Abstract
Snf1 protein kinase containing the β subunit Gal83 is localized in the cytoplasm during growth of Saccharomyces cerevisiae cells in abundant glucose and accumulates in the nucleus in response to glucose limitation. Nuclear localization of Snf1-Gal83 requires activation of the Snf1 catalytic subunit and depends on Gal83, but in the snf1Δ mutant, Gal83 exhibits glucose-regulated nuclear accumulation. We show here that the N terminus of Gal83, which is divergent from those of the other β subunits, is necessary and sufficient for Snf1-independent, glucose-regulated localization. We identify a leucine-rich nuclear export signal in the N terminus and show that export depends on the Crm1 export receptor. We present evidence that catalytically inactive Snf1 promotes the cytoplasmic retention of Gal83 in glucose-grown cells through its interaction with the C terminus of Gal83; cytoplasmic localization of inactive Snf1-Gal83 maintains accessibility to the Snf1-activating kinases. Finally, we characterize the effects of glucose phosphorylation on localization. These studies define roles for Snf1 and Gal83 in determining the nucleocytoplasmic distribution of Snf1-Gal83 protein kinase.
Snf1 protein kinase of Saccharomyces cerevisiae is a member of the Snf1/AMP-activated protein kinase family and has broad roles in cellular responses to carbon source limitation and other stresses. The kinase is heterotrimeric, comprising the Snf1 (α), Snf4 (γ), and β subunits. Multiple mechanisms appear to control the catalytic activity and subcellular localization of Snf1 protein kinase. Three upstream kinases, Sak1 (formerly Pak1), Tos3, and Elm1, are each capable of phosphorylating and activating the Snf1 catalytic subunit (11, 22, 24), and protein phosphatase 1 (Reg1-Glc7) dephosphorylates it (20, 21). The signaling mechanism is not known, but AMP does not appear to play a major role (30), and Snf1 activity is still inhibited by glucose when a heterologous mammalian kinase is responsible for its activation (12).
The subcellular localization of the kinase is also regulated and depends on the β subunit, which has three isoforms, Sip1, Sip2, and Gal83. During exponential growth of cells in glucose, Snf1 and all β subunits are cytoplasmic and excluded from the nucleus. In response to glucose depletion or growth on nonfermentable carbon sources, Snf1 protein kinase containing Gal83, called Snf1-Gal83, accumulates in the nucleus, Snf1-Sip1 relocalizes to the vacuolar membrane, and Snf1-Sip2 remains cytoplasmic (27). Snf4 is found in both the cytoplasm and the nucleus in both glucose-grown and glucose-limited cells (27). Protein kinase A controls the localization of Snf1-Sip1 but not Snf1-Gal83 (10, 27). The β subunits contain conserved C-terminal sequences that interact with Snf1 and Snf4 (14); however, their N termini are divergent and, in the case of Sip1, the N terminus confers localization to the vacuole (10).
The nuclear localization of Snf1-Gal83 is of particular interest because Snf1 protein kinase has major roles in transcriptional control, notably in response to glucose depletion. Snf1 affects the expression of a wide array of genes (33) and interacts with various nuclear transcription factors, chromatin, and the transcriptional apparatus (16, 18, 19, 23, 25, 26, 33, 34). Nuclear localization serves to permit the interaction of Snf1-Gal83 with nuclear proteins; conversely, nuclear exclusion of Snf1 in glucose-grown cells limits such interactions, serving as an additional layer of regulatory control. The localization of Snf1-Gal83 may differentially affect the expression of subsets of genes, dependent on the ability of the relevant transcription factors to shuttle in and out of the nucleus.
Previous work indicated that regulation of the nucleocytoplasmic distribution of Snf1-Gal83 is complex. Nuclear accumulation of the Snf1 catalytic subunit is strongly dependent on Gal83 in the S288C background (27) and completely dependent on Gal83 in W303 (9), which has been used for subsequent studies. Activation of Snf1 is required for nuclear localization of Snf1-Gal83, as is the Snf1-activating kinase Sak1; alteration of the activation-loop threonine (T210A) or the ATP-binding site (K84R) of Snf1 or mutation of SAK1 inhibited nuclear accumulation (9). However, activation of Snf1 by a heterologous kinase did not rescue the localization defect of the sak1Δ mutant, suggesting an additional role for Sak1 (12). In the absence of Snf1, moreover, Gal83 exhibits glucose-regulated localization and Sak1 is dispensable (9), indicating that a Snf1- and Sak1-independent mechanism also regulates localization. Finally, previous analyses of hexose kinase-deficient mutants and the effects of 2-deoxyglucose, which can be phosphorylated but not metabolized, were interpreted as evidence for the phosphorylation of glucose as a signal for nuclear exclusion of Gal83 (27); however, the requirement of Snf1 activation for nuclear accumulation raised the possibility that the effects on localization reflected effects on Snf1 activity.
Here, we have examined the roles of the Snf1 and Gal83 subunits in regulating localization of the kinase. We first characterized the divergent N-terminal sequence of Gal83 and showed that it is necessary and sufficient for Snf1-independent, glucose-regulated localization. We identified a Crm1-dependent nuclear export signal (NES) in this region. We further showed that inactive Snf1 contributes to the cytoplasmic retention of Gal83 in glucose-grown cells through its interaction with the C terminus of Gal83. Finally, we have revisited the effects of glucose phosphorylation on localization.
MATERIALS AND METHODS
Strains and genetic methods.
