Abstract
The Ste12 transcription factor of Saccharomyces cerevisiae regulates transcription programs controlling two different developmental fates. One is differentiation into a mating-competent form that occurs in response to mating pheromone. The other is the transition to a filamentous-growth form that occurs in response to nutrient deprivation. These two distinct roles for Ste12 make it a focus for studies into regulatory mechanisms that impart biological specificity. The transient signal characteristic of mating differentiation led us to test the hypothesis that regulation of Ste12 turnover might contribute to attenuation of the mating-specific transcription program and restrict activation of the filamentation program. We show that prolonged pheromone induction leads to ubiquitin-mediated destabilization and decreased amounts of Ste12. This depletion in pheromone-stimulated cultures is dependent on the mating-pathway-dedicated mitogen-activated protein kinase Fus3 and its target Cdc28 inhibitor, Far1. Attenuation of pheromone-induced mating-specific gene transcription (FUS1) temporally correlates with Ste12 depletion. This attenuation is abrogated in the deletion backgrounds (fus3Δ or far1Δ) where Ste12 is found to persist. Additionally, pheromone induces haploid invasion and filamentous-like growth instead of mating differentiation when Ste12 levels remain high. These observations indicate that loss of Ste12 reinforces the adaptive response to pheromone and contributes to the curtailing of a filamentation response.
Cellular responses to extracellular stimuli often entail alterations in transcriptional programs that are mediated by target transcription factors. Ste12 is one such transcription factor that directly regulates two different developmental gene expression programs in Saccharomyces cerevisiae. One is required for the transition to a mating-competent state upon exposure of haploid cells of one cell type to mating pheromone from the opposite cell type (7, 57). The other occurs in response to certain starvation conditions and is required for the transition to a filamentous form, which occurs during pseudohyphal growth in diploid cells and invasive growth in haploid cells (6, 41). Because of this dual function, studies focused on Ste12 are expected to provide insights into regulatory mechanisms that impart biological specificity.
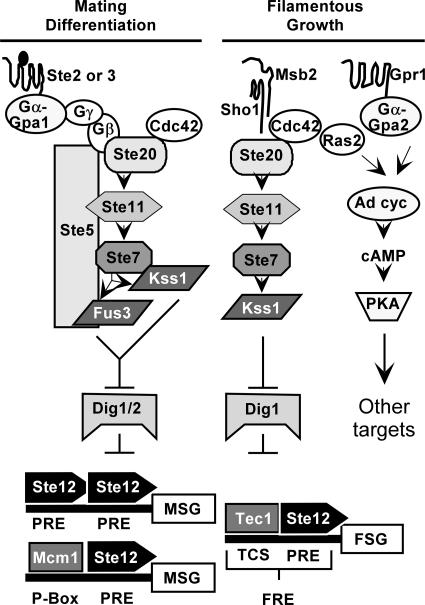
Combinatorial use of transcription factors distinguishes mating-specific genes from filamentation-specific genes (Fig. 1). Ste12 activates the promoters of mating-specific genes either as a homomultimer, by binding at reiterated pheromone response elements (PREs), or as a heterodimer with Mcm1, by binding to PREs that are adjacent to a P-box site (21, 26, 54). By contrast, Ste12 activates the promoters of some filamentation-specific genes as a heterodimer with Tec1 (4, 40). The heterodimer can act through binding either at Tec1 consensus binding sites (TCS) or at filamentation response elements, which are comprised of a PRE and an adjacent TCS element. In the former configuration the Ste12-Tec1 heterodimer is tethered to the DNA by virtue of the Tec1-TCS binding interaction, while in the latter configuration both Ste12 and Tec1 contact the DNA in a cooperative binding interaction (4, 13, 40). In both cases Tec1 confers the DNA binding specificity while Ste12 provides the activation function for transcription (4, 13).
FIG. 1.
Signal transduction pathways with common components mediate two life cycle transitions in haploid S. cerevisiae. See the text for an explanation.
Though the promoter configurations for Ste12 acting at mating- and filamentation-specific genes are distinct, it remains unclear how the separate transcriptional programs are regulated. Achieving specificity in this system is complicated because the same regulators of Ste12 activity function for both programs. In the absence of signal, the transcriptional inhibitors Dig1 and Dig2 (Rst1 and Rst2) suppress the ability of Ste12 to activate transcription of mating genes while Dig1 alone suppresses activation at filamentation genes (3, 13, 15, 50, 65). Mitogen-activated protein kinase (MAPK) cascades with shared components mediate the signal-dependent events that reverse Dig1-Dig2 suppression and promote Ste12 activation (Fig. 1).
Ste12 activation for mating differentiation occurs when pheromone (a- or α-factor) secreted from one cell type binds to its G-protein-coupled receptor (Ste3 or Ste2, respectively) on the opposite cell type. The activated G protein stimulates a MAPK cascade comprised of the p21 (Cdc42)-associated kinase Ste20, the MAPKKK Ste11, the MAPKK Ste7, and two MAPKs, Fus3 and Kss1 (Fig. 1) (see reference 19 for a review). The scaffold Ste5 organizes the core enzymes of this pathway by directly binding to Ste11, Ste7, and Fus3 (19). Kss1 does not bind directly to Ste5 but is part of this complex through binding to Ste7 (7).
