Abstract
Skp1p is an essential component of SCF-type E3 ubiquitin ligase complexes and associates with these through binding to F-box proteins. Skp1p also binds F-box proteins in a number of non-SCF complexes. The Skp1p-associated yeast protein Soi3p/Rav1p (hereafter referred to as Rav1p) is a component of the RAVE complex required for regulated assembly of vacuolar ATPase (V-ATPase). Rav1p is also involved in transport of TGN proteins and endocytic cargo between early and late endosomes. To evaluate the role of Skp1p in the RAVE complex, we made use of the fact that overexpression of Rav1p is toxic because it sequesters Skp1p from essential interactions. We isolated a separation of function allele of SKP1, skp1(Asn108Tyr), that completely abrogated the Rav1p interaction but allowed Skp1p to perform other essential cellular functions. Cells containing the skp1(Asn108Tyr) allele as the sole source of Skp1p exhibited normal V-ATPase assembly and activity. However, in the skp1(Asn108Tyr) mutant strain, the membrane-associated pool of Rav1-green fluorescent protein was increased, suggesting that Skp1p is important for the release of Rav1p from endosomal membranes where it functions in V-ATPase assembly. Thus, although part of the RAVE complex, Skp1p does not appear to be involved in V-ATPase assembly but instead in the cycling of the complex off membranes. This work also provides a generalizable approach to defining the roles of interactions of Skp1p with individual F-box proteins through the isolation of special alleles of SKP1.
The processing protease Kex2p is a type I transmembrane protein necessary for cleaving pro-α-factor and other proproteins in the secretory pathway of the budding yeast Saccharomyces cerevisiae (11). Steady-state localization of Kex2p to the trans-Golgi network (TGN) depends on cycling of the protein between the TGN and the late endosome/prevacuolar compartment (PVC) (6). Kex2p localization is mediated by two TGN localization signals (TLSs) in the Kex2p cytosolic tail (6, 41). TLS1 promotes retrieval of Kex2p from the PVC to the TGN; whereas, TLS2 slows Kex2p delivery to the PVC, either through retention in the TGN or by promoting an alternative pathway through the early endosome (6, 15). Disruption of either TLS1 or TLS2 results in increased trafficking of Kex2p to the vacuole, where it is degraded (6, 41).
The SOI3 gene was identified by a mutation (soi3-1) that suppressed mislocalization of Kex2p bearing a Tyr713Ala mutation in TLS1 (27). Deletion of SOI3 disrupted trafficking of TGN membrane proteins Kex2p and A-ALP, a fusion protein consisting of the cytosolic domain of dipeptidyl aminopeptidase A fused to the transmembrane and lumenal domains of the vacuolar protein alkaline phosphatase (24) but not the vacuolar sorting receptor, Vps10p, between the TGN and PVC (34). The soi3Δ mutation also delayed delivery of the plasma membrane-localized a-factor receptor, Ste3p, to the vacuole at a step after endocytic internalization (34). Although most of the protein is cytosolic, a fraction of tagged Soi3p was associated with dense membranes and localized to peripheral punctate structures (34). These data argued that Soi3p is required for transport between the early endosome and PVC.
A clue to the molecular function of Soi3p in early endosome-to-PVC trafficking comes from the fact that the protein was independently isolated as Rav1p, a Skp1p-interacting protein that forms part of a complex termed RAVE (regulator of the H+-ATPase of the vacuolar and endosomal membranes), which includes a third polypeptide, Rav2p (33) (Soi3p will hereafter be referred to as Rav1p). RAVE is associated with the V1 subcomplex (“sector”) of the vacuolar ATPase (V-ATPase), a multisubunit proton-translocating ATPase responsible for the acidification of intracellular compartments (13, 38). In yeast grown on a poor carbon source, V1 is largely cytosolic but can be induced, upon addition of glucose, to assemble with the integral membrane Vo sector to form an active holoenzyme (17). Rav1p is important for efficient assembly and for activation of V-ATPase (33, 36). We hypothesized that the role of Rav1p in early endosome-to-PVC transport reflects a requirement for assembly and activation of V-ATPase at the early endosome (34). This may be a conserved function in that V-ATPase activity is required for early endosome maturation in mammalian cells (8) and Rav1p has homologues in both Drosophila and humans (18, 19).
The Rav1p-Skp1 interaction is intriguing because Skp1p is a component of SCF (Skp1, Cdc53/Cullin, F-box protein)-type E3 ubiquitin-protein ligases. SCF complexes are responsible for ubiquitination of numerous substrates (10). The specificity of SCF complexes is achieved, in part, through the interchangeable F-box proteins that contain a Skp1p-binding motif (the F-box sequence) and a substrate recognition domain often consisting of WD-40 or leucine-rich repeats. The F-box protein thus acts as an adaptor between the ubiquitin transferase and its substrate (25). Although not an identified F-box protein, Rav1p does contain potential F-box motifs. Rav1p and the RAVE complex are hypothesized to be a non-SCF complex since additional components of the SCF complex cannot be coimmunoprecipitated (33).
Here we demonstrate that overexpression of Rav1p is toxic and that Skp1p acts as a multicopy suppressor of this toxicity. Taking advantage of the Rav1p overexpression phenotype, we designed a screen to identify skp1 mutants that would not interact with Rav1p but would maintain other essential interactions. One such mutant completely abrogated the Rav1p interaction and allowed us to dissect the role of the Rav1p-Skp1p interaction in Kex2p trafficking and V-ATPase assembly. We determined that the Rav1p-Skp1p interaction is not required for V-ATPase assembly/activation. Moreover, loss of the interaction alters Kex2p trafficking, but it does so differently than a rav1-null mutation. Finally, we found that loss of Skp1p-Rav1p binding alters localization of a Rav1-green fluorescent protein (GFP) fusion, suggesting that Skp1p functions in release of Rav1p from early endosomes.
MATERIALS AND METHODS
Reagents, strains, plasmids, and media.
Unless otherwise noted, chemicals were purchased from Sigma Chemical Co. (St. Louis, MO) and used as received. Antibodies are described where used. Yeast media were as described previously (31) except as noted. SGC-Leu plates were equivalent to SC-Leu dropout plates but contained 2% (wt/vol) galactose in place of glucose. YPAD, pH 5.6, plates were buffered with 50 mM Na-morpholineethanesulfonic acid (MES), pH 5.6, and 50 mM Na-morpholinepropanesulfonic acid (MOPS), pH 5.6. YPAD, pH 7.5, plates were buffered with 100 mM Tris-HCl, pH 7.5. YPAD zinc plates were supplemented with 5 mM ZnCl2 and buffered with 50 mM Na-MOPS, pH 5.6, and 50 mM Na-MES, pH 5.6. YPAD cobalt plates were supplemented with 750 μM CoCl2.
