Fungi are important pathogens of plants and cause more significant yield losses than bacteria or viruses. However, bacteria and viruses are more important than fungi as pathogens of animals; indeed, whether or not a fungus even becomes pathogenic on an animal often depends on the immune status of the host. Until the rapid rise of opportunistic fungal infections in humans, pathogenicity mechanisms in plant pathogens were better understood than those in animal pathogens. Increased research activity in medical mycology has coincided with the development of molecular genetic and genomic resources, which are being exploited to develop a detailed understanding of fungal pathogenesis in both animals and plants. Several constraints and peculiarities govern the types of information that can be derived from such studies. For instance, analyses of human-pathogenic fungi generally rely on cell lines and experimental animal models, in contrast to plant pathogens, which can be studied directly on their hosts. Many more fungal species infect plants than animals, and thus, more plant-fungus systems than animal-fungus systems are studied. This is mainly because there are far more plant hosts than animal hosts that are of economic importance, with the obvious exception of human disease, and because plants can be manipulated without the ethical issues associated with animal experimentation.
Pathogenesis involves the interaction of two partners with input from the environment, a concept described as the “disease triangle” in plant pathology. A more recent concept developed for animal pathogens is the “damage-response” framework which emphasizes that the outcome of an interaction is determined by the amount of damage incurred by the host (16). These concepts are useful reminders of the complexity of the interaction, as opposed to focusing on just the individual partners (host and pathogen). Surprisingly, there are few reports of fungal pathogenesis that describe commonalities of mechanisms for plants and animals. A commentary published nearly 10 years ago discussed issues such as degradation of the host, pathogen differentiation, regulatory genes, and signal transduction (37). These topics are still relevant, and an abundance of new information about them is available. This review describes parallels in fungal pathogenesis in plant and animal hosts, focusing on ascomycetes, as members of this phylum are generally well characterized. In general, four classes of ascomycetes are rich in plant pathogens (Dothideomycetes, Leotiomycetes, Sordariomycetes, and Taphrinomycetes), while animal pathogens generally belong to another two classes (Chaetothyriomycetes and Eurotiomycetes) that contain few plant pathogens (10). However, several ascomycete species can infect both animal and plant hosts, thus making it easier to identify commonalities of disease mechanisms. Other fungal phyla, particularly Basidiomycota, include important, well-characterized plant and animal pathogens (e.g., Ustilago maydis and Cryptococcus neoformans). These are reviewed extensively elsewhere (28, 41). Far less is known about the diseases caused by fungi from other phyla, including zygomycete diseases of humans and chytrid diseases of amphibians, as these are uncommon and are also challenging systems for experimental work. Although we emphasize the parallels in fungal pathogenesis in plant and animal hosts, in some cases we highlight a feature that has as yet been discovered only in either animal or plant pathogens but that may apply to both and thus lead to an enhanced understanding of disease mechanisms.
DEFENSE SYSTEMS OF PLANTS AND ANIMALS
The plant and animal host kingdoms have both innate and inducible/adaptive defense responses that are very different. These defense systems are generally effective in that the majority of fungi in the environment cannot cause disease. The immune system of mammals involves the innate complement system, circulating cells such as phagocytes that can internalize and destroy pathogen cells, and adaptive antibody-mediated defenses. The complement pathway, involving soluble factors and corresponding receptors, can lead to the formation of a pore complex in accessible pathogen membranes and subsequent lysis and opsonization, whereby proteins form a coating on antigens, pathogen cells, or host cells infected by the pathogen. This process “tags” them for clearance by the immune system and can trigger proinflammatory stimulation of chemotaxis (71, 77). Many serious fungal infections of mammals occur in immunocompromised hosts, suggesting that mammalian defense systems are usually very effective against fungi. The severity of fungal diseases ranges from serious infections (histoplasmosis, blastomycosis, and coccidioidomycosis) (21) requiring hospitalization to superficial cutaneous infections (e.g., tinea) which are extremely common and caused by fungi such as Candida and Trichophyton spp.
In contrast, fungal pathogens of plants have developed many mechanisms to evade or overcome healthy host plant defenses. Although plants do not have circulating or phagocytic cells, their cells have a thick, complex wall that acts as a barrier to invasion. Plants display innate pathogen-specific resistance, genetically controlled via resistance genes. Additionally, plants display inducible systemic acquired resistance, which occurs when previous exposure to a pathogen activates signaling pathways acting via molecules such as jasmonate, ethylene, and salicylic acid. These small and sometimes volatile molecules spread throughout the plant or even the plant population. This triggers responses such as the expression of “pathogenesis-related proteins,” including chitinases or glucanases, which can lead to the increased resistance of the whole plant against a subsequent pathogen attack (66). This outcome is analogous to that resulting from immunization or preexposure to a pathogen in animals, where the defense system is primed to improve resistance to subsequent challenge by the pathogen. The jasmonate (or lipoxygenase) pathway mentioned above involves oxygenation of fatty acids, and a similar pathway known as the eicosanoid (e.g., prostaglandins and leukotrienes) pathway is present in mammals. Oxylipins, end products of these pathways, are implicated in host defense and stress responses. These molecules are also present in fungi and involved in signaling and development (74); their roles in host-pathogen interactions are discussed later in this review.
Commonalities of the defense systems of different hosts against fungal pathogens include programmed cell death and oxidative burst response (55, 56). Animal- and plant-pathogenic microbes (fungal and bacterial) release molecules with pathogenicity-associated molecular patterns. Determinants on fungus-derived polysaccharides and proteins are recognized (usually indirectly) by conserved receptors in animals and plants and elicit a defense response. These receptors are often transmembrane proteins with leucine-rich repeat domains and are manifested as resistance gene products in plants and Toll-interleukin receptors in animal and insect cells (8). This appears to be either an ancient conserved eukaryotic pathway (57) or the result of convergent evolution whereby similar motifs have been recruited for defense in different systems (3).
APPROACHES TO ANALYSIS OF FUNGAL PATHOGENESIS
During the last 5 years, complete genome sequences, banks of tagged mutants, and large numbers of expressed sequence tags have become available for fungi. So far, most of the knowledge about fungal pathogenesis comes from the characterization of loss-of-function mutants generated by either random or targeted insertion of a selectable marker. Candidate genes for targeted mutagenesis are identified by comparative genome analysis between pathogens and nonpathogens and/or by large-scale transcriptome analyses. Recently, an internet database named PHI-base (Pathogen Host Interactions; http://www.phi-base.org/) that catalogues the phenotypes resulting from mutations in defined genes of both plant and animal pathogens has been developed (91). This database is becoming a useful resource for the discovery of candidate targets in agriculturally and medically important fungi. Since fungal genomes are relatively compact compared to those of their hosts, it is often possible to identify virulence factors by synteny. One important example of this is the prediction of genes involved in the biosynthesis of secondary metabolite toxins, as such genes are usually clustered in the fungal genome. Diagnostic genes for secondary metabolite production include those encoding nonribosomal peptide synthases or polyketide synthases, as well as those responsible for modifications of the core moiety (a peptide or polyketide), such as genes encoding methyltransferases, acetyltransferases, prenyltransferases, oxidoreductases, and cytochrome P450 monooxygenases. Such genes are much more common in pathogenic fungi than in nonpathogenic fungi (25, 93) and generally reflect the diverse spectrum of secondary metabolites produced by pathogens.
Genomic resources are now available for some plant hosts (rice and Arabidopsis thaliana), and much knowledge about pathogenesis mechanisms has been derived from disease studies where the model plant A. thaliana is the host. For animal diseases, the complete mouse genome sequence and the ability to create mouse gene knockout lines provide further potential for understanding host-pathogen interactions. Another approach for elucidating such mechanisms is to exploit nonmammalian hosts such as Drosophila melanogaster and Caenorhabditis elegans (32). Genes involved in virulence can be identified by screening mutagenized fungi in these model host organisms.
