Abstract
All eukaryotes carry out glycolysis, interestingly, not all using the same enzymes. Anaerobic eukaryotes face the challenge of fewer molecules of ATP extracted per molecule of glucose due to their lack of a complete tricarboxylic acid cycle. This may have pressured anaerobic eukaryotes to acquire the more ATP-efficient alternative glycolytic enzymes, such as pyrophosphate-fructose 6-phosphate phosphotransferase and pyruvate orthophosphate dikinase, through lateral gene transfers from bacteria and other eukaryotes. Most studies of these enzymes in eukaryotes involve pathogenic anaerobes; Monocercomonoides, an oxymonad belonging to the eukaryotic supergroup Excavata, is a nonpathogenic anaerobe representing an evolutionarily and ecologically distinct sampling of an anaerobic glycolytic pathway. We sequenced cDNA encoding glycolytic enzymes from a previously established cDNA library of Monocercomonoides and analyzed the relationships of these enzymes to those from other organisms spanning the major groups of Eukaryota, Bacteria, and Archaea. We established that, firstly, Monocercomonoides possesses alternative versions of glycolytic enzymes: fructose-6-phosphate phosphotransferase, both pyruvate kinase and pyruvate orthophosphate dikinase, cofactor-independent phosphoglycerate mutase, and fructose-bisphosphate aldolase (class II, type B). Secondly, we found evidence for the monophyly of oxymonads, kinetoplastids, diplomonads, and parabasalids, the major representatives of the Excavata. We also found several prokaryote-to-eukaryote as well as eukaryote-to-eukaryote lateral gene transfers involving glycolytic enzymes from anaerobic eukaryotes, further suggesting that lateral gene transfer was an important factor in the evolution of this pathway for denizens of this environment.
Glycolysis is the linchpin of eukaryotic metabolism. All eukaryotes carry out the glycolytic pathway, and many of the cell's anabolic and catabolic processes funnel through it in some way. The standard eukaryotic glycolytic pathway, the Embden-Meyerhof-Parnas (EMP) pathway (Fig. 1), consists of 10 enzymes, most of which catalyze reversible steps (5). Two enzymes, phosphofructokinase (PFK) and pyruvate kinase (PK), catalyze reactions requiring a high Gibbs free energy (G°) and are thus irreversible (25). The EMP pathway results in a net yield of two ATP molecules for each glucose molecule. In aerobic eukaryotes, pyruvate, the end product of the pathway, then enters the tricarboxylic acid cycle, resulting in a final yield of up to 36 ATP molecules for each glucose after one full cycle. Oxygen is required for the tricarboxylic acid cycle to result in such a high yield. However, this compound is unavailable to eukaryotes living in anaerobic environments.
FIG. 1.
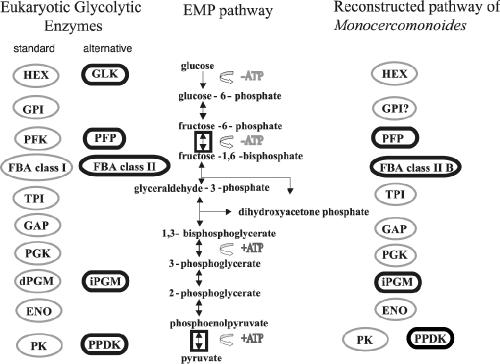
Schematic of the eukaryotic glycolytic pathway and the reconstructed pathway of Monocercomonoides. Gray ovals show the standard EMP pathway enzymes; black rounded squares show the alternative versions. Monocercomonoides possesses several alternative enzymes, including PFP, FBA class II, iPGM, and PPDK. GPI is predicted in Monocercomonoides but has not yet been found, and, thus, a question mark is appended to its GPI icon. Boxed double-headed arrows denote the two glycolytic steps which are irreversible in the standard pathway but for which reversible PPi-hydrolyzing enzymes exist.
One way in which anaerobes may increase ATP yield is through the use of alternative glycolytic enzymes for the steps performed by PFK and PK (Fig. 1). Although catalyzing the same steps, these enzymes do so through different biochemical reactions (10). Of the distantly related types of PFK that may coexist in both prokaryotes and eukaryotes (3, 24), the ATP-dependent PFK catalyzes the irreversible phosphorylation of fructose-6-phosphate, representing the first committed step of glycolysis (25). However, pyrophosphate-fructose-6-phosphate phosphotransferase (PFP) or inorganic pyrophosphate (PPi)-dependent PFK performs the same task using PPi as the phosphate donor; this reaction is reversible due to a much lower G° (25). Similarly, in the final step of the glycolytic pathway, several enzymes exist that catalyze the conversion of phosphoenolpyruvate to pyruvate (26). The reaction catalyzed by PK, the enzyme found in most eukaryotes and prokaryotes (26), is irreversible due to its high free energy demand. However, pyruvate phosphate dikinase (PPDK) is the energy-conserving, reversible alternative to PK (26, 34). Since both PFP and PPDK rely on PPi as the phosphate donor, they can thus increase the overall ATP yield of glycolysis by using a by-product of the cell's anabolic processes as an energy source rather than hydrolyzing ATP. Because these reactions are reversible, the enzymes can also participate in the regulation of the glycolytic pathway (25). In the case of PFK and PFP, both enzymes can coexist in the same organism, with examples found in both prokaryotes and eukaryotes (3). Similarly, PPDK does not necessarily replace, but rather may be expressed in addition to, PK (34).
Since all eukaryotes follow the EMP pathway of glycolysis (16), it might be reasonable to expect that they would all possess the same enzymes derived from the same source: the last common eukaryotic ancestor. This, however, is not the case. The phylogenetic picture of this complex situation has thus far deterred comprehensive reviews of the topic. However, studies of the individual steps have identified the above-described alternative glycolytic enzymes in eukaryotes and additional phylogenetic analyses suggest that other gene transfers occurred after the split from the last common eukaryotic ancestor (16, 30, 34, 37).
At least three other alternative enzymes have been found in the eukaryotic glycolytic pathway. The first step of glycolysis, the conversion of glucose to glucose-6-phosphate, may be performed by glucokinase or its strictly eukaryotic relative hexokinase (16). The third step in glycolysis may be performed by two unrelated enzymes: fructose-bisphosphate aldolase (FBA) classes I and II. FBA class I is found in most eukaryotes, while FBA class II is found in both prokaryotes and eukaryotes. FBA class II may be further divided into types A and B (17, 30), with type B being a subfamily characteristic of anaerobic eukaryotes. Finally, the conversion of 3-phosphoglycerate to 2-phosphoglycerate may be performed by either the standard dependent phosphoglycerate mutase (dPGM), which depends on 2,3-bisphosphoglycerate for activation, or the analogous enzyme, 2,3-bisphosphoglycerate independent phosphoglycerate mutase (iPGM), which does not (7, 11). While dPGM is found mostly among eukaryotes, iPGM is shared between prokaryotes and some eukaryotes; in rare cases, the two enzymes coexist in the same organism (7, 12).