Starvation-induced filamentous growth involves integration of at least two signaling pathways (Fig. 1). The glucose-sensing G-protein-coupled receptor Gpr1 stimulates one branch of the network that leads to activation of protein kinase A and its target transcriptional regulators (see reference 52 for a review). The Sho1 and Msb2 transmembrane proteins are upstream regulators of the MAPK-mediated branch of the network (17, 18, 51). With the exception of Fus3, the same kinases used in the mating pathway comprise the filamentous-growth MAPK activation module (52). Another notable difference is that activation of the MAPK cascade enzymes during starvation-induced filamentous growth takes place independently of the scaffold Ste5 (58).
Although Fus3 and Kss1 are closely related MAPKs, they have somewhat specialized roles for mating differentiation and filamentous growth. In most laboratory strains, mating differentiation is more dependent on Fus3 than on Kss1 (24). This more important role is now attributed to phosphoproteins essential for mating that are preferential substrates for Fus3 rather than for Kss1 (7, 29). Two examples of Fus3-specific targets are Far1, which is a bifunctional protein required for pheromone-induced cell cycle arrest and morphogenesis, and Sst2, which is a regulator of G protein signaling (7, 30). By contrast, active Kss1 is the principal MAPK required for promoting filamentous growth, and active Fus3 antagonizes this response (2, 3, 16, 42, 58, 61).
Genome-wide analysis of Ste12 promoter occupancy has suggested a mechanism for the negative role that Fus3 has on the filamentation transcriptional program. This analysis revealed that Fus3 activity inhibits Ste12 binding to genes assumed to be important for filamentation. Further, this inhibitory function is dependent on the Ste12 binding partner Tec1 (74). More recently it was shown that Fus3 specifies Ste12 promoter selectivity under pheromone-induced conditions through phosphorylation of Tec1 and its subsequent Skip-Cullin-F-box (SCF)-catalyzed ubiquitination and degradation (1, 9, 12, 70).
There are also temporal considerations that distinguish the filamentation and mating programs. Mating differentiation involves transient MAPK activation and requires arrest in the G1 phase of the cell cycle. Pheromone imposes G1 arrest by inducing the expression and activation of Far1, which functions as a Cdc28-Cln inhibitor (11, 53). The transience of the mating response is evident in that cells reenter the cell cycle to resume vegetative growth whether or not mating occurs. Accordingly, the responsiveness of cells to persistent pheromone induction diminishes with time. This phenomenon occurs through several desensitization or adaptation mechanisms that impinge on different components of the signaling pathway (see reference 19 for a review). Rapid attenuation of MAPK activation is reinforced through pheromone-dependent mechanisms that directly affect the MAPK module enzymes themselves. Examples include dephosphorylation of the MAPKs by phosphatases, one of which is induced by pheromone; ubiquitin-dependent turnover of Ste11 that apparently depletes activated Ste11 from the Ste5-organized MAPK activation complex; and ubiquitination of Ste7 upon prolonged pheromone exposure (20, 27, 67-70, 75). By contrast, filamentous growth requires persistent signaling through successive rounds of cell division. Therefore, it appears that the cell cycle arrest and transient signaling, which are characteristic of mating differentiation, are incompatible with continued growth in a filamentous form. The more-transient requirements associated with mating differentiation than with filamentous growth led us to speculate that regulation of Ste12 turnover might contribute to attenuation of the mating-specific transcription program and restrict activation of the filamentation program. The studies described herein were designed to test this hypothesis.
MATERIALS AND METHODS
Recombinant DNA procedures and plasmid constructions.
Bacterial transformations, bacterial DNA preparation, plasmid constructions, and DNA restriction enzyme digestions were performed by standard methods (62).
pNC739 carries a STE12-GST allele with a kanr selectable marker for integration at the STE12 genomic locus. The allele was constructed in several steps and involves two intermediate plasmids. pNC489 was constructed by ligating a SacI fragment encompassing the 657-bp GST coding sequence to pLB1214, which had been linearized by partial digestion with SacI. pLB1214 is a genomic clone that encompasses the entire STE12 locus (26). The GST fragment for this ligation reaction was generated by PCR amplification using primers 261 (5′-CCCGGAGCTCTCATGTCCCC) and 262 (5′-CGCCGAGCTCCCGATTTTGG) with pGEX-2T (Amersham Biosciences, Piscataway, NJ) as template DNA, followed by digestion with SacI. (Restriction enzyme recognition sites within oligonucleotides used in these studies are highlighted by underlined nucleotides.) The resulting STE12-GST fusion allele has the complete GST coding sequence inserted into the STE12 coding sequence at the SacI site (nucleotide 2007) that is 58 bp upstream from the TGA stop codon. (All nucleotide positions of STE12 given in this description are with respect to the A of the ATG initiation codon.) pNC736 was constructed by inserting a 291-bp SacI-EcoRI STE12 fragment from the 3′ noncoding region into the SacI-EcoRV sites that follow the kanMX4 region in pFA6a-kanMX4. This fragment (STE12 nucleotides 2149 to 2440) was generated by PCR amplification using oligonucleotide primers 321 (5′-CGTGCTGAGCTCCAACAAGGAC) and 322 (5′-CGGTCCGATATCGGCAATACTACG) with the plasmid pLB205 (26) as template DNA. pNC739 was then generated by inserting a 1,493-bp SalI-BglII fragment, encompassing the STE12-GST-STE12 sequences derived from pNC489, at the SalI-BglII sites preceding the kanMX4 region in pNC736. The fragment, with SalI and BglII ends, was generated by PCR amplification using oligonucleotide primers 323 (5′-CACACCGTCGACGCCGTATAATCC) and 324 (5′-CGCTGCAGATCTTCTGGCCCGCA) with pNC489 as template DNA.