HOST SPECIFICITY
Pathogenic fungi can have very broad or narrow host ranges. Some plant-pathogenic fungi such as the stem rot pathogen Sclerotinia sclerotiorum and the gray mold fungus Botrytis cinerea can infect hundreds of plant species (13, 84). Much is known about the host ranges of pathogens of agriculturally important plants, as resistance breeding is a major strategy for disease control. The specificities of interactions between many plants and pathogens are governed by resistance genes in the host and complementary avirulence (effector) genes in the pathogen (30). Of the 14 fungal avirulence genes from ascomycetes described so far, 6 encode small globular cysteine-rich proteins that have secretion signals, thus allowing these proteins to be accessible for recognition by the plant. These genes do not have sequence similarity even in closely related fungal species. In contrast, plant resistance genes contain domains that are homologous among even distantly related plants and with Toll-interleukin receptors in other eukaryotes, as mentioned above (94).
Other interactions with host molecules may contribute to a pathogen's host range. For example, the fatty alcohol fraction of the surface wax of avocado fruit, a host for the anthracnose fungus Colletotrichum gloeosporioides, induces spore germination and formation of melanized infection structures, termed appressoria. In contrast, waxes from nonhost plants do not induce appressorium formation in this fungus (67). For mammalian systems, there is not much literature on host specificity and recognition with hosts, but analogous mechanisms whereby host species-specific molecules trigger stages of the infection processes of particular fungal pathogens may exist.
Some fungi grow saprophytically in soil or dead matter and produce airborne spores which can cause pulmonary disease when inhaled by animals. Indeed, almost any fungus that produces spores small enough to reach alveoli, that can grow at 37°C, and that can evade host immune responses threatens human health. Fortunately, such infections usually do not have major consequences for healthy people. A few fungi can infect both plants and animals. The soilborne fungus Chaetomium globosum can infect plant roots endophytically without inducing disease symptoms and can control infection by some plant-pathogenic fungi (63). The airborne spores of this fungus can cause invasive disease with life-threatening symptoms, such as pneumonia, in immunocompromised patients (64). The best-studied fungal isolate that can affect both animals and plants is Fusarium oxysporum f. sp. lycopersici, which can kill both immunodepressed mice and tomato plants (58). Studies of this fungus highlight commonalities and differences in the mechanisms of pathogenicity on animal and plant hosts. For example, a mitogen-activated protein (MAP) kinase gene, Fmk1, of F. oxysporum is not required for virulence in mice but is essential for virulence in tomatoes. In contrast, the zinc finger transcription factor gene PacC is necessary for full virulence in mice but not in tomatoes (58). PacC is important for virulence in a range of other plant-specific and animal-specific fungi, as it mediates the environmental pH signal, which in turn alters gene expression appropriately. The regulation of responses to pH is discussed in more detail below. Another soilborne fungus, Aspergillus flavus, can infect animals, insects, and plants, particularly seeds of corn, peanuts, cotton, and nut trees. This fungus, like several other Aspergillus species, produces highly toxic, carcinogenic aflatoxins. Several strains isolated from humans and insects can also cause disease in corn (78). Different nutritional pathways may be important for the virulence of this fungus on different hosts, since a cysteine and methionine auxotroph of A. flavus has reduced conidiation in vitro and on plant hosts, but this auxotroph can still complete a disease cycle on insect hosts (73).
DEVELOPMENTAL REGULATION OF PATHOGENESIS
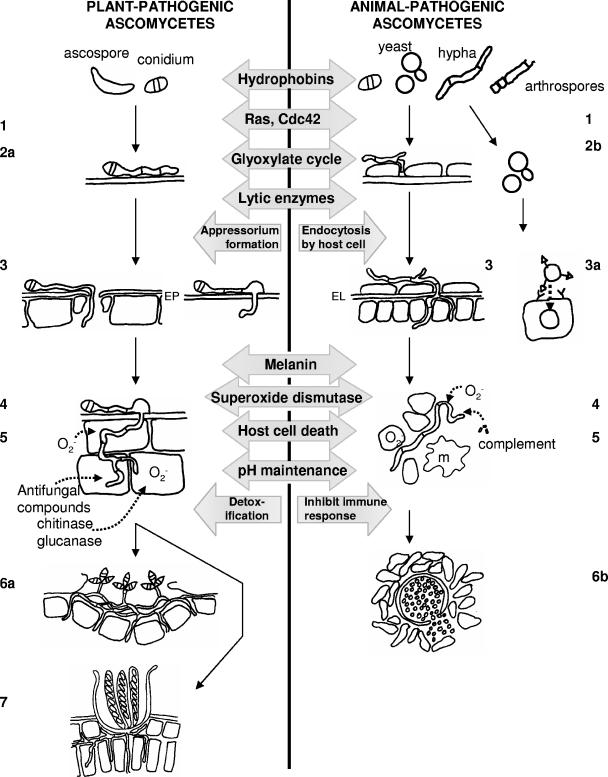
To be a pathogen, an organism must be able to cause disease and complete its life cycle on a host. Thus, pathogen spores must germinate on a host surface or in the alveoli of the lungs if inhaled, and the resulting hyphae must penetrate the host tissue, colonize, alter host physiology, and cause disease. Finally, the fungus must reproduce and disperse. These developmental steps are sequential, although each step often involves common mechanisms. For instance, pH changes can be important for both regulating penetration and creating a favorable environment for the subsequent colonization of a host. The types of genes essential for pathogenesis depend on the lifestyle and infection process of the fungus. Signaling cascades occur throughout these processes. Fungal genes or molecules involved in each of these developmental steps are discussed and those common to both animal and plant pathogens are emphasized in the sections below. The infection process for plants and animals is depicted in Fig. 1. This is a generalized scheme, and details of these processes vary from one host-fungus interaction to another.
FIG. 1.
Comparison of infection mechanisms used by ascomycete pathogens of plants and animals. Stage 1, attachment of conidia, ascospores (plant pathogens), yeast cells, hyphae, or arthrospores to a surface and recognition of the host. Stage 2a, germination of ascospores (plant pathogens), conidia, or arthrospores. Stage 2b, dimorphic switching of animal pathogens from a yeast phase to a pathogenic hyphal stage or from hyphae to a pathogenic yeast phase. Stage 3, penetration of the host surface or host cells may involve mechanical pressure, such as that produced by appressoria in some plant pathogens; lytic enzymes, such as proteases; and additional cell wall-degrading enzymes, including cutinases, cellulases, pectinases, and xylanases in plant pathogens. Natural openings in the host, such as stomata in plants or wounds in animals and plants, are also entry points for pathogenic fungi. Stage 3a, some animal pathogens (e.g., Histoplasma capsulatum) use receptors on host cells to bind and facilitate endocytosis as a means of penetrating the host cells. Stage 4, avoidance of host defenses. Pathogenic fungi may detoxify oxidative molecules such as superoxide and antifungal compounds and synthesize protective molecules such as melanin. Animal-pathogenic ascomycetes often avoid or inhibit animal immune system components. Plant pathogens may avoid exposure to fungal wall-degrading enzymes by causing little host cell damage when undergoing intercellular biotrophic growth or by producing inhibitors of these plant enzymes. Stage 5, colonization of the host environment. Colonization often results in host cell death, may require specific nutritional mechanisms such as those for iron uptake in animal hosts, and can produce other changes in host physiology, such as the pH level, to create a more favorable environment for the pathogen. Stage 6a, asexual reproduction often results in conidia emerging from lesions on the host surface of plants. Stage 6b, spore formation in the host is less common in animal pathogens, and direct host-to-host transmission is rare. Coccidioides immitis produces endospores in host tissue by numerous mitotic divisions inside a spherule. Stage 7, sexual reproduction. Mating and meiotic division produce ascospores during the disease cycle of plant-pathogenic fungi. This can result in recombinant offspring if mating occurs with a genetically different individual (obligatory in heterothallic but not in homothallic fungi). Sexual reproduction is not reported to occur in animal-pathogenic fungi, with a few exceptions, such as Pneumocystis spp. EP, plant epidermis/cuticle; EL, endothelium; m, macrophage.