In the course of characterizing these alternate glycolytic enzymes, the mosaic evolutionary nature of the pathway has come to light, with lateral gene transfer (LGT) clearly having played a significant role in the distribution of glycolytic enzymes, particularly to anaerobic eukaryotes. Glucokinase in diplomonads and parabasalids seems to be of eubacterial origin (16). While the diplomonad version is specifically related to cyanobacterial homologs, no strong support has been established for the monophyly of diplomonads and parabasalids for this enzyme (16). On the other hand, a study of glucose-6-phosphate isomerase (GPI) strongly indicated common ancestry for the diplomonad and parabasalid homologs to the exclusion of other eukaryotes as well as suggesting cyanobacteria as the origin of their GPI (16). Another LGT from eubacteria involved FBA class II, type B, found in diplomonads, parabasalids, and amoebae, though the analysis suggested separate acquisitions of the gene for these organisms (30). Finally, from its patchy distribution in eukaryotes and from the lack of monophyly of eukaryotic homologs of the enzyme, PPDK has been proposed to have arisen in eukaryotes via multiple gene transfers from bacteria, with oxymonads and diplomonads, but not parabasalids or kinetoplastids, linked with moderate support (34).
Much of the study of anaerobic glycolysis has focused on parasitic organisms in either the Amoebozoa or the Excavata, with the notable exception of the free-living amoeboflagellate Mastigamoeba balamuthi. The Amoebozoa and the Excavata represent two of the six recently established eukaryotic supergroups (1), with the Excavata being particularly controversial. Although this supergroup as a whole has been proposed based on a combination of inferred relationships between protists possessing the excavate feeding groove and lineages linked to them by molecular phylogenetics (31), there has never been a molecular phylogenetic result uniting all excavate taxa. Nonetheless, resolution has been established to the point where the 10 proposed excavate lineages can be robustly grouped into two major clades and an orphan lineage (Malawimonas) (32). Hampl et al. (14) did show representatives of the two major groups united into a clade in their concatenated analysis, but without node support (14), while Giardia, Trichomonas, and the kinetoplastids were united with good support in a recent concatenated protein analysis (28).
We have chosen to examine the glycolytic pathway of Monocercomonoides sp., an oxymonad. As excavate taxa (8, 33), oxymonads are thought to be related to the grouping of parabasalids (including Trichomonas vaginalis) and diplomonads (including Giardia intestinalis) (14). However, beyond the close affiliation with the excavate flagellate Trimastix (8), the phylogenetic position of oxymonads within the Excavata is far from fully resolved. Since Monocercomonoides is also a nonpathogenic organism living in low abundance in the gut fauna of some vertebrates (22, 23), it represents a phylogenetic sampling for enzymes of the anaerobic glycolytic pathway that is evolutionarily and ecologically distinct from the majority of systems investigated to date. The one study examining a glycolytic enzyme of an oxymonad (Streblomastix strix) demonstrated the presence of the alternative, PPi-hydrolyzing enzyme PPDK (34), suggesting that the anaerobic pathway of oxymonads could be a fruitful source of information about the evolution and mechanism of this pathway.
Here we report the molecular biological and phylogenetic characterization of enzymes from the oxymonad Monocercomonoides sp. for 9 of the 10 steps of glycolysis. This allows us to reconstruct its glycolytic pathway, including the presence and relative expression of several of the alternate glycolytic enzymes. Phylogeny of enolase produces moderately to well-supported monophyly of kinetoplastids, oxymonads, and Giardia, representing some of the best single-gene support to date for the unity of the two major excavate clades. Shared LGT events between oxymonads and both Giardia and Trichomonas further support the relationship of the Metamonada sensu Cavalier-Smith (6). Finally, the prevalence of LGT events in the glycolytic pathway of this nonpathogenic eukaryote, including a putative oxymonad-Entamoeba event, further reinforces the major role of LGT in the evolution of anaerobic glycolysis and suggests that it is selection for ATP efficiency and not pathogenicity that drives this phenomenon.
MATERIALS AND METHODS
Isolation of clones and sequencing.
Monocercomonoides cDNA clones encoding glycolytic enzyme homologs were identified by using the automated annotation program MAGPIE (http://magpie.ucalgary.ca/) as part of our ongoing expressed sequence tag survey of this organism (J. Dacks and V. Hampl, unpublished data). In some cases, the sequence was assembled from reads of multiple clones, with all sequence derived from a minimum of 2× coverage. Subsequent to initial transcript assembly using Paracel Transcript Assembler (Paracel, Inc., Carlsbad, CA), clones were deemed to carry the same gene if they could be assembled using the program Sequencher (Gene Codes Corp., Ann Arbor, MI) with a cutoff of 98% nucleotide identity. These criteria also provided us with the relative expression levels of the glycolytic enzymes because the library used for this study had not been normalized to remove highly abundant transcripts and thus was representative of mRNA expression at the time of isolation. In the cases where multiple clones were not available, clones were sequenced on both strands using T3 or T7 primers that primed within the cloning vector. In a few cases, walking primers were designed in order to obtain the full open reading frame. For hexokinase, the forward primer 440SF1 (5′-CGA GCG ATC ATA TGC TG-3′) and reverse primer 440SR1 (5′-GCA ATG GCT GAA AGA CG-3′) were designed; 1279SF1 (5′-ACT GGA ACA TTG TAC GC-3′) and 1279SR1 (5′-TCA GTG ACT TCT TTG GC-3′) for phosphoglycerate mutase and PFK-SF1 (5′-GTT TGA GGT TGC TGG AC-3′) for phosphofructokinase were also designed. Sequencing was performed by the University of Calgary Core DNA Services. Preliminary identification of the enzymes was established by a BLASTp search (2) of the conceptually translated coding regions against the “nr” protein database at the NCBI. For each Monocercomonoides homolog, the top five BLAST-retrieved sequences were compared in order to ensure consistency of annotation. In all cases, the top hit had an E value of at least 1e-40 (Table 1).
TABLE 1.
Glycolytic enzyme homologues from Monocercomonoides sp.
