Abstract
In the fission yeast Schizosaccharomyces pombe, the pgr1+ gene encoding glutathione (GSH) reductase (GR) is essentially required for cell survival. Depletion of GR caused proliferation arrest at the G1 phase of the cell cycle under aerobic conditions. Multicopy suppressors that restore growth were screened, and one effective suppressor was found to be the trx2+ gene, encoding a mitochondrial thioredoxin. This suggests that GR is critically required for some mitochondrial function(s). We found that GR resides in both cytosolic and organellar fractions of the cell. Depletion of GR lowered the respiration rate and the activity of oxidation-labile Fe-S enzymes such as mitochondrial aconitase and cytosolic sulfite reductase. Trx2 did not reverse the high ratio of oxidized glutathione to GSH or the low respiration rate observed in GR-depleted cells. However, it brought the activity of oxidation-labile Fe-S enzymes to a normal level, suggesting that the maintenance of Fe-S enzymes is a critical factor in the survival of S. pombe. The activity of succinate dehydrogenase, an oxidation-insensitive Fe-S enzyme, however, was not affected by GR depletion, suggesting that GR is not required for the biogenesis of the Fe-S cluster. The total iron content was greatly increased by GR depletion and was brought to a nearly normal level by Trx2. These results indicate that the essentiality of GR in the aerobic growth of S. pombe is derived from its role in maintaining oxidation-labile Fe-S enzymes and iron homeostasis.
The abundant tripeptide glutathione (GSH) (l-γ-glutamyl-l-cysteinylglycin) is one of the most prevalent reducing thiols in vivo, with diverse functions in many biological processes such as protein and DNA synthesis, transport, modulation of enzyme activity, maintenance of the intracellular redox state, and detoxification of damaging molecules (34, 35). GSH is also an electron donor for antioxidant enzymes such as glutaredoxins (Grx) and glutathione-dependent peroxidases. GSH is oxidized through reactions with disulfides, metals, and reactive oxygen species (ROS), yielding oxidized glutathione (GSSG) (8). The reduction of GSSG to GSH is efficiently mediated by glutathione reductase (GR) using NADPH specifically as a reducing equivalent (7). GR, a member of the pyridine nucleotide-disulfide oxidoreductase family, is conserved in a broad spectrum of organisms, from bacteria to higher eukaryotes.
In addition to glutathione, several peptide thiols such as thioredoxin (Trx) and glutaredoxin, containing redox-active dithiol or monothiol groups, provide a reducing environment to the cell (22, 32, 55). Thioredoxins reduce disulfide bonds in a variety of substrate proteins and provide electrons to thioredoxin-dependent peroxidases. They also serve as electron donors for several enzymes such as ribonucleotide reductase, methionine sulfoxide reductase, and 3′-phosphoadenosyl-5′-phosphosulfate reductase and regulate activities of transcription factors such as NF-κB, AP-1, and ASK1. Trx is reduced by Trx reductase using NADPH (32, 55).
As thiol buffer molecules, GSH, Grx, and Trx share some overlapping functions. However, in each organism, the specific role and contribution could be different. In Escherichia coli, a lack of GSH does not cause sensitivity toward oxidative stress (18). Furthermore, GR-deficient mutants maintain a highly reduced glutathione pool, suggesting that GSSG can be reduced independently of GR (52), most likely by Grx or Trx systems (45). In Saccharomyces cerevisiae, GSH is critically required for aerobic survival, and GR is required to protect cells against oxidative stress (16). However, the GLR1 gene encoding GR in Saccharomyces cerevisiae is not essentially required for normal growth (12, 17). Since the glr1 trx1 trx2 triple mutant is not viable, the Trx system is thought to provide the overlapping function shared by GR (39). In Schizosaccharomyces pombe, glutathione biosynthetic mutants show glutathione auxotrophy (10). Unexpectedly, the pgr1+ gene encoding GR is absolutely required for cell proliferation (28). Therefore, S. pombe could provide a good model system to disclose the role of GR in cell proliferation.
In addition to their central task of ATP generation, mitochondria are the main source for both ROS and the antioxidative defenses (5, 53). ROS are generated during aerobic respiration and cause damage in various cellular components (6, 49). They also act as mediators of an early event of the apoptotic signaling pathway and induction of the permeability transition in mammalian cells (13, 15, 36). The mitochondrial defense system against ROS includes a number of antioxidant enzymes such as Mn-superoxide dismutase (SOD), Trx, Grx, glutathione peroxidase, and thioredoxin peroxidase (40, 53). GSH acts as an antioxidant molecule not only in the cytosol but also in mitochondria. Since little or no de novo synthesis of GSH occurs in mitochondria, it must be imported from the cytosol and translocated across the inner membrane of mitochondria by an anion carrier (11, 31). However, it is difficult for the GSSG formed in mitochondria to exit this organelle (42). Therefore, GR is required to maintain reduced GSH in mitochondria. In mammalian cells GR activity has been detected in both the cytosol and mitochondria (33), and in Saccharomyces cerevisiae, GLR1 encodes both the cytosolic and mitochondrial forms of GR using alternative start codons (43). However, not much is known about the functions of GR in mitochondria and hence its potential role in maintaining cell survival.
One of the essential processes occurring in the mitochondrial matrix is the generation of iron-sulfur (Fe-S) clusters. Fe-S clusters are versatile prosthetic groups found in all living organisms, being involved in electron transport, biochemical catalysis, environmental sensing, and regulation of gene expression (3, 4). They could be sensitive to oxidative stress and liberate free iron that could further ROS production. Biogenesis of Fe-S clusters is a conserved process from bacteria to higher eukaryotes (30). Mitochondria can assemble their endogenous Fe-S proteins and export Fe-S clusters to assemble cytosolic Fe-S proteins (26). In Saccharomyces cerevisiae, Grx5, a mitochondrial monothiol glutaredoxin, is required for the activity of Fe-S proteins, especially in a step after cluster synthesis, and its absence affects cellular iron homeostasis (38, 44). It has been reported that GSH depletion does not affect the activities of mitochondrial Fe-S proteins but impairs the maturation of cytosolic Fe-S proteins, and the defect is not caused by oxidative stress (46).
In this work, we demonstrate that GR is required for the mitochondrial functions of S. pombe, such as efficient respiration and maintenance of the Fe-S cluster. This finding was made through the discovery of mitochondrial thioredoxin that compensated for the growth defect of GR-deficient cells.
MATERIALS AND METHODS
Yeast strains and culture media.
S. pombe strains used in this study were 972 (h− wild type), ED665 (h− ade6-M210 leu1-32 ura4-D18), ED668 (h+ ade6-M216 leu1-32 ura4-D18), JL38 (h− ade6-M210 leu1-32 ura4+), and JL36 (h− ade6-M210 leu1-32 ura4+ nmt-pgr1 Δpgr1) (28). Growth and maintenance of all the strains were generally done as described previously by Moreno et al. (37) and Alfa et al. (1). Cells were grown in Edinburgh minimal medium (EMM) with appropriate supplements as described previously by Alfa et al. (1). E. coli strain DH5α was used for plasmid construction and preparation.
Construction of the S. pombe genomic library and screening of the multicopy suppressor.