Plasmids and strains are outlined in Tables 1 and 2. Plasmid pSOI3.HA.HIS6-SL was created by replacing the SacI site in pSOI3.HA.HIS6, in which sequences encoding a triple hemagglutinin (HA) epitope tag followed by six His codons are fused to the 3′ end of the wild-type RAV1 gene (34), with a SalI site by linker ligation. Plasmid p413.ADH.SOI3.HA.HIS6, in which the RAV1 fusion is under the control of the ADH1 promoter on CEN6 ARS4 HIS3 plasmid p413 (22), was created by ligating the NarI/SalI fragment of pSOI3.HA.HIS6 containing the C terminus of SOI3.HA.HIS6 into p413ADH.SOI3 cleaved by NarI and SalI. Plasmid p416.ADH.SOI3.GFP was created by inserting an EcoRI/SalI fragment containing the SOI3.GFP fragment from p413.ADH.SOI3.GFP (34) into plasmid p416.ADH cleaved by EcoRI and SalI. Plasmid p413GAL.SOI3 was constructed by PCR amplifying the RAV1 structural gene with primers containing EcoRI sites and inserting the resulting fragment into the EcoRI site of p413.GAL.
TABLE 1.
Yeast strains
| Strain | Relevant genotype | Parental strain |
|---|---|---|
| CRY1 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | W303-1A |
| CRY2 | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | W303-1B |
| DC14 | MATahis1 | |
| KRY18-1A | MATα kex2Δ::TRP1 | CRY2 |
| GSY11-2A | MATα soi3Δ::Kanr | CRY2 |
| GSY11-r1 | MATα soi3Δ::Kanrkex2Δ::TRP1 | CRY2 |
| EBY23 | MATa Kanr::PGAL1-SOI3 | CRY1 |
| EBY72-2 | MATα vma2Δ::Kanr | CRY2 |
| EBY79-12 | MATα doa4Δ::HIS3 soi3Δ::Kanr | CRY2 |
| EBY94 | MATα vma2Δ::LEU2 | CRY2 |
| EBY116-4 | MATα skp1Δ::HIS3 soi3Δ::Kanr [pskp1-Asn108Tyr] | CRY2 |
| EBY116-6 | MATα skp1Δ::HIS3 [pskp1(Asn108Tyr)] | CRY2 |
| EBY127-2 | MATα kex2Δ::TRP1 skp1Δ::HIS3 | CRY2 |
| LPY01-4 | MATα soi3Δ::Kanrskp1Δ::URA3 [pskp1(Asn108Tyr)] | CRY2 |
| LPY02-4 | MATα soi3Δ::Kanrskp1Δ::URA3 [pCB6] | CRY2 |
TABLE 2.
Plasmids
| Plasmid name | Relevant characteristic(s) | Source or reference |
|---|---|---|
| P413.GAL | Galactose-regulated expression plasmid | D. Thiele |
| P413.GAL.SOI3 | SOI3 under GAL1 promoter | This study |
| YEp13 | 2μm plasmid (library backbone) | 42 |
| YEp96 | Wild-type ubiquitin under CUP1 promoter | Ellison and Hochstrasser |
| pLib.SKP1 | Library vector containing SKP1 | This study |
| pskp1(Asn108Tyr) | Soi3p-resistant skp1 allele | This study |
| p413ADH.SOI3 | SOI3 under ADH promoter | 34 |
| p413.ADH.SOI3.HA.HIS6 | SOI3.HA.HIS6 under ADH promoter | This study |
| p413.ADH.SOI3.GFP | SOI3.GFP under ADH promoter (HIS3) | 34 |
| p416.ADH.SOI3.GFP | SOI3.GFP under ADH promoter (URA3) | This study |
| pCWKX20 | Wild-type KEX2 under GAL1 promoter | 41 |
| pCWKX21 | Y713A-KEX2 under GAL1 promoter | 41 |
| pCWKX20-I718 | I718tail-KEX2 under GAL1 promoter | 41 |
| pCWKX21-I718 | Y713A, I718tail-KEX2 under GAL1 promoter | 41 |
| pCB6 | Wild-type ubiquitin under CUP1 promoter | 4 |
| pCB6-Xho1Δ | XhoI digest and blunt ligation of pCB6 | This study |
All yeast strains were derived from the W303 background (28) (Table 1). Gene disruptions were created by homologous recombination of a marker into the desired gene locus and confirmed by PCR (14), and subsequent strains were created by standard genetic crosses. The GAL1 promoter was integrated upstream of RAV1 using the Kanr marker to create EBY23 (21).
Identification of Skp1p and isolation of skp1(Asn108Tyr).
EBY23 was transformed with a multicopy YEp13 library (42) and plated on SGC-Leu plates after a 4-h outgrowth in YPAD media. Transformants that grew on galactose-containing plates were isolated, and the resulting plasmids were retested for suppression of the toxicity of Rav1p overexpression. Inserts of confirmed suppressor plasmids were partially sequenced to identify the genomic insert.
A library of SKP1 mutants was created by PCR mutagenesis of wild-type SKP1 in plasmid pCB6 (4). Plasmid pCB6-XhoIΔ was created by digesting pCB6 with XhoI and blunting the ends with Klenow polymerase before ligation. Taq polymerase was used under standard conditions to maximize the yield of single-point mutations. Mutagenized PCR products were cotransformed into EBY23 along with pCB6-XhoIΔ, digested with AvaI/NsiI, to generate pCB6 containing mutagenized SKP1 inserts by homologous recombination. Transformants were plated on SGC-Leu plates after a 4-h outgrowth in liquid YPAD medium. Suppressors of Rav1p toxicity were isolated, and the phenotypes were confirmed. Confirmed plasmids were sequenced to identify the mutations.
Rav1p-Skp1p coimmunoprecipitation.
Cell extracts were prepared by gentle freeze-thaw lysis of yeast spheroplasts as described by Baker et al. and Sipos and Fuller (5, 35). S13 fractions were obtained from the supernatant by centrifugation of thawed spheroplasts, diluted in 800 μl of immunoprecipitation buffer containing protease inhibitors (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.1 mM EDTA, 0.1% Triton X-100, and Complete Mini protease inhibitor cocktail tablets [Roche]), at 13,000 × g for 5 min at 4°C. Extracts from 10 optical densities (OD) of cells (equivalent to 10 ml of cells grown in log phase to an A600 of 1.0; ∼1 × 108 cells) were immunoprecipitated with 0.25 μl of 12CA5 (5 mg/ml, anti-HA from Roche), 2 μl of rabbit anti-mouse (2 mg/ml; Jackson Laboratory), and 10 μl of Pansorbin (Calbiochem) for 2 h at 4°C. Immunoprecipitates were washed three times with immunoprecipitation buffer. Samples were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE, 11%), and gels were subsequently blotted to nitrocellulose (175 mA, 2 h). Nitrocellulose filters, probed with either anti-HA (1:5,000, 12CA5 from Roche) and sheep anti-mouse horseradish peroxidase (HRP, 1:2,000; Amersham) or rabbit anti-Skp1p antiserum (1:2,500; gift of Stephen Elledge, Harvard University) and protein A-HRP (1:5,000; Zymed), were developed using the ECL plus kit (Amersham).