| Enzyme (Monocercomonoides) | GenBank accession number | No. of clones at 98% identity | Size (aa) | Top BLAST hit | E value | Size (aa) |
|---|---|---|---|---|---|---|
| Hexokinase 1 | DQ665855 | 1 | 463 | AAG22926.1 hexokinase-t2 (Drosophila melanogaster) | 9e-52 | 453 |
| Hexokinase 2 | DQ665856 | 1 | 517 | CAA57681.1 hexokinase (Entamoeba histolytica) | 1e-40 | 445 |
| Pyrophosphate fructose phosphotransferase | DQ665857 | 1 | 461 | YP_460806.1 pyrophosphate-fructose 6-phosphate 1-phosphotransferase (Syntrophus aciditrophicus SB) | 4e-114 | 434 |
| Fructose bisphosphate aldolase | DQ665853 | 3 | 321 | XP_779406.1 fructose-1,6-bisphosphate aldolase (Giardia lamblia ATCC 50803) | 5e-116 | 323 |
| Triosephosphate isomerase | DQ665863 | 1 | 274 | CAA83533.1 triosephosphate isomerase (Secale cereale) | 2e-74 | 298 |
| Glyceraldehyde-3-phosphate dehydrogenase | DQ665854 | 16 | 357 | CAD72746.1 glyceraldehyde 3-phosphate dehydrogenase (Rhodopirellula baltica SH1) | 5e-116 | 342 |
| Phosphoglycerate kinase | DQ665858 | 13 | 402 | CAA48479.1 phosphoglycerate kinase (Spinacia oleracea) | 1e-98 | 433 |
| Phosphoglycerate mutase | DQ665859 | 6 | 565 | EAL48794.1 phosphoglycerate mutase, 2,3-bisphosphoglycerate-independent, putative (Entamoeba histolytica HM-1:IMSS) | 0.0 | 555 |
| Enolase | DQ665852 | 9 | 430 | EAN97849.1 enolase, putative (Trypanosoma cruzi) | 3e-156 | 429 |
| Pyruvate kinase | DQ665860 | 2 | 517 | ABA20118.1 Pyruvate kinase (Anabaena variabilis ATCC 29413) | 1e-97 | 476 |
| Pyruvate, phosphate dikinase 1 | DQ665861 | 15 | 879 | ABB90247.1 pyruvate phosphate dikinase 2 (Streblomastix strix) | 0.0 | 782 |
| Pyruvate, phosphate dikinase 2 | DQ665862 | 12 | 879 | ABB90247.1 pyruvate phosphate dikinase 2 (Streblomastix strix) | 0.0 | 782 |
Sequence sampling strategy and alignment.
In order to ensure representation from all of the major prokaryotic and eukaryotic groups possessing homologs of the enzymes of interest, to allow taxonomic comparability between phylogenetic analyses of the various enzymes, to avoid issues of hidden paralogy, and to maintain a taxon cardinality in the analyses amenable to rigorous computational analysis, an a priori sampling and phylogenetic strategy was employed. Homologous amino acid sequences of the glycolytic proteins were retrieved from the main representatives of the major groups of Eukaryota, Bacteria, and Archaea, listed below. The majority of sequences were retrieved from NCBI (http://www.ncbi.nlm.nih.gov); in the case of exceptions, the URLs are given in brackets. A table listing the accession numbers for proteins used for the phylogenetic analyses can be found in Table SA1 in the supplemental material.
In alphabetical order, with eukaryotes denoted by “(E)” after the strain name, the taxa sampled were: Aeropyrum pernix K1, Aquifex aeolicus VF5, Arabidopsis thaliana (E), Archaeoglobus fulgidus DSM 2661, Bacillus subtilis subsp. subtilis 168, Bordetella parapertussis 12822, Borrelia burgdorferi B31, Bradyrhizobium japonicum USDA 110, Burkholderia mallei ATCC 23344, Chlamydomonas reinhardtii (E) (http://genome.jgi-psf.org/cgi-bin/runAlignment?db=Chlre3&advanced=1), Chlamydophila caviae GPIC, Cryptococcus neoformans (E) Cyanidioschyzon merolae (E) (http://merolae.biol.s.u-tokyo.ac.jp/), Cytophaga hutchinsonii, Desulfovibrio desulfuricans G20, Dictyostelium discoideum (E), Drosophila melanogaster (E), Entamoeba histolytica (E), Escherichia coli O157:H7 EDL933, Fusobacterium nucleatum subsp. nucleatum ATCC 25586, Giardia intestinalis (E), Haemophilus influenzae Rd KW20, Halobacterium sp. strain NRC-1, Helicobacter pylori 26695, Homo sapiens (E), Leishmania major (E) (http://www.genedb.org/), Methanocaldococcus jannaschii DSM 2661, Mycobacterium tuberculosis H37Rv, Nostoc sp. strain PCC7120, Phytophthora sojae (E) (http://genome.jgi-psf.org/cgi-bin/runAlignment?db=sojae1&advanced=1), Plasmodium falciparum (E), Pyrobaculum aerophilum IM2, Rhodopirellula baltica SH1, Rickettsia prowazekii Madrid E, Saccharomyces cerevisiae (E), Streptococcus pyogenes MGAS315, Streptomyces coelicolor A3(2), Sulfolobus solfataricus P2, Synechocystis sp. strain PCC6, Tetrahymena thermophila (E) (http://tigrblast.tigr.org/er-blast/index.cgi?project=ttg), Thermotoga maritima MSB8, Trichomonas vaginalis (E) (http://tigrblast.tigr.org/er-blast/index.cgi?project=tvg), and Trypanosoma brucei (E). The genomes above were last sampled in March 2006.
The amino acid sequences were aligned using T-Coffee, version 1.37 (27), and manually adjusted. Only unambiguously aligned regions of homology were selected for final analyses. All alignments are available on request.
Phylogenetic analyses.
For each enzyme, several alignments were constructed and analyses were performed. An initial phylogenetic analysis was performed in order to identify lineage-specific paralog sets and to identify long branch sequences, which were likely to contribute artifact to subsequent analyses. This initial analysis involved topology determination by either maximum-likelihood (ML) or Bayesian phylogeny and support values produced by ML-corrected distance and either ML or Bayesian analyses. However, due to their large taxon cardinality, the initial data sets of PFK and PK were analyzed using only ML for the topology and ML-corrected distances to provide node support values. The long branch or lineage-specific duplicated sequences were then removed from the alignment, and a new mask was created, allowing for more alignment positions to be analyzed and for more intense phylogenetic analysis involving Bayesian, ML, and ML-corrected distance methodologies to be performed. Further subalignments were created and analyzed as additional taxon sampling was required, as described in Results. In cases where the M. balamuthi or S. strix homologs were available in the nr protein database, these were added to the alignment after the initial a priori sampling and lineage-specific duplicate phylogenies were performed.
The model of sequence evolution for each data set was determined using Tree-Puzzle, version 5.2 (35), based on initial neighbor-joining trees and incorporating either an eight-category gamma or a gamma plus invariant correction for rates among sites. Trees were then built using MrBayes, version 3.1.2 (29), for Bayesian analysis to determine optimal tree topology and posterior probability (PP) values for the nodes, with 500,000 Markov chain Monte Carlo generations and the burn-in value determined graphically by removing trees before the plateau; PHYML, version 2.4.4 (13), was used to obtain maximum-likelihood bootstrap values; and Fitch or Neighbor, version 3.6a3 from the PHYLIP package (9), was used to obtain ML-corrected distance bootstrap values from distance matrices generated by Tree-Puzzle based on 1,000 (for neighbor joining) and 100 (Fitch) pseudoreplicate data sets. Nodes with a posterior probability greater than 0.95 and bootstrap support better than 80% were considered robust, although on tree figures, all nodes with support values greater than 0.80 for posterior probability and 50% for bootstrap are shown. The figures illustrating the results of all preliminary analyses and analyses stated as “data not shown” are available upon request.
Nucleotide sequence accession numbers.