Germination.
The germination of spores has been well characterized in saprophytes such as Neurospora crassa and Aspergillus nidulans, and unsurprisingly, the fundamentals are similar to those in animal and plant pathogens. These include a requirement for specific nutrients and moisture; morphological changes, including swelling and polarized growth; and metabolic changes (59). In plant pathogens, the germination of spores on a host may require additional triggers related to surface recognition and hydrophobicity (for instance, the avocado waxes described above) or other molecules (Fig. 1, diagram 2a). Lipids such as prostaglandin E2 can affect germ tube formation in the human pathogen Candida albicans (43) in a way somewhat analogous to that of waxes in some plant pathogens. In some cases, spore germination involves host-specific molecules. For instance, macroconidia of the pea pathogen Nectria haematococca germinate in response to the addition of flavonoids, including those that induce nod gene expression in pea-specific rhizobial species (4). This effect appears to be modulated via the inhibition of cyclic AMP (cAMP) phosphodiesterase, which affects cAMP levels. Additionally, pisatin, an antimicrobial pea isoflavonoid (phytoalexin), induces spore germination; however, this effect does not appear to be mediated via cAMP phosphodiesterase (4).
Temperature is often a key trigger for germination of and infection by animal pathogens, particularly for fungi that undergo dimorphic switching whereby their life cycles alternate between yeast and filamentous phases (Fig. 1, diagrams 2a and 2b). Conidia of Penicillium marneffei germinate at 25°C into a nonpathogenic filamentous phase and at 37°C proceed into the pathogenic yeast phase. The germination processes at the two temperatures are initially indistinguishable, with conidia swelling and forming germ tubes and then hyphae. However, at 37°C, the hyphae are highly branched and then proceed to arthroconidiation, where septation and nuclear division produce the single cells of the pathogenic yeast phase (2). In addition to temperature, other conditions such as pH and the presence of serum can regulate germination. Signaling components such as MAP kinase pathways, histidine kinases, and G proteins regulate germination in plant pathogens such as Botrytis cinerea. These factors also control other developmental processes in animal pathogens such as Candida albicans, including growth on the mucosal membrane of the gastrointestinal tract, biofilm formation, thigmotropism, and invasion (26, 46).
Many animal-pathogenic fungi undergo dimorphic switching. In some cases, the yeast form is more pathogenic than the hyphal phase, while in others the reverse is true. Fungi that grow almost exclusively in the yeast form in the host include Histoplasma capsulatum, Paracoccidioides brasiliensis, and Blastomyces dermatitidis; those that grow exclusively in the hyphal form in the host include Aspergillus species; and those that grow in both forms in the host include Candida albicans. This switching in C. albicans requires the regulation of genes including Cdc42 and Cdc24 (7). Additional signals that trigger this switch operate via MAP kinase- and cAMP-dependent protein kinase pathways and include elevated temperature, alkaline pH, serum, and the presence of N-acetylglucosamine (6, 7). During dimorphic switching, the composition of the cell wall may change; for example, in P. brasiliensis it alters from predominantly β-1,3-glucan to α-1,3-glucan (14). The ability to switch between yeast and hyphal morphologies is thought to be an important pathogenicity mechanism for escaping immune responses and establishing infection. For instance, in C. albicans yeast phase, a β-glucan is exposed that is recognized by the mammalian dectin-1 receptor and leads to the activation of macrophage defenses. However, during the hyphal phase, this β-glucan is not exposed (34). Such dimorphic switching is not a feature of plant-pathogenic ascomycetes, although it occurs in the basidiomycete corn pathogen U. maydis.
Specific nutritional mechanisms are activated via complex regulatory networks that include genes involved in germination, such as those encoding cAMP-dependent kinases, G α proteins, and MAP kinases, as well as Ras proteins, which are a family of small monomeric GTPases involved in a variety of cellular signaling pathways (26, 29, 59). In the alfalfa pathogen Colletotrichum trifolii, Ras regulates spore germination and pathogenic development via genes including Cdc42, which is required for growth and development (20). Cdc42 is involved in germ tube formation and invasive hyphal growth in C. albicans, and ectopic expression leads to loss of virulence in mice (6). Signal transduction cascades represent a clear example of conservation in determinants for disease development by plant and animal pathogens (26, 48, 54). There is an abundance of literature concerning signaling in both animal and plant pathogens, and this aspect is not discussed further here.
Until the fungus enters its host, it must rely on its own nutrition. The glyoxylate cycle plays an important role in the nutrition of both plant- and animal-pathogenic fungi. These include plant pathogens Magnaporthe grisea (88), Leptosphaeria maculans (42), and Stagonospora nodorum (76) and animal pathogens such as C. albicans (51) and Aspergillus fumigatus (27). Fungal mutants with mutations in the glyoxylate cycle enzymes isocitrate lyase or malate synthase have low levels of germination and are not infectious. The pathogenicity of an isocitrate lyase mutant of L. maculans on Brassica napus (canola) is restored by the addition of glucose to the inoculum (42). This finding supports the hypothesis that due to growth in a sugar-limited environment, fungi rely on fatty acid metabolism for growth before penetration of the host.
Common protective molecules are produced by plant- and animal-pathogenic fungi. The surfaces of many fungal spores are covered by a rodlet layer consisting of molecules including hydrophobins, which are low-molecular-weight, cysteine-rich, hydrophobic proteins. In plant pathogens such as M. grisea, particular hydrophobins are needed for a variety of processes during infection (44, 79). For instance, mutations in hydrophobin MHP1 have pleotropic effects on fungal morphogenesis, including reductions in conidiation, spore germination, and appressorium formation and infectious growth (44), while mutations in hydrophobin MPG1, which has about 20% sequence similarity to MHP1, significantly reduce appressorium formation on the host but not on other surfaces. Conidial germination and infectious growth are not altered (79). Hydrophobins in conidial walls protect the animal pathogen A. fumigatus from being killed by macrophages during initial invasion events (62). Although these A. fumigatus hydrophobins have structural features similar to those of M. grisea, they have very low levels of sequence similarity. Novel roles for hydrophobins in cell adhesion, protection, and protein-protein interactions in fungal pathogens may yet be discovered. Other fungal cell wall molecules, such as melanin, have crucial roles in protecting the pathogen during the initial stage of growth in the host environment. Melanin is essential for the viability of spores of Pneumocystis species (40), and melanized yeast forms of P. brasiliensis are more resistant to phagocytosis by macrophages than nonmelanized forms (24). An increased melanin content of the conidial cell wall in A. fumigatus causes hypervirulence and faster germination, as well as increased resistance to reactive oxygen species (52). At least five plant-pathogenic fungal mutants with mutations affecting melanin biosynthesis and resulting in the loss or reduction of pathogenicity are listed in the PHI-base database (91). This phenotype is commonly due to a defect at the penetration stage, as melanin is produced during appressorial formation (65). Fungicides targeting melanin biosynthesis have been developed (49). Other fungal wall components, such as chitin and β-glucan, are also important for the growth and virulence of several animal and plant pathogens. Ironically, some of these protective molecules also trigger defense responses, such as the synthesis of pathogenesis-related protein in plant hosts and proinflammatory responses in animal hosts. Accordingly, these molecules are potential targets for antifungal drugs and vaccines (17, 75, 89).
