Abstract
Galactose metabolism is essential for the survival of Trypanosoma brucei, the etiological agent of African sleeping sickness. T. brucei hexose transporters are unable to transport galactose, which is instead obtained through the epimerization of UDP-glucose to UDP-galactose catalyzed by UDP-glucose 4′-epimerase (galE). Here, we have characterized the phenotype of a bloodstream form T. brucei galE conditional null mutant under nonpermissive conditions that induced galactose starvation. Cellular levels of UDP-galactose dropped rapidly upon induction of galactose starvation, reaching undetectable levels after 72 h. Analysis of extracted glycoproteins by ricin and tomato lectin blotting showed that terminal β-d-galactose was virtually eliminated and poly-N-acetyllactosamine structures were substantially reduced. Mass spectrometric analysis of variant surface glycoprotein confirmed complete loss of galactose from the glycosylphosphatidylinositol anchor. After 96 h, cell division ceased, and electron microscopy revealed that the cells had adopted a morphologically distinct stumpy-like form, concurrent with the appearance of aberrant vesicles close to the flagellar pocket. These data demonstrate that the UDP-glucose 4′-epimerase is essential for the production of UDP-galactose required for galactosylation of glycoproteins and that galactosylation of one or more glycoproteins, most likely in the lysosomal/endosomal system, is essential for the survival of bloodstream form T. brucei.
The flagellated protozoan parasite Trypanosoma brucei, transmitted by the bite of the tsetse fly, is the etiological agent of African sleeping sickness in humans and the related disease nagana in cattle. T. brucei organisms are able to survive in the blood of the host by virtue of a dense surface coat of variant surface glycoprotein (VSG) that protects the parasite from the complement pathway and undergoes antigenic variation to evade specific immune responses, making the production of a vaccine unfeasible (4). Sleeping sickness is invariably fatal if untreated, and it is estimated that up to 500,000 people are currently infected in sub-Saharan Africa (22). Current drugs are expensive, toxic, and difficult to administer, leaving an urgent need for new therapeutic agents.
The bloodstream form of T. brucei is rich in galactose-containing glycoproteins, most notably the abundant VSG (11), and components of the flagellar pocket and lysomal/endosomal system, including the transferrin receptor (1, 13). T. brucei hexose transporters are unable to transport galactose (5, 18), which is instead obtained through the epimerization of UDP-glucose (UDP-Glc) to UDP-galactose (UDP-Gal) via the NADH-dependent oxidoreductase UDP-Glc 4′-epimerase (galE) (17). Construction of conditional null mutant cell lines has demonstrated that galE is an essential gene for both bloodstream and procyclic form T. brucei, and the phenotype of procyclic form cells upon induction of galactose starvation has been reported (16, 17).
In this paper, we examine the phenotype of the bloodstream form T. brucei conditional galE null mutant (galE-cKO) under nonpermissive conditions that induce galactose starvation. We show that galE is required to maintain the cellular levels of UDP-Gal and that the decreased UDP-Gal levels lead to the loss of galactose from glycoproteins and changes in cell morphology and metabolism.
MATERIALS AND METHODS
Cell culture.
Culture-adapted strain 427 monomorphic bloodstream form T. brucei (variant 221) genetically modified to express T7 polymerase and the tetracycline repressor protein, referred to here as the wild type, were cultured in HMI-9 medium containing 2.5 μg/ml G418 at 37°C in a 5% CO2 incubator as described by Wirtz et al. (21). Bloodstream form T. brucei conditional galE null mutant cells (galE-cKO) derived from the same cell line (17) were cultured as described above with the addition of 1 μg/ml tetracycline to ensure expression of galE. Galactose starvation was induced by washing cells three times in tetracycline-free HMI-9 prior to culturing in tetracycline-free HMI-9.
Sugar nucleotide analysis.
Approximately 5 × 107 accurately counted bloodstream form T. brucei cells were washed in phosphate-buffered saline (PBS), lysed in 300 μl of chloroform-methanol-water (2:4:1) containing 10 pmol GDP-glucose internal standard (Sigma), and centrifuged at 13,000 × g for 10 min. The supernatant was dried under a stream of nitrogen, and the products were partitioned between 200 μl water and 400 μl butan-1-ol. The aqueous phase was then reextracted twice with another 400 μl of water-saturated butan-1-ol, dried under a stream of nitrogen, and redissolved in 1 ml of 10 mM ammonium bicarbonate, and the sugar nucleotides were extracted using Envi-Carb columns (Supelco) (14). The eluant was freeze-dried overnight and stored at −80°C prior to analysis.
High-performance liquid chromatography conditions were adapted from Rabina et al. (14) as follows. The column (HiChrom C18, 1 by 250 mm) was equilibrated in 0.5% acetonitrile, 20 mM triethylammonium acetate (TEAA) buffer (pH 6.0) at 20 μl/min, and then eluted using 0.5% acetonitrile, 20 mM TEAA buffer for 20 min, followed by a linear gradient of 0.5 to 4.0% acetonitrile, 20 mM TEAA buffer over 20 min at 25 μl/min. The column was then washed for 20 min in 4% acetonitrile, 20 mM TEAA buffer prior to reequilibration. Sugar nucleotides were detected by negative ion electrospray mass spectrometry (ES-MS) using a Micromass Ultima triple quadrupole instrument and multiple-reaction monitoring of the transition from the [M-H]−1 precursor ion to a nucleotide product ion (UMP, UDP, or GDP) for each class of sugar nucleotide. Quantification of the sugar nucleotides was achieved via the GDP-glucose internal standard using empirically determined molar relative response factors for commercially available sugar nucleotide standards (Sigma).
Lectin Western blots.