The genomic DNA from strain 972 was prepared according to a method described previously by Moreno et al. (37). It was partially digested with HindIII, and DNA fragments of 9 to 12 kb in size were collected by electroelution. These fragments were ligated with the pWH5 vector cut with HindIII and amplified in E. coli. To screen for multicopy suppressors of the growth defect caused by a depletion of GR, JL36 (nmt1-pgr1 Δpgr1) cells were transformed with the library (37), and the transformants that grew on EMM plates with 10 μM thiamine were isolated. Total DNA was prepared from the transformants and was introduced into E. coli cells by electroporation, followed by preparation of the plasmids containing the multicopy suppressor gene. The nucleotide sequence was determined, and the results were analyzed by using the S. pombe BLAST server at the Sanger Institute (http://www.sanger.ac.uk).
Cloning of the thioredoxin genes.
The 1.5-kb DNA fragment that contains the trx2+ gene was amplified by PCR using primers TRX-F1 (5′-CAAAACTTCTGCAGTACAACATCG-3′ [the PstI site is underlined]) and TRX-R (5′-TACGAATGGGGATCCCAATATCCC-3′ [the BamHI site is underlined]). The PCR fragment was cut with PstI and BamHI and cloned into pREP1 to overproduce Trx2 in S. pombe. A 2.2-kb XbaI/EcoRI fragment containing the trx1+ gene was cloned into the XbaI/EcoRI site of pUC19, and from this plasmid, the 1.4-kb PstI/NdeI fragment was obtained and cloned into vector pREP1 to produce pREP1-trx1, which overproduces Trx1. To obtain mitochondrially targeted Trx1, the mitochondrial leader sequence of the trx2+ gene was fused to the N terminus of the trx1+ open reading frame (ORF) by PCR using primers Trx1-ML (5′-TTAAAGCCGCATATGAGATCTTTTGCTCTGCGCAGAAGTTTTACTAGCTCTCGTATTTTGAGAATGGTGAAACAAGTC-3′ [the NdeI site is underlined, and the mitochondrial leader sequences are in italics]) and Trx1-C (5′-ACTATCAGGATCCGTAAATTAGAGG-3′ [the BamHI site is underlined]). The amplified DNA was cut with NdeI and BamHI and cloned into pREP1-trx2 cut with BamHI and NdeI to remove the Trx2 ORF, resulting in pREP1-Mito-trx1.
Construction of pgr1 mutants.
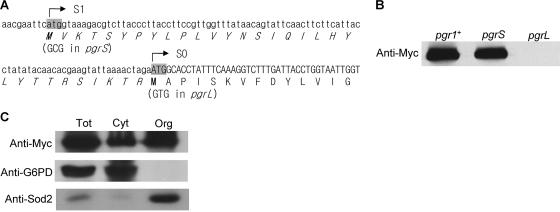
To determine the translational initiation site and the localization of GR within subcellular fractions, a Pgr1-Myc chimeric protein with a C-terminally fused nine-Myc tag was constructed. The 3.5-kb DNA fragment containing the pgr1+ gene was amplified by PCR using primers pgr-F1 (5′-CCTGTAAGCTTTTGCTGCAGTATTTGTTTC-3′ [the PstI site is underlined]) and pgr-HPM (5′-AATAAAAGACCCCGGGTAAACTAAAGTAAC-3′ [the SmaI site is underlined]). The PCR fragment was cut with PstI and SmaI and cloned into pRIP1-HPM (C-terminal nine-Myc tagging vector) through PstI and SmaI site to create pPgrF. The Pgr1 variants (pPgrS and pPgrL) that used defined ATG start sites were constructed by site-directed mutagenesis of pPgrF using pgr-S1 (5′-CATAACGAATTCGCGATAAAGACGTC-3′) and pgr-S2 (complementary sequence of pgr-S1) to remove the upstream start site (S1) (see Fig. 4A) or pgr-L3 (5′-GTATTAAAACTAGAGTGGCACCTATTTC-3′) and pgr-L4 (complementary sequence of pgr-L1) to remove the original start site (S0). The pPgrF, pPgrS, and pPgrL plasmids were linearized at the BglII site in the pgr1 promoter region and transformed into JL36 to promote integration into the chromosome. The correctly integrated transformants were selected by colony PCR and confirmed by Southern hybridization.
FIG. 4.
Determination of the translational start site and subcellular localization of GR. (A) The nucleotide sequence around the previously reported translational start site of the pgr1+ gene (S0) is shown with its amino acid sequence. The deduced amino acid sequence translated from another in-frame ATG site (S1) is indicated with italics. The two potential translational start codons were mutated to create pgrS (S1 Met to Ala) and pgrL (S0 Met to Val) variants. (B) The nine-Myc-tagged wild-type pgr1+ ORF and its translation initiation mutants (pgrS and pgrL) were integrated into the chromosome in JL36 cells. The cells were incubated in EMM containing thiamine and harvested at an OD595 of 0.5. Total cell extracts were prepared, and Pgr1-Myc protein was detected by Western blotting. (C) Cell extracts were prepared from the total (Tot), cytosolic (Cyt), and organellar (Org) fractions of cells containing the integrated nine-Myc-tagged wild-type pgr1+ ORF. In each fraction, Pgr1-Myc, G6PD, and Sod2 (mitochondrial Mn-SOD) were detected by Western blotting.
Flow cytometry.
Procedures for flow cytometry described previously by Alfa et al. (1) were used. For the control cells of 1C (a single copy of the genome) or 2C DNA content, wild-type cells starved for the N source or FYC5 (cdc25-22) cells grown at restrictive temperature (36°C) for 4 h were used, respectively. Exponentially growing cells also served as a 2C-type control, being indistinguishable from FYC5 at the restrictive temperature. Cells were fixed with ethanol, stained with propidium iodide (PI), and analyzed with a flow cytometer (FACScan; Becton Dickinson) using LYSIS II software.
Organelle fractionation.
The cytosol and organelle fractions were prepared according to a method described previously by Skoneczny et al. (47), with some modifications. Approximately 10 g of washed cells was resuspended in 50 ml of 0.1 M Tris-HCl (pH 9.0) containing 2.5 mM dithiothreitol and incubated for 20 min at 30°C with shaking. Cells were harvested, washed with digestion buffer (1.3 M sorbitol, 1 mM EDTA, 5 mM MOPS [morpholinepropanesulfonic acid], pH 7.2), resuspended in 20 ml of digestion buffer containing 10 mg lyticase (Sigma) and 0.05% (vol/vol) 2-mercaptoethanol, and incubated for 1 h at 30°C with gentle shaking. Protoplasts were pelleted and washed twice with digestion buffer and finally resuspended in 30 ml of chilled homogenization buffer (0.65 M sorbitol, 0.5 mM EDTA, 2.5 mM MOPS, pH 7.2). Following homogenization with a pestle for 20 strokes, the homogenate was centrifuged at 3,000 × g for 10 min. The supernatants were pooled and centrifuged at 65,000 × g (27,000 rpm with a Ti50 rotor) in a Beckman ultracentrifuge for 30 min. The supernatant was taken for the cytosol fraction, and the pellet was resuspended in 1 ml of homogenization buffer for the crude organelle fraction, followed by sonication.