Invasion.
Some fungal pathogens enter the host via natural openings (stomata of plants or air passages of animals) or even wounds, whereas others secrete toxins and/or enzymes, apply mechanical force, or subvert cellular processes of the host (Fig. 1, diagram 3). Often entry by force (mechanical pressure and enzymatic degradation of host tissue) is preceded by a morphological change in the fungus. For instance, some plant-pathogenic fungi produce invasive hyphae and/or infection structures such as appressoria, which sequester high concentrations of glycerol, producing high turgor pressure and thus allowing the fungus to puncture the leaf and gain entry (81). Appressorial formation involves a range of molecules and signals, including tetraspanins, P-type ATPases, and cyclophilins in M. grisea (5, 85, 86). Tetraspanins are integral membrane proteins involved in a variety of cellular functions such as differentiation and adhesion in animal cells and are necessary for virulence in plant-pathogenic fungi such as B. cinerea and M. grisea due to their role in the formation of the penetration pegs which emerge from appressoria and enter the host tissue (22). A homolog is present in the saprophytic fungus N. crassa, and there are hypothetical protein homologs in the animal-pathogenic fungus Coccidioides immitis and in the multihost pathogen Chaetomium globosum. However, whether the tetraspanin genes play a role in the infection processes of these pathogens is not known. Interestingly, appressoria have not been described for animal-pathogenic fungi (except for a few similarly shaped structures arising at the swollen hyphal tips of C. albicans) (45). However, other mechanisms, such as the binding of specific receptors, are used to gain entry to host cells. For instance, Histoplasma capsulatum binds via Hsp60 to β2 integrin receptors on macrophages, is then internalized (Fig. 1, diagram 3a), and survives intracellularly in a modified phagosome compartment (92).
Many plant pathogens need hydrolytic enzymes such as cutinases, pectinases, and cellulases to degrade the cuticle (waxy layer) and the plant cell wall. These enzymes are usually encoded by multigene families, and pathogenicity cannot be attributed to one particular member. One class of hydrolytic enzymes, the aspartyl proteases, is expressed by both animal- and plant-pathogenic ascomycetes during infection. C. albicans has at least 10 aspartyl proteinases, which assist in the infection process by degrading host cell surface molecules and membranes, facilitating adhesion, and degrading host molecules for nutrition (72). Two of these, Sap9 and Sap10, are cell surface associated and involved in adhesion to epithelial cells (1). Plant pathogens Sclerotinia sclerotiorum and Botrytis cinerea express aspartyl proteases during plant infection, but the importance of these proteases in plant disease has not been demonstrated (69, 80).
Colonization and alteration of host physiology.
To survive in and colonize its host, a pathogen must adapt itself or else modify the host environment (Fig. 1, diagrams 4 and 5). Molecules such as oxalic acid, which lowers pH, are virulence factors against plant, animal, and insect hosts. Oxalic acid produced by S. sclerotiorum affects stomatal opening and closing and suppresses the oxidative burst of the host defense response. This activity regulates the production of many enzymes (e.g., polygalacturonases) and developmental processes (e.g., formation of sclerotia, which are thick-walled resting structures) (18, 36). As mentioned previously, PacC is involved in controlling these processes and is required for virulence in tomato and Arabidopsis spp., as well as for sclerotial development (70). Furthermore, PacC is required for the full virulence of F. oxysporum and Aspergillus nidulans in immunocompromised mice, and its overexpression enhances fungal virulence in the pulmonary aspergillosis model (11, 58).
Oxylipins have complex pleiotropic effects in animals, plants, and fungi, and considerable efforts are under way to understand their roles in the interactions between fungi, particularly Aspergillus species, and animal and plant hosts (15). Oxylipins from A. fumigatus are involved in virulence in invasive pulmonary aspergillosis in mice. Fungal strains unable to produce these molecules are hypervirulent and have increased tolerance to hydrogen peroxide stress relative to that of the wild type. This suggests that the fungal oxylipins may activate mammalian immune responses, thus contributing to enhanced resistance to fungi (82). Oxylipin-deficient mutants of Aspergillus nidulans are less able than wild-type isolates to colonize seeds (83). Such fungal oxylipins, as well as those from seeds, stimulate sporulation and mycotoxin production in several Aspergillus species. Trihydroxy oxylipins from plants can inhibit the growth of phytopathogenic fungi (87). Indeed, there may even be some overlap in lipid signaling responses by animals and plants to fungal pathogens, especially since fungi also produce oxylipins.
Another way in which fungi may alter host physiology to facilitate colonization is by inducing or directly causing the death of host cells. As mentioned above, oxidative burst and programmed cell death are rapid defense responses of plants and animals to infection. However, some plant-pathogenic fungi that are necrotrophic (relying on dead host tissue to colonize their hosts) subvert these responses to provide nutrition for their growth and colonization (84). Such fungi must protect themselves from the oxidative defense response of their hosts, and both plant and animal pathogens produce enzymes such as superoxide dismutase, which degrades reactive oxygen species such as hydrogen peroxide generated by the invaded host. Disruption of superoxide dismutase genes in C. albicans leads to reduced growth and virulence and increased susceptibility to macrophage attack (39). Hydrogen peroxide inhibits the infection of plants by various fungi (53), and plant pathogens also have superoxide dismutase genes, but whether these are required for full virulence has not been reported. Proline protects the plant pathogen Colletotrichum trifolii against oxidative stress and other environmental stresses such as UV, salt, and heat. This molecule may be a universal protection mechanism in fungi, as it also protects Saccharomyces cerevisiae from damage by reactive oxygen species (19).
Once within the host, many fungi make virulence factors such as toxins that kill host tissue, thus providing nutrition. Toxins can be host specific or non-host specific, although, interestingly, there are no reports of host-specific toxins in fungal pathogenesis in animals. While some toxins are small peptides, many are secondary metabolites, which are low-molecular-weight molecules with a diverse range of structures. These secondary metabolites are dispensable for the fungus but presumably confer some selective advantage (38). Epipolythiodioxopiperazines (ETPs) are a class of secondary metabolites that are thought to contribute to virulence in animals and plants. These molecules mediate their toxicity via generation of reactive oxygen by redox cycling and by cross-linking to proteins with free thiols (35). The ETP gliotoxin is immunosuppressive and has been proposed to be associated with aspergillosis in humans caused by Aspergillus fumigatus. This latter assumption has been tested recently by two research groups who disrupted the peptide synthetase gene in the biosynthetic pathway and studied the effect of these non-gliotoxin-producing mutants in vitro and in vivo in animal models. The mutants showed reduced cytotoxic activity on cells such as mast cells or macrophages but did not affect the survival of mice with invasive aspergillosis (23, 47). Thus, gliotoxin does not appear to be required for the pathogenicity of the fungus in immunocompromised mice. However, secondary metabolite toxins do contribute significantly to the virulence of this fungus in mice, as shown by mutational and overexpression studies with the global transcriptional regulator LaeA of A. fumigatus (12). Another ETP, sirodesmin PL, is a non-host-selective toxin produced by L. maculans, a pathogen of Brassica napus. This toxin has antibacterial and antiviral properties and causes chlorotic lesions on leaves (35). A sirodesmin-deficient mutant with a disrupted peptide synthetase in the biosynthetic pathway made lesions on the cotyledons and primary leaves of B. napus that were similar in size to those made by the wild type. However, this mutant colonized stem tissue half as effectively as the wild type, thus implicating sirodesmin as a pathogenicity determinant in the late stages of canola plant infection (C. E. Elliott and B. J. Howlett, unpublished data).
Reproduction and transmission.
