Abstract
In the yeast Saccharomyces cerevisiae, the Snf1 protein kinase of the Snf1/AMP-activated protein kinase (AMPK) family regulates a wide range of responses to stress caused by glucose deprivation. The stress signal is relayed via upregulation of Snf1, which depends on phosphorylation of its activation loop Thr210 residue by upstream kinases. Although Snf1 is also required for coping with various stresses unrelated to glucose deprivation, some evidence suggests a role for low-level basal activity of unphosphorylated Snf1, rather than a specific signaling function. We previously found that Snf1 is required for diploid pseudohyphal differentiation, a developmental response to nitrogen limitation. Here, we present evidence that Snf1 is directly involved in nitrogen signaling. First, genetic analyses suggest that pseudohyphal differentiation depends on the stimulatory phosphorylation of Snf1 at Thr210. Second, immunochemical data indicate that nitrogen limitation improves Thr210 phosphorylation. Analyses of pseudohyphal differentiation in cells with catalytically inactive and hyperactive Snf1 support the role of Snf1 activity. Finally, we show that Snf1 is negatively regulated by the rapamycin-sensitive TOR kinase which plays essential roles in signaling nitrogen and amino acid availability. This and other evidence implicate Snf1 in the integration of signals regarding nitrogen and carbon stress. TOR and Snf1/AMPK are highly conserved in evolution, and their novel functional interaction in yeast suggests similar mechanisms in other eukaryotes.
The Snf1/AMP-activated protein kinase (AMPK) family is highly conserved in eukaryotes, and its members are involved in effecting responses to cellular stress. In mammalian cells, AMPK is activated by increased AMP:ATP ratios and controls responses to stimuli that affect the cellular energy supply. Evidence implicates the AMPK pathway in type 2 diabetes, obesity, cardiac disorders, and tumorigenesis (for reviews, see references 6, 18, 19, and 32). In the yeast Saccharomyces cerevisiae, the Snf1 protein kinase is required for multiple aspects of transcriptional and metabolic adaptation to reduced levels of available glucose, the preferred source of carbon and energy (7, 15). Snf1 is not simply required for growth on alternative carbon sources but plays a direct role in glucose signaling, as its function is regulated by glucose availability. Maximal catalytic activation of Snf1 requires phosphorylation of its conserved activation loop Thr210 residue (14) by upstream kinases, and cellular levels of phospho-Thr210-Snf1 increase dramatically upon glucose deprivation (44). Three Snf1 protein kinase kinases, Sak1 (Pak1), Tos3, and Elm1, have been identified and are related to the mammalian tumor suppressor kinase LKB1 and Ca2+/calmodulin-dependent protein kinase kinases, which activate AMPK by phosphorylation of the cognate Thr172 residue (21, 22, 25, 26, 28, 45, 46, 64, 74). Dephosphorylation and downregulation of Snf1 depend on type 1 protein phosphatase Glc7 in association with its specific targeting protein, Reg1 (44, 66, 67). The exact mechanisms by which glucose modulates the levels of Thr210 phosphorylation remain unclear.
The Snf1 protein kinase functions as a heterotrimeric complex containing the catalytic α subunit Snf1, the stimulatory γ subunit Snf4, and one of three alternative β subunits, Sip1, Sip2, or Gal83, which define three forms of the Snf1 complex (30, 75). All three forms of the complex are catalytically activated on limiting glucose and perform overlapping and distinct functions (1, 23, 36, 43, 58, 68, 70, 71).
Although Snf1 is also required for coping with a number of stresses unrelated to glucose limitation, its involvement does not automatically indicate the existence of a specific Snf1 signaling cascade. As with glucose limitation, activation of Snf1 by Thr210 phosphorylation was observed under conditions of sodium stress, suggesting a signaling mechanism (44). By contrast, evidence suggests a role for basal activity of unphosphorylated Snf1 in providing resistance to hydroxyurea and hygromycin B (13, 50). We previously found that Snf1 is required for diploid pseudohyphal (PH) differentiation (34), a filamentous-growth response to nitrogen limitation (16). The requirement of Snf1 for a nitrogen-regulated phenotype suggested a role in nitrogen signaling, but it remained possible that Snf1 contributes at a basal level of activity.
Signaling mechanisms that regulate PH differentiation have been extensively studied and involve the function of several protein kinase pathways. The cyclic AMP-dependent protein kinase (PKA) and mitogen-activated protein kinase (MAPK) pathways are required for PH differentiation under conditions of limiting nitrogen (40, 49, 53), and the Srb10-Srb11 (Cdk8-cyclin C) kinase functions to inhibit PH differentiation under nitrogen-rich conditions (47). Important roles are played by the rapamycin-sensitive TOR protein kinase pathway. The TOR protein kinases are highly conserved in evolution and function to signal nutrient availability and promote growth (for reviews, see references 41 and 56). S. cerevisiae has two homologs, Tor1 and Tor2 (collectively referred to as TOR), either of which can be employed by the rapamycin-sensitive TOR complex called TORC1 (37). Inhibition of yeast TOR with rapamycin broadly mimics the effects of nitrogen and amino acid deprivation, including inhibition of translation and ribosome biogenesis as well as greatly increased transcription of genes regulated by nitrogen catabolite repression (for reviews, see references 17, 51, and 54). Some of the regulators negatively controlled by TOR in nitrogen-rich conditions, such as the transcriptional activator Gln3, protein kinase Npr1, and Mep2 ammonium permease/sensor, are known to play critical positive roles in PH differentiation (2, 5, 20, 39, 57).
Here, we have further examined the role of Snf1 in the regulation of PH differentiation and present evidence that Snf1 is directly involved in nitrogen signaling controlled by Thr210 phosphorylation. We also show that Thr210 phosphorylation is negatively regulated by TOR.