Membrane fractionation.
Cell extracts were prepared as described above except that thawed spheroplasts were homogenized with 15 strokes in a Dounce homogenizer prior to centrifugation. Sucrose gradients contained steps of 750 μl of 60% sucrose (in 20 mM Na-HEPES, pH 6.8) and 750 μl of 15% sucrose (in 20 mM Na-HEPES, pH 6.8) in 11- by 34-mm polyallomer centrifuge tubes (Beckman). S13 fractions from 125 OD of cells were loaded onto gradients and centrifuged at 200,000 × g for 4 h using a TLX centrifuge and TLS-55 swinging bucket rotor (Beckman). Membranes were collected from the interface of the two steps. S13 fractions were not examined because Rav1-HAp was unstable in this fraction (E. J. Brace and R. S. Fuller, unpublished data). The Golgi protein Mnn1p was used as a loading control (29). Membranes from 3.3 OD of cells were subjected to SDS-PAGE (8%), and gels were blotted to nitrocellulose (200 mA, 2 h). Blots were probed with either mouse monoclonal anti-GFP (1:100; Clontech) and sheep anti-mouse HRP (1:2,000; Amersham) or rabbit anti-Mnn1p (1:500; gift of Todd Graham, Vanderbilt University) and donkey anti-rabbit HRP (1:2,000; Amersham).
Microscopy.
Microscopy was performed using a Nikon Eclipse 800 microscope and an ORCA2 charge-coupled device camera (Hamamatsu). ESEE Software (Inovision, Raleigh, NC) was used for image capture. Soi3-GFP images were from cells grown in log phase in minimal medium at 25°C. Quinacrine staining of yeast vacuoles was as described previously (30).
RESULTS
Overexpression of Rav1p is toxic.
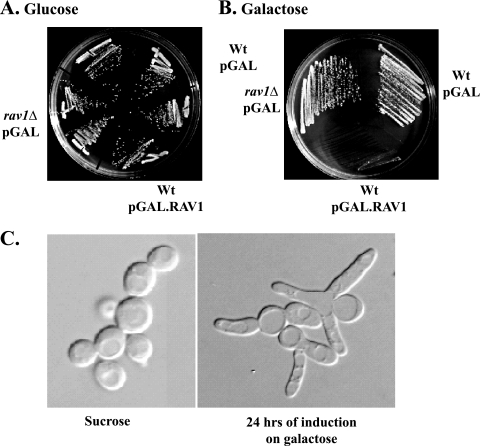
During the cloning of SOI3/RAV1, we observed that strains expressing RAV1 from strong promoters exhibited poor growth (E. J. Brace and R. S. Fuller, unpublished data). To test specifically whether high levels of Rav1p expression were toxic to yeast cells, RAV1 was placed under the galactose-inducible GAL1 promoter. Strains expressing Rav1p from the GAL1 promoter were able to grow on plates containing glucose but were unable to grow on plates containing galactose (Fig. 1A and B). Following a shift from sucrose to galactose, strains grew normally for approximately 10 h before exhibiting poor growth (E. J. Brace and R. S. Fuller, unpublished data), eventually arresting with an elongated, multibudded phenotype (Fig. 1C).
FIG. 1.
Overexpression of Rav1p is toxic. (A and B) CRY1 (wild type) cells were transformed either with p413GAL or p413GAL.RAV1, and GSY11-1A (rav1Δ) was transformed with p413GAL. The p413GAL plasmid possesses the galactose-inducible GAL1 promoter. Strains were tested on SC-His (glucose) and SGC-His (galactose) for the ability to grow when Rav1p was overexpressed. (C) CRY1 (wild type) cells transformed with p413GAL.RAV1 were grown in log phase in either SSC-His (Sucrose) or SGC-His (galactose). Strains were visualized under differential interference contrast microscopy after 24 h.
Overexpression of Skp1p suppresses Rav1p toxicity.
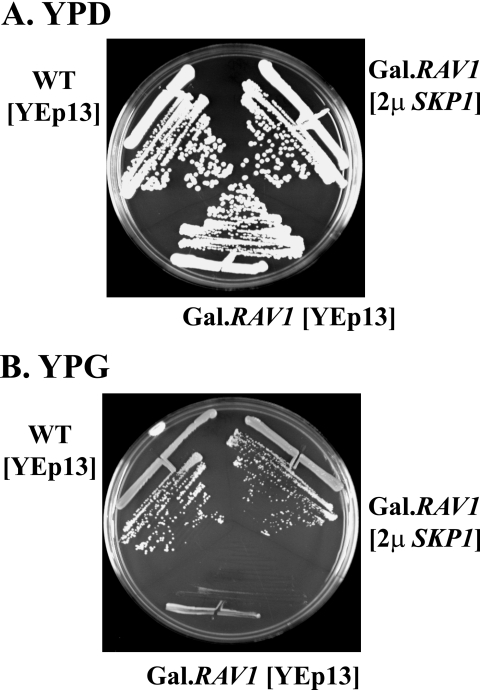
Rav1p is predicted to possess several WD-40 repeats that likely mediate interactions with other proteins (37). As an approach to identify possible Rav1p-interacting proteins, we attempted to isolate multicopy suppressors of the slow-growth phenotype caused by Rav1p overexpression. The GAL1-RAV1 strain was transformed with a 2μm plasmid multicopy library and plated on galactose. Twenty-one clones that grew on galactose were selected for characterization. By sequence analysis, each contained only the SKP1 gene in common. Figure 2 shows suppression by one of these plasmids, YEp-SKP1, of the toxicity of Rav1p overexpression.
FIG. 2.
Overexpression of Skp1p suppresses Rav1p toxicity. (A and B) CRY1 cells (wild type) were transformed with a YEp13 plasmid (2μm plasmid marked by LEU2), and EBY23 cells (integrated GAL.RAV1) were transformed with either the YEp13 plasmid or one of the isolated YEp13 library plasmids containing SKP1 (YEp13-SKP1, “2μ SKP1” in figure). Cells were grown on YPD (glucose) or YPG (galactose) to demonstrate that Skp1p is a multicopy suppressor of Rav1p overexpression.