pNC777 carries a C-terminal fusion of GFP to STE12 with a kanr selectable marker for integration at the STE12 genomic locus. It was constructed by the strategy described for pNC739 except that a 703-bp fragment encompassing the green fluorescent protein (GFP) coding region was used for the in-frame fusion at the SacI site of STE12. This GFP-encoding fragment was generated by PCR amplification using primers 398 (5′-GGCGGAGCTCTGAGTAAAGGAGAAG) and 399 (5′-GGTGAGCTCGCTCATCCATGCC) with pYSL17 as template DNA. pYSL17 was provided by A. Dranginis (St. Johns University, New York, NY). It is identical to pGFLO (37) except that it contains enhanced GFP instead of GFP.
pNC999 carries the erg6Δ::His3MX6 allele for replacement of the ERG6 coding region. It was constructed by subcloning a 396-bp BamHI-HindIII fragment and a 389-bp SacI-EcoRV fragment from the ERG6 3′ and 5′ noncoding regions, respectively, into the corresponding restriction sites of pFA6a-HIS3MX6. The flanking sequences were generated by PCR amplification using S288C DNA as template and primers 725 (5′-GATCAATAGGATCCAAATAAAGCG) and 726 (5′-AACCAAAGCTTCACCTTTCTTAATC) for the 3′ region and primers 721 (5′-CGATATCTGCCGTTTCCGATAA) and 722 (5′-GTTTGAGCTCATCTTATGCTGCCT) for the 5′ region.
pYES-8HisUbi is a 2μm plasmid with a URA3 selectable marker used for expression of ubiquitin with a polyhistidine tag (8His-Ubi) from the GAL1 promoter. A fragment with the 8His-UBI coding sequence was generated by PCR amplification using YEp120 as template DNA and the following two primers: 5′-CGGAATTCCAGAATGCATCATCACCATCATCACCATCACGGTATGCAGATCTTCGTCAAGACGTTAACCGGT and 5′-GGGGTACCGGATCCGCGTTAACCACCTCTTAGTCTTAAGACAAGATG. The amplified DNA was cloned into the TOPO site of the yeast expression vector pYES2.1/V5-His-TOPO (Invitrogen) to generate pYES-8HisUbi. YEp120 was provided by M. Hochstrasser, Yale University) and carries a synthetic ubiquitin gene cassette that was originally described by Ecker et al. (23).
Yeast strains, media, and genetic manipulations.
Unless otherwise specified, standard media and genetic procedures were used (63). Yeast transformations were carried out by the method of Ito et al. (34). Homologous replacements with the different DNA fragments were made by the method of Rothstein (60) or by the one-step PCR-mediated technique described by Longtine et al. (38). All gene integrations and replacements were verified by PCR analysis of genomic DNA.
Table 1 gives the genotypes and sources of strains used in these studies. E929-6C-92, E929-6C-95, E929-6C-96, E929-6C-97, BB151, and BB153 express Ste12 with a fusion to glutathione S-transferase (GST) from the endogenous STE12 locus. These strains were constructed by gene replacement with an EcoRV-BamHI fragment from the plasmid pNC739. Strains E929-6C-94 and E929-6C106 express Ste12 with a fusion to GFP from the endogenous Ste12 locus. These strains were constructed by gene replacement with an EcoRV-BamHI fragment from the plasmid pNC777. Strain Z1315 expresses Ste12 with an N-terminal fusion to 18 copies of an myc epitope tag (18-myc-Ste12) from the endogenous STE12 locus (56). Strains YW001 and YW002 were derived from Z1315 and an untagged Ste12 reference strain (BY4741) by gene replacement with the EcoRV-BamHI fragment that carries the erg6Δ::HIS3MX6 allele from pNC999.
TABLE 1.
Yeast strains
| Straina | Parent | Genotype | Source or reference |
|---|---|---|---|
| E929-6C* | MATa cyc1 CYC7-H2 can1 leu2-3,112 trp1-Δ1 ura3-52 STE12 | 14 | |
| E929-6C-6* | E929-6C | MATa cyc1 CYC7-H2 can1 leu2-3,112 trp1-Δ1 ura3-52 ste12Δ::LEU2 | 14 |
| E929-6C-30* | E929-6C | MATa cyc1 CYC7-H2 can1 leu2-3,112 trp1-Δ1 ura3-52 STE12 fus3Δ6::LEU2 | 76 |
| E929-6C-48* | E929-6C | MATa cyc1 CYC7-H2 can1 leu2-3,112 trp1-Δ1 ura3-52 STE12 kss1Δ::URA3 | 76 |
| E929-6C-49* | E929-6C | MATa cyc1 CYC7-H2 can1 leu2-3,112 trp1-Δ1 ura3-52 STE12 far1Δ::HisG | 76 |
| E929-6C-92* | E929-6C | MATa cyc1 CYC7-H2 can1 leu2-3,112 trp1-Δ1 ura3-52 STE12-GST::kanMX6 | This work |
| E929-6C-94* | E929-6C | MATa cyc1 CYC7-H2 can1 leu2-3,112 trp1-Δ1 ura3-52 STE12-GFP::kanMX6 | This work |
| E929-6C-95* | E929-6C-30 | MATa cyc1 CYC7-H2 can1 leu2-3,112 trp1-Δ1 ura3-52 fus3Δ6::LEU2 STE12-GST::kanMX6 | This work |
| E929-6C-96* | E929-6C-49 | MATa cyc1 CYC7-H2 can1 leu2-3,112 trp1-Δ1 ura3-52 far1Δ1::HisG STE12-GST::kanMX6 | This work |
| E929-6C-97* | E929-6C-48 | MATa cyc1 CYC7-H2 can1 leu2-3,112 trp1-Δ1 ura3-52 kss1Δ::URA3 STE12-GST::kanMX6 | This work |
| E929-6C-106* | E929-6C-30 | MATa cyc1 CYC7-H2 can1 leu2-3,112 trp1-Δ1 ura3-52 fus3Δ6::LEU2 STE12-GFP::kanMX6 | This work |
| 15Daub† | MATa ura3ΔNS ade1 his2 leu2-3,112 bar1Δ STE12 CDC28 | 44 | |
| D13au† | MATa ura3ΔNS ade1 his2 leu2-3,112 bar1Δ STE12 cdc28-13 | 44 | |
| BB151† | 15Daub | MATa ura3ΔNS ade1 his2 leu2-3,112 bar1Δ CDC28 STE12-GST::kanMX6 | This work |
| BB153† | D13au | MATa ura3ΔNS ade1 his2 leu2-3,112 bar1Δ cdc28-13 STE12-GST::kanMX6 | This work |
| Z1315 | MATa ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3 STE12::18myc-STE12 | 56 | |
| YW001 | Z1315 | MATa ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3 erg6Δ::HIS3MX6 STE12::18myc-STE12 | This work |
| BY4741 | MATa leu2Δ met15Δ ura3Δ | 69 | |
| YW002 | BY4741 | MATa leu2Δ met15Δ ura3Δ erg6Δ::HIS3MX6 STE12 | This work |
Immunoblot analysis of Ste12 protein abundance.