MATERIALS AND METHODS
Strains and genetic methods.
The S. cerevisiae strains used in this study are listed in Table 1. The strains were in the Σ1278b genetic background and were descendants of wild-type strains MY1401 (MATα ura3Δ leu2Δ his3Δ) and MY1402 (MATa ura3Δ leu2Δ trp1Δ) of the Sigma2000 series (Microbia, Cambridge, Mass.). Derivatives carrying snf1::LEU2, sip1Δ::KanMX6, and reg1Δ::URA3 have been described previously (34, 71). To generate snf1Δ::KanMX6 and reg1Δ::KanMX6, the KanMX6 sequence (38) was amplified by PCR with primers flanking the corresponding open reading frames (72). The alleles sip2Δ3::LEU2 (75), gal83Δ::TRP1 (68), and snf1Δ::KanMX6 were introduced into wild-type haploids by transformation; all yeast transformations were performed using standard methods (55). The reg1Δ::KanMX6 allele was first introduced into a wild-type diploid, with subsequent recovery of haploid segregants by tetrad analysis. Deletions were confirmed by PCR analysis of genomic DNA and by mutant phenotypes. Strains carrying deletion combinations were constructed by genetic crossing, and homozygous mutant diploids were obtained by mating appropriate mutant haploids (55).
TABLE 1.
S. cerevisiae strains
| Strain | Genotype | Source |
|---|---|---|
| MCY4472 | MATa/MATα ura3Δ/ura3Δ leu2Δ/leu2Δ HIS3/his3Δ TRP1/trp1Δ | 34 |
| MCY4473 | MATa/MATα ura3Δ/ura3Δ leu2Δ/leu2Δ HIS3/his3Δ TRP1/trp1Δ snf1::LEU2/snf1::LEU2 | 34 |
| KY38 | MATaura3Δ leu2Δ his3Δ snf1Δ::KanMX6 | This study |
| KY40 | MATa/MATα ura3Δ/ura3Δ leu2Δ/leu2Δ HIS3/his3Δ TRP1/trp1Δ snf1Δ::KanMX6/snf1Δ::KanMX6 | This study |
| KY43 | MATa/MATα ura3Δ/ura3Δ leu2Δ/leuΔ HIS3/his3Δ TRP1/trp1Δ reg1Δ::KanMX6/reg1Δ::URA3 | This study |
| KY55 | MATa/MATα ura3Δ/ura3Δ leu2Δ/leu2Δ HIS3/his3Δ TRP1/trp1Δ sip1Δ::KanMX6/sip1Δ::KanMX6 sip2Δ3::LEU2/sip2Δ3::LEU2 | This study |
| KY58 | MATa/MATα ura3Δ/ura3Δ leu2Δ/leu2Δ HIS3/his3Δ trp1Δ/trp1Δ sip1Δ::KanMX6/sip1Δ::KanMX6 gal83Δ::TRP1/gal83Δ::TRP1 | This study |
| KY61 | MATa/MATα ura3Δ/ura3Δ leu2Δ/leu2Δ HIS3/his3Δ trp1Δ/trp1Δ sip2Δ3::LEU2/sip2Δ3::LEU2 gal83Δ::TRP1/gal83Δ::TRP1 | This study |
| KY64 | MATa/MATα ura3Δ/ura3Δ leu2Δ/leu2Δ HIS3/his3Δ trp1Δ/trp1Δ sip1Δ::KanMX6/sip1Δ::KanMX6 sip2Δ3::LEU2/sip2Δ3::LEU2 gal83Δ::TRP1/gal83Δ::TRP1 | This study |
| KY65 | MATaura3Δ leu2Δ his3Δ snf1Δ::KanMX6 TOR1-S1972R | This study |
Construction of the chromosomal TOR1-S1972R allele.
First, a 430-bp DNA fragment corresponding to nucleotides 5515 to 5944 of the TOR1 open reading frame was generated by PCR with Platinum Taq DNA Polymerase High Fidelity (Invitrogen), wild-type yeast genomic DNA as a template, and primers F (5′-GACACGTTGAGGTTATTGACTC-3′) and M (5′-CTATGTTATGTTCAACGAAAAATTGGCGGcgcGCATCTTCCAGTCCTTCATAACCATAATTC-3′); in primer M, the mismatching nucleotides (lowercase type) encode the Ser1972-to-Arg substitution and create a silent diagnostic BssHII site (underlined). The fragment was then gel purified and used as a primer in the second PCR with the second primer, R (5′-GGCCTTTGCTTCGAAGAGATCAC-3′), and wild-type yeast genomic DNA as a template, to generate a more extended DNA fragment corresponding to nucleotides 5515 to 6317 of the TOR1 open reading frame. The obtained 803-bp DNA fragment was gel purified, confirmed to contain the BssHII site, and used to transform a wild-type haploid, with selection on yeast extract-peptone-dextrose agar plates (55) containing 200 ng/ml rapamycin (Sigma-Aldrich). Two independent rapamycin-resistant isolates were colony purified. The presence of the TOR1-S1972R allele at the correct chromosomal location was confirmed by the presence of the diagnostic BssHII site following PCR amplification of the corresponding chromosomal region. To confirm the dominance of the introduced mutation, the rapamycin-resistant isolates were crossed to a wild-type strain. In contrast to the control wild-type diploid (TOR1/TOR1), the obtained diploids displayed resistance to 200 ng/ml rapamycin, as anticipated. Strain KY65 (snf1Δ::KanMX6 TOR1-S1972R) is a segregant from a cross between one of the rapamycin-resistant isolates and a snf1Δ::KanMX6 (TOR1) strain. Rapamycin hyperresistance segregated in a 2:2 manner in all nine tetrads from this cross, and two of the tetrads were used to confirm its cosegregation with the diagnostic BssHII site.