S. cerevisiae strains used in this work are listed in Table 1. All MCY strains were in the W303 background. Selective synthetic complete (SC) media or rich yeast extract-peptone (YEP) medium contained the indicated carbon source.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| W303-1A | MATaade2 can1 his3 leu2 trp1 ura3 | R. Rothstein |
| W303-1B | MATα ade2 can1 his3 leu2 trp1 ura3 | R. Rothstein |
| MCY4034 | W303-1A reg1Δ::HIS3 | Our lab |
| MCY4093 | MATα gal83Δ::TRP1 ade2 can1 his3 leu2 trp1 ura3 | Our lab |
| MCY4908 | W303-1A snf1Δ10 | Our lab |
| MCY5115 | W303-1B sak1Δ::kanMX4 | Our lab |
| LDY937 | MATacrm1-1 ade2 ura3 trp1 leu2 his3 | 31 |
| LDY940 | MATacrm1-3 ade2 ade3 ura3 trp1 leu2 his7 | 31 |
| WAY.78-1 | MATα hxk1Δ::HIS3 hxk2Δ::LEU2 glk1Δ::LEU2 leu2-3,112 ura3-52 trp1-289 MAL2-8 MAL3 SUC3 | P. Sanz |
| ENY-WA-1A | MATα leu2-3,112 ura3-52 trp1-289 MAL2-8 MAL3 SUC3 | P. Sanz |
| DH63-0 | MATα trp1 leu2 ura3-52 pgi1Δ::URA3 | 13 |
Plasmids.
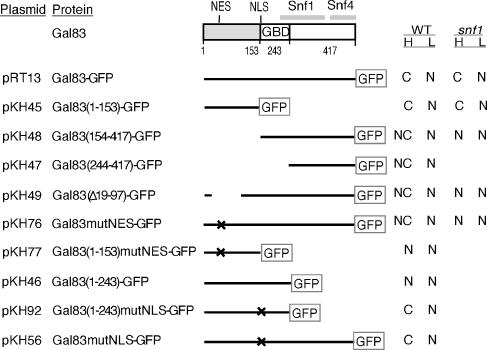
Proteins expressed from plasmids are diagrammed in Fig. 1. Centromeric plasmids pRT13, pRT12, and pOV72 express Gal83-green fluorescent protein (GFP) from the native promoter (27). To construct pKH45 and pKH84 expressing Gal83(1-153)-GFP, we amplified a PCR product from template pRT13 using a primer in the promoter of GAL83 and a second primer containing nucleotides 436 to 459 of GAL83 and N-terminal GFP coding sequence. The product was used to transform MCY4093, together with NheI/BclI-cut pRT13 or pOV72, and pKH45 and pKH84, respectively, were generated by recombination in vivo. pKH46 was constructed like pKH45 except that the second primer contained nucleotides 703 to 729 of GAL83 and GFP sequence. pKH48 and pKH47 were constructed similarly, by recombination between NheI/BclI-cut pRT13 and a PCR product amplified with a primer containing sequences 5′ of and including the start codon of GAL83 followed by nucleotides 460 to 483 and nucleotides 730 to 754, respectively, and a primer with GFP sequence. pKH49 was constructed by religating the vector-containing NheI fragment of pRT13. pKH76 and pKH77 were derived from pRT13 and pKH45, respectively, by site-directed mutagenesis using a primer containing nucleotide substitutions altering L39, F43, M46, and V48 to alanines. This primer and a GFP sequence primer were used to amplify a mutant PCR product, which was then used together with a GAL83 promoter primer to yield a new PCR product. NheI-cut pRT13 and pKH45 and their respective PCR products were used to transform MCY4093, and pKH76 and pKH77 were generated by recombination. pKH56, expressing a protein with altered residues K155A, K156A, and R158G, was similarly constructed by site-directed mutagenesis, except that NheI/NsiI-cut pRT13 was used. pKH92 was derived from pKH56 by using the same primers as for pKH46. All mutations were confirmed by sequencing.
FIG. 1.
Structure and localization of Gal83 proteins. Shaded region, Gal83 sequence that is divergent from that of other β subunits. GBD, glycogen-binding domain (residues 161 to 243) (29). Gray bars, regions mapped by deletion analysis as sufficient for interaction with Snf1 and Snf4, as indicated (designated KIS and ASC regions [14]). Lines, Gal83 sequence fused to GFP. Gal83 includes other potential NESs and NLSs. X, positions of the mutations L39A, F43A, M46A, and V48A (mutNES) and K155A, K156A, and R158G (mutNLS). All indicated plasmids carry the LEU2 marker. pRT12 and pOV72 are HIS3- and URA3-marked versions of pRT13, respectively. pKH84 is a URA3-marked version of pKH45. Localization in wild-type (WT) and snf1Δ mutant cells during growth in high (H) glucose or after a shift to low (L) glucose is indicated. N, nuclear; C, cytoplasmic; NC, some nuclear accumulation but less pronounced relative to cytoplasm than for cases described as N.
Microscopy.
Cultures were grown to mid-log phase in selective SC medium with 2% glucose except where noted otherwise. For shifts to limiting glucose, cells were collected by brief centrifugation (9 s) and resuspended in selective SC medium with 0.05% glucose. Nuclei were stained for 5 min by the addition of 4′,6′-diamidino-2-phenylindole (DAPI; 0.8 μg/ml). Cells (1 ml) were collected by brief centrifugation, resuspended in residual medium, and placed on a microscope slide. Cells were viewed using a Nikon Eclipse E800 fluorescence microscope. Images were taken with an Orca100 (Hamamatsu) camera by using Open Lab (Improvision) software and were processed with Adobe Photoshop 5.5. At least 500 cells were examined in each case, and the selected images were representative of the population.
Assay of Snf1 protein kinase activity by phosphorylation of a synthetic peptide.