Skp1p has multiple essential roles in yeast. We hypothesized that overexpression of Rav1p is toxic because excess Rav1p sequesters Skp1p, preventing Skp1p from participating in essential SCF complexes. One likely candidate is the SCF complex containing Cdc4p, SCFCdc4, because cdc4ts mutants arrest with a multibudded terminal phenotype similar to that seen with Rav1p overexpression (1, 16).
The skp1(Asn108Tyr) mutant performs essential functions of Skp1p but does not bind Rav1p.
To characterize specifically the role of Skp1p in Rav1p function, we screened for skp1 mutants that provided resistance to Rav1p overexpression but could support yeast cell growth. Presumably, such mutants would fail to bind Rav1p but would retain the ability to interact with other essential F-box proteins. After PCR mutagenesis, a single skp1 allele was identified that suppressed the toxic phenotype of Rav1p overexpression and could support growth in single copy in the absence of wild-type SKP1. The mutant, skp1(Asn108Tyr), was phenotypically distinct from other characterized skp1 point mutations (skp1-1, -2, -3, -11, and -12) (4, 9) in that none of these suppressed Rav1p overexpression at 25°C, 30°C, or 33°C (E. J. Brace and R. S. Fuller, unpublished data).
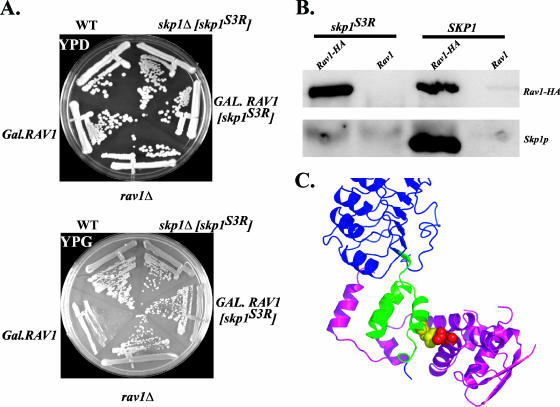
To determine directly whether the Rav1p-Skp1p interaction was disrupted by the skp1(Asn108Tyr) mutation, the ability of Rav1p to interact with Skp1p(Asn108Tyr) was tested by coimmunoprecipitation. Protein extracts of strains expressing Rav1-HAp and either Skp1p or Skp1p(Asn108Tyr) were immunoprecipitated with anti-HA under native conditions and probed with either anti-HA or anti-Skp1 antibodies. Rav1-HA coimmunoprecipitated wild-type Skp1p, but not Skp1p(Asn108Tyr) (Fig. 3B). This result confirmed that Skp1(Asn108Tyr) did not interact with Rav1p and supports both the conclusion that toxicity of Rav1p overexpression is caused by titration of Skp1p and that the Skp1p-Rav1p interaction is direct. Furthermore, because the allele could support growth as the lone copy of SKP1 (Fig. 3A, top), the essential interactions of Skp1p must remain intact.
FIG. 3.
Skp1p(Asn108Tyr) performs essential functions of Skp1p but does not bind Rav1p. A skp1 mutant [skp1(Asn108Tyr)] was identified through a genetic screen that suppressed the toxicity of Rav1p overexpression. (A) CRY1 (wild type), EBY23 (GAL.RAV1), GSY11-1A (rav1Δ), EBY23 [pskp1(Asn108Tyr)], and EBY116-6 (skp1Δ) [pskp1(Asn108Tyr)] were streaked out on YPD (glucose) and YPG (galactose) plates, demonstrating that skp1(Asn108Tyr) can both suppress Rav1p overexpression and exist as the sole copy of Skp1p in the cell. (B) Rav1p does not coimmunoprecipitate Skp1p(Asn108Tyr). Strains LPY01-4 {rav1Δ skp1Δ [pskp1(Asn108Tyr)]} and LPY02-4 (rav1Δ skp1Δ [pCB6]) were transformed with p413.ADH.RAV1.HA.HIS6 or p413.ADH.RAV1. The strains were grown at 25°C, and 10 ODs of S13 extract from yeast spheroplasts were immunoprecipitated under native conditions with 12CA5 antibody (anti-HA). Immunoprecipitations were separated on SDS-PAGE and transferred to nitrocellulose where they were probed with anti-HA (12CA5) or anti-Skp1 antibody to characterize the extent of Rav1p-Skp1 interactions. (C) The crystal structure of the mammalian Skp1-Skp2 complex is depicted with the skp1(Asn108Tyr) mutation Asn108Tyr modeled (Skp1, magenta; leucine-rich repeats of Skp2, blue; F-box of Skp2, green; Skp2-Leu116, yellow; Skp1-Asn108, red) (32). Graphic representation was made using MacPyMol (DeLano Scientific, LLC).
The skp1(Asn108Tyr) sequence contained an AAC-to-TAC transversion in codon 108, corresponding to a Tyr-for-Asn substitution. Asn108 is conserved in human Skp1 and lies in helix five, adjacent to the F-box binding site. In the three-dimensional structure of the human Skp1-Skp2 (32), Asn108 of human Skp1 contacts residue Leu116 in the F-box domain of Skp2 (Fig. 3C) and yet is at the periphery of the interface between the two proteins. In a compilation of F-box sequences, the position equivalent to Skp2 Leu116 varies between Leu, Val, and Ile (25). Thus, Skp1p Asn108 is in a position where substitutions could selectively alter affinity for F-box proteins depending on the identity of the contacting F-box residues. Although the precise Skp1p binding domain of Rav1p has not been identified, it has been noted previously that Rav1p contains several sequences with similarity to F-box domains (34).
skp1(Asn108Tyr) does not affect V-ATPase function.