Plasmids.
Vector pSK134HA is a derivative of pSK134 (69) and contains the triple hemagglutinin (HA) epitope tag-encoding sequence and polycloning site from pWS93 (63). pMO18 expresses N-terminal triple HA-tagged Snf1 (HA-Snf1) from the yeast ADH1 promoter of vector pSK134HA; the SNF1 sequence was from pSK119 (65). pMO19 expresses HA-Snf1-T210A (Thr210 to Ala) and was constructed using pSK134HA by inserting the snf1-T210A sequence from pRJ217 (35). pSK119, pSK120, and pIT517 express HA-Snf1, HA-Snf1-K84R, and HA-Snf1-G53R, respectively, from the ADH1 promoter of vector pWS93 (63, 65).
PH differentiation assays.
Solid synthetic low-ammonia plus 2% dextrose (SLAD) medium containing 50 μM ammonium sulfate as the sole nitrogen source was used for standard PH differentiation assays (16). SMAD medium was identical to SLAD except that it contained 500 μM ammonium sulfate as the sole nitrogen source (48). Diploids were streaked to single cells, and the plates were incubated at 30°C for 3 to 4 days. Colonies were photographed using a Nikon Eclipse 50i microscope (10× objective), a CoolSnap ES camera (Photometrics), and MetaMorph software (Universal Imaging Corporation).
Immunoblot assays of Thr210 phosphorylation.
For nitrogen-rich conditions, cells were grown at 30°C to mid-log phase (optical density at 600 nm of 0.5 to 0.7) in synthetic complete medium with plasmid selection (55). For nitrogen-limiting conditions, the cells were shifted to liquid SLAD medium containing 50 μM ammonium sulfate as the sole nitrogen source (16) for 60 min; abundant (2%) glucose was present under both conditions. For rapamycin treatment experiments, cells were grown in nitrogen-rich conditions, and rapamycin (Sigma-Aldrich) was added to a final concentration of 200 ng/ml; a 1 mg/ml stock solution of rapamycin was prepared in the drug vehicle, 90% ethanol plus 10% Tween 20 (10); control cultures were treated with the drug vehicle alone. Protein extracts were prepared by vortexing with glass beads essentially as described previously (33), except that to arrest Snf1 in its corresponding Thr210 phosphorylation state (73), the cultures were placed in a boiling water bath for 3 min, followed by cooling and harvesting. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and Thr210-phosphorylated HA-Snf1 was detected by immunoblotting with anti-phospho-Thr172-AMPK (Cell Signaling Technologies). Total levels of HA-Snf1 proteins were determined by reprobing the blots with monoclonal HA antibody 12CA5. Antibodies were detected by enhanced chemiluminescence using ECL and ECLPlus (Amersham Biosciences).
RESULTS
Nonphosphorylable Snf1 does not support PH differentiation.
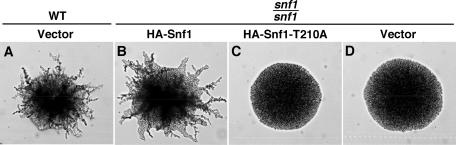
We first examined the ability of a nonphosphorylable mutant kinase (HA-Snf1-T210A), which contains a Thr210-to-Ala substitution previously used for studies of basal Snf1 function (13, 50), to complement the PH differentiation defect of a diploid deleted for both genomic copies of the SNF1 gene. A snf11/snf1 diploid was transformed with plasmids expressing HA-Snf1 or HA-Snf1-T210A or with the parent vector, and immunoblot analyses indicated that both Snf1 proteins are expressed (see below). The resulting strains were tested on SLAD plates for PH differentiation. The expression of HA-Snf1 restored PH development in the snf1/snf1 recipient (Fig. 1A and B). By contrast, HA-Snf1-T210A was ineffective and conferred no phenotypic improvement over the vector control (Fig. 1C and D). Thus, these results indicate that PH differentiation requires Thr210, suggesting that basal activity of unphosphorylated Snf1 is not sufficient.
FIG. 1.
HA-Snf1-T210A does not support PH differentiation. Strain KY40 (snf1Δ/snf1Δ) was transformed with pMO18 expressing HA-Snf1, pMO19 expressing HA-Snf1-T210A, or with the parent vector pSK134HA (Vector). The wild-type control strain MCY4472 (WT) carried the empty vector pSK134HA. All transformants also carried pRS316 (62), a centromeric vector with URA3, to provide prototrophy. The strains were examined on SLAD plates for PH differentiation. Colonies were photographed after 4 days at 30°C.
Nitrogen limitation improves Thr210 phosphorylation.
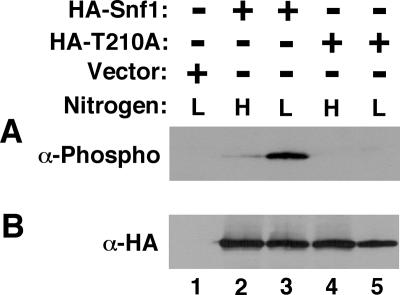
We therefore tested whether nitrogen availability regulates Thr210 phosphorylation. The above-described snf1/snf1 cells, expressing HA-Snf1, expressing HA-Snf1-T210A, or carrying the empty vector, were cultured under nitrogen-rich or nitrogen-limiting conditions in the presence of abundant glucose (2%). Thr210 phosphorylation was assayed by immunoblot analysis with an anti-phospho-Thr172-AMPK antibody (Thr172 of AMPK corresponds to Thr210 of Snf1), an approach used previously for analyses of yeast Snf1 (64). Thr210 phosphorylation of HA-Snf1 under nitrogen-limiting conditions was considerably increased relative to the nitrogen-rich conditions (Fig. 2A, lanes 2 and 3). As expected, anti-phospho-Thr172-AMPK did not detect HA-Snf1-T210A, regardless of growth conditions (Fig. 2A, lanes 4 and 5). Reprobing the blot with anti-HA indicated that the levels of the HA-tagged proteins were comparable to one another (Fig. 2B). Thus, nitrogen limitation improves Thr210 phosphorylation.