Novel sequence data reported in this paper have been deposited in GenBank under accession numbers DQ665852 to DQ665863. Details of sequence accession numbers and annotation are given in Table 1.
RESULTS
Sequence characterization and expression levels.
From our ongoing expressed sequence tag survey of Monocercomonoides (J. Dacks and V. Hampl, unpublished), we were able to identify cDNA clones putatively encoding glycolytic enzyme homologs. The nucleotide sequence of each gene was obtained either by assembling independent sequencing reads from multiple clones of the same gene (defined as sharing 98% or greater nucleotide identity) or, if multiple clones were not available, by double-strand sequencing of the clone of interest. Preliminary identification of the gene was determined by BLASTp.
As detailed in Table 1, enzyme homologs for 9 of the 10 steps in glycolysis were found to be present. Although we did not identify a GPI homolog, we did identify both PK and PPDK homologs. In comparing the predicted conceptual amino acid sequences of the oxymonad enzymes to that of the top returned BLASTp hit for each respective homolog, the oxymonad enzymes were as long as or longer than the predicted sequences in all but two cases and, thus, it is likely that we obtained the entire, or at least most of the, open reading frame for the oxymonad genes.
From the number of randomly obtained clones encoding each enzyme homolog (Table 1), we were able to deduce two points regarding the relative expression of the enzymes in the glycolytic pathway of oxymonads. Firstly, the enzymes for the last five steps of the pathway are more abundant than those for the first five steps (except GPI, about which we have no information) and, secondly, we found that there were 10 times more clones encoding PPDK than PK (Table 1).
Phylogenetic analyses.
While BLASTp analysis was sufficient in many cases to identify the Monocercomonoides sequence to its protein family and the step in glycolysis with which it is likely associated, phylogenetic analyses were required to more specifically classify the genes to the class or subclass level and to investigate their evolutionary histories. Therefore, phylogenetic analyses were performed on all 10 enzyme homologs that we obtained, corresponding to steps 1 and 3 through 10 of the glycolytic pathway.
Step 1: hexokinase.
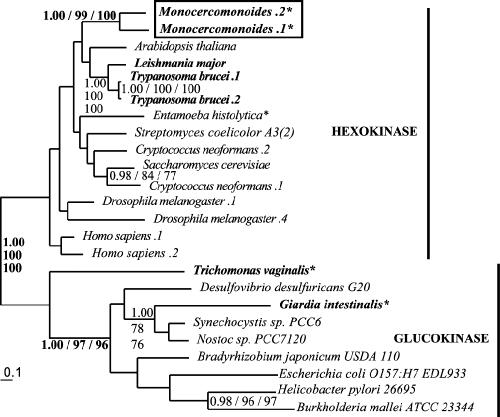
At least two different glucokinases can be found in eukaryotes, and two proposed relatives of oxymonads, Giardia and Trichomonas, are thought to have a shared LGT of glucokinase from cyanobacteria (16). Preliminary analyses using ML and ML-corrected distance methods and with bootstrap partitions (BP), given in that order, show three distinct clades: the eukaryote-specific hexokinases (BP, 100 and 100), the bacterial glucokinases also found in Trichomonas and Giardia (BP, 73 and 70), and the ROK sugar kinases (BP, 71 and 40). After removal of long branch sequences and lineage-specific duplicates, the monophyly of the ROK kinases was supported by 73% BP in ML analyses and 70% BP in ML-corrected distance analyses (data not shown). Since this class of transcription regulators, homologous to glucokinase and hexokinase, is specific to Bacteria and Archaea (15) and thus irrelevant to the question of oxymonad hexokinase evolutionary affinity, the ROK kinases were removed from subsequent phylogenies.
The resulting analyses (Fig. 2) robustly resolved the glucokinases and hexokinases with a PP of 1.00 and bootstrap support of 100% with both ML and ML-corrected distance methods (all subsequent values are given in the same order). The two Monocercomonoides homologs are resolved together as lineage-specific duplicates and both are clearly embedded within the hexokinase clade (Fig. 2). This is in contrast to the homolog from Giardia intestinalis, a glucokinase specifically related to the cyanobacterial homologs (PP, 1.00; BP, 78 and 76).
FIG. 2.
Phylogeny of glucokinase and hexokinase. This phylogeny shows the robust separation of glucokinase and hexokinase sequences into separate clades (demarked by the vertical bars). Monocercomonoides possesses two homologs of hexokinase; Giardia has glucokinase. Support values for these three results are in bold. In this figure and all subsequent phylogenetic analyses shown, the best Bayesian tree topology is presented. Posterior probability values, PHYML bootstrap values, and ML-corrected distance bootstrap values are provided in that order for all nodes with values better than 0.80 for posterior probability and 50% for ML and ML-corrected distance bootstrap values. Sequences from putative excavate taxa are presented in bold, Monocercomonoides sequences are boxed, and anaerobic eukaryotes are denoted by an asterisk. Multiple sequences from the same organism are denoted by numbers preceded by a period (.1, .2, etc.). Accession numbers for all sequences used in the analyses are provided in Table SA1 in the supplemental material; accession numbers for Monocercomonoides sequences are shown in Table 1.
Step 3: phosphofructokinase.
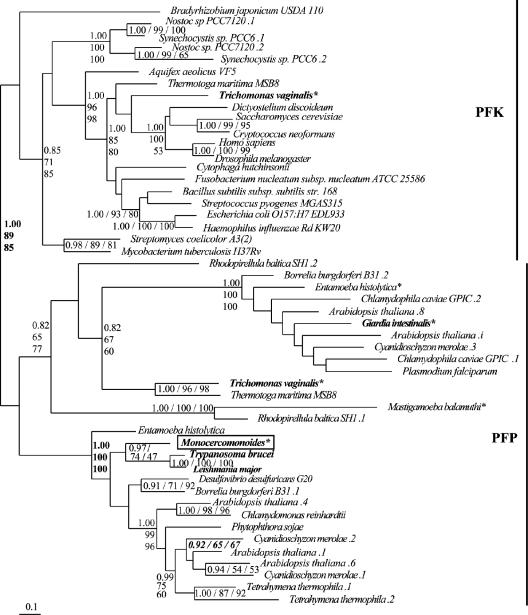
One particularly interesting question, raised by Slamovits and Keeling (34) regarding the glycolytic complement of oxymonads, is whether the second step in the pathway is performed by the irreversible PFK enzyme or the reversible, PPi-hydrolyzing PFP. We obtained only a single homolog for an enzyme encoding this step. After the preliminary analysis to remove long branch taxa and lineage-specific duplicates, analysis of the reduced data set resolved the PFP from the PFK homologs with Monocercomonoides homolog appearing as a PFP (Fig. 3). The division between the PFP and PFK clades is strong (PP, 1.00; BP, 89 and 85), with the PFP subdivided further into two groups. The Monocercomonoides homolog is robustly nested within one of these (PP, 1.00; BP, 100 and 100).
FIG. 3.