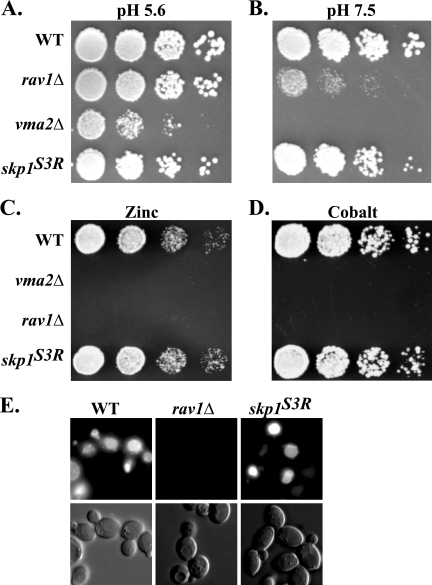
The skp1(Asn108Tyr) allele made it possible to determine whether the Skp1p-Rav1p interaction is important for V-ATPase assembly and activity. Mutations in V-ATPase subunit genes, such as VMA2, which encodes the B subunit of the ATPase hexamer, result in growth that is sensitive to high pH, zinc, and cobalt (3, 23) (E. J. Brace and R. S. Fuller, unpublished data). rav1 mutants, which are defective in ATPase assembly/activation, exhibit similar phenotypes (33, 34, 36). As shown in Fig. 4A and B, whereas vma2Δ and rav1Δ strains grew at pH 5.6 but not at pH 7.5, the skp1(Asn108Tyr) strain grew well at both low and high pH. Also, unlike rav1Δ and vma2Δ mutants, the skp1(Asn108Tyr) mutant did not show sensitivity to 5 mM ZnCl2 or 750 μM CoCl2 (Fig. 4C and D). V-ATPase and rav1Δ mutants are defective for vacuolar accumulation of the fluorescent dye quinacrine (33, 36). In contrast, the skp1(Asn108Tyr) strain exhibited normal quinacrine accumulation (Fig. 4E). These results demonstrate that binding of Skp1p to Rav1p is not required for V-ATPase assembly/activation.
FIG. 4.
skp1(Asn108Tyr) does not affect V-ATPase function. (A to D) Serial dilutions of strains CRY2 (wild type), GSY11-2A (rav1Δ), EBY72-2 (vma2Δ), and EBY116-6 [skp1Δ pskp1(Asn108Tyr)] were pronged onto the indicated media and incubated for 2 days. (A and B) YPAD plates, pH 5.6 or pH 7.5, grown at 37°C; (C) YPAD with 5 mM ZnCl2 (pH 5.6) grown at 30°C; (D) YPAD with 750 μM CoCl2 at 30°C. (E) Strains CRY2 (wild type), GSY11-2A (rav1Δ), and EBY116-6 [skp1Δ pskp1(Asn108Tyr)] were tested for their ability to accumulate quinacrine in their vacuoles. Cells were allowed to accumulate quinacrine for 10 min at 30°C and cooled on ice for 5 min. Cells were then visualized immediately by differential interference contrast and fluorescence microscopy.
skp1(Asn108Tyr) and rav1Δ alter Kex2p trafficking in distinct ways.
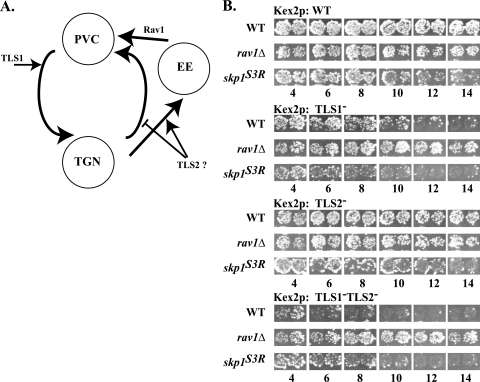
rav1Δ mutants exhibit defects in the trafficking of Kex2p between the early endosome and PVC (34). The effect of rav1Δ or other mutations on Kex2p localization can be monitored in vivo using the “onset of impotence” assay (27). Mislocalization of Kex2p disrupts processing of pro-α-factor and thus impairs production of the peptide mating pheromone α-factor, which is required for mating. When Kex2p expression, under control of the glucose-repressible GAL1 promoter, is shut off by shifting cells to glucose medium, the rate of loss of mating competence over time reflects the loss of preexisting enzyme from the pro-α-factor processing compartment (Fig. 5A). A Y713A mutation in TLS1 in the Kex2p cytosolic tail causes more rapid loss of mating competence because of failure to retrieve Kex2p from the PVC to the TGN. Loss of TLS2 function results in more rapid loss of mating competence because of increased transport to the PVC (6).
FIG. 5.
Skp1p affects trafficking of Kex2p. (A) Model of TLS1 and TLS2 function in Kex2p trafficking between the TGN and endosomal system. (B) Strains KRY18-1A (kex2Δ), GSY11-r1 (kex2Δ rav1Δ), and EBY127-2 [kex2Δ skp1Δ pskp1(Asn108Tyr)] were transformed with pCWKX20 (GAL1-KEX2; wild type [WT]), pCWKX21 (GAL1-KEX2 Y713A; TLS1−), pCWKX20-I718tail (GAL1-KEX2-I718 tail; TLS2−), and pCWKX21-I718tail (GAL1-KEX2 Y713A-I718 tail; TLS1− TLS2−). Strains were grown at 25°C, shifted from galactose to glucose medium for the indicated times, and assessed for mating competence as described previously (27).
In the onset of impotence assay, the effects of the skp1(Asn108Tyr) mutation were distinct from effects of the rav1Δ mutation with all forms of Kex2p tested (Fig. 5B). Whereas the rav1Δ mutation suppressed effects of both the TLS1 mutation alone and in combination with the TLS2 mutation, the skp1(Asn108Tyr) mutation did not. The skp1(Asn108Tyr) mutation did have effects in the assay, causing more rapid loss of mating competence compared to the wild-type strain background with wild-type (by 10 h) (Fig. 5B, top panel) and TLS2 mutant (by 6 h) (Fig. 5B, third panel) Kex2p and had a slight effect on TLS1 mutant Kex2p (Fig. 5B, second panel). These effects of the skp1(Asn108Tyr) allele were not due to general defects in mating because the skp1(Asn108Tyr) mutant demonstrated mating indistinct from wild-type strains in a quantitative mating assay (E. J. Brace and R. S. Fuller, unpublished data). Paradoxically, skp1(Asn108Tyr) improved mating over time with the TLS1− TLS2− double mutant (Fig. 5B, bottom panel). Although the effects of skp1(Asn108Tyr) in this assay are complex, the key result, that the skp1(Asn108Tyr) and rav1Δ mutants behave differently, is completely consistent with the conclusion reached above that the Skp1p-Rav1p interaction is not required for the function of Rav1p in V-ATPase assembly/activation.
Of note, the skp1(Asn108Tyr) mutation did not disrupt either the trafficking of the vacuolar protein sorting receptor, Vps10p, as measured by secretion of carboxypeptidase Y or endocytosis, as measured by FM4-64 uptake (E. J. Brace and R. S. Fuller, unpublished results). This points out one similarity between the rav1Δ and skp1(Asn108Tyr) mutations: both affect Kex2p but not Vps10p trafficking. This may reflect the fact that Kex2p reaches the PVC both by direct TGN-PVC transport and by trafficking through the early endosome, whereas Vps10p follows only the direct pathway (34).