Bloodstream form T. brucei organisms were hypotonically lysed in the presence of 0.1 μM 1-chloro-3-tosylamido-7-amino-2-heptone and 1 μg/ml leupeptin. Cell ghosts were centrifuged at 13,000 × g for 10 min, and high-molecular-mass glycoproteins were extracted by resuspension of the pellets in boiling 8 M urea and 2% sodium dodecyl sulfate (SDS) (1). The extracts were subjected to electrophoresis (2.5 × 107 cell equivalents/lane) on a NuPAGE bis-Tris 4 to 12% gradient acrylamide gel under reducing conditions and either stained with Coomassie blue or transferred to a nitrocellulose membrane. Membranes were probed with either a ricin-horseradish peroxidase conjugate (0.2 μg/ml; Sigma), with and without 3 mg/ml galactose to block ricin binding, or with biotinylated tomato lectin (1 μg/ml; Vector Labs), with and without 3 mg/ml chitin hydrolysate (Vector Labs) to block tomato lectin binding, followed by ExtraAvidin-horseradish peroxidase conjugate (0.1 μg/ml; Sigma). In both cases, the blots were developed by chemiluminescent detection (ECLplus; Amersham).
sVSG analysis.
Soluble-form VSG (sVSG) was isolated from ∼2 × 108 T. brucei cells by hypotonic lysis in the presence of 0.1 μM 1-chloro-3-tosylamido-7-amino-2-heptone, 1 μg/ml leupeptin, and 1 μg/ml aprotinin and purified on a DE52 anion-exchange column, as described previously (7). Samples were concentrated and diafiltered with water on a YM-10 spin concentrator (Microcon), and ∼100 μg of sVSG was recovered in a final volume of 100 μl water. Half of each sample was subjected to digestion with 2 μl of 1 mg/ml pronase (Sigma) in 100 mM ammonium bicarbonate and 5 mM calcium acetate for 3 h at 37°C. Digests were acidified with an equal volume of 0.2 M acetic acid and centrifuged for 1 min at 13,000 × g, and the supernatant was passed though a mixed-bed column consisting of 50 μl Chelex 100 (Na+) over 100 μl of Dowex AG50 (H+). The column was washed four times with 0.1 M acetic acid (150 μl), and the flowthrough and washes were combined and lyophilized.
Intact and pronase-digested sVSG were diluted to ∼0.05 μg/μl in 50% methanol, 1% formic acid, loaded into Micromass type-F nanotips, and analyzed by positive-ion ES-MS using an Applied Biosystems Q-StarXL instrument. The intact sVSG masses were calculated using the Bayesian protein reconstruction algorithm (ABI Analyst software). The identities of pronase glycopeptide ions were confirmed by ES-MS/MS (5).
Quantification of VSG copy number.
Bloodstream form T. brucei organisms were hypotonically lysed in the presence of 0.1 μM 1-chloro-3-tosylamido-7-amino-2-heptone and 1 μg/ml leupeptin, subjected to electrophoresis (2.5 × 105 cell equivalents/lane) on a 10% acrylamide gel under reducing conditions, and stained with Coomassie blue. The ratio of VSG (55 kDa) to β-tubulin (49 kDa) was determined by laser densitometry scanning.
Complement sensitivity.
Human serum was obtained by clotting blood in a glass vial at 37°C for 1 h, spinning at 800 × g for 10 min, and storing the supernatant at −80°C until use. Bloodstream and procyclic form T. brucei cells were washed twice in trypanosome dilution buffer (TDB) (5 mM KCl, 80 mM NaCl, 1 mM MgSO4, 20 mM Na2HPO4, 2 mM NaH2PO4, 20 mM glucose, pH 7.4) and resuspended in TDB supplemented with 1 mM CaCl2, 1 mM MgCl2, and 1 mM MnCl2 at 2 × 107 cells/ml. The complement sensitivity of galE-cKO cells with or without Tet was assessed by incubating cells with 50% serum at 37°C and recording the number of viable cells at hourly intervals, using procyclic and wild-type (WT) bloodstream form cells as positive and negative controls, respectively.
Dihydrolipoamide dehydrogenase assay.
The activity of dihydrolipoamide dehydrogenase was determined from the initial rate of oxidation of dihydrolipoamide to lipoic acid by monitoring NADH formation at 340 nm as described by Breidbach et al. (2). Stumpy cells were obtained by incubating wild-type bloodstream form T. brucei cells with 1 mM 8-(4-chlorophenylthio)-cyclic AMP for 72 h to induce stumpy formation (2).
Fluorescence microscopy.
T. brucei cell cultures were harvested by centrifugation at 8,000 × g for 10 min, washed twice in ice-cold TDB, and resuspended in TDB at 2 × 107 cells/ml. The parasites were air dried on coverslips (13 mm round) for 15 min and then fixed in 4% paraformaldehyde in trypanosome dilution buffer for 1 h at room temperature. The fixed cells were washed three times in PBS, incubated with PBS plus 0.5% bovine serum albumin (PBA) for 1 h to block nonspecific binding, and incubated with biotinylated tomato lectin (5 μg/ml; Vector labs) in PBA for 1 h. The cells were washed three times in PBA, incubated with streptavidin-fluorescein isothiocyanate (5 μg/ml; Vector labs), and 4′,6′-diamidino-2-phenylindole (DAPI) (2 μg/ml) for 1 h in the dark. The coverslips were washed a further three times in PBA before mounting on slides in Hydromount. Data were collected on a Zeiss Axiovert 200 M microscope using a 100× oil lens.
Scanning electron microscopy.
T. brucei cell cultures were fixed directly in HMI-9 medium supplemented with 2.5% glutaraldehyde for 30 min at room temperature before collection on 1-μm membrane filters (Nuclepore). Filters were washed twice in 0.1 M sodium cacodylate buffer (pH 7.2) for 30 min, followed by incubation in 1% aqueous osmium tetroxide for 1 h. The filters were washed twice in distilled water for 15 min, dehydrated through a graded ethanol series, and dried in a Bal-Tec 030 critical point drier. Specimens were mounted on stubs and coated with 20-nm gold/palladium using a Cressington 208HR sputter coater. Samples were examined using a Philips XL30 FEG environmental scanning electron microscope operating at an accelerating voltage of 15 kV.
Transmission electron microscopy.