FIG. 2.
Nitrogen levels regulate Thr210 phosphorylation. The transformants of KY40 (snf1Δ/snf1Δ) analyzed in Fig. 1 expressing HA-Snf1, expressing nonphosphorylable HA-Snf1-T210A (HA-T210A), or carrying the empty vector pSK134HA (Vector) were grown in nutrient-rich synthetic medium to mid-log phase (high nitrogen, H) and shifted for 1 h to SLAD medium (low nitrogen, L). All transformants carried pRS316 to provide prototrophy. (A) Thr210 phosphorylation of HA-Snf1 was assessed by immunoblotting with anti-phospho-Thr172-AMPK (α-Phospho). (B) Total levels of HA-Snf1 proteins were determined by reprobing the blot with anti-HA (α-HA).
Effects of inactivation and hyperactivation of Snf1.
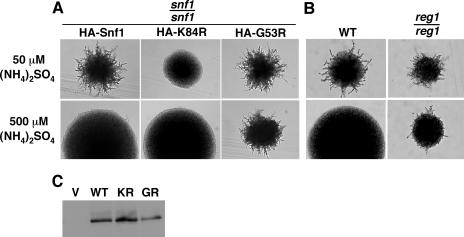
To address the role of Snf1 kinase activity, we first tested PH differentiation in cells (snf1/snf1) expressing HA-Snf1-K84R, which is catalytically inactive because the conserved lysine in the ATP-binding site is replaced with arginine (14). PH differentiation was defective, indicating that Snf1 catalytic activity is required (Fig. 3A). To examine the effects of increased Snf1 activity, we similarly expressed HA-Snf1-G53R, a catalytically hyperactive Gly53-to-Arg mutant identified by a function-based screening (14). In the presence of nitrogen levels 10-fold elevated relative to the optimal PH differentiation-inducing conditions (500 μM instead of 50 μM ammonium sulfate as the sole nitrogen source), wild-type HA-Snf1 was insufficient, as anticipated (48), whereas HA-Snf1-G53R still supported PH differentiation (Fig. 3A). In another approach, we examined cells lacking Reg1, in which Snf1 is constitutively activated (26, 44). Unlike the wild-type diploid, which displayed PH differentiation on 50 μM but not 500 μM ammonium sulfate medium, the reg1/reg1 diploid formed filaments even under the nonpermissive conditions (Fig. 3B). These findings indicate that upregulation of Snf1 results in improved PH differentiation.
FIG. 3.
Effects of inactive and hyperactive Snf1 on PH differentiation. (A) A snf1/snf1 diploid, MCY4473, was transformed with plasmids pSK119, pSK120, or pIT517 expressing HA-Snf1, catalytically inactive HA-Snf1-K84R (HA-K84R), and hyperactive HA-Snf1-G53R (HA-G53R), respectively. Immunoblot analyses indicated that all HA-tagged proteins are expressed (panel C). Strains were examined for PH differentiation on SLAD and SMAD media containing 50 μM and 500 mM ammonium sulfate as the sole nitrogen source, respectively; colonies were photographed after 3 days at 30°C. Cells expressing no Snf1 protein (carrying the corresponding empty vector pWS93) failed to undergo PH differentiation under either condition (not shown). (B) Wild-type diploid MCY4472 (WT) and KY43 (reg1/reg1) were examined for PH differentiation as described above. Both strains carried pLCLG-Staf (31), a centromeric plasmid with LEU2 and URA3 to confer prototrophy. (C) Cells analyzed in panel A were grown to mid-log phase in nutrient-rich synthetic medium with plasmid selection and shifted to liquid SLAD medium for 2 h, and levels of the HA-Snf1 proteins were compared by immunoblotting with anti-HA. V, vector pWS93; WT, HA-Snf1; KR, HA-Snf1-K84R; GR, HA-Snf1-G53R.
Collectively, our immunochemical and genetic analyses strongly suggest that the regulation of Snf1 is a major mechanism by which nitrogen levels control PH differentiation and strongly implicate Snf1 in nitrogen signaling.
Requirement for the β subunits.
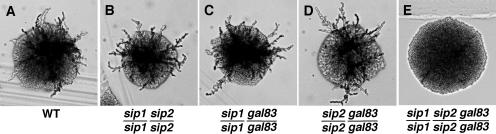
There are three distinct forms of the Snf1 complex, as defined by the associated alternate β subunit. Given a new signaling role of Snf1, it was of interest to assess the individual roles of these complexes, which are known to have not only overlapping but also distinct functions (1, 23, 36, 43, 58, 68, 70, 71). An examination of diploids expressing individual β subunits indicated that Sip1, Sip2, and Gal83 can each support PH differentiation, and only the loss of all three resulted in a strong phenotypic defect (Fig. 4). These results suggest that all three forms of the Snf1 complex can respond to signaling and make functionally equivalent contributions to PH differentiation. However, we cannot rule out the possibility that the different complexes also have distinct functions in this and other nitrogen-regulated processes.
FIG. 4.
Requirement for the β subunits. Diploids with the indicated genotypes were grown on SLAD plates for 4 days at 30°C, and colonies were photographed. WT, wild type. All strains carried pLCLG-Staf to confer prototrophy (see legend to Fig. 3).