Rav1-GFP is recruited to membranes in skp1(Asn108Tyr) and doa4Δ mutants.
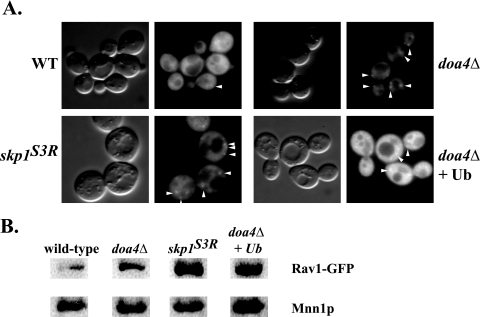
In yeast cells, Rav1p is present both in a soluble pool and as a peripheral protein associated with a high-density membrane compartment suggestive of early endosomes (34). The bulk of Rav1-GFP fluorescence was cytosolic, but image deconvolution made it possible to identify discrete Rav1-GFP puncta (34). The skp1(Asn108Tyr) mutant was examined to determine whether Skp1p affects recruitment of Rav1-GFP to membranes. Surprisingly, in the skp1(Asn108Tyr) mutant, Rav1-GFP localized more extensively to puncta that were visualized even without the aid of image deconvolution (Fig. 6A). To determine if total Rav1p protein levels were altered in the skp1(Asn108Tyr) or doa4Δ mutants, Rav1-GFP levels were determined by Western blotting (L. P. Parkinson and R. S. Fuller, unpublished results). Rav1-GFPp levels were unchanged in the skp1(Asn108Tyr) mutant but were slightly lower in the doa4Δ mutant. The latter effect may account for the lower cytosolic Rav1-GFP fluorescence seen in the doa4Δ strain. These data suggest that Rav1-GFP accumulation on membranes is not due to increased levels of Rav1-GFP protein but is due to altered localization of the protein. Sucrose step gradients were used to provide an alternative way of assessing Rav1-GFP localization to membranes in the skp1(Asn108Tyr) background. S13 fractions (13,000 × g supernatant) of yeast extracts were further fractionated by equilibrium centrifugation at 200,000 × g using a two-step sucrose gradient. Samples were tested for the accumulation of Rav1-GFP at the 15%-60% sucrose interface that separates membrane fractions from large protein complexes that pellet and cytosolic proteins that sediment slowly and fractionate above the 15% step. Rav1-GFP more readily accumulated at the 15%-60% interface of the gradient in a skp1(Asn108Tyr) mutant, suggesting that the mutant is defective in releasing Rav1p from membranes (Fig. 6B).
FIG. 6.
Rav1-GFP is localized to membranes in mutants. (A) Strains GSY11-2A (rav1Δ), EBY116-4 [rav1Δ skp1Δ skp1(Asn108Tyr)], EBY79-12 (rav1Δ doa4Δ), and EBY79-12 (YEp96; wild-type [WT] ubiquitin) were transformed with p416.ADH.RAV1-GFP. For simplicity, the rav1Δ is not marked on the figure, since it is covered by the RAV1 plasmid. Strains were grown at 25°C in minimal media (including 100 μM CuSO4 for the ubiquitin experiment) and maintained in log phase. Differential interference contrast and fluorescence microscopy were performed using an E800 Nikon microscope and ISEE digital imaging software. All images were obtained with the same exposure times and gain. Punctate staining can be seen in each of the mutants pictured (white arrowheads). (B) Rav1-GFP accumulates in the membrane fraction. Strains GSY11-1A (rav1Δ), EBY116-4 [rav1Δ skp1Δ pskp1(Asn108Tyr)], EBY79-11 (rav1Δ doa4Δ), and EBY79-11 (rav1Δ doa4Δ pYEp96) were transformed with p416ADH.RAV1-GFP. S13 was created from 125 ODs of yeast spheroplasts and loaded on top of a sucrose step gradient consisting of 15% and 60% layers. To measure Rav1-GFP localization to membranes, membrane fractions were collected at the 15 to 60% interface and separated by SDS-PAGE before being transferred to nitrocellulose. Rav1-GFP was not visible in whole-cell extracts because of apparent degradation. The nitrocellulose was probed with anti-GFP or anti-Mnn1p, which was used as a loading control.
Although Rav1p-Skp1p complexes do not contain other components typically found in SCF complexes (Cdc32p, Rbx1p, and Cdc53p) (33), it remains possible that ubiquitin plays a role in Rav1p function or localization. DOA4, which encodes a deubiquitinating enzyme, is localized to endosomes and is involved in the multivesicular body pathway (2). Mutations in DOA4 result in lower levels of free ubiquitin in the cells, which often leads to the impairment of cellular functions requiring ubiquitin (39). Therefore, Rav1-GFP localization was tested in a doa4Δ mutant to determine if Rav1p localization depended on ubiquitin. Indeed, Rav1-GFP demonstrated increased membrane localization in a doa4Δ mutant, as shown both by fluorescence microscopy and by the sucrose step gradient fractionation (Fig. 6A and B). However, ubiquitin overexpression did not rescue the effect of the doa4Δ mutant, suggesting that Doa4p may be directly involved in Rav1p localization.
DISCUSSION
We isolated the SKP1 gene as a multicopy suppressor of the toxicity caused by Rav1p overexpression. Because Skp1p is essential, we designed a screen to identify a novel SKP1 allele that would specifically abrogate the Rav1p-Skp1 interaction but would not disrupt other, essential Skp1p interactions. This approach allowed us to identify a single point mutation in Skp1p that was adjacent to the F-box binding domain and that blocked the Rav1p-Skp1p interaction but maintained essential protein-protein interactions of Skp1p. This separation of function allele, skp1(Asn108Tyr), allowed us to characterize the role of the Rav1p-Skp1p interaction in Rav1p function and localization.