Cultures of nonconditional strains were grown to early log phase (1 × 107 to 2 × 107 cells/ml) in complete medium at 30°C. Cultures of temperature-sensitive mutant strains were grown at the permissive temperature (22°C) to early log phase and shifted to the nonpermissive temperature (37°C) for 30 min prior to initiation of the pheromone induction time courses. After removal of the initial samples for protein extract preparation (time zero), incubation of the cultures continued at 30°C or 37°C, as specified, with pheromone (3 μM α-factor) for induction of mating differentiation. Parallel cultures were maintained at the corresponding temperatures in the absence of pheromone as references. Whole-cell protein extracts were prepared by the method of Mattison et al. (46). Protein concentration determinations for extracts were done with the Bio-Rad Lowry protein assay reagent kit (Bio-Rad DC protein assay reagents 500-0113, 500-0114, and 500-0115).
For determination of the Ste12 protein half-life under uninduced and pheromone-induced conditions, the protein synthesis inhibitor cycloheximide (20 μM final concentration) was added to the cultures with or without pheromone immediately after removal of the initial samples at time zero. Samples of the cultures taken for extract preparation were adjusted to keep the cell numbers constant.
Protein extracts from the Ste12 strain (50 μg) were fractionated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and those from the various Ste12-GST strains (40 μg) were fractionated by 7.5% SDS-PAGE, with a Mini-PROTEAN (Bio-Rad) gel apparatus. The corresponding amount of extract from a Ste12-deficient (ste12Δ) strain was included as a control for nonspecific cross-reacting proteins. The fractionated proteins were transferred to nitrocellulose membranes (Schleicher & Schuell) by either a semidry transfer apparatus (LKB) or a Mini-Trans-Blot (Bio-Rad) transfer cell. Detection of Ste12 was carried out with affinity-purified rabbit anti-Ste12 polyclonal antibodies (G. Ammerer, University of Vienna, Vienna, Austria) at a dilution of 1:1,000 followed by a donkey anti-rabbit immunoglobulin G (IgG) antibody conjugated to horseradish peroxidase (HRP) (Amersham) at a dilution of 1:6,500. Detection of the Ste12-GST fusion protein used a mouse monoclonal anti-GST antibody (Santa Cruz Biotechnology) at 0.2 μg/ml followed by a goat anti-mouse IgG antibody conjugated to alkaline phosphatase (AP) (Promega) at 0.17 μg/ml. Detection of Tub1 used a rat anti-tubulin monoclonal antibody at 0.17 μg/ml followed by rabbit anti-rat IgG antibody conjugated to AP (Sigma) at a dilution of 1:10,000. Visualization of HRP-conjugated complexes and AP-conjugated complexes was accomplished with the Amersham ECL Western blotting system and Promega Protoblot immunoblot system, respectively.
The signal intensities for anti-Ste12, anti-GST (Ste12-GST), and anti-tubulin reacting proteins were quantified from scanned blots with NIH Image J software. The normalized amount of Ste12-GST or Ste12 in each sample was calculated from the ratio of the Ste12-GST or Ste12 signal to that of the corresponding internal reference signal for that sample. The signal from a nonspecific anti-Ste12 cross-reacting protein was used as an internal reference for the samples on the anti-Ste12 blots. The signal from anti-tubulin was used as the internal reference for the samples on the anti-GST blots. Relative amounts of Ste12 or Ste12-GST in samples at different times after addition of pheromone are the normalized ratio relative to the normalized ratio for the sample at time zero on the same blot.