T. brucei cell cultures were harvested by centrifugation at 8,000 × g for 10 min and fixed in 1 pellet volume of 4% paraformaldehyde in 0.1 M piperazine-N,N′-bis(2-ethanesulfonic acid) for 30 min at room temperature. Fixed cells were spun at 13,000 × g for 10 min, the supernatant was removed, and the pellet was resuspended in 1% aqueous osmium tetroxide (to fix and stain lipids), dehydrated, and set in Durcupan epoxy resin (Sigma). Sections were cut using a Leica Ultracut UCT system and analyzed using a Philips Tecnai 12 transmission electron microscopy instrument.
RESULTS
GalE is essential to maintain cellular UDP-galactose levels.
In our previous study demonstrating the essentiality of galE to bloodstream form T. brucei, we generated a galE conditional knockout (cKO) cell line containing a tetracycline-inducible ectopic copy of galE and both allelic copies replaced with drug resistance genes (17). In the presence of tetracycline (+Tet) the galE-cKO cells express sufficient GalE to grow normally, but withdrawal of tetracycline from the culture (−Tet) results in cessation of cell division after 3 to 4 days, followed by cell death (Fig. 1A). Northern blot analysis showed that galE mRNA was more abundant in the galE-cKO +Tet than in wild-type cells but became undetectable within 8 h of tetracycline removal (17).
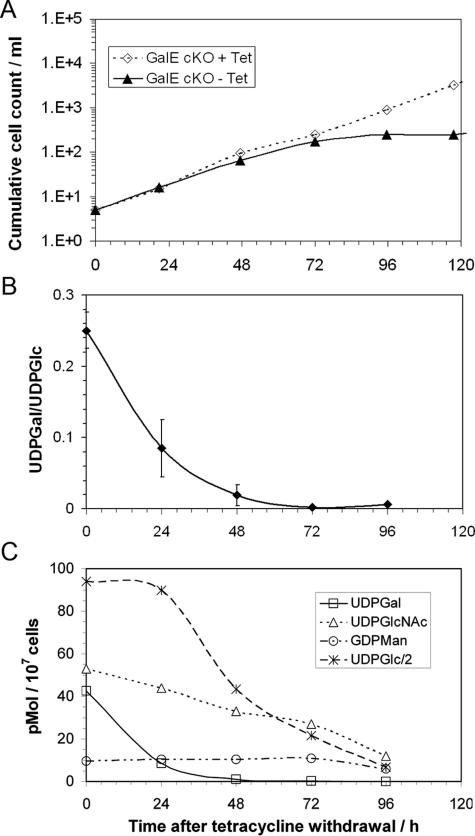
FIG. 1.
Effect of galactose starvation on cell growth and cellular sugar nucleotide content. (A) Growth of galE-cKO with and without Tet; (B) ratio of UDP-Gal/UDP-Glc in the galE-cKO −Tet; (C) sugar nucleotide levels of the galE-cKO −Tet cells.
Analysis of the cellular sugar nucleotide levels by liquid chromatography-MS showed that the galE-cKO +Tet cells contained quantities of the major cellular sugar nucleotides UDP-GlcNAc, GDP-Man, and UDP-Glc similar to those of wild-type cells but contained a slightly lower amount of UDP-Gal, reducing the UPD-Gal/UDP-Glc ratio from 0.30 ± 0.03 in wild-type cells to 0.25 ± 0.025 in galE-cKO +Tet cells. Upon removal of tetracycline from the culture, the level of UDP-Gal of the galE-cKO dropped substantially (80%) within 24 h, while the level of UDP-Glc was unaffected, leading to a rapid decrease in the UDP-Gal/UDP-Glc ratio (Fig. 1B). The lack of increase in the cellular level of UDP-Glc in response to the decreasing UDP-Gal level indicates that the cellular level of UDP-Glc is regulated by an independent mechanism. Subsequently, the quantity of UDP-Glc dropped, decreasing by ∼50% after 48 h (Fig. 1C), perhaps indicating general metabolic stress. The level of UDP-Gal became too low to be accurately detected after 72 h, concurrent with cessation of cell division and subsequent cell death. The other major cellular sugar nucleotides UDP-GlcNAc and GDP-Man also decreased after 72 h, probably reflecting the decreasing health of the cells undergoing galactose starvation.
Galactose starvation reduces the binding of lectins specific to galactosylated glycoproteins.
To determine the effect of galactose starvation on the level of galactose present in glycoproteins, the galE-cKO cell line was grown in the presence or absence of tetracycline for 72 h and the glycoproteins were extracted and analyzed. High-molecular-weight and membrane-bound glycoproteins were solubilized in 8 M urea and 2% SDS (1). The extracts were either stained with Coomassie blue to confirm equal protein loading (Fig. 2A) or subjected to Western blotting with lectins (Fig. 2 B to E). Blots were probed either with ricin to detect the presence of terminal β-d-galactose or with tomato lectin to detect the presence of galactose contained in linear poly-N-acetyllactosamine structures.
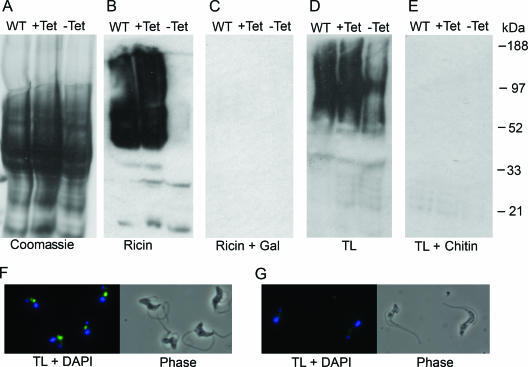
FIG. 2.