Cells were grown in the indicated medium to an optical density at 600 nm of 1 and then exposed to different conditions. In all cases, cells were collected by filtration. When collected for preparation of extracts, cells were frozen immediately in liquid nitrogen. Extracts were prepared from at least two independent cultures, and assays of phosphorylation of the synthetic peptide HRMSAMSGLHLVKRR (SAMS peptide) (4) were done as previously described (9). Kinase activity is expressed as nanomoles of phosphate incorporated into the peptide per minute per milligram of protein.
RESULTS
N terminus of Gal83 suffices for Snf1-independent, glucose-regulated nuclear localization.
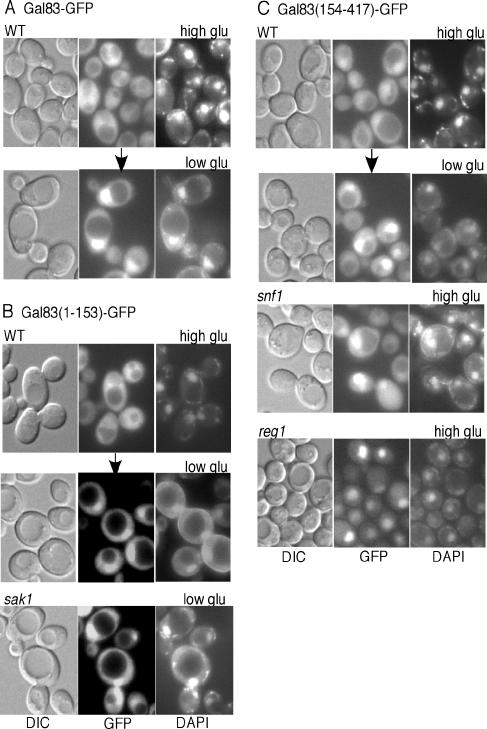
The sequence of Gal83 N-terminal to the conserved glycogen-binding domain (29) is divergent from that of the other β subunits. To examine the role of this divergent sequence in determining the localization of Gal83, we expressed residues 1 to 153, fused to GFP, from the GAL83 promoter on a centromeric plasmid (Fig. 1). The resulting protein, designated Gal83(1-153)-GFP, does not contain the C-terminal sequences that interact with Snf1 and Snf4 (14) and does not provide β-subunit function. Gal83(1-153)-GFP was expressed at higher levels than Gal83-GFP, as judged from fluorescence and immunoblot analyses (data not shown). In wild-type cells, Gal83(1-153)-GFP was cytoplasmic during growth in glucose and relocalized to the nucleus when cells were shifted to limiting glucose or glycerol plus ethanol (Fig. 2B and data not shown). Similar results were observed with gal83Δ cells, and Gal83(1-153)-GFP also accumulated in the nucleus when cells were grown in glycerol plus ethanol (data not shown). These patterns are the same as those exhibited by full-length Gal83-GFP (Fig. 2A) (9, 27). In accord with the absence of Snf1- and Snf4-interacting sequences, Gal83(1-153)-GFP showed the same localization patterns in snf1Δ, snf4Δ, and sak1Δ mutant cells as in the wild type during growth in glucose and in response to glucose limitation (Fig. 2B and data not shown). Thus, the N-terminal 153 residues of Gal83 are sufficient for glucose-regulated localization. These findings are consistent with previous results that Gal83(1-90)-GFP exhibits partially regulated localization (27).
FIG. 2.
Nucleocytoplasmic distribution of full-length and truncated Gal83-GFP. (A and B) Transformants of W303-1A (WT) or MCY5115 (sak1Δ) expressed Gal83-GFP or Gal83(1-153)-GFP from plasmids, as indicated. Cells were grown in selective SC medium plus 2% glucose (high glu) and shifted to SC medium plus 0.05% glucose (low glu) for 10 min. (C) WT, snf1Δ, and reg1Δ cells expressed Gal83(154-417)-GFP. Cells were grown in selective SC medium plus 2% glucose, and WT cells were also shifted to 0.05% glucose for 10 min. GFP fluorescence, DAPI fluorescence, and differential interference contrast (DIC) images are shown. Arrows indicate a shift to a different medium. Similar results were observed for Gal83(1-153)-GFP and Gal83(154-417)-GFP when WT cells were shifted to 2% glycerol plus 3% ethanol for 10 min.
Cytoplasmic retention of the C terminus of Gal83 by inactive Snf1 in glucose-grown cells.
To determine if the N terminus is necessary for regulated localization, we expressed Gal83(154-417)-GFP from the native promoter (Fig. 1). In glucose-grown wild-type or gal83Δ cells, Gal83(154-417)-GFP was present in the cytoplasm but also accumulated in the nucleus (Fig. 2C and data not shown), whereas Gal83-GFP was nuclear excluded (Fig. 2A). When cells were shifted to limiting glucose, Gal83(154-417)-GFP became more strongly nuclear (Fig. 2C). Similar results were observed with Gal83(244-417)-GFP, although it was not well expressed (data not shown).
Gal83(154-417)-GFP and Gal83(244-417)-GFP conferred growth on raffinose in sip1Δ sip2Δ gal83Δ cells (data not shown), indicating that they both provide at least minimal β-subunit function, consistent with the presence of Snf1- and Snf4-interacting regions C-terminal to the glycogen-binding domain (residues 161 to 243). Deletion analysis mapped the Snf1-interacting sequence (designated the KIS domain) between residues 198 and 343, which is now known to include part of the glycogen-binding domain, and mapped the Snf4-interacting sequence (ASC domain) distal to residue 343 (14).