Surprisingly, the skp1(Asn108Tyr) mutant exhibited phenotypes that were distinct from those of both rav1Δ and vma2Δ mutants, suggesting that the Skp1p-Rav1p interaction is not essential for the function of Rav1p in V-ATPase assembly/activation. Unlike rav1Δ and vma2Δ mutants, the skp1(Asn108Tyr) mutant exhibited no defects in quinacrine accumulation in the vacuole or in growth on zinc-containing, cobalt-containing, or high-pH media. In a previous study, the temperature-sensitive skp1-12 mutant exhibited growth that was sensitive to high pH as well as a defect in vacuolar quinacrine accumulation (33). However, skp1-12 did not suppress Rav1p overexpression (E. J. Brace and R. S. Fuller, unpublished data), suggesting that the Skp1p-Rav1p interaction is not disrupted by skp1-12. The effects of the skp1-12 mutation may therefore be indirect, resulting from the nonspecific nature of the allele. skp1-12 was identified as a mutant that caused temperature sensitivity for growth and which, therefore, disrupted at least one essential Skp1p function (4). In contrast, the skp1(Asn108Tyr) mutant specifically disrupts the Skp1p-Rav1p interaction but leaves intact the Skp1p interactions that are essential for growth. Synthesis of these findings suggests that the skp1-12 mutation may affect pH sensitivity and quinacrine accumulation not by loss of the specific Skp1p-Rav1p interaction but by loss of one or more other Skp1p interactions. We conclude, therefore, that the Skp1p-Rav1p interaction is not required for V-ATPase assembly/activation. A second line of evidence that Skp1p function is distinct from that of Rav1p is the fact that although the skp1(Asn108Tyr) mutation did alter Kex2p localization, as judged by the onset of impotence assay, the effects were quite different than those of the rav1Δ mutation. The effects of the skp1(Asn108Tyr) allele may be a consequence of alterations in Rav1p localization.
Whereas skp1(Asn108Tyr) did not affect Rav1p function in V-ATPase assembly and activation, it did have a clear effect on Rav1p localization. In contrast to wild-type cells, in the skp1(Asn108Tyr) mutant, Rav1-GFP was readily apparent in cytoplasmic puncta even without image deconvolution, presumably reflecting both an increase in membrane localization and a decrease in the cytosolic pool. Increased membrane localization was confirmed by biochemical fractionation. Interestingly, although Rav1-GFP showed increased localization to membranes in the skp1(Asn108Tyr) mutant, microscopic examination showed that sites of localization corresponded to punctate organelles rather than to vacuolar membranes. This is consistent with the proposed role for the RAVE complex in V-ATPase assembly/activation at the early endosome (34). A link between the roles of Rav1p in trafficking and V-ATPase activity is suggested by the fact that bafilomycin A1, a potent and specific inhibitor of V-ATPase, disrupts traffic between the early and late endosome in mammalian cells (8). Mutations in V-ATPase cause defects in the internalization of the fluorescent dye FM4-64 from the plasma membrane to the vacuole in yeast (26). Additionally, V-ATPase mutants exhibit defects in Kex2p trafficking (E. J. Brace and R. S. Fuller, unpublished data).
Loss of Doa4p also resulted in increased membrane association and punctate localization of Rav1-GFP. Because this effect was not overcome by ubiquitin overexpression, it was not an indirect result of ubiquitin depletion in the cells. These results suggest that both the Skp1p interaction with Rav1p and a deubiquitination event mediated by Doa4p are required for release of Rav1p from endosomal membranes. Although the RAVE complex has been classified as a non-SCF type Skp1p complex because of the absence of Cdc53p, Rbx1p, and Cdc34p (33), an intriguing but highly speculative possibility is that Skp1p may participate transiently in a ubiqitination reaction that is important for release of the RAVE complex from endosomal membranes. In this view, Skp1p and Doa4p might be required for a cycle of ubiquitination and deubiquitination that regulate Rav1p association with the endosomal compartment.
There are two other known Skp1p-containing non-SCF complexes in yeast, the Rcy1p-Skp1p complex (12) and the kinetochore complex CBF3 (9). Notably, the F-box protein Rcy1p functions at the early endosome to regulate recycling of internalized proteins from the cell surface (12, 20, 40). Mutations in RCY1 have recently been shown to affect Kex2p trafficking (7). Rav1p and Rcy1p may govern distinct pathways out of the early endosome. Thus, it is plausible that Skp1p may function as a regulator of both processes.
Acknowledgments
We are grateful to the following for generously providing reagents and antisera and for helpful discussions: Steve Elledge, Todd Graham, and Dennis Thiele. Additional thanks to Damian Krysan, Jen Blanchette, and other members of the Fuller Lab for helpful discussions.
This work was supported in part by NIH grants GM50915 and GM39697 to R.S.F. and P30 CA46592 to the University of Michigan Comprehensive Cancer Center.
Footnotes
Published ahead of print on 13 October 2006.
REFERENCES
- 1.Adams, A. E., and J. R. Pringle. 1984. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J. Cell Biol. 98:934-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amerik, A. Y., J. Nowak, S. Swaminathan, and M. Hochstrasser. 2000. The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol. Biol. Cell 11:3365-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachhawat, A. K., M. F. Manolson, D. G. Murdock, J. D. Garman, and E. W. Jones. 1993. The VPH2 gene encodes a 25 kDa protein required for activity of the yeast vacuolar H(+)-ATPase. Yeast 9:175-184. [DOI] [PubMed] [Google Scholar]
- 4.Bai, C., P. Sen, K. Hofmann, L. Ma, M. Goebl, J. W. Harper, and S. J. Elledge. 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86:263-274. [DOI] [PubMed] [Google Scholar]
- 5.Baker, D., L. Hicke, M. Rexach, M. Schleyer, and R. Schekman. 1988. Reconstitution of SEC gene product-dependent intercompartmental protein transport. Cell 54:335-344. [DOI] [PubMed] [Google Scholar]
- 6.Brickner, J. H., and R. S. Fuller. 1997. SOI1 encodes a novel, conserved protein that promotes TGN-endosomal cycling of Kex2p and other membrane proteins by modulating the function of two TGN localization signals. J. Cell Biol. 139:23-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, S. H., S. Chen, A. A. Tokarev, F. Liu, G. Jedd, and N. Segev. 2005. Ypt31/32 GTPases and their novel F-box effector protein Rcy1 regulate protein recycling. Mol. Biol. Cell 16:178-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clague, M. J., S. Urbe, F. Aniento, and J. Gruenberg. 1994. Vacuolar ATPase activity is required for endosomal carrier vesicle formation. J. Biol. Chem. 269:21-24. [PubMed] [Google Scholar]