We also note that these findings distinguish diploid PH differentiation from two related processes in haploids, surface adhesion and filamentation, which are activated by glucose limitation (11, 52, 71): only Gal83 can support haploid surface adhesion, and haploid filamentation can be provided by Gal83 or Sip2 but not by Sip1 (71).
Snf1 is negatively regulated by TOR.
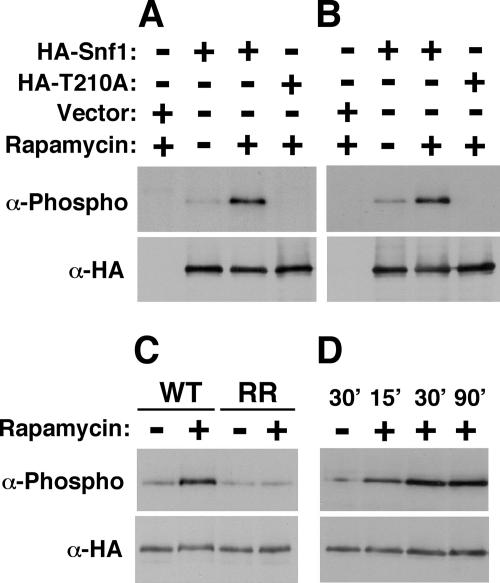
The involvement of Snf1 in nitrogen signaling suggested that Snf1 might function downstream of the rapamycin-sensitive TOR kinase. First, under nitrogen-rich conditions, TOR negatively regulates numerous functions required for coping with nitrogen limitation, some of which are known to be essential for PH differentiation. Second, previous evidence implicated Snf1 as an effector of rapamycin toxicity (3). We therefore examined whether treating the cells with rapamycin would affect Thr210 phosphorylation. Diploid cells expressing HA-Snf1 were grown in nutrient-rich conditions and treated with rapamycin or with the drug vehicle alone as a control. Rapamycin treatment resulted in a significant improvement of Thr210 phosphorylation (Fig. 5A). A similar result was obtained for haploid cells (Fig. 5B), suggesting that this regulatory effect is not ploidy specific. Rapamycin treatment did not improve Thr210 phosphorylation in cells carrying a TOR1-S1972R allele (Fig. 5C), which encodes a rapamycin-resistant Ser1972-to-Arg Tor1 protein (4, 24), confirming that rapamycin elicits its effect on Snf1 via inhibition of TOR. Finally, time-course experiments showed that a detectable response develops within 15 to 30 min of rapamycin addition (3, 42), arguing against grossly indirect effects (Fig. 5D). Thus, we conclude that the rapamycin-sensitive TOR negatively regulates Snf1.
FIG. 5.
Rapamycin-sensitive TOR negatively regulates Snf1. Cells (snf1Δ/snf1Δ or snf1Δ) expressing HA-Snf1 from pMO18, expressing HA-Snf1-T210A (HA-T210A) from pMO19, or carrying the empty vector pSK134HA (Vector) were grown in nutrient-rich synthetic complete medium lacking Leu to mid-log phase and treated with 200 ng/ml rapamycin (+) or with the corresponding amount (1 μl per 5-ml culture) of drug vehicle (−) for 90 min (A to C) or for the indicated times (D). Thr210 phosphorylation was analyzed as described for Fig. 2. (A) Results for a diploid strain, KY40. (B) Results for a haploid strain, KY38. (C) Results for strains KY38 and KY65 carrying the wild-type TOR1 allele (WT) or a rapamycin-resistant TOR1-S1972R allele (RR), respectively, expressing HA-Snf1. (D) Cells of KY40 expressing HA-Snf1 were treated with rapamycin (+) or with the drug vehicle (−) for the indicated times (min).
DISCUSSION
We have investigated the requirement of the Snf1 protein kinase for diploid PH differentiation in S. cerevisiae and present evidence that Snf1 is directly involved in nitrogen signaling. First, nonphosphorylable Snf1-T210A does not support PH differentiation, suggesting a requirement for maximal activation. Second, nitrogen limitation leads to improved Thr210 phosphorylation, indicating that Snf1 responds to a nitrogen signal. Furthermore, we show that Thr210 phosphorylation is negatively regulated by the rapamycin-sensitive TOR kinase, which plays essential roles in signaling nitrogen and amino acid availability.
Evidence that the three alternative β subunits can each support PH differentiation suggests that all three forms of the Snf1 complex are responsive to regulation by nitrogen. All three forms are also controlled by glucose (43, 58) and, collectively, these findings implicate Snf1 in broad integration of signals regarding nitrogen and carbon stress. Further studies are required to decipher how such integration occurs and whether it relies on differential roles of the known regulators, such as the three partially redundant Snf1-activating kinases (Sak1, Tos3, Elm1) or involves other mechanisms. In this regard, an interesting possibility raised by our findings is that at least some signal integration upstream of Snf1 in fact occurs at TOR. Indeed, besides its broad involvement in nitrogen signaling, TOR has also been implicated in responses to carbon and salt stress (2, 9, 10, 27, 59).
Snf1 extends the list of known positive regulators of PH differentiation that are negatively regulated by TOR (2, 5, 20, 39, 57), further supporting the involvement of TOR in the negative control of this developmental process. Interestingly, however, the net effect of rapamycin on PH differentiation is inhibitory rather than stimulatory (12), and our experiments confirm this conclusion (data not shown). On the one hand, this result indicates that TOR also has a positive role, likely reflecting the fact that PH differentiation still requires a signal(s) confirming the presence of nitrogen, albeit in limiting amounts or in a nonpreferred form (12). On the other hand, its involvement in both positive and negative control of PH differentiation suggests that TOR can distinguish between different physiological (non-rapamycin) situations to allow the activation of one set of responses but not the other. This possibility is in line with evidence that TOR can function as a “multichannel processor” that couples specific upstream inputs to specific downstream events (10, 59).