The amount of extract used for quantification of Ste12 and Ste12-GST in these experiments was empirically determined to be in the linear range for detection by these methods. The same strategy for this determination was used for Ste12 and Ste12-GST but is described only for the latter. Calibration standards were made by mixing decreasing amounts of extract from an uninduced STE12-GST strain with increasing amounts of extract from a ste12Δ strain (micrograms of STE12-GST to micrograms of ste12Δ: 100:0, 67:33, 33:67, 17:83, and 0:100). This method provides standards with decreasing amounts of Ste12-GST while keeping the total protein constant. Therefore, any potential interference from other proteins on either blotting efficiency or detection of Ste12-GST is controlled. Plots of the anti-GST signal for Ste12-GST on immunoblots versus micrograms of STE12-GST extract indicated that 40-μg samples have a signal within the linear range. Also, the dynamic range for this amount of extract was adequate to reveal either increases or deceases in Ste12-GST abundance that might occur during pheromone induction. To validate the inferences from the calibration plots, three independent experiments were performed, with standardized samples being examined on blots developed in parallel with those for the samples from the pheromone induction time courses.
Isolation of 18myc-Ste12-ubiquitin conjugated to His-tagged ubiquitin.
We used erg6Δ strains that are sensitive to the proteasome inhibitor MG132 (Sigma), GAL1-expressed polyhistidine-tagged ubiquitin, and poly-myc-tagged Ste12 expressed from the endogenous STE12 locus (36, 48). Strain YW001 (erg6Δ 18Myc-STE12), with either pYES-8HisUbi (GAL1-8His-ubiquitin, 2 μm URA3) or vector (pYES2.1/V5-His-TOPO), and reference strain YW002 (erg6Δ STE12), with pYES-8HisUbi, were grown in synthetic galactose (2%, wt/vol) medium without uracil. MG132 (50 μM) was added to inhibit proteasome function when cultures reached a density of 1 × 107 to 1.2 × 107 cells/ml. After 90 min in the presence of MG132, each culture was divided into two portions. One portion was treated with pheromone (3 μM), and the other was maintained as a noninduced reference. After 60 min, 109 cells from each culture were harvested by centrifugation.
Extracts were prepared from these pellets for metal affinity purification of His-tagged ubiquitin and conjugates by methods that were adapted from Muratani et al. (48). Each cell pellet was suspended in 1 ml of buffer A2 (6 M guanidine-HCl, 100 mM Na2HPO4/NaH2PO4 [pH 8.0], 10 mM imidazole, 250 mM NaCl, 0.5% NP-40, 2 mM N-ethylmaleimide, and 1 pellet of complete EDTA-free protease inhibitor from Roche for every 50 ml of buffer). Suspensions were subjected to eight cycles of glass bead beating of 30 seconds each. The lysates were solubilized by mixing at 4°C for 1 h and clarified by two rounds of centrifugation at maximum speed. The first was for 5 min, and the second was for 25 min, at 4°C. The resulting supernatants were incubated with TALON Superflow metal affinity resin (BD Biosciences, Clontech) for 2 h at room temperature with rocking. Following this incubation, the resin was washed twice with buffer A2, twice with buffer A2/T2 (1 volume of buffer A2 and 3 volumes of buffer T2 [50 mM Na2HPO4/NaH2PO4 {pH 8.0}, 250 mM NaCl, 20 mM imidazole, 0.5% NP-40), and twice with buffer T2. Resin was then washed with buffer T2 containing 50 mM histidine to reduce the retention of proteins that nonspecifically bind to the resin. Finally, 8His-ubiquitin and its conjugated proteins were eluted with 2× SDS loading buffer containing 250 mM imidazole. Samples of the metal affinity-purified proteins and total extract were fractionated by SDS-PAGE with 6.5% gels and transferred to nitrocellulose membranes (Bio-Rad). Detection of 18-myc-Ste12 was carried out with mouse Myc1-9E10 monoclonal antibodies (28) at 0.2 μg/ml followed by goat anti-mouse IgG-HRP (Santa Cruz Biotechnology). Detection of 8His-ubiquitin used rabbit anti-ubiquitin antibodies at a 1:200 dilution followed by goat anti-rabbit IgG-HRP (Santa Cruz Biotechnology) at a 1:6,000 dilution. Visualization of HRP-conjugated complexes was accomplished by the enhanced chemiluminescence Reagent Plus from Perkin-Elmer.
RNA blot analysis.
Strains E929-6C-92 (wild type), E929-6C-95 (fus3Δ), and E929-6C-96 (far1Δ) were grown to 1 × 107 to 2 × 107 cells/ml in complete medium (YPD) at 30°C. A 20-ml aliquot from each culture was removed for analysis of steady-state mRNAs in uninduced cultures. Mating pheromone (α-factor, 3 μM) was added to the remaining portion of each culture, and samples (20 ml) were removed for analysis at the indicated times. Cells in each aliquot were harvested by centrifugation, and total RNA was prepared by the glass bead lysis method (8). RNA was glyoxal denatured, fractionated on 1% agarose gels in 10 mM phosphate buffer (pH 7.0), and transferred to nylon membranes (Immobilon-Ny+; Millipore, Bedford, MA).
Radiolabeled probes for hybridization to the mRNA blots were prepared from purified DNA fragments by random-primed synthesis reactions (Boehringer Mannheim, Indianapolis, IN). The FUS1 mRNA probe was synthesized with the 1.5-kb EcoRI fragment of pSL589 as template DNA (G. Sprague, Jr., University of Oregon, Eugene). The ACT1 mRNA probe was synthesized with the 2.2-kb EcoRI-HindIII fragment of pYACT1 as template DNA (49). Blots were hybridized to the FUS1 radiolabeled probe, stripped, and then hybridized to the ACT1 radiolabeled probe to provide an internal loading reference. Conditions for hybridization to radiolabeled probes, washing, and stripping of blots were as described by Cameron et al. (10). The hybridization signals were detected by direct scanning with a Molecular Dynamics STORM 860 PhosphorImager (Amersham, Piscataway, NJ) and quantified with the Molecular Dynamics ImageQuant 5.0 software package (Amersham, Piscataway, NJ). Amounts of mRNA in each sample are specified as the ratio of the signal for the query mRNA to that of the ACT1 mRNA reference.