Previous evidence indicated that kinase-dead Snf1 inhibits the nuclear localization of Gal83-GFP (9), suggesting that, in glucose-grown cells, the presence of inactive Snf1 inhibits nuclear localization. To test this idea, we first examined snf1Δ mutant cells. Gal83(154-417)-GFP was strongly nuclear localized under high- and low-glucose conditions (Fig. 2C and data not shown). We next examined reg1Δ mutant cells, in which Snf1 is constitutively active; the Reg1-Glc7 protein phosphatase 1 is required for dephosphorylation and inactivation of Snf1 in glucose-grown cells (12, 21). In reg1Δ cells, Gal83(154-417)-GFP was nuclear in high glucose (Fig. 2C), whereas Gal83-GFP and Gal83(1-153)-GFP were not (reference 27 and data not shown). These findings indicate that the presence of catalytically inactive Snf1 was responsible for the partial cytoplasmic retention of Gal83(154-417)-GFP in glucose-grown wild-type cells. When Snf1 was active or absent, Gal83(154-417)-GFP was constitutively nuclear. Thus, inactive Snf1 contributes to the cytoplasmic retention of Gal83 in glucose-grown cells through its interaction with C-terminal sequences of Gal83, and the N-terminal sequence of Gal83 is necessary for the Snf1-independent mechanism that regulates the localization of Gal83 in response to glucose signals.
N-terminal leucine-rich nuclear export signal is required for glucose-dependent nuclear exclusion of Gal83.
We next took advantage of conveniently located NheI sites to remove residues 19 to 97 from Gal83-GFP (Fig. 1). In snf1Δ and reg1Δ mutant cells, Gal83(Δ19-97)-GFP showed nuclear accumulation in both high and low glucose (Fig. 3A and data not shown). In wild-type cells, the mutant protein was not excluded from the nucleus; it was, in some cases, enriched in the nucleus during growth in high glucose, and it became strongly nuclear in response to glucose limitation (Fig. 3A). These findings indicate that sequences residing within the deleted region are required for the Snf1-independent, glucose-dependent nuclear exclusion of Gal83.
FIG. 3.
Glucose-dependent nuclear exclusion of Gal83 requires an NES and Crm1. (A) snf1Δ, reg1Δ, and WT (W303-1A) cells expressing Gal83(Δ19-97)-GFP were grown selectively in SC medium plus 2% glucose (high glu). WT cells were also shifted to 0.05% glucose (low glu) for 10 min. (B) WT cells expressing Gal83(1-153)mutNES-GFP were grown in 2% glucose and shifted to 0.05% glucose as in panel A. (C) crm1-3 mutant cells expressing Gal83(1-153)-GFP were grown in 2% glucose. GFP fluorescence, DAPI fluorescence, and DIC images are shown. Arrows indicate a shift to a different medium.
The N terminus of Gal83 contains the sequence LAYTFSQMNV (residues 39 to 48), which matches the leucine-rich NES consensus ΦX2-3ΦX2-3ΦXΦ (Φ = L, I, V, F, M) (7, 17, 28). We introduced mutations altering the four critical residues in Gal83-GFP to alanines (L39A, F43A, M46A, V48A) and refer to the quadruply mutant protein as Gal83mutNES-GFP (Fig. 1). In wild-type, snf1Δ, and reg1Δ cells, Gal83mutNES-GFP showed the same patterns of localization as Gal83(Δ19-97)-GFP (data not shown). The alteration S38A, L39A also caused the same pattern, whereas S38A alone had no effect (wild-type cells were examined; data not shown). The truncated Gal83(1-153)mutNES-GFP showed similar nuclear accumulation in both high and low glucose even in wild-type cells (Fig. 3B), consistent with its inability to interact with Snf1. We also introduced the alterations T42A and S44A to eliminate potential phosphorylation, but they had no effect (data not shown). Thus, the NES LX3FX2MXV is required for the Snf1-independent nuclear export of Gal83 in glucose-grown cells.
Nuclear export of Gal83 requires the Crm1 export receptor.
Crm1, a member of the importin-β family of nuclear transport receptors, mediates nuclear export through binding to leucine-rich NESs (8). To determine whether Gal83 depends on Crm1 for export, we examined the localization of Gal83-GFP and Gal83(1-153)-GFP in strains carrying two different point mutations, crm1-1 and crm1-3, that cause defects in nuclear export (31). Both mutant strains showed strong nuclear localization of Gal83(1-153)-GFP during growth in high glucose (Fig. 3C), thereby implicating Crm1 in the export of Gal83. In accord with these results, the localization of Gal83(1-153)-GFP was not affected by the absence of the Msn5 transport receptor, which is required for the glucose-regulated export of the Mig1 repressor but does not recognize a leucine-rich NES (5) (data not shown).
Identification of NLSs.
We next examined the N terminus of Gal83 for basic nuclear localization signals (NLSs). The only clustered basic residues are KH at position 28 and RHK at position 61; the latter is also a potential protein kinase A site (RHKSS) and resembles the Snf1 consensus site (3). Mutations altering the basic residues of either or both clusters to alanines had no effect on the localization of Gal83(1-153)-GFP; alteration of the serines also had no effect (data not shown). Nuclear localization of Gal83(1-153)-GFP may be conferred through an import signal that we have not recognized or through interactions with another protein containing an NLS.
The full-length Gal83 has additional potential NLSs. We altered one of these, KKGR at position 155, to AAGG. This alteration alone, or in combination with alteration of both N-terminal basic clusters, did not impair nuclear localization of Gal83-GFP (data not shown). However, KKGR affected the localization of Gal83(1-243)-GFP (Fig. 1) which, as expected, does not provide β-subunit function. Gal83(1-243)-GFP is constitutively nuclear in both wild-type and gal83Δ cells, and alteration of KKGR abolished its nuclear localization in glucose-grown cells, restoring glucose-regulated localization (data not shown). Thus, this sequence appears to be recognized as an NLS in the context of this truncated protein, although we cannot rule out more complicated models. The localization patterns observed for Gal83(1-153), Gal83(1-243), and Gal83 and their mutant derivatives would then simply reflect the different balance between NLS and NES elements.