Measurement of respiration rate.
The respiration rate was measured polarographically at 25°C using a YSI5300 Biologcal Oxygen Monitor Micro system (Yellow Spring Instrument). After collection by centrifugation at 3,000 × g, cells were suspended in 50 mM potassium phosphate (pH 6.5) buffer with 0.1 M glucose at 0.1 g (wet weight) of cells/ml. Sixty microliters of the cell suspension was introduced into the buffer-filled chamber, and the amount of oxygen consumed was recorded.
Enzyme assays and measurements of intracellular glutathione and iron.
Cell extracts were prepared as described previously (29). GR activity was measured according to a method described previously by Smith et al. (48) by monitoring the reduction of 5′-dithiobis(2-nitrobenzoic acid) by GSH, which is produced by GR. The amount of total and oxidized glutathione was determined using GR and 2-vinylpyridine as described previously by Griffith (19). Enzyme activities of aconitase and succinate dehydrogenase in organellar extracts and sulfite reductase in the cytosolic fraction were measured as described previously (9, 20, 21). To determine the total amount of intracellular iron, approximately 200 mg of dried cells was analyzed by using an ICP-atomic emission spectrometer (ICP-AES) (ICPS-1000IV; SHIMADZU) at the National Center for Inter-University Research Facilities at Seoul National University. All enzyme assays and measurements in this study were carried out at least three times independently.
Western immunoblot analysis.
The JL36 variants integrated with pPgrF, pPgrS, and pPgrL were harvested at an optical density at 595 nm (OD595) of 0.5. Their cell extracts, prepared from total cells or the cytosolic or organellar fraction, were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subjected to Western analysis using anti-Myc monoclonal antibody (Santa Cruz Biotech, Santa Cruz, CA) to detect Pgr1-Myc protein. Immunoblotting with mouse polyclonal antibodies against cytosolic glucose-6-phosphate dehydrogenase (G6PD) and mitochondrial Mn-SOD (Sod2) (24) was done to confirm a clean separation of the fractions. The secondary antibody (goat anti-mouse IgGAM; Cappell) conjugated with horseradish peroxidase was used at a 10−5 dilution, and the signal was detected using an ECL system (Amersham).
GFP fluorescence microscopy.
The wild-type cell (ED668) containing an integrated copy of the red fluorescent protein (RFP)-tagged sdh4+ gene encoding a subunit of the succinate dehydrogenase complex as a mitochondrial marker protein was constructed using pRIP1-RFP (created from pFA6a-RFP-kanMX6) with a C-terminal RFP tag. The green fluorescent protein (GFP)-tagged trx2+ gene was introduced into the sdh4+-RFP cells on a multicopy pREP42EGFP-C plasmid (14) with an enhanced GFP tag fused in frame to the C terminus. Cells were grown in EMM to the stationary phase without thiamine to allow the expression of the fused genes from the intermediate-strength nmt promoter. Fluorescence microscopy was performed at ×100 magnification, and the images were captured by an LSM510 MLD confocal microscope (Carl-Zeiss). GFP and RFP signals were detected at 488 nm and 553 nm, respectively, and merged by using the Zeiss LSM image browser.
RESULTS
GR depletion causes a growth halt.
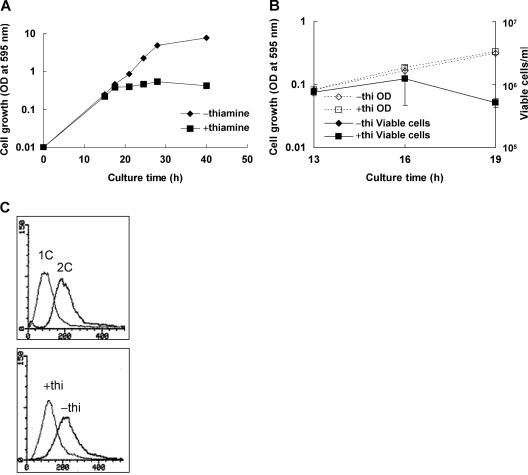
It has been previously reported that the pgr1+ gene encoding glutathione reductase is absolutely required for the proliferation of S. pombe (28). Growth of strain JL36, in which the expression of the pgr1 gene is driven by a thiamine-repressible nmt1 promoter, was halted at an OD595 of about 0.5 when inoculated and grown in EMM containing 10 μM thiamine (Fig. 1A). It did not form colonies on EMM plates containing thiamine under aerobic (air-exposed) conditions but grew as well as the wild-type cells under anaerobic conditions or on GSH-supplemented medium (data not shown). When freshly grown JL36 (nmt1-pgr1) cells were transferred to the thiamine-containing medium, GR activity decreased gradually, and by the time of the growth halt, at around 19 h postinoculation, activity was no longer detected in cell extracts (Table 1). Around this time, thiamine-repressed GR-deficient cells started to lose cell viability, as monitored by colony counts on EMM plates, whereas nonrepressed JL36 cells retained growth and viability (Fig. 1B). To determine whether the growth-halted cells were arrested at a specific cell stage, we analyzed cells with a fluorescence-activated cell sorter after staining nuclear DNA with PI. As demonstrated in Fig. 1C, the GR-deficient (thiamine-repressed) cells at growth halt contained 1C content of DNA, whereas nonrepressed cells showed the typical 2C content as predicted. This suggests that the growth halt was initially caused by an arrest at the G1 phase of cell cycle.
FIG. 1.
Effect of GR deficiency on the growth, viability, and cell cycle arrest in S. pombe. (A) A seed culture of JL36 (nmt1-pgr1) cells grown overnight was inoculated to an OD595 of 0.01 in EMM in the presence (square) or absence (diamond) of 10 μM thiamine. The growth was monitored at 30°C by measuring the OD595. Growth analyses were carried out at least three times independently for each strain, and a representative growth curve for each condition is shown. (B) At around the time of growth arrest (13, 16, and 19 h postinoculation), cells were diluted and plated onto EMM plates in triplicate to monitor viable cell counts. (C) At the arrested time point (16 h postinoculation), cells were harvested and fixed with ethanol. After removing RNA, the remaining nucleic acid was stained with PI and subjected to fluorescence-activated cell sorter analysis. x and y axes represent fluorescence (DNA content) and cell number, respectively. The upper panel represents patterns for control cells of 1C or 2C DNA content. The lower panel represents results for JL36 cells grown in the absence (−) and presence (+) of thiamine (thi).
TABLE 1.
Effect of Trx overproduction on GR activity and the GSH/GSSG ratio
| Straina | GR (mU/mg) ± SDb | Total glutathione (nmol/mg) ± SD | GSSG (nmol/mg) ± SD | GSH/GSSG ratio |
|---|---|---|---|---|
| JL38(pREP1) | 6.26 ± 1.75 | 109.9 ± 10.6 | 0.4 ± 0.06 | 261.24 |
| JL36(pREP1) | NDc | 136.5 ± 12.9 | 21.6 ± 1.37 | 4.33 |
| JL36(pREP1-trx2+) | ND | 119.1 ± 14.5 | 11.7 ± 1.99 | 8.15 |
| JL36(pREP1-trx1+) | ND | 147.8 ± 13.2 | 10.2 ± 3.57 | 12.48 |
Wild-type (JL38) and JL36 (nmt1-pgr1) cells harboring recombinant or parental (pREP1) plasmids were grown to an OD595 of 0.5 in EMM containing 10 μM thiamine. Enzyme activities and the amount of glutathione per mg protein in cell extracts were determined as described in Materials and Methods.