Both animal- and plant-infecting fungi reproduce mitotically within the host as part of the disease cycle (Fig. 1, diagram 6). A striking contrast is that while plant-to-plant transmission of fungal disease is very common, direct transmission of fungal pathogens between mammalian hosts is not usually a regular part of the disease cycle. In plant pathogens, conidium production usually occurs after infection has been established and lesions have developed. Conidia are spread from plant to plant by wind or in water droplets. Similarly, spores of the mammalian pathogen Pneumocystis jirovecii are thought to be spread in droplets coughed from the lungs of an infected host. However, many pathogens of mammals (e.g., Coccidioides immitis) are transmitted by inhalation as hyphae or as arthroconidia, in dust, wind, or soil. Increasingly, biofilms, in which a community of microorganisms attaches to a solid surface, mediate the dispersal of fungi in hospital environments.
Various genetic factors and regulatory pathways control conidiation and spore dispersal. These pathways can be triggered in response to light, pH, and nutrient availability. In A. fumigatus, a particular Ras gene, rasA, interferes with the mitotic cycle when introduced as a constitutively active form, causing less conidiation and malformed conidiophores, whereas the mutant expressing an inactive form of RasA displays problems with conidial germination. RasB also affects conidiation as well as germination (31). A Ras-related protein, RhbA, is involved in signaling for growth and asexual development, possibly via a nutrient-sensing pathway. Additionally, when this gene is disrupted, the resultant mutant has reduced virulence in mice (60). Hydrophobins, as well as being important for spore germination, are necessary for spore dispersal and survival. For instance, in the tomato pathogen Cladosporium fulvum, hydrophobins assist conidial dispersal on the surfaces of water droplets (90).
Many animal fungal pathogens do not appear to complete a sexual cycle. In contrast, many ascomycete plant pathogens commonly undergo meiosis in the late stages of disease, producing tough resting structures (e.g., pseudothecia and perithecia) that survive until the next growing season and release ascospores (Fig. 1, diagram 7). Either homothallism (selfing) or heterothallism (outcrossing) occur in many plant pathogens, with heterothallism requiring the presence of opposite mating types. This creates new genotypes and thus theoretically facilitates the potential to adapt to selection pressures such as host resistance. Mating-type genes are well characterized in many saprophytic and plant-pathogenic ascomycetes (68). A few animal-pathogenic ascomycetes, such as Pneumocystis jirovecii, Candida lusitaniae, and Candida guilliermondii, are thought to undergo a sexual cycle. These Candida species are haploid and undergo meiosis and sporulation, in contrast to diploid C. albicans, which is thought to be asexual in vivo. Interestingly, C. albicans maintains both mating-type loci, which may provide a selective advantage in virulence. This fungus can also undergo a parasexual cycle where a tetraploid cell forms, followed by chromosome loss leading to a diploid with genetic reshuffling (9, 50). Recently, the expression of pheromone and mating-type genes has been reported to occur in A. fumigatus, which was previously thought to lack a sexual cycle. Also, population studies indicate that recombination occurs in this fungus, further supporting the hypothesis that this pathogen can reproduce sexually (61). Discovering whether pathogenic fungi undergo sexual reproduction during the disease cycle has important implications for understanding the evolution of new strains of these pathogens and their potential to overcome host resistances and control mechanisms such as fungicides.
CONCLUSIONS AND FUTURE DIRECTIONS
A multitude of pathogenicity mechanisms has been uncovered in studies of a range of fungi. There appear to be few universal pathogenicity factors within and between plant- and animal-pathogenic fungi. A particular pathogenicity mechanism may have been described only for a few fungi, and it may or may not pertain to many others. As yet, there are not enough in-depth studies on a range of fungus-host interactions to deduce this. Analyses of fungal interactions with other hosts, such as insects (e.g., diseases caused by Metarhizium anisopliae), other fungi (e.g., mycoparasites such as Trichoderma spp., used as biocontrol agents for other fungi), and plants that host endosymbiont fungi, may provide important comparisons between pathogenic and nonpathogenic interactions. Despite the lack of universal virulence factors, many pathogens appear to utilize common signaling cascades and protective compounds during their development and pathogenesis. Lipids, such as oxylipins, play important roles in signaling in disease. There is a complex web of interactions involving these molecules, particularly since both partners in the interaction (host and pathogen) synthesize them; this web is just beginning to be untangled (74).
Recent technologies are providing opportunities to identify commonalities among and differences between pathogens in their disease mechanisms. The complement of genes in a fungal pathogen of plants is not so different from that of a fungal pathogen of animals in both content and synteny, as demonstrated by the recent genome-sequencing projects for many fungi (33). These genome projects also allow comparisons of nonpathogens with closely related pathogens; for example, Aspergillus nidulans and Aspergillus oryzae can be compared with the pathogens A. flavus, A. fumigatus, and Aspergillus terreus, or the nonpathogen Uncinocarpus reesii can be compared with the pathogen Coccidioides immitis. The genomes of A. oryzae and A. flavus, a food biotechnology organism and a pathogen, respectively, are extremely similar, so it will be interesting to discover whether the differences can be implicated in infection. Improvements in bioinformatic tools for gene annotations and comparative analyses will be needed, and the development of more fungal databases in this area will be extremely valuable (93). Global transcriptional analyses of multihost fungi, such as Fusarium oxysporum, that invade both animals and plants will highlight networks and signaling components that are regulated in response to two extremely different host types. The vibrant research activity centered on understanding fungal pathogenesis in animals will give clues to the mechanisms of pathogenesis in plants and vice versa. The next decade promises to be as exciting as the last.
Acknowledgments
We thank Candace Elliott, Alex Indurm, and three anonymous reviewers for helpful comments.
We thank the Australian Research Council and the Australian Grains Research and Development Corporation for supporting our research.
Footnotes
Published ahead of print on 13 October 2006.