The functional interaction between AMPK and mTOR in mammalian cells has been intensively studied in connection with its biomedical importance. Interestingly, it is AMPK that has been reported to negatively control mTOR, by phosphorylating and stimulating the tumor suppressor protein TSC2 which functions in mTOR inhibition (29). Upregulation of mTOR caused by defects in the LKB1-AMPK-TSC pathway has been implicated as an underlying molecular cause of inherited hamartomatous tumor syndromes, such as tuberous sclerosis complex and Peutz-Jeghers syndrome (8, 29, 60, 61).
Our findings for S. cerevisiae suggest that other eukaryotes may have mechanisms for the negative regulation of Snf1/AMPK by TOR. The existence of such a mechanism in mammals would suggest further clues to the molecular etiology and therapy of tumorigenesis and other diseases associated with deregulated energy metabolism. Regardless of whether the specific mechanisms are conserved, however, a form of functional antagonism between Snf1/AMPK and TOR clearly appears to be a common regulatory feature in eukaryotes from yeast to humans.
Acknowledgments
This work was supported by the UWM College of Letters and Science and a Graduate School Faculty Research Award.
We thank Mark McBride, Tom Schuck, Heather Owen, and Raymond Hovey for advice and technical assistance.
This work was initiated in the laboratory of M. Carlson and supported by NIH grant GM34095.
Footnotes
Published ahead of print on 15 September 2006.
REFERENCES
- 1.Ashrafi, K., S. S. Lin, J. K. Manchester, and J. I. Gordon. 2000. Sip2p and its partner Snf1p kinase affect aging in S. cerevisiae. Genes Dev. 14:1872-1885. [PMC free article] [PubMed] [Google Scholar]
- 2.Beck, T., and M. N. Hall. 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402:689-692. [DOI] [PubMed] [Google Scholar]
- 3.Bertram, P. G., J. H. Choi, J. Carvalho, T. F. Chan, W. Ai, and X. F. Zheng. 2002. Convergence of TOR-nitrogen and Snf1-glucose signaling pathways onto Gln3. Mol. Cell. Biol. 22:1246-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cafferkey, R., P. R. Young, M. M. McLaughlin, D. J. Bergsma, Y. Koltin, G. M. Sathe, L. Faucette, W. K. Eng, R. K. Johnson, and G. P. Livi. 1993. Dominant missense mutations in a novel yeast protein related to mammalian phosphatidylinositol 3-kinase and VPS34 abrogate rapamycin cytotoxicity. Mol. Cell. Biol. 13:6012-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cardenas, M. E., N. S. Cutler, M. C. Lorenz, C. J. Di Como, and J. Heitman. 1999. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 13:3271-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carling, D. 2004. The AMP-activated protein kinase cascade—a unifying system for energy control. Trends Biochem. Sci. 29:18-24. [DOI] [PubMed] [Google Scholar]
- 7.Carlson, M. 1999. Glucose repression in yeast. Curr. Opin. Microbiol. 2:202-207. [DOI] [PubMed] [Google Scholar]
- 8.Corradetti, M. N., K. Inoki, N. Bardeesy, R. A. DePinho, and K. L. Guan. 2004. Regulation of the TSC pathway by LKB1: evidence of a molecular link between tuberous sclerosis complex and Peutz-Jeghers syndrome. Genes Dev. 18:1533-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crespo, J. L., K. Daicho, T. Ushimaru, and M. N. Hall. 2001. The GATA transcription factors GLN3 and GAT1 link TOR to salt stress in Saccharomyces cerevisiae. J. Biol. Chem. 276:34441-34444. [DOI] [PubMed] [Google Scholar]
- 10.Crespo, J. L., T. Powers, B. Fowler, and M. N. Hall. 2002. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc. Natl. Acad. Sci. USA 99:6784-6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cullen, P. J., and G. F. Sprague, Jr. 2000. Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. USA 97:13619-13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cutler, N. S., X. Pan, J. Heitman, and M. E. Cardenas. 2001. The TOR signal transduction cascade controls cellular differentiation in response to nutrients. Mol. Biol. Cell 12:4103-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubacq, C., A. Chevalier, and C. Mann. 2004. The protein kinase Snf1 is required for tolerance to the ribonucleotide reductase inhibitor hydroxyurea. Mol. Cell. Biol. 24:2560-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Estruch, F., M. A. Treitel, X. Yang, and M. Carlson. 1992. N-terminal mutations modulate yeast SNF1 protein kinase function. Genetics 132:639-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gancedo, J. M. 1998. Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 62:334-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gimeno, C. J., P. O. Ljungdahl, C. A. Styles, and G. R. Fink. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68:1077-1090. [DOI] [PubMed] [Google Scholar]
- 17.Gingras, A. C., B. Raught, and N. Sonenberg. 2004. mTOR signaling to translation. Curr. Top. Microbiol. Immunol. 279:169-197. [DOI] [PubMed] [Google Scholar]
- 18.Hardie, D. G. 2005. New roles for the LKB1→AMPK pathway. Curr. Opin. Cell Biol. 17:167-173. [DOI] [PubMed] [Google Scholar]
- 19.Hardie, D. G., D. Carling, and M. Carlson. 1998. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 67:821-855. [DOI] [PubMed] [Google Scholar]