One milliunit (mU) is defined as the activity which produces 1 nmol of TNB per min.
ND, not detected.
Overexpression of the trx2+ gene encoding a mitochondrial thioredoxin suppressed the growth defect of GR-deficient cells.
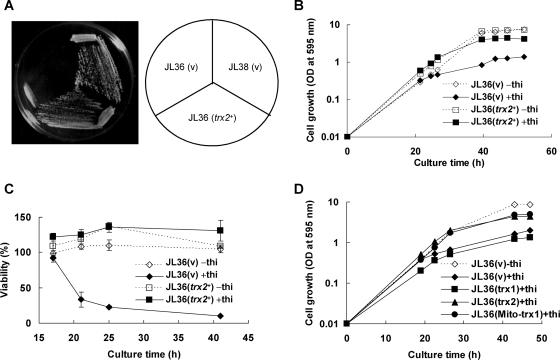
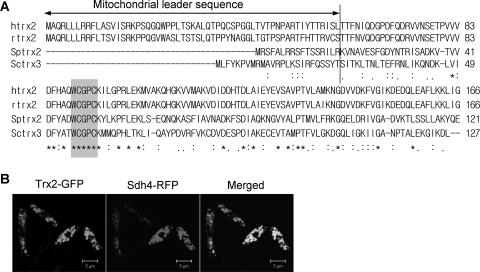
In order to find a clue for the reason why GR is absolutely required for the growth of S. pombe, multicopy suppressor genes were screened. JL36 (nmt1-pgr1+) cells were transformed with the S. pombe genomic DNA library, and the transformants that grew on thiamine-containing plates were selected. Transformants provided with the wild-type pgr1+ gene or regulator genes such as thi1+, which could fully or partially derepress the nmt1 promoter even in the presence of thiamine, were excluded by PCR, Southern blotting, or Northern analyses, since even a tiny amount of GR activity that would hardly be detectable by enzyme assay could cause survival. Among the candidates with various degrees of a suppressive effect, a plasmid containing a 9-kb fragment from chromosome II restored almost normal growth in thiamine-containing medium without restoring GR activity. One of the five ORFs in the plasmid (SPBC12D12.07c) encodes a putative thioredoxin of 121 amino acids, similar to Saccharomyces cerevisiae TRX2, with 40% identities and 68% similarities. When a multicopy plasmid containing the subcloned 1.5-kb fragment covering the promoter and the entire coding region of the putative thioredoxin gene, named trx2+, was introduced into JL36, it restored the cell viability by suppressing the growth defect caused by a GR deficiency (Fig. 2). Trx2-overproducing JL36 cells formed colonies on EMM plates with thiamine and showed a growth behavior similar to that of nonrepressed cells in liquid medium containing thiamine (Fig. 2A and B). Consistent with the growth restoration, Trx2-overproducing JL36 cells restored viability monitored by colony counts on plates (Fig. 2C). The putative thioredoxin gene trx2+ shared many conserved residues with thioredoxins from Saccharomyces cerevisiae and other eukaryotes, including the conserved active-site sequence WCGPC (Fig. 3A). The N-terminal 17-amino-acid sequence of Trx2 was predicted to be a mitochondrial signal peptide by PSORT II (http://psort.nibb.ac.jp) and MitoProtII (http://www.mips.biochem.mpg.de). To examine its subcellular localization, we produced a Trx2-GFP fusion protein using the pREP42EGFP-C-based multicopy plasmid in S. pombe. The Trx2-GFP recombinant plasmid was introduced into the cell that has a single copy of the RFP-tagged sdh4+ gene encoding a subunit of the mitochondrial succinate dehydrogenase complex, as described in Materials and Methods. The GFP fluorescence signal was detected at the same position as the red mitochondrial marker protein Sdh4 (Fig. 3B), indicating that Trx2 is indeed localized to mitochondria.
FIG. 2.
Suppression of GR deficiency by multicopy trx2+. (A) The trx2+ gene was subcloned into pREP1. JL38 cells harboring the control vector (pREP1) (v) and JL36 (nmt1-pgr1) cells harboring the control vector or pREP1-trx2+ were streaked onto an EMM plate containing 10 μM thiamine and incubated for 4 days at 30°C. (B) JL36 cells containing plasmid pREP1 (v) or pREP1-trx2+ (trx2+) were incubated in EMM in the presence (filled marks and solid lines) or absence (open marks and dotted lines) of 10 μM thiamine as described in the legend of Fig. 1. The growth was monitored by measuring the OD595. Growth analyses were carried out at least three times independently for each strain, and the growth curves from a single experiment are shown. (C) Cells were diluted and plated onto EMM plates in triplicate to monitor viable cell counts. Viability was presented as the percentage of viable cells compared to the total cell number calculated from the OD595. (D) To examine the effect of Trx1, JL36 cells containing plasmid pREP1 (v), pREP1-trx1+ (trx1), pREP1-trx2+ (trx2), or pREP1-Mito-trx1 (Mito-trx1) were incubated in EMM in the presence (filled marks and solid lines) or absence (open marks and dotted lines) of 10 μM thiamine as described in the legend of Fig. 1. The growth was monitored by measuring the OD595. Growth analyses were carried out at least three times, and representative growth curves from a single experiment are shown.
FIG. 3.
Subcellular localization of Trx2. (A) Amino acid sequences of mitochondrial thioredoxins from human, rat, S. pombe, and Saccharomyces cerevisiae were compared by CLUSTAL W. The positions of identical and similar amino acids are marked with asterisks and dots, respectively. The active site is shaded. The vertical line represents the plausible cleavage site for the mitochondrial leader peptide. (B) Cells containing an integrated copy of the RFP-tagged sdh4+ gene for mitochondrial succinate dehydrogenase were transformed with a pREP42EGFP-C-based recombinant plasmid containing the GFP-tagged trx2+ gene. Cells were grown in EMM to the stationary phase. Fluorescent images were visualized by using an LSM510 MLD confocal microscope (Carl-Zeiss).
A search for Trx homologues in the S. pombe genome revealed two candidate genes: trx1+, which encodes a putative cytosolic thioredoxin, and trx2+. When we examined the effect of trx1+ on suppressing the growth inhibition caused by a GR deficiency, we found that it did not restore the growth of GR-deficient cells as trx2+ did (Fig. 2D). However, when the predicted mitochondrial targeting sequence of trx2+ was fused to the N terminus of the trx1+ ORF, the resulting chimera (Mito-Trx1) on a multicopy plasmid (pREP1) suppressed the growth defect of GR-deficient cells (Fig. 2D). This confirms the proposal that mitochondrial function was damaged by a GR depletion, and the damaged function could be restored by thioredoxins.
Determination of start codon and subcellular localization of GR.