REFERENCES
- 1.Albrecht, A., A. Felk, I. Pichova, J. Naglik, M. Schaller, P. de Groot, D. Maccallum, F. Odds, W. Schafer, F. Klis, M. Monod, and B. Hube. 2006. Glycosylphosphatidylinositol-anchored proteases of Candida albicans target proteins necessary for both cellular processes and host-pathogen interactions. J. Biol. Chem. 281:688-694. [DOI] [PubMed] [Google Scholar]
- 2.Andrianopoulos, A. 2002. Control of morphogenesis in the human fungal pathogen Penicillium marneffei. Int. J. Med. Microbiol. 292:331-347. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. 2005. Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol. 6:973-979. [DOI] [PubMed] [Google Scholar]
- 4.Bagga, S., and D. Straney. 2000. Modulation of cAMP and phosphodiesterase activity by flavonoids which induce spore germination of Nectria haematococca MP VI (Fusarium solani). Physiol. Mol. Plant Pathol. 56:51-61. [Google Scholar]
- 5.Balhadere, P. V., and N. J. Talbot. 2001. PDE1 encodes a P-type ATPase involved in appressorium-mediated plant infection by the rice blast fungus Magnaporthe grisea. Plant Cell 13:1987-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bassilana, M., J. Blyth, and R. A. Arkowitz. 2003. Cdc24, the GDP-GTP exchange factor for Cdc42, is required for invasive hyphal growth of Candida albicans. Eukaryot. Cell 2:9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bassilana, M., J. Hopkins, and R. A. Arkowitz. 2005. Regulation of the Cdc42/Cdc24 GTPase module during Candida albicans hyphal growth. Eukaryot. Cell 4:588-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belkhadir, Y., R. Subramaniam, and J. L. Dangl. 2004. Plant disease resistance protein signaling: NBS-LRR proteins and their partners. Curr. Opin. Plant Biol. 7:391-399. [DOI] [PubMed] [Google Scholar]
- 9.Bennett, R., and A. Johnson. 2003. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 22:2505-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berbee, M. L. 2001. The phylogeny of plant and animal pathogens in the Ascomycota. Physiol. Mol. Plant Pathol. 59:165-187. [Google Scholar]
- 11.Bignell, E., S. Negrete-Urtasun, A. M. Calcagno, K. Haynes, H. N. Arst, Jr., and T. Rogers. 2005. The Aspergillus pH-responsive transcription factor PacC regulates virulence. Mol. Microbiol. 55:1072-1084. [DOI] [PubMed] [Google Scholar]
- 12.Bok, J. W., S. A. Balajee, K. A. Marr, D. Andes, K. F. Nielsen, J. C. Frisvad, and N. P. Keller. 2005. LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryot. Cell 4:1574-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolton, M. D., B. Thomma, and B. D. Nelson. 2006. Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 7:1-16. [DOI] [PubMed] [Google Scholar]
- 14.Borges-Walmsley, M. I., D. Chen, X. Shu, and A. R. Walmsley. 2002. The pathobiology of Paracoccidioides brasiliensis. Trends Microbiol. 10:80-87. [DOI] [PubMed] [Google Scholar]
- 15.Brodhagen, M., and N. Keller. 2006. Signalling pathways connecting mycotoxin production and sporulation. Mol. Plant Pathol. 7:285-301. [DOI] [PubMed] [Google Scholar]
- 16.Casadevall, A., and L. A. Pirofski. 2003. The damage-response framework of microbial pathogenesis. Nat. Rev. Microbiol. 1:17-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casadevall, A., and L. A. Pirofski. 2006. Polysaccharide-containing conjugate vaccines for fungal diseases. Trends Mol. Med. 12:6-9. [DOI] [PubMed] [Google Scholar]
- 18.Cessna, S., V. Sears, M. Dickman, and P. Low. 2000. Oxalic acid, a pathogenicity factor for Sclerotinia sclerotiorum, suppresses the oxidative burst of the host plant. Plant Cell 12:2191-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen, C., and M. B. Dickman. 2005. Proline suppresses apoptosis in the fungal pathogen Colletotrichum trifolii. Proc. Natl. Acad. Sci. USA 102:3459-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen, C., Y.-S. Ha, J.-Y. Min, S. D. Memmott, and M. B. Dickman. 2006. Cdc42 is required for proper growth and development in the fungal pathogen Colletotrichum trifolii. Eukaryot. Cell 5:155-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu, J. H., C. Feudtner, K. Heydon, T. J. Walsh, and T. E. Zaoutis. 2006. Hospitalizations for endemic mycoses: a population-based national study. Clin. Infect. Dis. 42:822-825. [DOI] [PubMed] [Google Scholar]
- 22.Clergeot, P. H., M. Gourgues, J. Cots, F. Laurans, M. P. Latorse, R. Pepin, D. Tharreau, J. L. Notteghem, and M.-H. Lebrun. 2001. PLS1, a gene encoding a tetraspanin-like protein, is required for penetration of rice leaf by the fungal pathogen Magnaporthe grisea. Proc. Natl. Acad. Sci. USA 98:6963-6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cramer, R. A., Jr., M. P. Gamcsik, R. M. Brooking, L. K. Najvar, W. R. Kirkpatrick, T. F. Patterson, C. J. Balibar, J. R. Graybill, J. R. Perfect, S. N. Abraham, and W. J. Steinbach. 2006. Disruption of a nonribosomal peptide synthetase in Aspergillus fumigatus eliminates gliotoxin production. Eukaryot. Cell 5:972-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.da Silva, M. B., A. Marques, J. D. Nosanchuk, A. Casadevall, L. R. Travassos, and C. P. Taborda. 2006. Melanin in the dimorphic fungal pathogen Paracoccidioides brasiliensis: effects on phagocytosis, intracellular resistance and drug susceptibility. Microbes Infect. 8:197-205. [DOI] [PubMed] [Google Scholar]
- 25.Dean, R., N. J. Talbot, D. J. Ebbole, M. L. Farman, T. K. Mitchell, M. J. Orbach, M. Thon, R. Kulkarni, J. R. Xu, H. Pan, N. D. Read, Y. H. Lee, I. Carbone, D. Brown, Y. Y. Oh, N. Donofrio, J. S. Jeong, D. M. Soanes, S. Djonovic, E. Kolomiets, C. Rehmeyer, W. Li, M. Harding, S. Kim, M. H. Lebrun, H. Bohnert, S. Coughlan, J. Butler, S. Calvo, L. J. Ma, R. Nicol, S. Purcell, C. Nusbaum, J. E. Galagan, and B. W. Birren. 2005. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434:980-986. [DOI] [PubMed] [Google Scholar]
- 26.Doehlemann, G., P. Berndt, and M. Hahn. 2006. Different signalling pathways involving a Galpha protein, cAMP and a MAP kinase control germination of Botrytis cinerea conidia. Mol. Microbiol. 59:821-835. [DOI] [PubMed] [Google Scholar]
- 27.Ebel, F., M. Schwienbacher, J. Beyer, J. Heesemann, A. A. Brakhage, and M. Brock. 2006. Analysis of the regulation, expression, and localisation of the isocitrate lyase from Aspergillus fumigatus, a potential target for antifungal drug development. Fungal Genet. Biol. 43:476-489. [DOI] [PubMed] [Google Scholar]
- 28.Feldbrugge, M., J. Kamper, G. Steinberg, and R. Kahmann. 2004. Regulation of mating and pathogenic development in Ustilago maydis. Curr. Opin. Microbiol. 7:666-672. [DOI] [PubMed] [Google Scholar]
- 29.Fillinger, S., M. K. Chaveroche, K. Shimizu, N. Keller, and C. d'Enfert. 2002. cAMP and ras signalling independently control spore germination in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 44:1001-1016. [DOI] [PubMed] [Google Scholar]
- 30.Flor, H. H. 1971. Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9:275-296. [Google Scholar]
- 31.Fortwendel, J. R., J. Panepinto, A. E. Seitz, D. S. Askew, and J. C. Rhodes. 2004. Aspergillus fumigatus rasA and rasB regulate the timing and morphology of asexual development. Fungal Genet. Biol. 41:129-139. [DOI] [PubMed] [Google Scholar]