Pheromone-induced filamentation assays.
Halo assays for pheromone-induced invasion were done with a lawn of cells (105) from the MATa haploid strains E929-6C-92 (wild type), E929-6C-95 (fus3Δ), E929-6C-96 (far1Δ), and E929-6C-6 (ste12Δ) spread onto the surface of YPD agar plates. Then, 2 μl of α-factor (10 μg/μl) or solvent (ethanol) was applied to a 4-mm-diameter disc (made from Whatman 3-mm paper) placed in the center of the lawns. Plates were incubated for 48 h at 30°C. Plates were photographed to record the halo size, which is an indicator of the extent of pheromone-induced growth arrest. After a wash under running tap water, plates were again photographed to record the zones where cells invaded the agar substrate.
For comparison of Ste12 stability in wild-type and fus3Δ cells undergoing pheromone-induced mating differentiation or haploid filamentation, we assessed the stability of Ste12-GFP in individual living cells by imaging analysis. These studies used cultures of the MATa haploid strains E929-6C-92 (wild type) and E929-6C-106 (fus3Δ) grown to early log phase, harvested by centrifugation, and suspended at 1 × 107 to 2 × 107 cells/ml in synthetic dextrose and sonicated briefly to disrupt clumps. Standard 3- by 1-in. glass slides were prepared by pipetting 1 ml of synthetic dextrose agar containing a high concentration of pheromone (3 μM α-factor) to induce mating differentiation (shmoo formation) in the wild-type strain or haploid filamentation in the fus3Δ strain. For reference, a low concentration of pheromone (30 nM α-factor) was used to induce haploid filamentation in the wild-type strain. Five microliters of each cell suspension was allowed to settle onto the solidified agar surface, and coverslips were applied. In preliminary experiments we established the timing of shmoo versus filament formation under the specified experimental conditions. Developmental morphologies and Ste12 abundance were monitored by visualizing a different field of cells at 60-min intervals, but images were captured only at time zero, at the time when shmoo or filamentous morphologies first became evident, and 2 h thereafter. (Images were captured from different fields of cells at each time interval so that photobleaching would not be an issue for Ste12-GFP quantification.)
Ste12-GFP quantification was based on the average fluorescence intensity within the nucleus of individual cells measured from captured images by Metamorph software. The area used for each measurement was a circle with dimensions contained within the nuclear boundary. The raw fluorescence intensity within the circle was measured. The background intensity for an identical area was then measured by dragging the same circle to the background adjacent to the cell. The average fluorescence intensity specific to the nucleus was obtained after subtracting the average background intensity from the raw fluorescence measurement. The individual cells in these images were scored to have either mating projections (shmoos), filamentous morphology, or vegetative morphology. Only cells with a length-to-width ratio of 1.6 or greater were classified as filamentous. The reported quantification of developmental morphologies and Ste12-GFP signal intensity included cells scored in two independent trials.
Microscopy was performed with a Nikon Eclipse E600 FN using a 100× 1.4 NA Plan Apochromatic objective. Images were generated by a Hamamatsu OrcaII Progressive Scan Interline-cooled charge-coupled-device digital camera (model c4742-98). GFP fluorescence was detected with the 8200 V2 filter set from Chroma Technology Corp. (Rockingham, VT). Exposure times for differential interference contrast (DIC) and fluorescence images were 20 ms and 1,000 ms, respectively. Microscope automation, image acquisition, and analysis were accomplished with the Metamorph 5.0r7 software package (Universal Imaging Corp., Downingtown, PA).
RESULTS
The amount of Ste12 protein declines following pheromone induction.
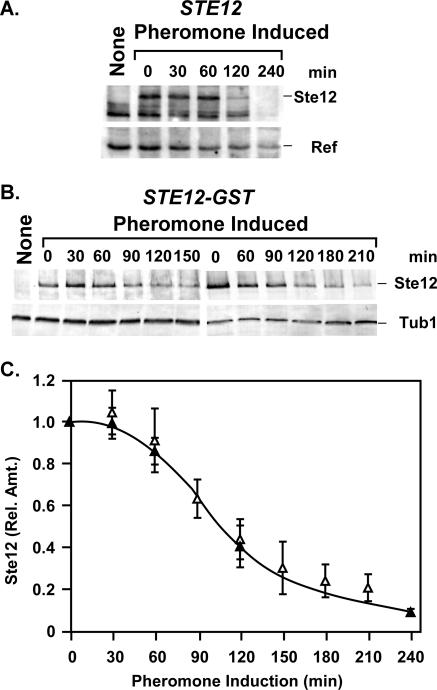
We examined the amount of Ste12 protein present in haploid cells before and at different times during exposure to pheromone. Whole-cell protein extracts were prepared from culture samples, fractionated by SDS-PAGE, and transferred to nitrocellulose for detection of Ste12 protein by anti-Ste12 antibodies (Fig. 2A). This analysis revealed that the amount of Ste12 protein begins to decline after 60 min of exposure to mating pheromone. This decline occurs even though the amounts and polysome profiles of STE12 mRNA are sustained under these conditions (39).
FIG. 2.