2-Deoxyglucose inhibits Snf1 activity but not nuclear accumulation of Gal83(1-153)-GFP.
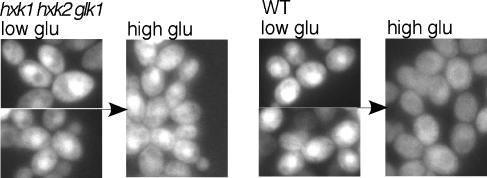
Previous evidence suggested that phosphorylation of glucose is necessary and sufficient for the glucose-dependent nuclear exclusion of Gal83-GFP, and glucose-6-phosphate was proposed as a candidate signal (27). As it is now evident that inhibition of Snf1 activity affects Gal83-GFP localization, we have revisited this issue. We first examined the localization of Gal83(1-153)-GFP in hxk1Δ hxk2Δ glk1Δ cells, which lack hexose phosphorylating activity; localization of Gal83-GFP was normal in hxk1Δ hxk2Δ cells but defective in the triple mutant (27). Cultures were grown on glycerol plus ethanol (the mutant cannot utilize glucose), shifted to 0.05% glucose for 10 min, and then adjusted to 2% glucose for 10 min. Gal83(153)-GFP remained nuclear, whereas in wild-type cells, it translocated to the cytoplasm (Fig. 4).
FIG. 4.
Glucose-dependent nuclear exclusion of Gal83(1-153)-GFP is defective in the hxk1 hxk2 glk1 mutant. Gal83(1-153)-GFP was expressed in hxk1Δ hxk2Δ glk1Δ cells and the corresponding WT cells (strains WAY.78-1 and ENY-WA-1A). Cells were grown in selective SC medium plus 2% glycerol and 3% ethanol and were shifted to 0.05% glucose (low glu) for 10 min (for consistency with other experiments). Arrows indicate the addition of glucose to a level of 2% (high glu) for 10 min. Nuclei were identified by DAPI staining (not shown). GFP fluorescence is shown.
We next examined the effects of 2-deoxyglucose, which can be phosphorylated but not metabolized. Cells were grown in 2% glucose and shifted to 0.05% glucose for 10 min, and then 0.02% 2-deoxyglucose was added. In wild-type cells, Gal83(1-153)-GFP remained nuclear after 10 min and 1 h (Fig. 5A and data not shown), whereas Gal83-GFP rapidly relocated to the cytoplasm, as reported previously (27) (Fig. 5B). In snf1Δ and reg1Δ cells, however, Gal83-GFP remained nuclear (Fig. 5B). Thus, the effect of 2-deoxyglucose on the localization of Gal83 is mediated by Snf1.
FIG. 5.
Effects of 2-deoxyglucose on localization of Gal83 and activity of Snf1. (A and B) WT (W303-1A), snf1Δ, and reg1Δ cells expressed Gal83(1-153)-GFP or Gal83-GFP as indicated. Cells were grown in selective SC medium plus 2% glucose and shifted to 0.05% glucose (low glu) for 10 min, and then 2-deoxyglucose (2DG) was added to a final concentration of 0.02% (1.2 mM) for 10 min. Arrows indicate this addition. GFP fluorescence, DAPI fluorescence, and DIC images are shown. (C) Assay of Snf1 protein kinase activity. WT cells were grown in YEP medium plus 2% glucose and shifted to YEP medium plus 0.05% glucose (low glu) for 10 min. One aliquot of the culture was collected, and other aliquots were adjusted to 2% glucose (+glu), 0.02% 2-deoxyglucose (+2DG), or 0.02% 6-deoxyglucose (+6DG) and incubated for an additional 10 min. Cells were collected by filtration, extracts were prepared, and Snf1 was partially purified and assayed by phosphorylation of the SAMS peptide.
We next assayed Snf1 catalytic activity. Glucose-grown wild-type cells were shifted to limiting glucose for 10 min and then adjusted to 2% glucose, 0.02% 2-deoxyglucose, or 0.02% 6-deoxyglucose for 10 min. Extracts were prepared, and Snf1 was partially purified and assayed by phosphorylation of a synthetic substrate, the SAMS peptide (4). The addition of glucose or 2-deoxyglucose inhibited Snf1 activity, whereas 6-deoxyglucose did not (Fig. 5C); no inhibition was observed for hxk1Δ hxk2Δ glk1Δ cells (data not shown). The inhibition of Snf1 may be indirect, and it has been noted that intracellular phosphorylation of 2-deoxyglucose depletes cellular ATP (30).
In cells lacking phosphoglucose isomerase, the addition of glucose inhibits Snf1 activity but not nuclear localization of Gal83(1-153)-GFP.
We examined the localization of Gal83(1-153)-GFP and assayed Snf1 activity in the pgi1Δ mutant, which lacks phosphoglucose isomerase and cannot isomerize glucose-6-phosphate to fructose-6-phosphate (1). Gal83(1-153)-GFP was nuclear excluded in cells grown on 2% fructose plus 0.1% glucose and translocated to the nucleus upon a shift to medium with no added carbon source. The addition of 2% fructose resulted in relocalization to the cytoplasm, but the addition of 2% glucose did not (Fig. 6A). In contrast, Gal83-GFP relocalized to the cytoplasm upon addition of either fructose or glucose (Fig. 6B). Consistent with these results, Snf1 was activated when cells were shifted from fructose to carbon source-deficient medium, and the addition of 2% glucose or 0.02% 2-deoxyglucose inhibited Snf1 (Fig. 6C). These findings provide evidence that glucose-6-phosphate is not the signal responsible for the glucose-dependent nuclear exclusion of Gal83(1-153)-GFP.
FIG. 6.