- 32.Fuchs, B. B., and E. Mylonakis. 2006. Using non-mammalian hosts to study fungal virulence and host defense. Curr. Opin. Microbiol. 9:346-351. [DOI] [PubMed] [Google Scholar]
- 33.Galagan, J. E., S. Calvo, C. Cuomo, L. J. Ma, J. R. Wortman, S. Batzoglou, S. I. Lee, M. Basturkmen, C. C. Spevak, J. Clutterbuck, V. Kapitonov, J. Jurka, C. Scazzocchio, M. Farman, J. Butler, S. Purcell, S. Harris, G. H. Braus, O. Draht, S. Busch, C. D'Enfert, C. Bouchier, G. H. Goldman, D. Bell-Pedersen, S. Griffiths-Jones, J. H. Doonan, J. Yu, K. Vienken, A. Pain, M. Freitag, E. U. Selker, D. B. Archer, M. A. Penalva, B. R. Oakley, M. Momany, T. Tanaka, T. Kumagai, K. Asai, M. Machida, W. C. Nierman, D. W. Denning, M. Caddick, M. Hynes, M. Paoletti, R. Fischer, B. Miller, P. Dyer, M. S. Sachs, S. A. Osmani, and B. W. Birren. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105-1115. [DOI] [PubMed] [Google Scholar]
- 34.Gantner, B., R. Simmons, and D. Underhill. 2005. Dectin-1 mediates macrophage recognition of Candida albicans yeast but not filaments. EMBO J. 24:1277-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gardiner, D. M., P. Waring, and B. J. Howlett. 2005. The epipolythiodioxopiperazine (ETP) class of fungal toxins: distribution, mode of action, functions and biosynthesis. Microbiology 151:1021-1032. [DOI] [PubMed] [Google Scholar]
- 36.Guimaraes, R. L., and H. U. Stotz. 2004. Oxalate production by Sclerotinia sclerotiorum deregulates guard cells during infection. Plant Physiol. 136:3703-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamer, J. E., and D. W. Holden. 1997. Linking approaches in the study of fungal pathogenesis: a commentary. Fungal Genet. Biol. 21:11-16. [PubMed] [Google Scholar]
- 38.Howlett, B. J. 2006. Secondary metabolite toxins and nutrition of plant pathogenic fungi. Curr. Opin. Plant Biol. 9:371-375. [DOI] [PubMed] [Google Scholar]
- 39.Hwang, C. S., G. E. Rhie, J. H. Oh, W. K. Huh, H. S. Yim, and S. O. Kang. 2002. Copper- and zinc-containing superoxide dismutase (Cu/ZnSOD) is required for the protection of Candida albicans against oxidative stresses and the expression of its full virulence. Microbiology 148:3705-3713. [DOI] [PubMed] [Google Scholar]
- 40.Icenhour, C. R., T. J. Kottom, and A. H. Limper. 2006. Pneumocystis melanins confer enhanced organism viability. Eukaryot. Cell 5:916-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Idnurm, A., Y. Bahn, K. Nielsen, X. Lin, J. Fraser, and J. Heitman. 2005. Deciphering the model pathogenic fungus Cryptococcus neoformans. Nat. Rev. Microbiol. 3:753-764. [DOI] [PubMed] [Google Scholar]
- 42.Idnurm, A., and B. J. Howlett. 2002. Isocitrate lyase is essential for pathogenicity of the fungus Leptosphaeria maculans to canola (Brassica napus). Eukaryot. Cell 1:719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalo-Klein, A., and S. S. Witkin. 1990. Prostaglandin E2 enhances and gamma interferon inhibits germ tube formation in Candida albicans. Infect. Immun. 58:260-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim, S., I. Ahn, H. Rho, and Y. Lee. 2005. MHP1, a Magnaporthe grisea hydrophobin gene, is required for fungal development and plant colonization. Mol. Microbiol. 57:1224-1237. [DOI] [PubMed] [Google Scholar]
- 45.Kriznik, A., M. Bouillot, J. Coulon, and F. Gaboriaud. 2005. Morphological specificity of yeast and filamentous Candida albicans forms on surface properties. C. R. Biol. 328:928-935. [DOI] [PubMed] [Google Scholar]
- 46.Kumamoto, C., and M. Vinces. 2005. Alternative Candida albicans lifestyles: growth on surfaces. Annu. Rev. Microbiol. 59:113-133. [DOI] [PubMed] [Google Scholar]
- 47.Kupfahl, C., T. Heinekamp, G. Geginat, T. Ruppert, H. Hartl, H. Hof, and A. A. Brakhage. 2006. Deletion of the gliP gene of Aspergillus fumigatus results in loss of gliotoxin production but has no effect on virulence of the fungus in a low-dose mouse infection model. Mol. Microbiol. 62:292-302. [DOI] [PubMed] [Google Scholar]
- 48.Lengeler, K. B., R. C. Davidson, C. D'Souza, T. Harashima, W.-C. Shen, P. Wang, X. Pan, M. Waugh, and J. Heitman. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64:746-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liao, D. I., G. Basarab, A. A. Gatenby, and D. B. Jordan. 2000. Selection of a potent inhibitor of trihydroxynaphthalene reductase by sorting disease control data. Bioorg. Med. Chem. Lett. 10:491-494. [DOI] [PubMed] [Google Scholar]
- 50.Lockhart, S. R., W. Wu, J. B. Radke, R. Zhao, and D. R. Soll. 2005. Increased virulence and competitive advantage of a/alpha over a/a or alpha/alpha offspring conserves the mating system of Candida albicans. Genetics 169:1883-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lorenz, M. C., and G. R. Fink. 2002. Life and death in a macrophage: role of the glyoxylate cycle in virulence. Eukaryot. Cell 1:657-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maubon, D., S. Park, M. Tanguy, M. Huerre, C. Schmitt, M. C. Prevost, D. S. Perlin, J. P. Latge, and A. Beauvais. 2006. AGS3, an alpha(1-3)glucan synthase gene family member of Aspergillus fumigatus, modulates mycelium growth in the lung of experimentally infected mice. Fungal Genet. Biol. 43:366-375. [DOI] [PubMed] [Google Scholar]
- 53.Mellersh, D., I. Foulds, V. Higgins, and M. Heath. 2002. H2O2 plays different roles in determining penetration failure in three diverse plant-fungal interactions. Plant J. 29:257-268. [DOI] [PubMed] [Google Scholar]
- 54.Monge, R. A., E. Roman, C. Nombela, and J. Pla. 2006. The MAP kinase signal transduction network in Candida albicans. Microbiology 152:905-912. [DOI] [PubMed] [Google Scholar]
- 55.Mur, L. A., T. Carver, and E. Prats. 2006. NO way to live; the various roles of nitric oxide in plant-pathogen interactions. J. Exp. Bot. 57:489-505. [DOI] [PubMed] [Google Scholar]
- 56.Nascimento, F. R., V. Calich, D. Rodriguez, and M. Russo. 2002. Dual role for nitric oxide in paracoccidioidomycosis: essential for resistance, but overproduction associated with susceptibility. J. Immunol. 168:4593-4600. [DOI] [PubMed] [Google Scholar]
- 57.Nurnberger, T., F. Brunner, B. Kemmerling, and L. Piater. 2004. Innate immunity in plants and animals: striking similarities and obvious differences. Immunol. Rev. 198:249-266. [DOI] [PubMed] [Google Scholar]
- 58.Ortoneda, M., J. Guarro, M. P. Madrid, Z. Caracuel, M. I. G. Roncero, E. Mayayo, and A. Di Pietro. 2004. Fusarium oxysporum as a multihost model for the genetic dissection of fungal virulence in plants and mammals. Infect. Immun. 72:1760-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Osherov, N., and G. May. 2001. The molecular mechanisms of conidial germination. FEMS Microbiol. Lett. 199:153-160. [DOI] [PubMed] [Google Scholar]
- 60.Panepinto, J. C., B. G. Oliver, J. R. Fortwendel, D. L. H. Smith, D. S. Askew, and J. C. Rhodes. 2003. Deletion of the Aspergillus fumigatus gene encoding the Ras-related protein RhbA reduces virulence in a model of invasive pulmonary aspergillosis. Infect. Immun. 71:2819-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Paoletti, M., C. Rydholm, E. U. Schwier, M. J. Anderson, G. Szakacs, F. Lutzoni, J. P. Debeaupuis, J. P. Latge, D. W. Denning, and P. S. Dyer. 2005. Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus. Curr. Biol. 15:1242-1248. [DOI] [PubMed] [Google Scholar]