Effect of galactose starvation on lectin binding of glycoproteins. Detergent- and urea-solubilized glycoproteins from wild-type (WT), galE-cKO +Tet (+Tet), and galE-cKO −Tet for 72 h (−Tet) cells. (A) Coomassie blue-stained SDS-polyacrylamide gel electrophoresis, (B) ricin Western blot, (C) ricin Western blot blocked with 3 mg/ml galactose (Gal), (D) tomato lectin (TL) Western blot, (E) tomato lectin Western blot blocked with 3 mg/ml chitin hydrolysate, (F) tomato lectin immunofluorescence microscopy of WT, (G) tomato lectin immunofluorescence microscopy of galE-cKO −Tet for 96 h. Tomato lectin is shown in green; DAPI is shown in blue. Note, the inclusion of 8 M urea to effect the complete extraction of high-molecular-weight glycoproteins and the loading of sufficient material to see residual ricin binding in the −Tet sample resulted in some distortion in the gels shown in panels A to E.
Ricin detected a range of glycoproteins in wild-type or galE-cKO +Tet cells with apparent molecular masses of 50 to 180 kDa, whereas virtually no binding was seen with galE-cKO −Tet cells (Fig. 2B). All ricin binding was abolished by coincubation with galactose (Fig. 2C), confirming the specificity of binding. The majority of ricin-binding glycans are N-linked structures attached to glycoprotein components of the flagellar pocket and lysosomal/endosomal system (1). Galactose starvation clearly reduces the incorporation of galactose into all of the extracted glycoproteins such that none of them terminate with β-galactosidase (β-Gal).
Tomato lectin detected a range of glycoproteins in wild-type or galE-cKO +Tet cells with apparent molecular masses similar to those detected by ricin (Fig. 2D). The intensity of tomato lectin binding was significantly reduced in galE-cKO −Tet cells (Fig. 2D), although some binding still occurred, but was completely abolished for all cell types by coincubation with chitin hydrolysate (Fig. 2E), confirming the specificity of binding.
Immunofluorescence microscopy of wild-type cells with tomato lectin (Fig. 2F) demonstrated that binding was confined to components of the flagellar pocket and lysosomal/endosomal system, as previously described (13); however, tomato lectin binding was negligible in galE-cKO cells after 96 h without tetracycline (Fig. 2G), demonstrating significantly reduced levels of poly-N-acetyllactosamine structures in these cells.
The tomato lectin blots and immunofluorescence microscopy indicate that there is substantial turnover of the poly-N-acetyllactosamine-containing glycoproteins within 72 h of tetracycline withdrawal, and the ricin blots indicate that the galactosylation step becomes rate-limiting in poly-N-acetyllactosamine assembly such that no β-Gal termini can be detected.
Galactose starvation eliminates galactose from sVSG GPI anchors.
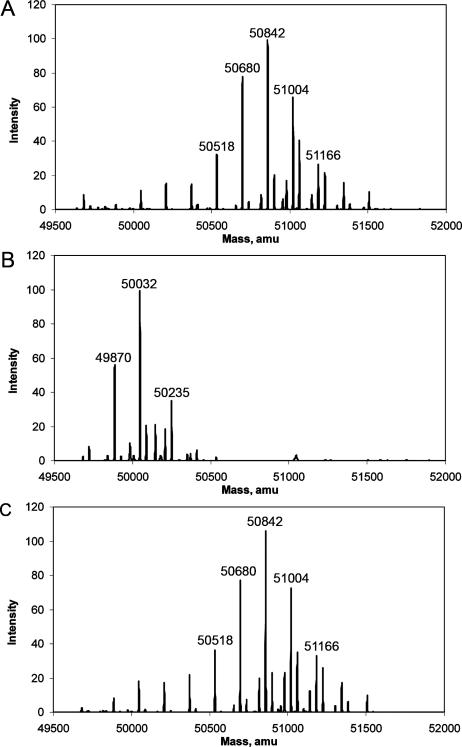
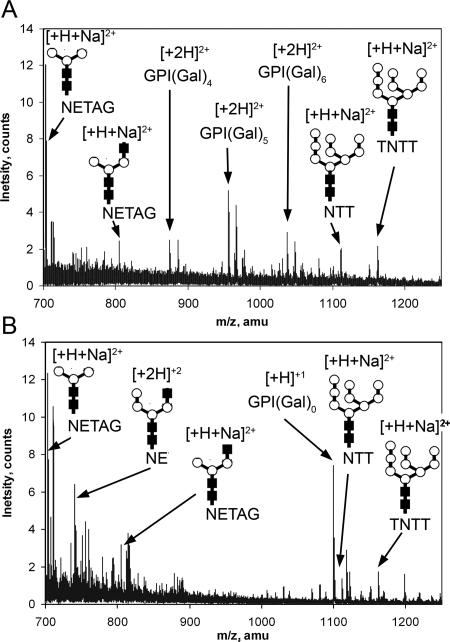
The effect of galactose starvation on the galactosylation of VSG221 glycosylphosphatidylinositol (GPI) anchors was also examined. sVSG variant 221 was isolated from the wild type and galE-cKO incubated with or without Tet for 72 h. Analysis of sVSG by positive-ion ES-MS showed that a shift to lower-molecular-weight species had occurred in the sVSG from galE-cKO −Tet compared to wild-type cells and galE-cKO +Tet (Fig. 3A to C). The wild-type sVSG profile (Fig. 3A) shows the range of different glycoforms that arise from known heterogeneity in the GPI anchor (10) and the N-glycan sites (23). The most abundant form of sVSG221 contains a C-terminal N-glycan of Man9GlcNAc2, an internal N-glycan of Man3GlcNAc2, and a GPI anchor of composition ethanolamine phosphate-(Gal5)Man3GlcN-PI (7). The sVSG from the galE-cKO −Tet (Fig. 3B) exhibits less heterogeneity, with the major species having lost mass equivalent to five hexose residues (Table 1). The sVSG isolated from galE-cKO +Tet shows a distribution of glycoforms identical to the wild-type profile (Fig. 3A and C). Aliquots of sVSG from wild-type and galE-cKO −Tet cells were digested with pronase and analyzed by positive-ion ES-MS, and the identities of putative glycopeptide ions were confirmed by analysis of the fragmentation pattern produced in ES-MS/MS product ion scans. Pronase-digested sVSG from wild-type cells contained characteristic glycopeptide fragments from the C-terminal N-glycosylation site (Asn486), the internal N-glycosylation site (Asn263), and the GPI anchor attached to Ser491 (Fig. 4A). The GPI anchor ions, observed as doubly charged species with and without a sodium adduct, contain four to six galactose residues embellishing the core GPI structure (8). Pronase-digested sVSG from galE-cKO −Tet contained the same C-terminal and internal N-glycans, but only a single GPI anchor fragment was observed, corresponding to a singly charged GPI anchor ion completely free of galactose (Fig. 4B). These data showed that galactose starvation prevented the embellishment of the GPI anchor with additional galactose residues.