The steady-state amount of Ste12 decreases during pheromone induction. (A) A representative immunoblot showing amounts of Ste12 at different times during pheromone induction of strain E929-6C (STE12). The signal from a nonspecific cross-reacting protein (Ref) serves as an internal reference for loading and blotting. (B) Representative immunoblots showing amounts of Ste12-GST at different times during pheromone induction of strain E929-6C-92 (STE12-GST). Tubulin (Tub1) serves as an internal reference for loading and blotting. The first lanes (None) in panels A and B show an extract from an isogenic ste12Δ strain (E929-6C-6) as a negative control. (C) Plots comparing the relative amounts of Ste12 versus time from STE12 (▴) and STE12-GST (▵) strains undergoing pheromone induction. The amount of Ste12 at each time point is the Ste12 or Ste12-GST signal intensity divided by the signal intensity for the corresponding internal reference. Relative amounts are normalized to the amount of Ste12 or Ste12-GST at time zero for each time course. Values are averages from three or more independent experiments. Bars show the average deviation for each point.
The suitability of using a tagged version of Ste12 for further studies was investigated because we had limited amounts of anti-Ste12 antibodies. A strain that expresses a Ste12 fusion to GST (Ste12-GST) from the endogenous locus was constructed. The Ste12-GST fusion functions identically to Ste12 in qualitative assays for mating, quantitative assays for pheromone-induced reporter gene expression, G1 arrest, and mating projection formation (data not shown). Before and at indicated times following pheromone treatment, whole-cell protein extracts were prepared from culture samples, fractionated by SDS-PAGE, and transferred to nitrocellulose filters for detection of Ste12-GST by anti-GST antibodies (Fig. 2B). The kinetics and extent of the decrease in Ste12 and Ste12-GST are identical, within experimental error, throughout the pheromone induction time course (Fig. 2C). We therefore used the Ste12-GST fusion for subsequent studies to further investigate regulation of Ste12 protein stability.
Pheromone induction promotes ubiquitin-dependent degradation of Ste12.
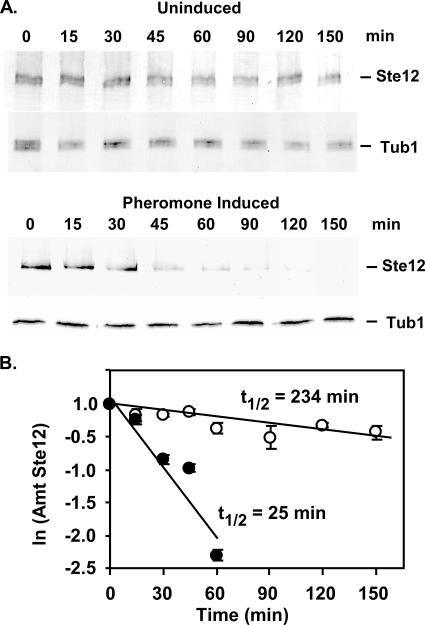
To test whether accelerated protein degradation contributes to the decline in the amount of Ste12 protein, we compared its half-life under uninduced and pheromone-induced conditions. For these determinations the amount of Ste12-GST before and at different times after inhibition of protein synthesis was quantified by analysis of protein extracts on immunoblots. A culture of a strain expressing Ste12-GST was grown to early log phase. One sample was removed immediately for extract preparation (time zero), and the remaining culture was divided in two. One portion was treated with pheromone, and the other was the uninduced reference. Cycloheximide was immediately added to both cultures to block new protein synthesis. Amounts of Ste12-GST and Tub1 in the sample extracts were detected by anti-GST and anti-tubulin antibodies, respectively. Whereas preexisting Ste12-GST and Tub1 from uninduced cultures have similar levels of persistence after protein synthesis inhibition, Ste12-GST is depleted much more rapidly than Tub1 under pheromone-induced conditions (compare the upper and lower panels in Fig. 3A). The half-lives under uninduced and pheromone-induced conditions were determined from least-squares fit of data to natural log plots of relative amounts of Ste12-GST versus time (Fig. 3B). The half-life of Ste12 for pheromone inducing conditions is estimated to be ∼25 min compared with ∼230 min for uninduced conditions. The ∼10-fold decrease in the Ste12 half-life under pheromone inducing conditions establishes that accelerated protein turnover contributes to the observed decline in Ste12 abundance.
FIG. 3.
Comparison of Ste12 half-lives under uninduced and pheromone-induced conditions. (A) Representative immunoblots showing amounts of Ste12-GST and Tub1 in extracts prepared from a STE12-GST strain (E929-6C-92) at different times after the addition of cycloheximide (20 μM) to cultures without and with pheromone. Tub1 serves as an internal reference to highlight the differences in Ste12-GST stability under the two conditions. (B) Plots comparing the natural log of the amount of Ste12-GST versus time. The amount of Ste12-GST at each time point is the signal intensity for Ste12-GST relative to the amount at time zero for each time course. Values for Ste12-GST from uninduced cultures (○) are averages of two independent experiments, and those for the pheromone-induced cultures (•) are averages of four independent experiments.