Localization of Gal83 and activity of Snf1 are differently affected by the lack of phosphoglucose isomerase. (A and B) pgi1Δ mutant cells expressed Gal83(1-153)-GFP or Gal83-GFP. Cells were grown in SC medium plus 2% fructose and 0.1% glucose and shifted to medium containing no added carbon source for 10 min. Fructose (fru) or glucose (glu) was then added to a final concentration of 2% for 20 min. GFP fluorescence, DAPI fluorescence, and DIC images are shown. (C) Assay of Snf1 protein kinase activity. pgi1Δ cells were grown in YEP medium plus 2% fructose and 0.1% glucose (fru) and shifted for 10 min to medium containing no added carbon source (none). Aliquots of the culture were then adjusted to 2% glucose (+glu) or 0.02% 2-deoxyglucose (+2DG) for an additional 20 min. Cells were collected by filtration, and Snf1 catalytic activity was assayed by phosphorylation of the SAMS peptide.
We note that the addition of glucose-6-phosphate (2 mM) to assays of Snf1 prepared from carbon source-deprived pgi1Δ cells had no inhibitory effect on Snf1 activity (data not shown), confirming that glucose-6-phosphate does not directly inhibit Snf1 protein kinase (30). Our experiment excludes the possibility that glucose-6-phosphate was metabolized by phosphoglucose isomerase present in the preparation.
Relationship between Snf1 activity and localization.
These findings indicate that the localization of Gal83, and thus that of Snf1-Gal83 protein kinase, is determined both by signals impinging directly on Gal83 and by the activation status of Snf1. During growth on abundant glucose, Snf1-Gal83 is largely inactive and cytoplasmic, whereas during growth on nonfermentable carbon sources or in response to acute carbon stress, Snf1-Gal83 is active and nuclear enriched (9, 10, 15, 27). We considered the possibility that, during growth on carbon sources that are neither very preferred or nonpreferred, the existence of multiple control mechanisms could result in the cytoplasmic localization of active Snf1-Gal83. The localization of Gal83-GFP in cells growing on some such carbon sources is highly strain dependent; however, Gal83-GFP was cytoplasmic in both W303 and an S288C-related strain during growth on sucrose (10, 27). The snf1 mutation was identified by its sucrose-nonfermenting phenotype, suggesting that this cytoplasmic Snf1-Gal83 was active.
To explore this further, we assayed Snf1 catalytic activity in sucrose-grown W303 cells. Cultures were grown to mid-log phase in synthetic complete medium containing 2% sucrose, an aliquot was taken for microscopic examination, and cells were collected for assays of Snf1 activity. Gal83-GFP was cytoplasmic, as was Gal83(1-153)-GFP; however, Snf1 activity was not significantly higher than that of cells grown in 2% glucose (0.63 ± 0.07 and 0.56 ± 0.13 nmol/min/mg, respectively). In control cultures shifted from glucose to sucrose for 10 min or grown on 2% glycerol plus 3% ethanol, Snf1 was activated (fivefold-higher activity) and Gal83(1-153)-GFP accumulated in the nucleus. The utilization of sucrose requires the Snf1-dependent expression of the SUC2 gene encoding invertase, a stable secreted enzyme that hydrolyzes sucrose; however, after adaptation, minimal Snf1 activity is apparently required for continued growth. Consistent with these findings, the snf1Δ mutant exhibits a severe growth defect on sucrose only under anaerobic (fermentative) conditions or in the presence of the respiratory inhibitor antimycin A. Although cytoplasmic Snf1 proved to be largely inactive in this case, it remains possible that other conditions result in cytoplasmic localization of active Snf1.
DISCUSSION
We have examined here the roles of the Snf1 and Gal83 subunits in regulating the nucleocytoplasmic distribution of Snf1-Gal83 protein kinase in response to glucose signals. We showed that the N terminus of Gal83, which is divergent from those of the other two β subunits, is necessary and sufficient for the Snf1-independent regulation of nuclear localization. Gal83(1-153)-GFP was cytoplasmic and excluded from the nucleus in high glucose conditions and translocated to the nucleus in response to glucose limitation. Localization of Gal83(1-153)-GFP was not affected by snf1Δ, reg1Δ, or sak1Δ mutations, indicating that it is controlled independently of Snf1.
We identified a leucine-rich NES in the N terminus (LX3FX2MXV, residues 39 to 48) that is required for the Snf1-independent nuclear exclusion of Gal83 in glucose-grown cells. Mutation of the NES resulted in nuclear accumulation during growth in high glucose for Gal83(1-153)mutNES-GFP in wild-type cells and for Gal83mutNES-GFP in reg1Δ or snf1Δ mutant cells. Mutation of the gene encoding the Crm1 export receptor also resulted in nuclear accumulation of Gal83(1-153)-GFP in high glucose. A leucine-rich NES, VX3VX2LXL, is conserved at a similar position in the N terminus (residues 44 to 53) of the β subunit Sip2, which is excluded from the nucleus (27). Gal83 also contains other possible NESs, notably LX3LX2VXL (residues 350 to 359), which is conserved in Sip2 and AMP-activated protein kinase β1 and β2; however, its functional role remains uncertain as Gal83(154-417)-GFP showed strong nuclear accumulation in the absence of Snf1. Although Gal83 includes various potential NLSs, including one at position 155 that affects the nuclear localization of Gal83(1-243)-GFP, we did not identify an NLS responsible for the import of Gal83(1-153)-GFP. This region may contain a nonclassical NLS, or import may depend on another unidentified protein with an NLS.