In Saccharomyces cerevisiae, GLR1 encodes both the cytosolic and mitochondrial forms of GR. It harbors two in-frame start codons, and both are used as translational initiation sites for two forms of GR (43). In S. pombe, pgr1+ was reported to encode a peptide of 465 amino acids (28). From the completed genome sequence database, we found that at 90 nucleotides upstream from the start codon, another in-frame ATG codon exists (Fig. 4A), and the extended sequences increases the probability of mitochondrial targeting from 23% to 58% as predicted by MitoProtII. To test whether S. pombe uses alternative start codons, the upstream codon (S1) as well as the originally reported downstream codon (S0) were mutated to GCG (Ala) and GTG (Val) to create the variant genes pgrS and pgrL, respectively. The wild-type and the mutant pgr genes with a linked promoter region were fused with a nine-Myc tag in the pRIP1-HPM vector and introduced into the JL36 chromosome as described in Materials and Methods. The fusion proteins were detected by Western blotting analysis using anti-Myc antibody. A mutation of the upstream S1 codon in pgrS produced a GR protein of the same size as the wild type, whereas an S0 mutation in pgrL resulted in no production of GR (Fig. 4B). This confirms the original proposal that GR is translated from the S0 start site (28). Consistent with this observation, the pgrS mutant suppressed the growth defect of JL36 (nmt1-pgr1) on thiamine-containing medium, whereas the pgrL variant did not (data not shown). Therefore, unlike Saccharomyces cerevisiae, S. pombe uses only one start codon for GR.
We then examined the subcellular localization of GR using the Myc-tagged pgr1+ construct integrated into the chromosome. Cell extracts were fractionated into cytosolic versus organellar fractions. Western blotting analysis using anti-Myc antibody revealed that GR resides not only in the cytosol but also in the organelle fraction (Fig. 4C). Parallel analyses with antibodies against G6PD and mitochondrial Mn-SOD (Sod2) (24) confirmed the relatively clean separation between the two fractions. Therefore, it appears that even without a predictable signal sequence, GR is distributed in organelles, most likely in mitochondria, as well as in the cytosol in S. pombe.
Trx2 overexpression does not increase the GSH/GSSG ratio.
Depletion of GR caused the accumulation of oxidized glutathione (GSSG) and dramatically reduced the GSH/GSSG ratio by more than 50-fold (Table 1). To examine the effect of Trx2 on the GSH/GSSG ratio, total glutathiones and the GSH/GSSG ratio were measured in Trx2-overproducing JL36 cells. As demonstrated in Table 1, Trx2 did not significantly change the total level of glutathione, nor did GSSG. Compared with the GR-deficient JL36 cells, the GSSG level was reduced by only about twofold. This level is still much lower than that of normal cells (JL38) by more than 30-fold. Introduction of multicopy trx1+, which did not restore the growth of GR-deficient cells, also decreased the level of oxidized glutathione by about twofold. Therefore, these results indicate that Trx2 restored cell growth not by reducing GSSG but through a different mechanism.
The respiration rate was decreased in GR-deficient cells.
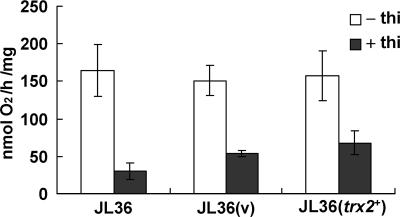
The observation that mitochondrial thioredoxin (Trx2), but not the cytosolic thioredoxin (Trx1), restored the growth of GR-deficient cells led us to examine the role of GR in various mitochondrial functions and whether an impairment of any of these functions causes growth inhibition that can be reversed by Trx2. As an initial trial, we monitored the rate of aerobic respiration. The oxygen consumption rate was measured for JL36 cells that were grown in thiamine-containing medium near the growth arrest point. GR depletion caused a reduction in the respiration rate by about three- or fourfold in JL36 cells with or without the control plasmid, respectively (Fig. 5). The trx2+-containing cells exhibited only a slightly increased rate of respiration compared with that of the vector-containing control. This suggests that GR serves to ensure the optimal respiratory function of mitochondria, but the reduced respiration in GR-deficient cells is not the cause of the growth defect that was reversed by Trx2.
FIG. 5.
Respiration rate of GR-deficient cells. JL36 cells with or without pREP1 (v) or pREP1-trx2+ (trx2+) were incubated in EMM in the presence (filled bars) or absence (open bars) of 10 μM thiamine as described in the legend of Fig. 1. Near the point of growth arrest (at an OD595 of about 0.5), cells were harvested, and the respiration rate was measured using an oxygen monitor. Average values with standard deviations from four independent experiments are presented.
GR is required to maintain oxidant-labile iron-sulfur enzymes and iron homeostasis.
Mitochondria play a central role in providing cellular Fe-S clusters. Reactive oxygen species such as superoxide or peroxinitrite inactivate Fe-S proteins by oxidizing the cluster, and a loss of antioxidative defense proteins causes a decrease in the activity of oxidant-labile Fe-S enzymes (25, 50). It has also been reported that glutaredoxins and glutathione contribute to the assembly of Fe-S proteins (38, 44, 46).
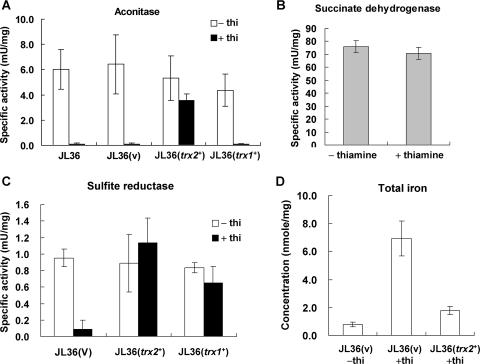
We therefore examined whether Fe-S enzyme activities have been affected by GR depletion and Trx2 overproduction. For this purpose, the activities of two mitochondrial Fe-S enzymes (aconitase and succinate dehydrogenase) and one cytosolic Fe-S enzyme (sulfite reductase) were measured. The Fe-S cluster of aconitase has previously been reported to be sensitive to oxidants, whereas succinate dehydrogenase is more stable (25, 50). Figure 6A demonstrates that upon GR depletion, the oxidant-sensitive aconitase activity disappeared to a nondetectable level. Trx2 overproduction, but not Trx1, restored the level of aconitase activity to near 70% of its normal level. On the contrary, succinate dehydrogenase activity is not affected by GR depletion (Fig. 6B). The cytosolic sulfite reductase activity was greatly reduced upon GR depletion but was fully restored by Trx2 overproduction (Fig. 6C). Its activity was also restored to about 80% by cytosolic Trx1 overproduction (Fig. 6C). These results suggest that (i) the primary reason for the growth defect caused by a GR depletion is the loss of (oxidation-sensitive) Fe-S protein activities not only in mitochondria but also in the cytosol and (ii) Trx2, when overproduced, is able to protect the labile Fe-S cluster against oxidative stress.
FIG. 6.