Phylogeny of PFK and PFP. This phylogeny shows the robust separation of PFK and PFP sequences into separate clades (demarked by the vertical bars). Monocercomonoides possesses a homolog of PFP, as do Trichomonas, Giardia, and the kinetoplastids. Trichomonas also possesses a homolog of PFK. Sequences from putative excavate taxa are presented in bold, Monocercomonoides sequences are boxed, and anaerobic eukaryotes are denoted by an asterisk. Multiple sequences from the same organism are denoted by numbers preceded by a period (.1, .2, etc.). Values separating PFP and PFK as well as embedding the Monocercomonoides sequence in a subclade of PFP are in bold.
Steps 4 to 7: fructose-bisphosphate aldolase class II, glyceraldehyde 3-phosphate dehydrogenase (GAP), triosephosphate isomerase (TPI), and phosphoglycerokinase.
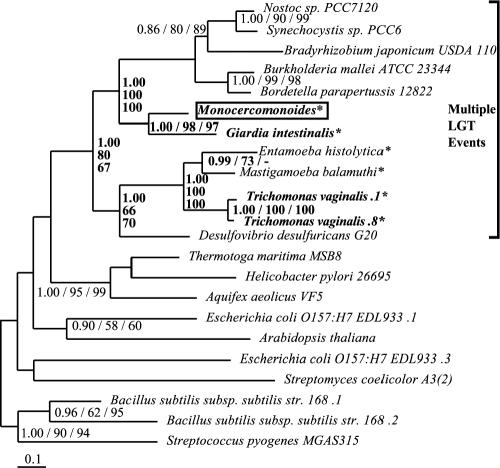
BLAST results indicated that the Monocercomonoides FBA homolog is of the class II family, while a preliminary phylogenetic analysis identified it specifically as a type B class II FBA. Therefore, a data set of FBA class II, type B homologs was assembled and analyzed (Fig. 4). Monocercomonoides appears as a sister taxon to G. intestinalis (PP, 1.00; BP, 98 and 97), the two grouping strongly with bacteria (PP, 1.00; BP, 100 and 100). Homologs from the two anaerobic amoebae, E. histolytica and M. balamuthi, grouped with the two T. vaginalis homologs (PP, 1.00; BP, 100 and 100).
FIG. 4.
Phylogeny of FBA, class II, type B. This phylogeny shows the excavates Monocercomonoides, Trichomonas, and Giardia as well as the amoebae strongly embedded within a bacterial clade. Multiple LGT events between T. vaginalis and the amoebae as well as between the oxymonads, diplomonads, and various prokaryotes must be invoked to account for the observed phylogeny (relevant nodes are shown in bold). Sequences from putative excavate taxa are presented in bold, Monocercomonoides sequences are boxed, and anaerobic eukaryotes are denoted by an asterisk. Multiple sequences from the same organism are denoted by numbers preceded by a period (.1, .2, etc.). A minus sign indicates that the node is not reconstructed in the ML-corrected distance bootstrap consensus tree.
Analyses of data sets of TPI, GAP, and phosphoglycerate kinase were largely unresolved (data not shown). While in the TPI analysis, the Monocercomonoides homolog appeared embedded within a robustly monophyletic eukaryotic clade (PP, 1.00; BP, 87 and 95), the phosphoglycerate kinase data set yielded little resolution for the placement of the oxymonad sequence. Likewise, the GAP phylogeny provided little overall resolution; however, the Monocercomonoides homolog did group specifically with T. vaginalis with strong support values (PP, 1.00; BP, 88 and 88).
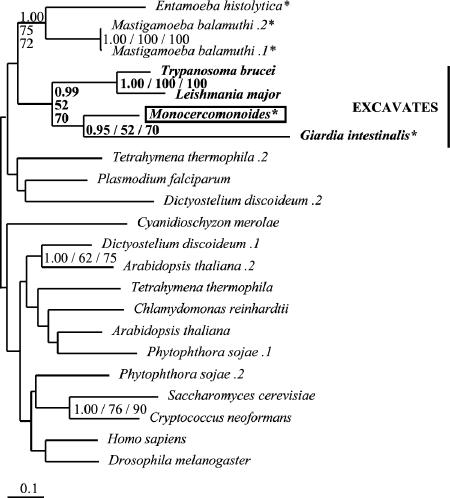
Step 8: phosphoglycerate-independent PGM.
Of the two unrelated classes of PGM, phosphoglycerate (or cofactor)-dependent dPGM and cofactor independent iPGM, BLASTp results (Table 1) identified the Monocercomonoides homolog as an iPGM. After preliminary phylogeny to facilitate removal of long branch taxa and lineage-specific duplicates, the analysis of the resulting reduced iPGM data set resolved two clades in the iPGM tree (PP, 1.00; BP, 100 and 99), separating most eukaryotes from prokaryotes (Fig. 5); however, a few eukaryotes, including T. vaginalis, appear within the prokaryotic clade. Monocercomonoides groups strongly (PP, 1.00; BP, 100 and 100) with E. histolytica; although G. intestinalis forms part of the eukaryotic clade, its position is unresolved.
FIG. 5.
Phylogeny of iPGM. This phylogeny shows that Monocercomonoides possesses a homolog of iPGM, along with Giardia, kinetoplastids, and Trichomonas. The Monocercomonoides and Entamoeba homologs are robustly placed as sisters, strongly suggestive of a eukaryote-to-eukaryote LGT (values are shown in bold). Sequences from putative excavate taxa are presented in bold, Monocercomonoides sequences are boxed, and anaerobic eukaryotes are denoted by an asterisk. Multiple sequences from the same organism are denoted by numbers preceded by a period (.1, .2, etc.).
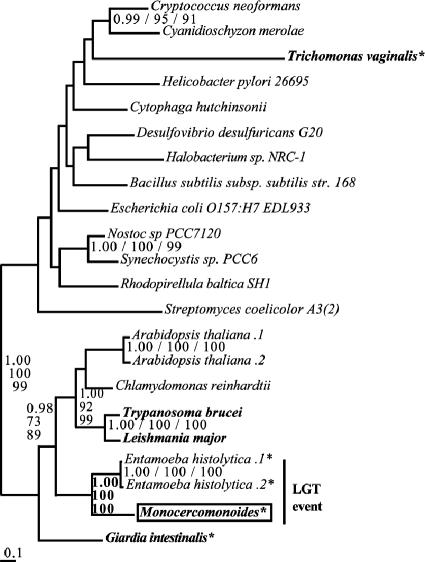
Step 9: enolase.
After the initial analysis of enolase homologs to identify long branch sequences and lineage-specific duplicates, our analysis with the long branch taxa removed separated eukaryotes from prokaryotes with moderate support values (PP, 1.00; BP, 65 and 58). Based on this analysis, and consistent with previous results (18, 21), a eukaryote-specific tree was made prior to the addition of the M. balamuthi enolase sequence, uniting representatives of the two major excavate clades with relatively strong support values (PP, 1.00; BP, 61 and 73). In order to avoid long branch attraction and to improve resolution, we removed long branches, including T. vaginalis; although it is an excavate, the enolase gene in this organism has been previously shown to have had a uniquely complex evolutionary history (4, 18, 20).