FIG. 3.
Mass spectrometric analysis of intact sVSG. (A) Wild-type cell sVSG; (B) galE-cKO −Tet for 72 h sVSG; (C) galE-cKO +Tet sVSG. Masses were calculated using a Bayesian protein reconstruction algorithm.
TABLE 1.
Isobaric forms of sVSG detected by mass spectrometrya
| Mol wt | sVSG composition
|
Abundance in cell lined:
|
||||||
|---|---|---|---|---|---|---|---|---|
| Proteinb | GlcN-Ino (cP)c | EtNPc | GlcNAc | Man+Gal | WT |
galE-cKO
|
||
| −Tet | +Tet | |||||||
| 49,708 | 1 | 1 | 1 | 4 | 13 | − | ++ | + |
| 49,870 | 1 | 1 | 1 | 4 | 14 | − | ++++ | − |
| 50,032 | 1 | 1 | 1 | 4 | 15 | + | +++++ | ++ |
| 50,073 | 1 | 1 | 1 | 5 | 14 | − | +++ | − |
| 50,194 | 1 | 1 | 1 | 4 | 16 | ++ | ++ | ++ |
| 50,235 | 1 | 1 | 1 | 5 | 15 | − | ++++ | − |
| 50,356 | 1 | 1 | 1 | 4 | 17 | ++ | + | ++ |
| 50,397 | 1 | 1 | 1 | 5 | 16 | − | ++ | − |
| 50,518 | 1 | 1 | 1 | 4 | 18 | +++ | + | +++ |
| 50,680 | 1 | 1 | 1 | 4 | 19 | ++++ | − | ++++ |
| 50,842 | 1 | 1 | 1 | 4 | 20 | +++++ | − | +++++ |
| 50,883 | 1 | 1 | 1 | 5 | 19 | ++ | − | ++ |
| 51,004 | 1 | 1 | 1 | 4 | 21 | ++++ | − | ++++ |
| 51,045 | 1 | 1 | 1 | 5 | 20 | +++ | − | +++ |
| 51,166 | 1 | 1 | 1 | 4 | 22 | +++ | − | +++ |
| 51,207 | 1 | 1 | 1 | 5 | 21 | ++ | − | ++ |
| 51,328 | 1 | 1 | 1 | 4 | 23 | ++ | − | ++ |
| 51,490 | 1 | 1 | 1 | 4 | 24 | + | − | + |
Intact sVSG was isolated from WT and galE-cKO cells grown in the presence (+Tet) or absence (−Tet) of tetracycline for 72 h.
The protein molecular weight of 46,284 is based on the VSG221 amino acid sequence with four disulfide bonds minus residues 1 to 27 (signal peptide) and residues 460 to 476 (GPI attachment signal peptide) (7).
GPI components common to all glycoforms. GlcN-Ino(cP), glucosamine-α1-6-myo-ionositol-1,2-cyclic phosphate; EtNP, ethanolamine phosphate.
The relative abundance of each isobaric form (Fig. 3) is scored from − (absent) to +++++ (most abundant).
FIG. 4.
Mass spectrometric analysis of pronase-digested sVSG. (A) Wild type; (B) galE-cKO −Tet for 72 h. The identities of the indicated glycopeptide ions were confirmed by ES-MS/MS. The m/z values of the parent ions and the product ion spectra are consistent with the following assignments: m/z 703.13 [Man3GlcNAc2-NETAG + H + Na]2+; m/z 729.19 [GlcNAcMan3GlcNAc2-NE + 2H]2+; m/z 804.74 [GlcNAcMan3GlcNAc2-NETAG + H + Na]2+; m/z 874.55 [GPI(Gal)4 + 2H]2+; m/z 955.55 [GPI(Gal)5 + 2H]2+; m/z 1,036.65 [GPI(Gal)6 + 2H]2+; m/z 1,100.10 [GPI(Gal)0 + H]+; m/z 1,111.49 [Man9GlcNAc2-NTT + H + Na]2+; m/z 1,162.05 [Man9GlcNAc2-TNTT + H + Na]2+, where GPI represents serine-ethanolaminephosphate-6Manα1-2Manα1-6Manα1-4GlcNα1-6-d-myo-inositol-1,2-cyclic phosphate and NETAG, NE, NTT, and TNTT are single-letter amino acid sequences for the internal and C-terminal N-glycosylation sites, respectively. In the glycopeptide cartoon structures, closed boxes represent GlcNAc residues and the open circles represent Man residues.
To determine if the loss of galactose from the VSG221 GPI anchor had any effect on the VSG coat, the complement sensitivity and VSG copy number of galE-cKO cells grown without Tet for 72 h were examined. The VSG copy number did not change significantly compared to galE-cKO +Tet or wild-type cells, as judged by laser densitometry scanning, and neither did the cell's sensitivity to lysis by human serum (Table 2), indicating that the change in GPI anchor had minimal effect on the integrity of the VSG coat.
TABLE 2.
T. brucei cell lysis in human serum
| Time (min) | % Cell lysis of cell typea:
|
|||
|---|---|---|---|---|
| WT | Procyclic | +Tet | −Tet | |
| 80 | <5 | 100 | <5 | <5 |
| 140 | 50 | 100 | 50 | 50 |
| 230 | >95 | 100 | >95 | >95 |
Incubation with 50% human serum.
Galactose starvation affects cellular morphology.