- 20.Hardwick, J. S., F. G. Kuruvilla, J. K. Tong, A. F. Shamji, and S. L. Schreiber. 1999. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. USA 96:14866-14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hawley, S. A., J. Boudeau, J. L. Reid, K. J. Mustard, L. Udd, T. P. Makela, D. R. Alessi, and D. G. Hardie. 2003. Complexes between the LKB1 tumor suppressor, STRADα/β and MO25α/β are upstream kinases in the AMP-activated protein kinase cascade. J. Biol. 2:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hawley, S. A., D. A. Pan, K. J. Mustard, L. Ross, J. Bain, A. M. Edelman, B. G. Frenguelli, and D. G. Hardie. 2005. Calmodulin-dependent protein kinase kinase-beta is an alternative upstream kinase for AMP-activated protein kinase. Cell Metab. 2:9-19. [DOI] [PubMed] [Google Scholar]
- 23.Hedbacker, K., R. Townley, and M. Carlson. 2004. Cyclic AMP-dependent protein kinase regulates the subcellular localization of Snf1-Sip1 protein kinase. Mol. Cell. Biol. 24:1836-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helliwell, S. B., P. Wagner, J. Kunz, M. Deuter-Reinhard, R. Henriquez, and M. N. Hall. 1994. TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol. Biol. Cell 5:105-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong, S.-P., F. C. Leiper, A. Woods, D. Carling, and M. Carlson. 2003. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc. Natl. Acad. Sci. USA 100:8839-8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong, S.-P., M. Momcilovic, and M. Carlson. 2005. Function of mammalian LKB1 and Ca2+/calmodulin-dependent protein kinase kinase α as Snf1-activating kinases in yeast. J. Biol. Chem. 280:21804-21809. [DOI] [PubMed] [Google Scholar]
- 27.Huang, J., H. Zhu, S. J. Haggarty, D. R. Spring, H. Hwang, F. Jin, M. Snyder, and S. L. Schreiber. 2004. Finding new components of the target of rapamycin (TOR) signaling network through chemical genetics and proteome chips. Proc. Natl. Acad. Sci. USA 101:16594-16599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hurley, R. L., K. A. Anderson, J. M. Franzone, B. E. Kemp, A. R. Means, and L. A. Witters. 2005. The Ca2+/calmodulin-dependent protein kinase kinases are AMP-activated protein kinase kinases. J. Biol. Chem. 280:29060-29066. [DOI] [PubMed] [Google Scholar]
- 29.Inoki, K., T. Zhu, and K. L. Guan. 2003. TSC2 mediates cellular energy response to control cell growth and survival. Cell 115:577-590. [DOI] [PubMed] [Google Scholar]
- 30.Jiang, R., and M. Carlson. 1997. The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol. Cell. Biol. 17:2099-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kartasheva, N. N., S. V. Kuchin, and S. V. Benevolensky. 1996. Genetic aspects of carbon catabolite repression of the STA2 glucoamylase gene in Saccharomyces cerevisiae. Yeast 12:1297-1300. [DOI] [PubMed] [Google Scholar]
- 32.Kemp, B. E., D. Stapleton, D. J. Campbell, Z. P. Chen, S. Murthy, M. Walter, A. Gupta, J. J. Adams, F. Katsis, B. van Denderen, I. G. Jennings, T. Iseli, B. J. Michell, and L. A. Witters. 2003. AMP-activated protein kinase, super metabolic regulator. Biochem. Soc. Trans. 31:162-168. [DOI] [PubMed] [Google Scholar]
- 33.Kuchin, S., I. Treich, and M. Carlson. 2000. A regulatory shortcut between the Snf1 protein kinase and RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. USA 97:7916-7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuchin, S., V. K. Vyas, and M. Carlson. 2002. Snf1 Protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol. Cell. Biol. 22:3994-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuchin, S., V. K. Vyas, E. Kanter, S. P. Hong, and M. Carlson. 2003. Std1p (Msn3p) positively regulates the Snf1 kinase in Saccharomyces cerevisiae. Genetics 163:507-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin, S. S., J. K. Manchester, and J. I. Gordon. 2003. Sip2, an N-myristoylated beta subunit of Snf1 kinase, regulates aging in Saccharomyces cerevisiae by affecting cellular histone kinase activity, recombination at rDNA loci, and silencing. J. Biol. Chem. 278:13390-13397. [DOI] [PubMed] [Google Scholar]
- 37.Loewith, R., E. Jacinto, S. Wullschleger, A. Lorberg, J. L. Crespo, D. Bonenfant, W. Oppliger, P. Jenoe, and M. N. Hall. 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10:457-468. [DOI] [PubMed] [Google Scholar]
- 38.Longtine, M. S., A. McKenzie, III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 39.Lorenz, M. C., and J. Heitman. 1998. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 17:1236-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madhani, H. D., and G. R. Fink. 1997. Combinatorial control required for the specificity of yeast MAPK signaling. Science 275:1314-1317. [DOI] [PubMed] [Google Scholar]
- 41.Martin, D. E., and M. N. Hall. 2005. The expanding TOR signaling network. Curr. Opin. Cell Biol. 17:158-166. [DOI] [PubMed] [Google Scholar]
- 42.Martin, D. E., A. Soulard, and M. N. Hall. 2004. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 119:969-979. [DOI] [PubMed] [Google Scholar]
- 43.McCartney, R. R., E. M. Rubenstein, and M. C. Schmidt. 2005. Snf1 kinase complexes with different beta subunits display stress-dependent preferences for the three Snf1-activating kinases. Curr. Genet. 47:335-344. [DOI] [PubMed] [Google Scholar]
- 44.McCartney, R. R., and M. C. Schmidt. 2001. Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J. Biol. Chem. 276:36460-36466. [DOI] [PubMed] [Google Scholar]
- 45.Momcilovic, M., S. P. Hong, and M. Carlson. 2006. Mammalian TAK1 activates Snf1 protein kinase in yeast and phosphorylates AMP-activated protein kinase in vitro. J. Biol. Chem. 281:25336-25343. [DOI] [PubMed] [Google Scholar]