- 9.Connelly, C., and P. Hieter. 1996. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell 86:275-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deshaies, R. J. 1999. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15:435-467. [DOI] [PubMed] [Google Scholar]
- 11.Fuller, R. S., R. E. Sterne, and J. Thorner. 1988. Enzymes required for yeast prohormone processing. Annu. Rev. Physiol. 50:345-362. [DOI] [PubMed] [Google Scholar]
- 12.Galan, J. M., A. Wiederkehr, J. H. Seol, R. Haguenauer-Tsapis, R. J. Deshaies, H. Riezman, and M. Peter. 2001. Skp1p and the F-box protein Rcy1p form a non-SCF complex involved in recycling of the SNARE Snc1p in yeast. Mol. Cell. Biol. 21:3105-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham, L. A., A. R. Flannery, and T. H. Stevens. 2003. Structure and assembly of the yeast V-ATPase. J. Bioenerg. Biomembr. 35:301-312. [DOI] [PubMed] [Google Scholar]
- 14.Gueldener, U., J. Heinisch, G. J. Koehler, D. Voss, and J. H. Hegemann. 2002. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 30:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ha, S. A., J. Torabinejad, D. B. DeWald, M. R. Wenk, L. Lucast, P. De Camilli, R. A. Newitt, R. Aebersold, and S. F. Nothwehr. 2003. The synaptojanin-like protein Inp53/Sjl3 functions with clathrin in a yeast TGN-to-endosome pathway distinct from the GGA protein-dependent pathway. Mol. Biol. Cell 14:1319-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnston, G. C., J. R. Pringle, and L. H. Hartwell. 1977. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp. Cell Res. 105:79-98. [DOI] [PubMed] [Google Scholar]
- 17.Kane, P. M., and A. M. Smardon. 2003. Assembly and regulation of the yeast vacuolar H+-ATPase. J. Bioenerg. Biomembr. 35:313-321. [DOI] [PubMed] [Google Scholar]
- 18.Kraemer, C., T. Enklaar, B. Zabel, and E. R. Schmidt. 2000. Mapping and structure of DMXL1, a human homologue of the DmX gene from Drosophila melanogaster coding for a WD repeat protein. Genomics 64:97-101. [DOI] [PubMed] [Google Scholar]
- 19.Kraemer, C., B. Weil, M. Christmann, and E. R. Schmidt. 1998. The new gene DmX from Drosophila melanogaster encodes a novel WD-repeat protein. Gene 216:267-276. [DOI] [PubMed] [Google Scholar]
- 20.Lafourcade, C., J. M. Galan, and M. Peter. 2003. Opposite roles of the F-box protein Rcy1p and the GTPase-activating protein Gyp2p during recycling of internalized proteins in yeast. Genetics 164:469-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 22.Mumberg, D., R. Muller, and M. Funk. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119-122. [DOI] [PubMed] [Google Scholar]
- 23.Nelson, H., and N. Nelson. 1990. Disruption of genes encoding subunits of yeast vacuolar H(+)-ATPase causes conditional lethality. Proc. Natl. Acad. Sci. USA. 87:3503-3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nothwehr, S. F., N. J. Bryant, and T. H. Stevens. 1996. The newly identified yeast GRD genes are required for retention of late-Golgi membrane proteins. Mol. Cell. Biol. 16:2700-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patton, E. E., A. R. Willems, and M. Tyers. 1998. Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet. 14:236-243. [DOI] [PubMed] [Google Scholar]
- 26.Perzov, N., V. Padler-Karavani, H. Nelson, and N. Nelson. 2002. Characterization of yeast V-ATPase mutants lacking Vph1p or Stv1p and the effect on endocytosis. J. Exp. Biol. 205:1209-1219. [DOI] [PubMed] [Google Scholar]
- 27.Redding, K., J. H. Brickner, L. G. Marschall, J. W. Nichols, and R. S. Fuller. 1996. Allele-specific suppression of a defective trans-Golgi network (TGN) localization signal in Kex2p identifies three genes involved in localization of TGN transmembrane proteins. Mol. Cell. Biol. 16:6208-6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Redding, K., C. Holcomb, and R. S. Fuller. 1991. Immunolocalization of Kex2 protease identifies a putative late Golgi compartment in the yeast Saccharomyces cerevisiae. J. Cell Biol. 113:527-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reynolds, T. B., B. D. Hopkins, M. R. Lyons, and T. R. Graham. 1998. The high osmolarity glycerol response (HOG) MAP kinase pathway controls localization of a yeast golgi glycosyltransferase. J. Cell Biol. 143:935-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts, C. J., C. K. Raymond, C. T. Yamashiro, and T. H. Stevens. 1991. Methods for studying the yeast vacuole. 194:644-661. [DOI] [PubMed]
- 31.Rose, M. D., F. Winston, and P. Heiter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 32.Schulman, B. A., A. C. Carrano, P. D. Jeffrey, Z. Bowen, E. R. Kinnucan, M. S. Finnin, S. J. Elledge, J. W. Harper, M. Pagano, and N. P. Pavletich. 2000. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408:381-386. [DOI] [PubMed] [Google Scholar]
- 33.Seol, J. H., A. Shevchenko, and R. J. Deshaies. 2001. Skp1 forms multiple protein complexes, including RAVE, a regulator of V-ATPase assembly. Nat. Cell Biol. 3:384-391. [DOI] [PubMed] [Google Scholar]
- 34.Sipos, G., J. H. Brickner, E. J. Brace, L. Chen, A. Rambourg, F. Kepes, and R. S. Fuller. 2004. Soi3p/Rav1p functions at the early endosome to regulate endocytic trafficking to the vacuole and localization of trans-Golgi network transmembrane proteins. Mol. Biol. Cell 15:3196-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sipos, G., and R. S. Fuller. 2002. Separation of Golgi and endosomal compartments. Methods Enzymol. 351:351-365. [DOI] [PubMed] [Google Scholar]
- 36.Smardon, A. M., M. Tarsio, and P. M. Kane. 2002. The RAVE complex is essential for stable assembly of the yeast V-ATPase. J. Biol. Chem. 277:13831-13839. [DOI] [PubMed] [Google Scholar]
- 37.Smith, T. F., C. Gaitatzes, K. Saxena, and E. J. Neer. 1999. The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24: 181-185. [DOI] [PubMed] [Google Scholar]
- 38.Stevens, T. H., and M. Forgac. 1997. Structure, function and regulation of the vacuolar (H+)-ATPase. Annu. Rev. Cell Dev. Biol. 13:779-808. [DOI] [PubMed] [Google Scholar]
- 39.Swaminathan, S., A. Y. Amerik, and M. Hochstrasser. 1999. The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol. Biol. Cell 10:2583-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiederkehr, A., S. Avaro, C. Prescianotto-Baschong, R. Haguenauer-Tsapis, and H. Riezman. 2000. The F-box protein Rcy1p is involved in endocytic membrane traffic and recycling out of an early endosome in Saccharomyces cerevisiae. J. Cell Biol. 149:397-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilcox, C. A., K. Redding, R. Wright, and R. S. Fuller. 1992. Mutation of a tyrosine localization signal in the cytosolic tail of yeast Kex2 protease disrupts Golgi retention and results in default transport to the vacuole. Mol. Biol. Cell 3:1353-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshihisa, T., and Y. Anraku. 1989. Nucleotide sequence of AMS1, the structure gene of vacuolar alpha-mannosidase of Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 163:908-915. [DOI] [PubMed] [Google Scholar]