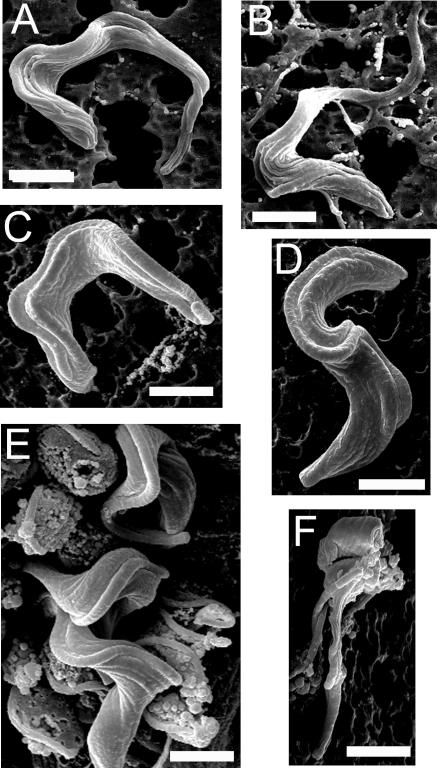
Scanning electron microscopy was used to examine the morphology the galE-cKO cells upon induction of galactose starvation. Culture-adapted wild-type T. brucei bloodstream form cells proliferate in a slender morphology, with an elongated body ∼20 μm long, flanked by a long flagellum joined to the body for almost its entire length. Pleomorphic wild-type T. brucei bloodstream form cells are able to differentiate into a nonproliferative stumpy form at high cell density (>2 × 106 cells/ml), with a shorter, wider body ∼10 μm long, flanked by a shortened flagellum (9, 15). However, such differentiation is not normally observed in culture-adapted monomorphic strains (2). The galE-cKO +Tet cells appear identical to slender wild-type cells (Fig. 5A) and appear unchanged after 48 h of galactose starvation (Fig. 5B), despite having cellular UDP-Gal levels reduced by >95%. After 96 h of galactose starvation, the morphology of the galE-cKO cells changed to a stumpy-like form (Fig. 5C and D), concurrent with cessation of cell growth, despite being maintained at low culture density (<1 × 106 cells/ml). After 144 h of galactose starvation, a large proportion of cells displayed gross morphological defects, deterioration of the cell surface, and lysis (Fig. 5E and F).
FIG. 5.
Scanning electron microscopy of galE-cKO −Tet. Effects on cellular morphology after galactose starvation for 0 h (A), 48 h (B), 96 h (C and D), and 144 h (E and F) are shown. Scale bars, 2 μm.
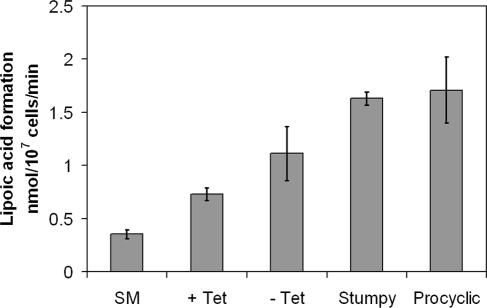
To further characterize the observed stumpy-like cells, we examined the activity of dihydrolipoamide dehydrogenase (DHLADH), the enzyme believed to be responsible for NADH diaphorase activity, a classical marker for differentiation to stumpy morphology (20). The DHLADH activity of galE-cKO +Tet cells was higher than that of wild-type cells and increased in the galE-cKO cells grown without Tet for 96 h, approaching the DHLADH activity seen for cyclic AMP (cAMP)-induced stumpy or procyclic cells (Fig. 6). The increased DHLADH activity of galE-cKO −Tet cells, in conjunction with the observed change in cell morphology and cessation of cell proliferation, suggests that the cells are undergoing galactose starvation-induced differentiation to a stumpy-like form. The increased DHLADH activity of the galE-cKO +Tet cells may result from the reduced cellular level of UDP-Gal of the conditional null mutant compared to the wild-type cells.
FIG. 6.
The activity of dihydrolipoamide dehydrogenase in cellular extracts. Wild type (WT), galE-cKO +Tet, and galE-cKO −Tet for 72 h compared with cAMP-induced stumpy and procyclic T. brucei.
Galactose starvation affects subcellular architecture.
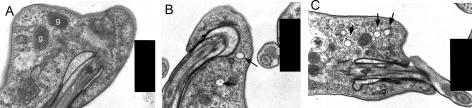
Transmission electron microscopy was used to investigate the subcellular architecture of galE-cKO cells upon galactose starvation. After 96 h of galactose starvation, the galE-cKO −Tet cells still have the same electron-dense cell surface coat as found on wild-type cells (Fig. 7), but a number of vesicles (∼120 nm diameter) consistently appear adjacent to the flagellar pocket.
FIG. 7.
Transmission electron microscopy of galE-cKO −Tet. Wild-type cells (A) and galE-cKO after 96 h of galactose starvation (B and C) are shown. The flagellum (f), flagellar pocket (fp), and glycosomes (g) are indicated in white type. Aberrant subcellular compartments are indicated with black arrows. Scale bars, 1 μm.
DISCUSSION
We previously demonstrated that galE is an essential gene for bloodstream form T. brucei (17). In this paper, we characterize the phenotype of bloodstream form T. brucei conditional galE null mutant cells undergoing galactose starvation. Withdrawal of tetracycline from galE-cKO for 24 h results in a substantial decrease (80%) in cellular UDP-Gal levels, with little or no effect on other sugar nucleotides. However, after 48 h, there is also a decrease in UDP-Glc levels and UDP-GlcNAc levels, perhaps indicating general metabolic stress. The levels of UDP-sugars continue to fall over time, with UDP-Gal levels becoming undetectable by 72 h. By this time, profound changes in parasite protein glycosylation are apparent. (i) The VSG GPI anchor, which normally contains an average of five side chain galactose residues, is essentially galactose free (though the mannose-, N-acetylglucosamine- and glucosamine-containing N-linked glycan and GPI anchor core structures are unchanged). (ii) The tomato lectin- and ricin-binding (i.e., poly-N-acetyllactosamine- and terminal β-Gal-containing) carbohydrate structures that normally fill the flagellar pocket and lysosomal/endosomal compartments are substantially reduced. Assuming that UDP-Gal is essentially unavailable after 24 h without tetracycline (Fig. 1), the lack of detectable galactose in the VSG GPI anchor after 72 h is broadly consistent with the combined effects of VSG turnover (half-life = 32 h) and the dilution effect of a 10-fold increase in cell numbers between 24 and 72 h, i.e., one would expect normally galactosylated VSG to represent maximally about 3% of the VSG glycoforms at the 72-h time point. The reduction in ricin and tomato lectin binding to the poly-N-acetyllactosamine-containing glycoproteins after 72 h appears to be >10-fold (Fig. 2B and D), suggesting that these glycoproteins are also undergoing turnover. However, there are insufficient data to estimate their turnover rates or to assess whether this is affected by galactose starvation.