- 46.Nath, N., R. R. McCartney, and M. C. Schmidt. 2003. Yeast Pak1 kinase associates with and activates Snf1. Mol. Cell. Biol. 23:3909-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nelson, C., S. Goto, K. Lund, W. Hung, and I. Sadowski. 2003. Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature 421:187-190. [DOI] [PubMed] [Google Scholar]
- 48.Pan, X., and J. Heitman. 1999. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:4874-4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan, X., and J. Heitman. 2002. Protein kinase A operates a molecular switch that governs yeast pseudohyphal differentiation. Mol. Cell. Biol. 22:3981-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Portillo, F., J. M. Mulet, and R. Serrano. 2005. A role for the non-phosphorylated form of yeast Snf1: tolerance to toxic cations and activation of potassium transport. FEBS Lett. 579:512-516. [DOI] [PubMed] [Google Scholar]
- 51.Powers, T., I. Dilova, C. Y. Chen, and K. Wedaman. 2004. Yeast TOR signaling: a mechanism for metabolic regulation. Curr. Top. Microbiol. Immunol. 279:39-51. [DOI] [PubMed] [Google Scholar]
- 52.Reynolds, T. B., and G. R. Fink. 2001. Bakers' yeast, a model for fungal biofilm formation. Science 291:878-881. [DOI] [PubMed] [Google Scholar]
- 53.Robertson, L. S., and G. R. Fink. 1998. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc. Natl. Acad. Sci. USA 95:13783-13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rohde, J. R., and M. E. Cardenas. 2004. Nutrient signaling through TOR kinases controls gene expression and cellular differentiation in fungi. Curr. Top. Microbiol. Immunol. 279:53-72. [DOI] [PubMed] [Google Scholar]
- 55.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- 56.Sarbassov, D. D., S. M. Ali, and D. M. Sabatini. 2005. Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 17:596-603. [DOI] [PubMed] [Google Scholar]
- 57.Schmidt, A., T. Beck, A. Koller, J. Kunz, and M. N. Hall. 1998. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 17:6924-6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmidt, M. C., and R. R. McCartney. 2000. β-Subunits of Snf1 kinase are required for kinase function and substrate definition. EMBO J. 19:4936-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shamji, A. F., F. G. Kuruvilla, and S. L. Schreiber. 2000. Partitioning the transcriptional program induced by rapamycin among the effectors of the Tor proteins. Curr. Biol. 10:1574-1581. [DOI] [PubMed] [Google Scholar]
- 60.Shaw, R. J., N. Bardeesy, B. D. Manning, L. Lopez, M. Kosmatka, R. A. DePinho, and L. C. Cantley. 2004. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell 6:91-99. [DOI] [PubMed] [Google Scholar]
- 61.Shaw, R. J., M. Kosmatka, N. Bardeesy, R. L. Hurley, L. A. Witters, R. A. DePinho, and L. C. Cantley. 2004. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc. Natl. Acad. Sci. USA 101:3329-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Song, W., and M. Carlson. 1998. Srb/mediator proteins interact functionally and physically with transcriptional repressor Sfl1. EMBO J. 17:5757-5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sutherland, C. M., S. A. Hawley, R. R. McCartney, A. Leech, M. J. Stark, M. C. Schmidt, and D. G. Hardie. 2003. Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae SNF1 complex. Curr. Biol. 13:1299-1305. [DOI] [PubMed] [Google Scholar]
- 65.Treitel, M. A., S. Kuchin, and M. Carlson. 1998. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:6273-6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tu, J., and M. Carlson. 1994. The GLC7 type 1 protein phosphatase is required for glucose repression in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:6789-6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tu, J., and M. Carlson. 1995. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J. 14:5939-5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vincent, O., and M. Carlson. 1999. Gal83 mediates the interaction of the Snf1 kinase complex with the transcription activator Sip4. EMBO J. 18:6672-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vincent, O., S. Kuchin, S. P. Hong, R. Townley, V. K. Vyas, and M. Carlson. 2001. Interaction of the Srb10 kinase with Sip4, a transcriptional activator of gluconeogenic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:5790-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vincent, O., R. Townley, S. Kuchin, and M. Carlson. 2001. Subcellular localization of the Snf1 kinase is regulated by specific beta subunits and a novel glucose signaling mechanism. Genes Dev. 15:1104-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vyas, V. K., S. Kuchin, C. D. Berkey, and M. Carlson. 2003. Snf1 kinases with different β-subunit isoforms play distinct roles in regulating haploid invasive growth. Mol. Cell. Biol. 23:1341-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259-265. [DOI] [PubMed] [Google Scholar]
- 73.Wilson, W. A., S. A. Hawley, and D. G. Hardie. 1996. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr. Biol. 6:1426-1434. [DOI] [PubMed] [Google Scholar]
- 74.Woods, A., K. Dickerson, R. Heath, S. P. Hong, M. Momcilovic, S. R. Johnstone, M. Carlson, and D. Carling. 2005. Ca2+/calmodulin-dependent protein kinase kinase-beta acts upstream of AMP-activated protein kinase in mammalian cells. Cell Metab. 2:21-33. [DOI] [PubMed] [Google Scholar]
- 75.Yang, X., R. Jiang, and M. Carlson. 1994. A family of proteins containing a conserved domain that mediates interaction with the yeast SNF1 protein kinase complex. EMBO J. 13:5878-5886. [DOI] [PMC free article] [PubMed] [Google Scholar]