- 62.Paris, S., J.-P. Debeaupuis, R. Crameri, M. Carey, F. Charlès, M. C. Prévost, C. Schmitt, B. Philippe, and J. P. Latgé. 2003. Conidial hydrophobins of Aspergillus fumigatus. Appl. Environ. Microbiol. 69:1581-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park, J.-H., G. J. Choi, K. S. Jang, H. K. Lim, H. T. Kim, K. Y. Cho, and J.-C. Kim. 2005. Antifungal activity against plant pathogenic fungi of chaetoviridins isolated from Chaetomium globosum. FEMS Microbiol. Lett. 252:309-313. [DOI] [PubMed] [Google Scholar]
- 64.Paterson, P. J., S. Seaton, T. Yeghen, T. D. McHugh, J. McLaughlin, A. V. Hoffbrand, and C. C. Kibbler. 2005. Molecular confirmation of invasive infection caused by Chaetomium globosum. J. Clin. Pathol. 58:334. [PMC free article] [PubMed] [Google Scholar]
- 65.Perpetua, N. S., Y. Kubo, N. Yasuda, Y. Takano, and I. Furusawa. 1996. Cloning and characterization of a melanin biosynthetic THR1 reductase gene essential for appressorial penetration of Colletotrichum lagenarium. Mol. Plant-Microbe Interact. 9:323-329. [DOI] [PubMed] [Google Scholar]
- 66.Pieterse, C. M., and Van Loon, L. 2004. NPR1: the spider in the web of induced resistance signaling pathways. Curr. Opin. Plant Biol. 7:456-464. [DOI] [PubMed] [Google Scholar]
- 67.Podila, G. K., L. M. Rogers, and P. E. Kolattukudy. 1993. Chemical signals from avocado surface wax trigger germination and appressorium formation in Colletotrichum gloeosporioides. Plant Physiol. 103:267-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Poeggeler, S. 2001. Mating-type genes for classical strain improvements of ascomycetes. Appl. Microbiol. Biotechnol. 56:589-601. [DOI] [PubMed] [Google Scholar]
- 69.Poussereau, N., S. Gente, C. Rascle, G. Billon-Grand, and M. Fevre. 2001. aspS encoding an unusual aspartyl protease from Sclerotinia sclerotiorum is expressed during phytopathogenesis. FEMS Microbiol. Lett. 194:27-32. [DOI] [PubMed] [Google Scholar]
- 70.Rollins, J. A. 2003. The Sclerotinia sclerotiorum pac1 gene is required for sclerotial development and virulence. Mol. Plant-Microbe Interact. 16:785-795. [DOI] [PubMed] [Google Scholar]
- 71.Roozendaal, R., and M. Carroll. 2006. Emerging patterns in complement-mediated pathogen recognition. Cell 125:29-32. [DOI] [PubMed] [Google Scholar]
- 72.Schaller, M., C. Borelli, H. C. Korting, and B. Hube. 2005. Hydrolytic enzymes as virulence factors of Candida albicans. Mycoses 48:365-377. [DOI] [PubMed] [Google Scholar]
- 73.Scully, L., and M. Bidochka. 2006. A cysteine/methionine auxotroph of the opportunistic fungus Aspergillus flavus is associated with host-range restriction: a model for emerging diseases. Microbiology 152:223-232. [DOI] [PubMed] [Google Scholar]
- 74.Shea, J. M, and M. Del Poeta. 2006. Lipid signaling in pathogenic fungi. Curr. Opin. Microbiol. 9:352-358. [DOI] [PubMed] [Google Scholar]
- 75.Shinya, T., R. Menard, I. Kozone, H. Matsuoka, N. Shibuya, S. Kauffmann, K. Matsuoka, and M. Saito. 2006. Novel beta-1,3-,1,6-oligoglucan elicitor from Alternaria alternata 102 for defense responses in tobacco. FEBS J. 273:2421-2431. [DOI] [PubMed] [Google Scholar]
- 76.Solomon, P., R. Lee, T. Wilson, and R. Oliver. 2004. Pathogenicity of Stagonospora nodorum requires malate synthase. Mol. Microbiol. 53:1065-1073. [DOI] [PubMed] [Google Scholar]
- 77.Speth, C., G. Rambach, C. Lass-Florl, M. P. Dierich, and R. Wurzner. 2004. The role of complement in invasive fungal infections. Mycoses 47:93-103. [DOI] [PubMed] [Google Scholar]
- 78.St. Leger, R. J., S. E. Screen, and B. Shams-Pirzadeh. 2000. Lack of host specialization in Aspergillus flavus. Appl. Environ. Microbiol. 66:320-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Talbot, N., M. J. Kershaw, G. E. Wakley, O. M. H. De Vries, J. G. H. Wessels, and J. Hamer. 1996. MPG1 encodes a fungal hydrophobin involved in surface interactions during infection-related development of Magnaporthe grisea. Plant Cell 8:985-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.ten Have, A., E. Dekkers, J. Kay, L. H. Phylip, and J. A. van Kan. 2004. An aspartic proteinase gene family in the filamentous fungus Botrytis cinerea contains members with novel features. Microbiology 150:2475-2489. [DOI] [PubMed] [Google Scholar]
- 81.Thines, E., R. Weber, and N. J. Talbot. 2000. MAP kinase and protein kinase A-dependent mobilization of triacylglycerol and glycogen during appressorium turgor generation by Magnaporthe grisea. Plant Cell 12:1703-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tsitsigiannis, D. I., J.-W. Bok, D. Andes, K. F. Nielsen, J. C. Frisvad, and N. P. Keller. 2005. Aspergillus cyclooxygenase-like enzymes are associated with prostaglandin production and virulence. Infect. Immunol. 73:4548-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tsitsigiannis, D. I., and N. P. Keller. 2006. Oxylipins act as determinants of natural product biosynthesis and seed colonization in Aspergillus nidulans. Mol. Microbiol. 59:882-892. [DOI] [PubMed] [Google Scholar]
- 84.van Kan, J. A. 2006. Licensed to kill: the lifestyle of a necrotrophic plant pathogen. Trends Plant Sci. 11:247-253. [DOI] [PubMed] [Google Scholar]
- 85.Veneault-Fourrey, C., K. Lambou, and M.-H. Lebrun. 2006. Fungal Pls1 tetraspanins as key factors of penetration into host plants: a role in re-establishing polarized growth in the appressorium? FEMS Microbiol. Lett. 256:179-184. [DOI] [PubMed] [Google Scholar]
- 86.Viaud, M. C., P. V. Balhadere, and N. J. Talbot. 2002. A Magnaporthe grisea cyclophilin acts as a virulence determinant during plant infection. Plant Cell 14:917-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Walters, D. R., T. Cowley, and H. Weber. 2006. Rapid accumulation of trihydroxy oxylipins and resistance to the bean rust pathogen Uromyces fabae following wounding in Vicia faba. Ann. Bot. 97:779-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang, Z. Y., C. R. Thornton, M. J. Kershaw, L. Debao, and N. J. Talbot. 2003. The glyoxylate cycle is required for temporal regulation of virulence by the plant pathogenic fungus Magnaporthe grisea. Mol. Microbiol. 47:1601-1612. [DOI] [PubMed] [Google Scholar]
- 89.Wheeler, R. T., and G. R. Fink. 2006. A drug-sensitive genetic network masks fungi from the immune system. PLoS Pathogens 2:e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Whiteford, J. R., and P. D. Spanu. 2001. The hydrophobin HCf-1 of Cladosporium fulvum is required for efficient water-mediated dispersal of conidia. Fungal Genet. Biol. 32:159-168. [DOI] [PubMed] [Google Scholar]
- 91.Winnenburg, R., T. K. Baldwin, M. Urban, C. Rawlings, J. Köhler, and K. E. Hammond-Kosack. 2006. PHI-base: a new database for pathogen host interactions. Nucleic Acids Res. 34:D459-D464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Woods, J. 2003. Knocking on the right door and making a comfortable home: Histoplasma capsulatum intracellular pathogenesis. Curr. Opin. Microbiol. 6:327-331. [DOI] [PubMed] [Google Scholar]
- 93.Xu, J. R., Y. L. Peng, M. B. Dickman, and A. Sharon. 2006. The dawn of fungal pathogen genomics. Annu. Rev. Phytopathol. 44:337-366. [DOI] [PubMed] [Google Scholar]
- 94.Young, N. 2000. The genetic architecture of resistance. Curr. Opin. Plant Biol. 3:285-290. [DOI] [PubMed] [Google Scholar]