After 72 h without tetracycline, cell division slows significantly, ceasing by 96 h, at which time the cells adopt a stumpy-like morphology and aberrant vesicular structures appear in the vicinity of the flagellar pocket. Differentiation to the stumpy form usually occurs in pleomorphic cell lines at high cell densities, thought to be triggered by an unidentified parasite-derived signal, but is not normally observed in momomorphic culture-adapted strains unless stimulated with cell-permeable cAMP analogues (2, 9, 15). In mammalian hosts, differentiation of the slender to the stumpy form is thought to prolong survival of the host and prepare the cells for synchronous differentiation into the procyclic form upon transmission to the tsetse fly (9). Slender-to-stumpy differentiation is accompanied by morphological, ultrastructural, and metabolic changes, including the elaboration of mitochondrial activity to provide an alternative energy source to glycolysis. We observed a significant increase in dihydrolipoamide dehydrogenase activity in the galactose-starved parasites, suggesting a stumpy-like elaboration of mitochondrial activity in addition to stumpy-like morphological changes and the cessation of cell division in these cells. However, another well-documented change during slender-to-stumpy differentiation is a significant increase in the level of a carbohydrate epitope recognized by the CB1 antibody (3). This antibody, like tomato lectin, recognizes the novel poly-N-acetyllactosamine-containing N-linked glycans found throughout the lysosomal/endosomal system (1). The synthesis of these Galβ1-4GlcNAc repeating structures is clearly prevented under galactose starvation. The appearance of aberrant vesicles close to the flagellar pocket and, ultimately, cell death may reflect this inability to appropriately glycosylate glycoproteins in the endocytic/exocytic recycling pathway, where protein glycosylation has been postulated to play a role (13). Thus, it is conceivable that the observed change to stumpy-like morphology is an attempt to escape metabolic starvation caused by the malfunctioning of its endosomal/lysomal system. Conversely, galactose starvation may trigger differentiation by some other mechanism and the inability of the cells to complete that program, because of their inability to synthesize poly-N-acetyllactosamine-containing N-linked glycans, may cause their ultimate demise.
We have suggested in the past, based on molecular modeling, that the galactose side chains attached to VSG GPI anchors might play a significant role in completing the macromolecular diffusion barriers characteristics of the surface coat (6). This does not appear to be the case, since there was no increase in complement-mediated lysis in trypanosomes that had lost all GPI galactose. Thus, although these galactose side chains are specific to T. brucei VSGs (11), their function(s) remains unknown.
The effects of galactose starvation in procyclic form T. brucei are somewhat different (16) than those reported here for bloodstream form T. brucei. In the procyclic form, galactose is found principally in the poly-N-acetyllactosamine-containing GPI anchor side chains of the predominant cell surface molecule, procyclin, and, to a lesser extent, on free GPI glycolipids (12, 19). Galactose starvation, induced in the same way with a galE conditional null mutant, caused the loss of the GPI-linked poly-N-acetyllactosamine side chains and resulted in a 10-fold increase in procyclin and the cessation of cell growth. However, although some cell death occurred, the effects were mainly cytostatic. Unlike bloodstream form T. brucei, the procyclic form parasites exhibited a partial phenotype (reduced galactosylation of procyclin) when one of the two galE alleles was replaced. Similarly, when one of the two galE alleles was replaced in Trypanosoma cruzi epimastigotes, the cells also exhibited a profound phenotype, losing their galactopyranose-containing GPI-anchored mucins (but maintaining their galactofuranose-containing free GPI glycolipids) and exhibiting cell agglutination and morphological defects (8). Taken together, these studies and the data reported here illustrate the significance of galactose metabolism to the physiology of T. brucei and T. cruzi.
In summary, we have demonstrated that galE is required to maintain the cellular level of UDP-Gal in bloodstream form T. brucei and that galactose starvation occurring in the absence of galE results in elimination of galactose from VSG and from the poly-N-acetyllactosamine-containing glycoproteins of the cell. This leads to the concomitant cessation of cell growth and differentiation to a stumpy-like form and, ultimately, cell death. The results demonstrate that galactosylation of one or more glycoprotein(s), most likely in the endosomal/lysosomal system, is essential for the survival of bloodstream form T. brucei and that galE-encoded epimerase and the downstream UDP-Gal transporters and UDP-Gal-dependent glycosyltransferases may be exploitable as drug targets.
Acknowledgments
We thank John James and Martin Kierans of the Centre for High Resolution Imaging and Processing at the University of Dundee for conducting the transmission and scanning electron microscopy, respectively, and M. Lucia Güther for provision of procyclic form T. brucei cells and helpful discussions.
This work was supported by a Programme grant from the Wellcome Trust (71463).