This final data set (Fig. 6) placed the excavate representatives into a single clade (PP, 0.99; BP, 52 and 70); within this clade the kinetoplastids group together (PP, 1.00, BP, 100 and 100), as do Monocercomonoides and G. intestinalis (PP, 0.95; BP, 52 and 70). Overall, the resolution of the two groups of excavates in this data set is among the more robust support seen for excavate monophyly to date in a single gene phylogeny.
FIG. 6.
Phylogeny of enolase. This phylogeny groups the putative excavates Monocercomonoides, Giardia, and kinetoplastids into a single clade. The putative excavate taxa and the values supporting their monophyly are in bold. Sequences from putative excavate taxa are presented in bold, Monocercomonoides sequences are boxed, and anaerobic eukaryotes are denoted by an asterisk. Multiple sequences from the same organism are denoted by numbers preceded by a period (.1, .2, etc.).
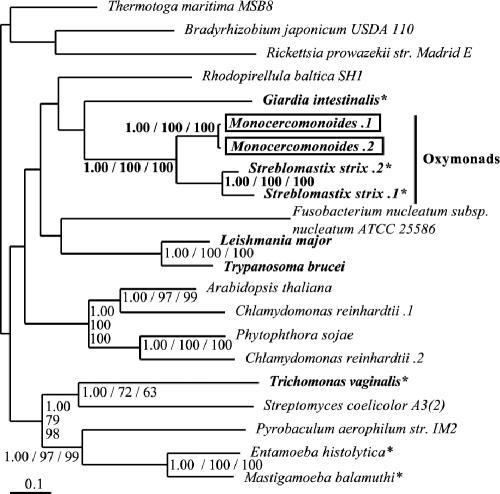
Step 10: pyruvate kinase and pyruvate phosphate dikinase.
Slamovits and Keeling (34) previously raised the question of whether PPDK and PK coexist in oxymonads. Our analysis identified homologs of both enzymes. The phylogeny of PK was generally poorly resolved. Although the PPDK tree has better resolution, the positions of the excavates are still unresolved (Fig. 7). The two oxymonads Streblomastix strix and Monocercomonoides appear together with very strong support values (PP, 1.00; BP, 100 and 100). For both oxymonad lineages, the two genes identified for each of the oxymonads appear to be lineage-specific duplicates (PP, 1.00; BP, 100 and 100).
FIG. 7.
Phylogeny of PPDK. This phylogeny groups the two oxymonads, Monocercomonoides and Streblomastix, into a single strongly supported clade. However, independent lineage-specific duplication events have occurred in both taxa. Relevant support values for these points are in bold. Sequences from putative excavate taxa are presented in bold, Monocercomonoides sequences are boxed, and anaerobic eukaryotes are denoted by an asterisk. Multiple sequences from the same organism are denoted by numbers preceded by a period (.1, .2, etc.).
DISCUSSION
Our analysis has allowed us to (i) reconstruct the glycolytic pathway of Monocercomonoides sp., (ii) identify both single-gene trees and shared derived LGT events that provide evidence for the monophyly of excavates and the Metamonada sensu Cavalier-Smith (6), and (iii) identify LGT events in the oxymonad glycolytic pathway, reinforcing previous conclusions that anaerobic glycolysis has been shaped by extensive lateral as well as vertical gene transfer.
Glycolytic enzymes of Monocercomonoides and their biological implications.
Based on the preliminary identification from the BLASTp and the phylogenetic analyses of the various sequences of glycolytic enzymes, it is possible to reconstruct the components of the glycolytic machinery in Monocercomonoides sp. (Fig. 1). While most of the standard eukaryotic versions of glycolytic enzymes were found, the alternative versions PFP, FBA class II (type B), iPGM, and PPDK were also present, making this pathway a mosaic similar to that of other anaerobes characterized to date (for examples, see references 26 and 36). Based on the relative number of clones for each of the glycolytic enzymes, the expression levels of the enzymes in the second half of the pathway are, on average, 10 times larger than those in the first half (Table 1). This result may reflect the fact that the pentose phosphate pathway feeds into the bottom half of the EMP pathway, requiring a higher expression of those enzymes as they do double duty in essentially two metabolic pathways (5).
Questions have also been raised in the past regarding the coexpression of PK and PPDK in the oxymonads and whether PPDK is accompanied by PFP (34); we found all three enzymes in Monocercomonoides as well as in all the other excavates sampled (T. vaginalis, G. intestinalis, and the kinetoplastids). T. vaginalis has both PFK and PFP enzymes. This highlights the possibility that, while the Monocercomonoides sequence that we identified was a PFP homolog, a PFK homolog may yet exist. We note that, while the fructokinases from kinetoplastids are evolutionarily directly homologous to PPi-dependent PFK (PFP), enzymatically they are ATP dependent (24). Therefore it remains to be determined by biochemical studies whether the related Monocercomonoides enzyme is PPi or ATP dependent.
Moreover, PPDK is expressed more than PK by an order of magnitude (Table 1). Because PPDK is more efficient than PK in terms of ATP conservation and reversibility, its high expression level relative to that of PK suggests that PPDK is an important enzyme in the glycolytic pathway of Monocercomonoides. That the ATP-conserving enzyme for this step in the pathway is expressed preferentially over the irreversible enzyme suggests that the drive for ATP efficiency may be acting at the cellular as well as the genomic level. We expect that, if PFK is found in this organism, its expression level would also be significantly below that of PFP. Enzymatic or genomic analyses will be useful in exploring the issue further. The presence of both PPi-utilizing enzymes in addition to the ATP-dependent PK underlines the potential importance of energy conservation to Monocercomonoides.
In examining the mosaic composition of the glycolytic complement of Monocercomonoides, we also identified numerous LGT events which have implications for both the evolution of oxymonads as a lineage and for the role of LGT in the evolution of anaerobic glycolysis.
Support for Excavata monophyly.
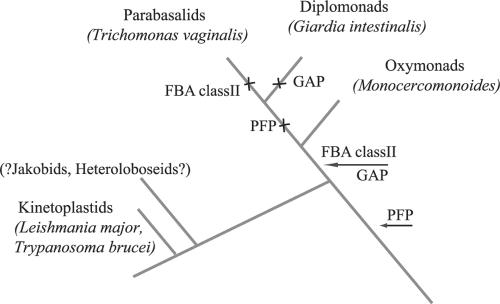
Although it is one of the six recognized eukaryotic supergroups (1), the composition and monophyly of the Excavata is still a matter of contention (19). While a sophisticated argument based on homologous cell structures and molecular phylogenetics has been used to knit together the various excavate taxa (31, 33), no analysis of single genes has yet been able to demonstrate excavate monophyly. Although a single concatenated analysis has recently united the kinetoplastids strongly with Trichomonas and Giardia (28), most analyses fail to find robust support for excavate monophyly (31). To the best resolution to date, the oxymonads were found to be the outgroup to a clade of parabasalids and diplomonads which form the Metamonada sensu Cavalier-Smith (6), representing one of the two major clades of excavates. The second clade is composed of the Euglenozoa, jakobids, and Heterolobosea (32).