Measurement of iron-sulfur enzymes and total iron. Cells were grown as described in the legend of Fig. 5. (A) Aconitase activity was measured for organellar cell extracts from JL36 cells with or without the parental pREP1 vector (v), the cloned trx2+ gene (trx2+), or the cloned trx1+ gene (trx1+) on pREP1. (B) Succinate dehydrogenase activity was measured for organellar extracts prepared from JL36 cells. (C) Sulfite reductase activity was measured using total cell extracts from JL36 cells containing the parental vector (v), the cloned trx2+ gene (trx2+), or the cloned trx1+ gene (trx1+). (D) Measurement of total iron concentration. Dried cells were prepared from JL36 cells containing either the parental vector (v) or the cloned trx2+ gene (trx2+). Total iron concentration was measured by ICP-AES. Average values with standard deviations from at least three independent experiments are presented.
Inactivation of components in Fe-S assembly system often results in the accumulation of iron in the cell or mitochondria (44, 46). We examined the change in total iron concentration in GR-deficient cells by ICP-AES. Consistent with its effect on Fe-S proteins, depletion of GR caused a drastic increase in the total iron concentration by more than sevenfold (Fig. 6D). The introduction of multicopy trx2+ reduced this increased level to about one-fourth in JL36 cells. These results coincide with the effect of GR and Trx2 on Fe-S protein activity and suggest that GR and high levels of Trx2 contribute to cellular iron homeostasis.
DISCUSSION
In S. pombe, GR activity is indispensable for normal proliferation (28). This contrasts with its dispensable role in E. coli or Saccharomyces cerevisiae, where its loss can be compensated for by Grx, Trx, or lipoamide and lipoamide reductase systems (12, 16, 17, 39, 45, 52). Upon GR depletion, S. pombe cells stop proliferation at the G1 phase and subsequently lose viability (Fig. 1). This study provides a clue for the role of GR in supporting various mitochondrial functions, and especially in providing Fe-S proteins, to sustain cell viability.
In Saccharomyces cerevisiae, the GR mutant grows under aerobic conditions when either one of the two cytosolic thioredoxin genes is active (39). This suggests that the function of GR partly overlaps with a thioredoxin system in the cytosol. In contrast, in S. pombe, we observed that not a cytosolic but a mitochondrial thioredoxin system suppressed growth inhibition caused by a GR deficiency. This indicates that the most critical damage caused by GR depletion results from mitochondrial dysfunction, even though not much has been understood about the role of GR in mitochondria.
GSH serves to maintain the thiol redox balance in the cytosol as well as in mitochondria. In mitochondria, GSH serves as an electron donor for GSH-dependent peroxidases to degrade hydroperoxide generated as a by-product in the process of electron transfer to oxygen (40). Mitochondrial GSH is imported from the cytosol by an energy-dependent process. However, GSSG formed in mitochondria is not exported to the cytosol unless it is reduced (11, 31, 42). Therefore, in order to maintain a reduced glutathione pool in mitochondria, glutathione reductase activity should reside in mitochondria. In Saccharomyces cerevisiae, GLR1 encodes both the cytosolic and the mitochondrial forms of GR using two different start codons located in frame (43). However, we showed in this study that S. pombe uses a single start codon and hence a single form of GR. This single form of GR is distributed in the cytosol as well as in organelles, most likely in mitochondria.
Upon GR depletion, we found that cells stopped proliferation at the G1 phase of the cell cycle. It can be simply postulated that the G1 arrest results from a decrease in ribonucleotide reductase activity that requires GSH or thioredoxin as an electron donor (32). However, there are reports that the mitochondrial dysfunction exemplified by the inhibition of mitochondrial protein synthesis, decreased ATP generation, and disruption of membrane potential causes cell cycle arrest at the G1 phase of mammalian cells (27, 54). Therefore, the G1 arrest observed by GR depletion in S. pombe could also be connected with mitochondrial dysfunction. Consistent with this proposal, a lack of dihydrolipoamide dehydrogenase, an E3 component of the α-ketoglutarate dehydrogenase complex in mitochondria, has been reported to cause cell cycle arrest at the G1 phase in S. pombe (23).
Mitochondria are the primary source of ROS generated during aerobic respiration and at the same place inhabit many oxidant-sensitive molecules. Some components of the respiratory chain complexes can be the target of ROS (41). In this study, we found that GR depletion caused a drastic decrease in the respiration rate, which was not recovered by Trx2 overproduction. Accumulation of GSSG (or a high GSSG/GSH ratio) in mitochondria could cause the oxidation of several thiol-containing proteins such as the permeability transition pore (13), respiratory chain molecules, or tricarboxylic acid (TCA) cycle enzymes. There are reports that GSH depletion causes a loss of complex I activity through thiol oxidation (41, 51). The redox-active thiols in complex I react with GSSG and form mixed disulfides in a Grx-dependent manner, thus causing the inhibition of complex I activity and an increase in ROS generation (2, 51). Supplementation with thioredoxin or GSH reduces the mixed disulfides but does not restore the activity of complex I. Respiratory dysfunction can result from the inactivation of enzymes in the TCA cycle as well. When NADH is not supplied efficiently from the TCA cycle, the electron flow in the respiratory chain is blocked. We found that aconitase, a key enzyme in the TCA cycle, lost activity in GR-deficient cells. It could have caused a decrease in the respiration rate. However, the restoration of aconitase activity by Trx2 failed to increase the respiration rate, suggesting that not the decrease in respiration but a function related with aconitase activity is what caused the growth defect in GR-deficient S. pombe cells.
Aconitase possesses an Fe-S cluster that is labile to oxidative attack. Since aconitase activity is greatly affected by GR deficiency, it can be hypothesized that either an increased ROS level or inefficient Fe-S maintenance results from a GR deficiency. Our preliminary observation revealed no change in protein carbonylation upon GR depletion, an indication of the oxidative environment in the cell, suggesting that the absence of GR does not increase the level of ROS but could impair Fe-S maintenance (data not shown). It is not plausible that the GR depletion impaired the generation of the Fe-S cluster, since succinate dehydrogenase activity was not affected by the GR depletion. Instead, the GR- and Trx2-dependent behavior of sulfite reductase as well as aconitase indicates that GR is required to maintain the Fe-S cluster in mitochondria as well as in the cytosol. The overproduction of cytosolic Trx1, which did not suppress the growth defect of GR-deficient cell, restored only the sulfite reductase activity, and the mitochondrially targeted trx1+ gene (Mito-trx1+) suppressed the growth defect of GR-deficient cells. These observations confirm this conclusion.
The change in the total iron concentration, an indication of change in Fe-S formation and maintenance, correlates well with changes in the activity of oxidation-labile Fe-S enzymes as well as the viability of cells, supporting the proposed role of GR and overproduced Trx2 in maintaining labile Fe-S enzymes. It can be postulated that GSH may contribute to stabilizing the Fe-S cluster or reactivating oxidized Fe-S clusters. This idea coincides with the observation in Saccharomyces cerevisiae that glutathione is required for the maturation of cytosolic Fe-S proteins and that ΔGSH1 accumulates high amounts of iron in mitochondria (46). GRX5, a mitochondrial thiol protein, is required for the later step of Fe-S cluster biogenesis (38). Our study adds GR and overproduced Trx2 in mitochondria to the group of proteins that maintain Fe-S cluster proteins. More studies are needed to determine the exact role of Trx2 in this process.