REFERENCES
- 1.Atrih, A., J. M. Richardson, A. R. Prescott, and M. A. J. Ferguson. 2005. Trypanosoma brucei glycoproteins contain novel giant poly-N-acetyllactoseamine carbohydrate chains. J. Biol. Chem. 280:865-871. [DOI] [PubMed] [Google Scholar]
- 2.Breidbach, T., E. Ngazoa, and D. Steverding. 2002. Trypansoma brucei: in vitro slender-to-stumpy differentiation of culture-adapted, monomorphic bloodstream forms. Exp. Parasitol. 101:223-230. [DOI] [PubMed] [Google Scholar]
- 3.Brickman, M. J., and A. E. Baber. 1990. Trypanosoma brucei rhodesianse bloodstream forms: surface ricin-binding glycoproteins are localised exclusively in the flagellar pocket and the flagellar adhesion zone. J. Protozool. 37:219-224. [DOI] [PubMed] [Google Scholar]
- 4.Cross, G. A. M. 1996. Antigenic variation in trypanosomes: secrets surface slowly. Bioessays 18:283-291. [DOI] [PubMed] [Google Scholar]
- 5.Eisenthal, R., S. Game, and G. D. Holman. 1989. Specificity and kinetics of hexose-transport in Trypanosoma brucei. Biochim. Biophys. Acta 985:81-89. [DOI] [PubMed] [Google Scholar]
- 6.Homans, S. W., and M. A. J. Ferguson. 1989. Solution structure of the glycosylphosphatidylinositol membrane anchor glycan of trypanosoma-brucei variant surface glycoprotein. Biochemistry 28:2881-2887. [DOI] [PubMed] [Google Scholar]
- 7.Jones, D. C., A. Mehlert, M. L. S. Guther, and M. A. J. Ferguson. 2005. Deletion of the glucosidase II gene in Trypanosoma brucei reveals novel N-glycosylation mechanisms in the biosynthesis of variant surface glycoprotein. J. Biol. Chem. 280:35929-35942. [DOI] [PubMed] [Google Scholar]
- 8.MacRae, J. I., S. O. Obado, D. C. Turnock, J. R. Roper, M. Kierans, J. M. Kelly, and M. A. J. Ferguson. 2006. The suppression of galactose metabolism in Trypanosoma cruzi epimastigotes causes changes in cell surface molecular architecture and cell morphology. Mol. Biochem. Parasitol. 147:126-136. [DOI] [PubMed] [Google Scholar]
- 9.Mathews, K. R. 2005. The developmental cell biology of Trypanosoma brucei. J. Cell Sci. 118:283-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mehlert, A., J. M. Richardson, and M. A. J. Ferguson. 1998. Structure of the glycosylphosphatidylinositol membrane anchor glycan of class-2 variant surface glycoprotein from Trypanosoma brucei. J. Mol. Biol. 277:379-392. [DOI] [PubMed] [Google Scholar]
- 11.Mehlert, A., N. Zitzmann, J. M. Richardson, A. Traumann, and M. A. J. Ferguson. 1998. The glycosylation of the variant surface glycoprotein and procyclic acidic repetitive proteins of Trypanosoma brucei. Mol. Biochem. Parasitol. 91:145-152. [DOI] [PubMed] [Google Scholar]
- 12.Nagamune, K., A. Acosta-Serrano, H. Uemura, R. Brun, C. Kunz-Renggli, Y. Maeda, M. A. J. Ferguson, and T. Kinoshita. 2004. Surface sialic acids taken from the host allow trypanosome survival in Tsetse fly vectors. J. Exp. Med. 199:1445-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nolan, D. P., M. Geuskens, and E. Pays. 1999. N-linked glycans containing linear poly-N-acetyllactosamine as sorting signals in endocytosis in Trypanosoma brucei. Curr. Biol. 9:1169-1172. [DOI] [PubMed] [Google Scholar]
- 14.Rabina, J., M. Maki, E. M. Savilahti, N. Jarvinen, L. Penttila, and R. Rekonen. 2002. Analysis of nucleotide sugars from cell lysate by ion-pair solid phase extraction and reverse phase HPLC. Glycoconj. J. 18:799-805. [DOI] [PubMed] [Google Scholar]
- 15.Reuner, B., E. Vassella, B. Yutzy, and M. Boshart. 1997. Cell density triggers slender to stumpy differentiation of Trypanosoma brucei bloodstream forms in culture. Mol. Biochem. Parasitol. 90:269-280. [DOI] [PubMed] [Google Scholar]
- 16.Roper, J. R., M. L. S. Guther, J. I. MacRae, A. R. Prescott, I. Hallyburton, A. Acosta-Serrano, and M. A. J. Ferguson. 2005. The suppression of galactose metabolism in procyclic form Trypanosoma brucei causes cessation of cell growth and alters procyclin glycoprotein structure and copy number. J. Biol. Chem. 280:19728-19736. [DOI] [PubMed] [Google Scholar]
- 17.Roper, J. R., M. L. S. Guther, K. G. Milne, and M. A. J. Ferguson. 2002. Galactose metabolism is essential for the African sleeping sickness parasite Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 99:5884-5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tetaud, E., M. P. Barrett, F. Bringaud, and T. Baltz. 1997. Kinetoplastid glucose transporters. Biochem. J. 325:569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Treumann, A., N. Zitzmann, A. Hulsmeier, A. R. Prescott, A. Almond, J. Sheehan, and M. A. J. Ferguson. 1997. Structural characterisation of two forms of procyclic acidic repetitive protein expressed by procyclic forms of Trypanosoma brucei. J. Mol. Biol. 269:529-549. [DOI] [PubMed] [Google Scholar]
- 20.Tyler, K. M., K. R. Matthews, and K. Gull. 1997. The bloodstream differentiation-division of Trypanosoma brucei studied using mitochondrial markers. Proc. R. Soc. Lond. B. 264:1481-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wirtz, E., S. Leal, C. Ochatt, and G. A. M. Cross. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89-101. [DOI] [PubMed] [Google Scholar]
- 22.World Health Organization. 2001. African trypanosomiasis, fact sheet number 259 ed. WHO Publications, Geneva, Switzerland.
- 23.Zamze, S. E., D. A. Ashford, E. W. Wooten, T. W. Rademacher, and R. A. Dwek. 1991. Structural characterization of the asparagine-linked oligosaccharides from Trypanosoma brucei type II and type III variant surface glycoproteins. J. Biol. Chem. 266:20244-20261. [PubMed] [Google Scholar]