Our analysis of glycolytic enzymes from Monocercomonoides uncovered evidence consistent with Metamonada and Excavata monophyly. Robust relationships were observed between excavate taxa in several of the phylogenies of glycolytic enzymes. While more complex scenarios of eukaryote-to-eukaryote LGT or phylogenetic artifact could be invoked, we interpret these phylogenetic results most parsimoniously as shared LGT events. Such evidence was found uniting Monocercomonoides and Giardia (FBA) (Fig. 4) as well as Monocercomonoides and Trichomonas (GAP). The hexokinase analysis is consistent either with a shared replacement from bacteria to the ancestor of Trichomonas and Giardia or with multiple replacement events (Fig. 2). Finally, it appears that the kinetoplastids and oxymonads share an LGT of their PFP enzymes (Fig. 3). This may be explained most simply by a replacement to the common ancestor of all excavates with subsequent replacements into Giardia and Trichomonas. The monophyly of excavates is further supported by the enolase tree, which places representatives of both major excavate clades (32) into one relatively well-supported clade (Fig. 6). Although, on their own, none of these pieces of data would be enough to claim Metamonada or Excavata monophyly, when taken with previous evidence (14, 31), these data are most consistent with a relationship of oxymonads with the other Metamonada, with this clade related to the other excavates and LGT continuing to both lay down and erase phylogenetic signal in this eukaryotic supergroup (Fig. 8). However, given the complex history of eukaryotic enolase and the possible alternate explanations of the phylogenies that we have interpreted as shared LGTs, additional support from either single-gene or multigene analyses will be needed before the excavate controversy is laid to rest. Nonetheless, since the analysis of glycolytic enzymes seems to have shed some light on the relationships within the proposed Excavata, studying the glycolytic pathway of Malawimonas may also help place this organism into its proper clade.
FIG. 8.
Cartoon illustrating proposed evolutionary history of glycolytic enzymes within the excavates. The arrows show gain or replacement of enzymes, and crosses represent enzyme loss after the split from the last common excavate ancestor. Kinetoplastids are proposed to have diverged first, followed by the oxymonads, with parabasalids and diplomonads diverging last. The various events are most parsimoniously interpreted as evidence consistent with the proposed relationships shown.
Role of LGT and selection in the evolution of anaerobic glycolysis.
Regardless of how one interprets the various LGT events observed in our analyses, it is clear that this phenomenon has had a significant role in shaping the glycolytic complement of anaerobic eukaryotes. From our research and previous investigations, both prokaryotic-to-eukaryotic and intraeukaryotic LGT must be invoked.
The simplest explanation for the FBA class II, type B phylogeny (Fig. 4) involves at least two separate LGTs from bacteria to anaerobic eukaryotes as well as a possible intraeukaryotic transfer between T. vaginalis and the amoebozoans E. histolytica and M. balamuthi. An LGT between two eukaryotes, E. histolytica and Monocercomonoides, is evident in the iPGM tree (Fig. 5). As eukaryote-to-eukaryote LGTs are rarely observed, it is also not impossible that these organisms received the same gene from the same bacterium, as opposed to from one of the eukaryotes to another.
There is a question as to whether the presence of ATP-conserving enzymes in certain anaerobic eukaryotes was a result of their pathogenicity or the anaerobic environment that they inhabit. We observed a number of shared LGT events to the common ancestor of two anaerobes, one of which is pathogenic and one that is not. One such example is the FBA class II tree (Fig. 4), which shows an LGT to the ancestor of the pathogenic Giardia and the commensal Monocercomonoides as well as an LGT to the ancestor of the pathogenic Entamoeba and the free-living Mastigamoeba. This, together with the presence and a higher expression level in Monocercomonoides of the ATP-conserving glycolytic homologs, suggests that it might be pressure towards energy conservation in a low-oxygen environment, rather than the parasitic nature of the anaerobes, that is driving the acquisition of alternate glycolytic enzymes via LGT.
Clearly, the evolution of anaerobic glycolysis is a muddy question. As more genomic data becomes available from diverse eukaryotic taxa, determining the evolutionary history of this pathway should be both fruitful and exciting.
Supplementary Material
Acknowledgments
The Monocercomonoides cDNA library was created in the lab of T. Martin Embley and we are grateful for his generous donation of these materials for analysis.
This work was supported by a Joint Canadian Institutes of Health Research/Wellcome Trust fellowship grant to J.B.D. and L.G. C.W.S. and P.G. are supported by Genome Alberta, in part through Genome Canada.
Footnotes
Published ahead of print on 27 October 2006.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Adl, S. M., A. G. Simpson, M. A. Farmer, R. A. Andersen, O. R. Anderson, J. R. Barta, S. S. Bowser, G. Brugerolle, R. A. Fensome, S. Fredericq, T. Y. James, S. Karpov, P. Kugrens, J. Krug, C. E. Lane, L. A. Lewis, J. Lodge, D. H. Lynn, D. G. Mann, R. M. McCourt, L. Mendoza, O. Moestrup, S. E. Mozley-Standridge, T. A. Nerad, C. A. Shearer, A. V. Smirnov, F. W. Spiegel, and M. F. Taylor. 2005. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52:399-451. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bapteste, E., D. Moreira, and H. Philippe. 2003. Rampant horizontal gene transfer and phospho-donor change in the evolution of the phosphofructokinase. Gene 318:185-191. [DOI] [PubMed] [Google Scholar]
- 4.Bapteste, E., and H. Philippe. 2002. The potential value of indels as phylogenetic markers: position of trichomonads as a case study. Mol. Biol. Evol. 19:972-977. [DOI] [PubMed] [Google Scholar]
- 5.Brown, J. R., and W. F. Doolittle. 1997. Archaea and the prokaryote-to-eukaryote transition. Microbiol. Mol. Biol. Rev. 61:456-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavalier-Smith, T. 2003. The excavate protozoan phyla Metamonada Grassé emend. (Anaeromonadea, Parabasalia, Carpediemonas, Eopharyngia) and Loukozoa emend. (Jakobea, Malawimonas): their evolutionary affinities and new higher taxa. Int. J. Syst. Evol. Microbiol. 53:1741-1758. [DOI] [PubMed] [Google Scholar]
- 7.Chevalier, N., D. J. Rigden, J. Van Roy, F. R. Opperdoes, and P. A. Michels. 2000. Trypanosoma brucei contains a 2,3-bisphosphoglycerate independent phosphoglycerate mutase. Eur. J. Biochem. 267:1464-1472. [DOI] [PubMed] [Google Scholar]
- 8.Dacks, J. B., J. D. Silberman, A. G. Simpson, S. Moriya, T. Kudo, M. Ohkuma, and R. J. Redfield. 2001. Oxymonads are closely related to the excavate taxon Trimastix. Mol. Biol. Evol. 18:1034-1044. [DOI] [PubMed] [Google Scholar]
- 9.Felsenstein, J. 1995. PHYLIP (Phylogeny Inference Package), 3.57 ed. Department of Genetics, University of Washington, Seattle, Wash.