We also present evidence that the C terminus of Gal83 is involved in the regulation of localization through its interaction with Snf1; previous studies showed that activation of Snf1 is required for nuclear accumulation of Snf1-Gal83 (9). During growth in glucose, Gal83(154-417)-GFP was partially cytoplasmic in wild-type cells but strongly nuclear in reg1Δ and snf1Δ cells. Similar patterns were observed with full-length Gal83 proteins lacking the NES. These findings indicate that inactive Snf1 contributes to the cytoplasmic retention of Gal83 in glucose-grown cells through its interaction with C-terminal sequences of Gal83.
Further studies are required to understand the mechanism by which Snf1 activity regulates localization. There are multiple possibilities; for example, a phosphorylation event could inhibit cytoplasmic retention by releasing interaction with an anchoring protein, or phosphorylation could be required for nuclear import of Snf1-Gal83 by unmasking an NLS or affecting interaction with an import factor. Gal83 is phosphorylated by Snf1 in vitro (32), but immunoblot analysis showed no differential modification in response to glucose availability in vivo (K. Hedbacker, unpublished results). The dependence of nuclear accumulation on the activation of Snf1 may be physiologically important because none of the Snf1-activating kinases is located in the nucleus (2, 9, 15); hence, the retention of inactive Snf1 in the cytoplasm maintains accessibility to the activating kinases.
Finally, we present evidence that the signal responsible for the glucose-dependent nuclear exclusion of Gal83(1-153)-GFP requires glucose phosphorylation but is not glucose-6-phosphate. While glucose had no effect in cells lacking hexose kinases, the addition of 2-deoxyglucose to glucose-limited wild-type cells, or the addition of glucose to pgi1Δ mutant cells lacking phosphoglucose isomerase, rapidly inhibited Snf1 activity but did not promote the cytoplasmic localization of Gal83(1-153)-GFP. Thus, previous evidence suggesting glucose-6-phosphate as a candidate signal regulating Gal83 localization (27) can be accounted for by inhibitory effects on Snf1 activity. These findings further suggest that Snf1 activity and localization of Gal83(1-153) are regulated by different signals; a caveat is that depletion of ATP may limit the phosphorylation of Snf1 by its activating kinases.
The signaling mechanisms that control localization of Snf1-Gal83 remain unclear. Nuclear accumulation of Snf1-Gal83 is defective in a sak1Δ mutant (9); one explanation is that Sak1 is the major kinase responsible for activating Snf1, but substantial activation of Snf1 by a heterologous kinase did not suppress this defect (12). We found that Sak1 is not required for nuclear localization of Gal83(1-153)-GFP, consistent with previous evidence that it is not required for localization of Gal83-GFP in the absence of Snf1. Protein kinase A regulates the localization of the Sip1 β subunit (10) but not that of Gal83-GFP (10, 27) or Gal83(1-153)-GFP (Hedbacker, unpublished). We have examined the localization of Gal83(1-153)-GFP and Gal83-GFP in an array of mutants lacking protein kinases that have been implicated in nutrient responses but did not identify another kinase that affects localization (Hedbacker, unpublished); functional redundancy may have precluded detection of a defect. We also mutated several potential CK2 sites in Gal83-GFP because CK2 catalytic subunits copurify with Sak1 (6); however, the substitution of alanine for serines 12 and 17, aspartates 94, 95, and 96, or serines 100 through 104 had no effect (Hedbacker, unpublished).
These studies show that regulation of the nucleocytoplasmic distribution of Snf1-Gal83 is complex, involving multiple regulatory mechanisms operating on Snf1 and Gal83. Under the growth conditions that we have examined, Snf1 activity correlated with the localization of Snf1-Gal83. Under other conditions, in particular those encountered during growth in natural environments, these multiple control mechanisms may serve to fine-tune the nucleocytoplasmic distribution of active Snf1-Gal83 and thus modulate the expression of subsets of its target genes.
Acknowledgments
We thank P. Roach, P. Sanz, and M. Rosbash for strains.
This work was supported by NIH grant GM34095 to M.C.
Footnotes
Published ahead of print on 27 October 2006.
REFERENCES
- 1.Aguilera, A. 1986. Deletion of the phosphoglucose isomerase structural gene makes growth and sporulation glucose dependent in Saccharomyces cerevisiae. Mol. Gen. Genet. 204:310-316. [DOI] [PubMed] [Google Scholar]
- 2.Bouquin, N., Y. Barral, R. Courbeyrette, M. Blondel, M. Snyder, and C. Mann. 2000. Regulation of cytokinesis by the Elm1 protein kinase in Saccharomyces cerevisiae. J. Cell Sci. 113:1435-1445. [DOI] [PubMed] [Google Scholar]
- 3.Dale, S., W. A. Wilson, A. M. Edelman, and D. G. Hardie. 1995. Similar substrate recognition motifs for mammalian AMP-activated protein kinase, higher plant HMG-CoA reductase kinase-A, yeast SNF1, and mammalian calmodulin-dependent protein kinase I. FEBS Lett. 361:191-195. [DOI] [PubMed] [Google Scholar]
- 4.Davies, S. P., D. Carling, and D. G. Hardie. 1989. Tissue distribution of the AMP-activated protein kinase, and lack of activation by cyclic-AMP-dependent protein kinase, studied using a specific and sensitive peptide assay. Eur. J. Biochem. 186:123-128. [DOI] [PubMed] [Google Scholar]
- 5.DeVit, M. J., and M. Johnston. 1999. The nuclear exportin Msn5 is required for nuclear export of the Mig1 glucose repressor of Saccharomyces cerevisiae. Curr. Biol. 9:1231-1241. [DOI] [PubMed] [Google Scholar]
- 6.Elbing, K., R. R. McCartney, and M. C. Schmidt. 2006. Purification and characterization of the three Snf1-activating kinases of Saccharomyces cerevisiae. Biochem. J. 393:797-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischer, U., J. Huber, W. C. Boelens, I. W. Mattaj, and R. Luhrmann. 1995. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82:475-483. [DOI] [PubMed] [Google Scholar]