Acknowledgments
This work was supported by a National Research Laboratory (NRL) grant (2004-02397) from the Ministry of Science and Technology to J.-H. Roe. J.-Y. Song and J. Cha were supported by a BK21 research fellowship from the Korean Ministry of Education and Human Resources.
Footnotes
Published ahead of print on 1 September 2006.
REFERENCES
- 1.Alfa, C., P. Fantes, J. Hyams, M. Mcleod, and E. Warbrick. 1993. Experiments with fission yeast: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Beer, S. M., E. R. Taylor, S. E. Brown, C. C. Dahm, N. J. Costa, M. J. Runswick, and M. P. Murphy. 2004. Glutaredoxin 2 catalyzes the reversible oxidation and glutathionylation of mitochondrial membrane thiol proteins: implications for mitochondrial redox regulation and antioxidant defense. J. Biol. Chem. 279:47939-47951. [DOI] [PubMed] [Google Scholar]
- 3.Beinert, H., R. H. Holm, and E. Münck. 1997. Iron-sulfur clusters: nature's modular, multipurpose structures. Science 277:653-659. [DOI] [PubMed] [Google Scholar]
- 4.Beinert, H., and P. J. Kiley. 1999. Fe-S proteins in sensing and regulatory functions. Curr. Opin. Chem. Biol. 3:152-157. [DOI] [PubMed] [Google Scholar]
- 5.Cadenas, E., and K. J. A. Davies. 2000. Mitochondrial free radical generation, oxidative stress, and aging. Free Radic. Biol. Med. 29:222-230. [DOI] [PubMed] [Google Scholar]
- 6.Cahill, A., X. Wang, and J. B. Hoek. 1997. Increased oxidative damage to mitochondrial DNA following chronic ethanol consumption. Biochem. Biophys. Res. Commun. 235:286-290. [DOI] [PubMed] [Google Scholar]
- 7.Carlberg, I., and B. Mannervik. 1975. Purification and characterization of the flavoenzyme glutathione reductase from rat liver. J. Biol. Chem. 250:5475-5480. [PubMed] [Google Scholar]
- 8.Carmel-Harel, O., and G. Storz. 2000. Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu. Rev. Microbiol. 54:439-461. [DOI] [PubMed] [Google Scholar]
- 9.Chang, E. C., and D. J. Kosman. 1990. O2-dependent methionine auxotrophy in Cu,Zn superoxide dismutase-deficient mutants of Saccharomyces cerevisiae. J. Bacteriol. 172:1840-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaudhuri, B., S. Ingavale, and A. K. Bachhawat. 1997. apd1+, a gene required for red pigment formation in ade6 mutants of Schizosaccharomyces pombe, encodes an enzyme required for glutathione biosynthesis: a role for glutathione and a glutathione-conjugate pump. Genetics 145:75-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, Z., D. A. Putt, and L. H. Lash. 2000. Enrichment and functional reconstitution of glutathione transport activity from rabbit kidney mitochondria: further evidence for the role of the dicarboxylate and 2-oxoglutarate carriers in mitochondrial glutathione transport. Arch. Biochem. Biophys. 373:193-202. [DOI] [PubMed] [Google Scholar]
- 12.Collinson, L. P., and I. W. Dawes. 1995. Isolation, characterization and overexpression of the yeast gene, GLR1, encoding glutathione reductase. Gene 156:123-127. [DOI] [PubMed] [Google Scholar]
- 13.Costantini, P., B. V. Chernyak, V. Petronilli, and P. Bernardi. 1996. Modulation of the mitochondrial permeability transition pore by pyridine nucleotides and dithiol oxidation at two separate sites. J. Biol. Chem. 271:6746-6751. [DOI] [PubMed] [Google Scholar]
- 14.Craven, R. A., D. J. Griffiths, K. S. Sheldrick, R. E. Randall, I. M. Hagan, and A. M. Carr. 1998. Vectors for the expression of tagged proteins in Schizosaccharomyces pombe. Gene 9:59-68. [DOI] [PubMed] [Google Scholar]
- 15.Esteve, J. M., J. Mompo, J. García de la Asunción, J. Sastre, M. Asensi, J. Boix, J. R. Viña, J. Viña, and F. V. Pallardó. 1999. Oxidative damage to mitochondrial DNA and glutathione oxidation in apoptosis: studies in vivo and in vitro. FASEB J. 13:1055-1064. [DOI] [PubMed] [Google Scholar]
- 16.Grant, C. M., F. H. MacIver, and I. W. Dawes. 1996. Glutathione is an essential metabolite required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae. Curr. Genet. 29:511-515. [DOI] [PubMed] [Google Scholar]
- 17.Grant, C. M., L. P. Collinson, J. H. Roe, and I. W. Dawes. 1996. Yeast glutathione reductase is required for protection against oxidative stress and is a target gene for yAP-1 transcriptional regulation. Mol. Microbiol. 21:171-179. [DOI] [PubMed] [Google Scholar]
- 18.Greenberg, J. T., and B. Demple. 1986. Glutathione in Escherichia coli is dispensable for resistance to H2O2 and gamma radiation. J. Bacteriol. 168:1026-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffith, O. W. 1980. Determination of glutathione and glutathione disulfide using glutathione reductase and 2-vinylpyridine. Anal. Biochem. 106:207-212. [DOI] [PubMed] [Google Scholar]
- 20.Hatefi, Y. 1978. Resolution of complex II and isolation of succinate dehydrogenase (EC 1.3.99.1). Methods Enzymol. 53:27-35. [DOI] [PubMed] [Google Scholar]
- 21.Hausladen, A., and I. Fridovich. 1996. Measuring nitric oxide and superoxide: rate constants for aconitase reactivity. Methods Enzymol. 269:37-41. [DOI] [PubMed] [Google Scholar]
- 22.Herrero, E., and J. Ros. 2002. Glutaredoxins and oxidative stress defense in yeast. Methods Enzymol. 348:136-146. [DOI] [PubMed] [Google Scholar]
- 23.Jang, Y. J., K. S. Chung, C. Park, and H. S. Yoo. 1997. Fission yeast dihydrolipoamide dehydrogenase gene is involved in G1/S cell cycle progression. Biochim. Biophys. Acta 1358:229-239. [DOI] [PubMed] [Google Scholar]
- 24.Jeong, J. H., E. S. Kwon, and J. H. Roe. 2001. Characterization of the manganese-containing superoxide dismutase and its gene regulation in stress response of Schizosaccharomyces pombe. Biochem. Biophys. Res. Commun. 283:908-914. [DOI] [PubMed] [Google Scholar]
- 25.Keyer, K., and J. A. Imlay. 1997. Inactivation of dehydratase [4Fe-4S] clusters and disruption of iron homeostasis upon cell exposure to peroxynitrite. J. Biol. Chem. 272:27652-27659. [DOI] [PubMed] [Google Scholar]
- 26.Kispal, G., P. Csere, C. Prohl, and R. Lill. 1999. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 18:3981-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuruvilla, S., C. W. Qualls, Jr., R. D. Tyler, S. M. Witherspoon, G. R. Benavides, L. W. Yoon, K. Dold, R. H. Brown, S. Sangiah, and K. T. Morgan. 2003. Effects of minimally toxic levels of carbonyl cyanide P-(trifluoromethoxy) phenylhydrazone (FCCP), elucidated through differential gene expression with biochemical and morphological correlations. Toxicol. Sci. 73:348-361. [DOI] [PubMed] [Google Scholar]