- 10.Fothergill-Gilmore, L. A., and P. A. Michels. 1993. Evolution of glycolysis. Prog. Biophys. Mol. Biol. 59:105-235. [DOI] [PubMed] [Google Scholar]
- 11.Fothergill-Gilmore, L. A., and H. C. Watson. 1989. The phosphoglycerate mutases. Adv. Enzymol. Relat. Areas Mol. Biol. 62:227-313. [DOI] [PubMed] [Google Scholar]
- 12.Fraser, H. I., M. Kvaratskhelia, and M. F. White. 1999. The two analogous phosphoglycerate mutases of Escherichia coli. FEBS Lett. 455:344-348. [DOI] [PubMed] [Google Scholar]
- 13.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 14.Hampl, V., D. S. Horner, P. Dyal, J. Kulda, J. Flegr, P. G. Foster, and T. M. Embley. 2005. Inference of the phylogenetic position of oxymonads based on nine genes: support for Metamonada and Excavata. Mol. Biol. Evol. 22:2508-2518. [DOI] [PubMed] [Google Scholar]
- 15.Hansen, T., and P. Schonheit. 2003. ATP-dependent glucokinase from the hyperthermophilic bacterium Thermotoga maritima represents an extremely thermophilic ROK glucokinase with high substrate specificity. FEMS Microbiol. Lett. 226:405-411. [DOI] [PubMed] [Google Scholar]
- 16.Henze, K., D. S. Horner, S. Suguri, D. V. Moore, L. B. Sanchez, M. Muller, and T. M. Embley. 2001. Unique phylogenetic relationships of glucokinase and glucosephosphate isomerase of the amitochondriate eukaryotes Giardia intestinalis, Spironucleus barkhanus and Trichomonas vaginalis. Gene 281:123-131. [DOI] [PubMed] [Google Scholar]
- 17.Henze, K., H. G. Morrison, M. L. Sogin, and M. Muller. 1998. Sequence and phylogenetic position of a class II aldolase gene in the amitochondriate protist, Giardia lamblia. Gene 222:163-168. [DOI] [PubMed] [Google Scholar]
- 18.Keeling, P. J. 2004. Polymorphic insertions and deletions in parabasalian enolase genes. J. Mol. Evol. 58:550-556. [DOI] [PubMed] [Google Scholar]
- 19.Keeling, P. J., G. Burger, D. G. Durnford, B. F. Lang, R. W. Lee, R. E. Pearlman, A. J. Roger, and M. W. Gray. 2005. The tree of eukaryotes. Trends Ecol. Evol. 20:670-676. [DOI] [PubMed] [Google Scholar]
- 20.Keeling, P. J., and J. D. Palmer. 2000. Parabasalian flagellates are ancient eukaryotes. Nature 405:635-637. [DOI] [PubMed] [Google Scholar]
- 21.Keeling, P. J., and J. D. Palmer. 2001. Lateral transfer at the gene and subgenic levels in the evolution of eukaryotic enolase. Proc. Natl. Acad. Sci. USA 98:10745-10750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulda, J., and E. Nohynková. 1978. Flagellates of the human intestine and of intestines of other species, p. 104-113. In J. P. Krieier (ed.), Parasitic protozoa, vol. II. Academic Press, San Diego, Calif. [Google Scholar]
- 23.Lee, J. J., G. F. Leedale, and P. Bradbury (ed.). 2002. The illustrated guide to the protozoa. Society of Protozoologists, Lawrence, Kans.
- 24.Lopez, C., N. Chevalier, V. Hannaert, D. J. Rigden, P. A. Michels, and J. L. Ramirez. 2002. Leishmania donovani phosphofructokinase. Gene characterization, biochemical properties and structure-modeling studies. Eur. J. Biochem. 269:3978-3989. [DOI] [PubMed] [Google Scholar]
- 25.Mertens, E. 1993. ATP versus pyrophosphate: glycolysis revisited in parasitic protists. Parasitol. Today 9:122-126. [DOI] [PubMed] [Google Scholar]
- 26.Nevalainen, L., I. Hrdy, and M. Muller. 1996. Sequence of a Giardia lamblia gene coding for the glycolytic enzyme, pyruvate, phosphate dikinase. Mol. Biochem. Parasitol. 77:217-223. [DOI] [PubMed] [Google Scholar]
- 27.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205-217. [DOI] [PubMed] [Google Scholar]
- 28.Rodríguez-Ezpeleta, N., H. Brinkmann, S. C. Burey, B. Roure, G. Burger, W. Loffelhardt, H. J. Bohnert, H. Philippe, and B. F. Lang. 2005. Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr. Biol. 15:1325-1330. [DOI] [PubMed] [Google Scholar]
- 29.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 30.Sánchez, L., D. Horner, D. Moore, K. Henze, T. Embley, and M. Muller. 2002. Fructose-1,6-bisphosphate aldolases in amitochondriate protists constitute a single protein subfamily with eubacterial relationships. Gene 295:51-59. [DOI] [PubMed] [Google Scholar]
- 31.Simpson, A. G. 2003. Cytoskeletal organization, phylogenetic affinities and systematics in the contentious taxon Excavata (Eukaryota). Int. J. Syst. Evol. Microbiol. 53:1759-1777. [DOI] [PubMed] [Google Scholar]
- 32.Simpson, A. G., Y. Inagaki, and A. J. Roger. 2006. Comprehensive multigene phylogenies of excavate protists reveal the evolutionary positions of “primitive” eukaryotes. Mol. Biol. Evol. 23:615-625. [DOI] [PubMed] [Google Scholar]
- 33.Simpson, A. G., R. Radek, J. B. Dacks, and C. J. O'Kelly. 2002. How oxymonads lost their groove: an ultrastructural comparison of Monocercomonoides and excavate taxa. J. Eukaryot. Microbiol. 49:239-248. [DOI] [PubMed] [Google Scholar]
- 34.Slamovits, C. H., and P. J. Keeling. 2006. Pyruvate-phosphate dikinase of oxymonads and parabasalia and the evolution of pyrophosphate-dependent glycolysis in anaerobic eukaryotes. Eukaryot. Cell 5:148-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strimmer, K., and A. von Haeseler. 1996. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13:964-969. [Google Scholar]
- 36.Suguri, S., K. Henze, L. B. Sanchez, D. V. Moore, and M. Muller. 2001. Archaebacterial relationships of the phosphoenolpyruvate carboxykinase gene reveal mosaicism of Giardia intestinalis core metabolism. J. Eukaryot. Microbiol. 48:493-497. [DOI] [PubMed] [Google Scholar]
- 37.Wu, G., K. Henze, and M. Muller. 2001. Evolutionary relationships of the glucokinase from the amitochondriate protist, Trichomonas vaginalis. Gene 264:265-271. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.