- 8.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 9.Hedbacker, K., S. P. Hong, and M. Carlson. 2004. Pak1 protein kinase regulates activation and nuclear localization of Snf1-Gal83 protein kinase. Mol. Cell. Biol. 24:8255-8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hedbacker, K., R. Townley, and M. Carlson. 2004. cAMP-dependent protein kinase regulates the subcellular localization of Snf1-Sip1 protein kinase. Mol. Cell. Biol. 24:1836-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hong, S.-P., F. C. Leiper, A. Woods, D. Carling, and M. Carlson. 2003. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc. Natl. Acad. Sci. USA 100:8839-8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong, S.-P., M. Momcilovic, and M. Carlson. 2005. Function of mammalian LKB1 and Ca2+/calmodulin-dependent protein kinase kinase α as Snf1-activating kinases in yeast. J. Biol. Chem. 280:21804-21809. [DOI] [PubMed] [Google Scholar]
- 13.Huang, D., W. A. Wilson, and P. J. Roach. 1997. Glucose-6-P control of glycogen synthase phosphorylation in yeast. J. Biol. Chem. 272:22495-22501. [DOI] [PubMed] [Google Scholar]
- 14.Jiang, R., and M. Carlson. 1997. The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol. Cell. Biol. 17:2099-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, M. D., S. P. Hong, and M. Carlson. 2005. Role of Tos3, a Snf1 protein kinase kinase, during growth of Saccharomyces cerevisiae on nonfermentable carbon sources. Eukaryot. Cell 4:861-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuchin, S., I. Treich, and M. Carlson. 2000. A regulatory shortcut between the Snf1 protein kinase and RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. USA 97:7916-7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.la Cour, T., R. Gupta, K. Rapacki, K. Skriver, F. M. Poulsen, and S. Brunak. 2003. NESbase version 1.0: a database of nuclear export signals. Nucleic. Acids Res. 31:393-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin, S. S., J. K. Manchester, and J. I. Gordon. 2003. Sip2, an N-myristoylated beta subunit of Snf1 kinase, regulates aging in Saccharomyces cerevisiae by affecting cellular histone kinase activity, recombination at rDNA loci, and silencing. J. Biol. Chem. 278:13390-13397. [DOI] [PubMed] [Google Scholar]
- 19.Lo, W. S., L. Duggan, N. C. Emre, R. Belotserkovskya, W. S. Lane, R. Shiekhattar, and S. L. Berger. 2001. Snf1—a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science 293:1142-1146. [DOI] [PubMed] [Google Scholar]
- 20.Ludin, K., R. Jiang, and M. Carlson. 1998. Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 95:6245-6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCartney, R. R., and M. C. Schmidt. 2001. Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J. Biol. Chem. 276:36460-36466. [DOI] [PubMed] [Google Scholar]
- 22.Nath, N., R. R. McCartney, and M. C. Schmidt. 2003. Yeast Pak1 kinase associates with and activates Snf1. Mol. Cell. Biol. 23:3909-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shirra, M. K., S. E. Rogers, D. E. Alexander, and K. M. Arndt. 2005. The Snf1 protein kinase and Sit4 protein phosphatase have opposing functions in regulating TATA-binding protein association with the Saccharomyces cerevisiae INO1 promoter. Genetics 169:1957-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sutherland, C. M., S. A. Hawley, R. R. McCartney, A. Leech, M. J. Stark, M. C. Schmidt, and D. G. Hardie. 2003. Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae SNF1 complex. Curr. Biol. 13:1299-1305. [DOI] [PubMed] [Google Scholar]
- 25.Treitel, M. A., S. Kuchin, and M. Carlson. 1998. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:6273-6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vincent, O., and M. Carlson. 1999. Gal83 mediates the interaction of the Snf1 kinase complex with the transcription activator Sip4. EMBO J. 18:6672-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vincent, O., R. Townley, S. Kuchin, and M. Carlson. 2001. Subcellular localization of the Snf1 kinase is regulated by specific β subunits and a novel glucose signaling mechanism. Genes Dev. 15:1104-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wen, W., J. L. Meinkoth, R. Y. Tsien, and S. S. Taylor. 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell 82:463-473. [DOI] [PubMed] [Google Scholar]
- 29.Wiatrowski, H. A., B. J. W. van Denderen, C. D. Berkey, B. E. Kemp, D. Stapleton, and M. Carlson. 2004. Mutations in the Gal83 glycogen-binding domain activate the Snf1/Gal83 kinase pathway by a glycogen-independent mechanism. Mol. Cell. Biol. 24:352-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson, W. A., S. A. Hawley, and D. G. Hardie. 1996. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr. Biol. 6:1426-1434. [DOI] [PubMed] [Google Scholar]
- 31.Yan, C., L. H. Lee, and L. I. Davis. 1998. Crm1p mediates regulated nuclear export of a yeast AP-1-like transcription factor. EMBO J. 17:7416-7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang, X., R. Jiang, and M. Carlson. 1994. A family of proteins containing a conserved domain that mediates interaction with the yeast SNF1 protein kinase complex. EMBO J. 13:5878-5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young, E. T., K. M. Dombek, C. Tachibana, and T. Ideker. 2003. Multiple pathways are co-regulated by the protein kinase Snf1 and the transcription factors Adr1 and Cat8. J. Biol. Chem. 278:26146-26158. [DOI] [PubMed] [Google Scholar]
- 34.Young, E. T., N. Kacherovsky, and K. Van Riper. 2002. Snf1 protein kinase regulates Adr1 binding to chromatin but not transcription activation. J. Biol. Chem. 277:38095-38103. [DOI] [PubMed] [Google Scholar]