- 28.Lee, J., I. W. Dawes, and J. H. Roe. 1997. Isolation, expression, and regulation of the pgr1+ gene encoding glutathione reductase absolutely required for the growth of Schizosaccharomyces pombe. J. Biol. Chem. 272:23042-23049. [DOI] [PubMed] [Google Scholar]
- 29.Lee, J., I. W. Dawes, and J. H. Roe. 1995. Adaptive response of Schizosaccharomyces pombe to hydrogen peroxide and menadione. Microbiology 141:3127-3132. [DOI] [PubMed] [Google Scholar]
- 30.Lill, R., and G. Kispal. 2000. Maturation of cellular Fe-S proteins: an essential function of mitochondria. Trends Biochem. Sci. 25:352-356. [DOI] [PubMed] [Google Scholar]
- 31.Martensson, J., C. K. James, and A. Meister. 1990. High-affinity transport of glutathione is part of a multicomponent system essential for mitochondrial function. Proc. Natl. Acad. Sci. USA 87:7185-7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masutani, H., and J. Yodoi. 2002. Thioredoxin. Methods Enzymol. 347:279-286. [DOI] [PubMed] [Google Scholar]
- 33.Mbemba, F., A. Houbion, M. Raes, and J. Remacle. 1985. Subcellular localization and modification with ageing of glutathione, glutathione peroxidase and glutathione reductase activities in human fibroblasts. Biochim. Biophys. Acta 838:211-220. [DOI] [PubMed] [Google Scholar]
- 34.Meister, A., and M. E. Anderson. 1983. Glutathione. Annu. Rev. Biochem. 52:711-760. [DOI] [PubMed] [Google Scholar]
- 35.Meister, A. 1988. Glutathione metabolism and its selective modification. J. Biol. Chem. 263:17205-17208. [PubMed] [Google Scholar]
- 36.Miñana, J. B., L. Gomez-Cambronero, A. Lloret, F. V. Pallardó, J. Del Olmo, A. Escudero, J. M. Rodrigo, A. Pellín, J. R. Viña, J. Viña, and J. Sastre. 2002. Mitochondrial oxidative stress and CD95 ligand: a dual mechanism for hepatocyte apoptosis in chronic alcoholism. Hepatology 35:1205-1214. [DOI] [PubMed] [Google Scholar]
- 37.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 38.Mühlenhoff, U., J. Gerber, N. Richhardt, and R. Lill. 2003. Components involved in assembly and dislocation of iron-sulfur clusters on the scaffold protein Isu1p. EMBO J. 22:4815-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muller, E. G. 1996. A glutathione reductase mutant of yeast accumulates high levels of oxidized glutathione and requires thioredoxin for growth. Mol. Biol. Cell 7:1805-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Netto, L. E. S., A. J. Kowaltowski, R. F. Castilho, and A. E. Vercesi. 2002. Thiol enzymes protecting mitochondria against oxidative damage. Methods Enzymol. 348:260-270. [DOI] [PubMed] [Google Scholar]
- 41.Nicholls, D. G. 2002. Mitochondrial function and dysfunction in the cell: its relevance to aging and aging-related disease. Int. J. Biochem. Cell Biol. 34:1372-1381. [DOI] [PubMed] [Google Scholar]
- 42.Olafsdottir, K., and D. J. Reed. 1988. Retention of oxidized glutathione by isolated rat liver mitochondria during hydroperoxide treatment. Biochim. Biophys. Acta 964:377-382. [DOI] [PubMed] [Google Scholar]
- 43.Outten, C. E., and V. C. Culotta. 2004. Alternative start sites in the Saccharomyces cerevisiae GLR1 gene are responsible for mitochondrial and cytosolic isoforms of glutathione reductase. J. Biol. Chem. 279:7785-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez-Manzaneque, M. T., J. Tamarit, G. Belli, J. Ros, and E. Herrero. 2002. Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol. Biol. Cell 13:1109-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Russel, M., and A. Holmgren. 1988. Construction and characterization of glutaredoxin-negative mutants of Escherichia coli. Proc. Natl. Acad. Sci. USA 85:990-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sipos, K., H. Lange, Z. Fekete, P. Ullmann, R. Lill, and G. Kispal. 2002. Maturation of cytosolic iron-sulfur proteins requires glutathione. J. Biol. Chem. 277:26944-26949. [DOI] [PubMed] [Google Scholar]
- 47.Skoneczny, M., A. Chelstowska, and J. Rytka. 1988. Study of the coinduction by fatty acids of catalase A and acyl-CoA oxidase in standard and mutant Saccharomyces cerevisiae strains. Eur. J. Biochem. 174:297-302. [DOI] [PubMed] [Google Scholar]
- 48.Smith, I. K., T. L. Vierheller, and C. A. Thorne. 1988. Assay of glutathione reductase in crude tissue homogenates using 5,5′-dithiobis(2-nitrobenzoic acid). Anal. Biochem. 175:408-413. [DOI] [PubMed] [Google Scholar]
- 49.Stadtman, E. R. 1993. Oxidation of free amino acids and amino acid residues in proteins by radiolysis and by metal-catalyzed reactions. Annu. Rev. Biochem. 62:797-821. [DOI] [PubMed] [Google Scholar]
- 50.Strain, J., C. R. Lorenz, J. Bode, S. Garland, G. A. Smolen, D. T. Ta, L. E. Vickery, and V. C. Culotta. 1998. Suppressors of superoxide dismutase (SOD1) deficiency in Saccharomyces cerevisiae. Identification of proteins predicted to mediate iron-sulfur cluster assembly. J. Biol. Chem. 273:31138-31144. [DOI] [PubMed] [Google Scholar]
- 51.Taylor, E. R., F. Hurrell, R. J. Shannon, T. K. Lin, J. Hirst, and M. P. Murphy. 2003. Reversible glutathionylation of complex I increases mitochondrial superoxide formation. J. Biol. Chem. 278:19603-19610. [DOI] [PubMed] [Google Scholar]
- 52.Tuggle, C. K., and J. A. Fuchs. 1985. Glutathione reductase is not required for maintenance of reduced glutathione in Escherichia coli K-12. J. Bacteriol. 162:448-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turrens, J. F. 2003. Mitochondrial formation of reactive oxygen species. J. Physiol. 552:335-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van den Bogert, C., G. van Kernebeek, L. de Leij, and A. M. Kroon. 1986. Inhibition of mitochondrial protein synthesis leads to proliferation arrest in the G1-phase of the cell cycle. Cancer Lett. 32:41-51. [DOI] [PubMed] [Google Scholar]
- 55.Vlamis-Gardikas, A., and A. Holmgren. 2002. Thioredoxin and glutaredoxin isoforms. Methods Enzymol. 347:286-296. [DOI] [PubMed] [Google Scholar]