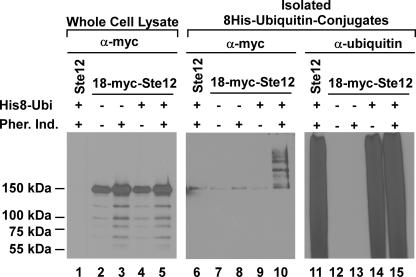
To test whether Ste12 turnover involves its ubiquitination and degradation by the proteasome, we exploited an affinity purification method for isolating His-tagged ubiquitin-conjugated proteins and strains (erg6Δ) that are sensitive to the proteasome inhibitor MG132 (36, 48). These strains carried either a high-copy-number plasmid that expresses poly-His-tagged ubiquitin (8His-Ubi) from the GAL1 promoter or vector as a control. For immune detection of Ste12, the strains also expressed an N-terminal poly-myc-tagged version of Ste12 (18myc-Ste12). 18-myc-Ste12 was used because the multiple epitopes enhance detection compared with Ste12-GST. A STE12 (untagged) erg6Δ strain expressing 8His-Ubi was included in the analysis for reference. Cultures were treated with MG132 for 90 min prior to pheromone induction to inhibit proteasome function and then divided into two portions. One portion was treated with pheromone for 60 min, and the other was maintained as an uninduced reference for the same interval. Extracts from each culture were prepared for affinity purification. Samples of the extracts and purified proteins were fractionated by SDS-PAGE and analyzed on immunoblots.
The 150-kDa species that is detected on the anti-myc blot of the whole-cell lysates specifically identifies 18-myc-Ste12 (Fig. 4; compare lanes 2 to 5 with lane 1). (The smaller species on this blot are attributed to degradation intermediates that accumulate because of diminished proteasome function resulting from treatment with MG132.) The population of 8His-Ubi-conjugated proteins specifically isolated by affinity purification is shown on the anti-ubiquitin blot (Fig. 4, lanes 11 and 14 to 15). The affinity-purified proteins visualized on the anti-myc blot with a size of >150 kDa identify 18-myc-Ste12 conjugated to one or more polyubiquitin chains of various lengths (Fig. 4, lane 10). These species are present in samples of the pheromone-treated culture coexpressing 8His-Ubi and 18-myc-Ste12 but not from samples of the corresponding uninduced culture or from those of the vector and untagged Ste12 reference cultures (Fig. 4, lanes 6 to 9). The minor 150-kDa species present in the control samples is attributed to anti-myc cross-reacting material that is enriched by nonspecific binding to the resin (Fig. 4, lanes 6 to 8). This analysis shows that pheromone induction promotes ubiquitination of Ste12.
FIG. 4.
Covalent ubiquitin-Ste12 conjugates are intermediates in proteasome degradation of Ste12. Cultures of STE12 8His-UBI erg6Δ (YW002 pYES-8HisUbi), 18myc-STE12 His8-UBI erg6Δ (YW001 pYES-8HisUbi), and 18-myc-STE12 erg6Δ (YW001 vector) strains were treated with the proteasome inhibitor MG132 (50 μM) for 90 min prior to the addition of pheromone (3 μM α-factor) for induction (+) or continued incubation without pheromone for the uninduced reference (−), as indicated. Immunoblots compare the amounts of 18-myc-Ste12 in samples from the different cultures by using anti-myc antibodies (lanes 1 to 5). Proteins covalently conjugated to 8His-ubiquitin from the indicated samples were isolated by metal affinity purification and analyzed on duplicate immunoblots. 18-myc-Ste12 conjugated to 8His-Ubi was detected on one blot by using anti-myc antibodies (lanes 6 to 10), and the total cellular population of 8His-Ubi-conjugated proteins was detected on the other by using anti-ubiquitin antibodies (lanes 11 to 15).
Depletion of Ste12 protein following pheromone induction is dependent on Fus3 and Far1.
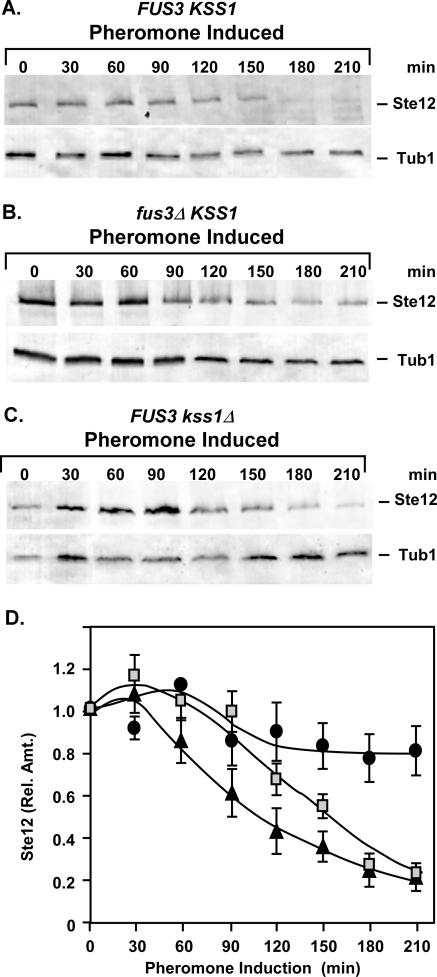
Protein phosphorylation often precedes and targets proteins for subsequent ubiquitination. Ste12 is a known substrate for both Fus3 and Kss1, which are the MAPK family members that are activated in response to mating pheromone (5, 35, 64). Therefore, we tested whether Ste12 protein degradation is dependent on either Fus3 or Kss1. Amounts of Ste12-GST during pheromone induction were monitored as before in strains expressing both (FUS3 KSS1) or only one of the MAPKs (fus3Δ KSS1 and FUS3 kss1Δ) (Fig. 5A to C). In the absence of Kss1, the kinetics of Ste12 degradation are delayed but the extent of depletion is the same as observed in the strain expressing both MAPKs (Fig. 5A, C, and D). By contrast, pheromone-induced depletion of Ste12 is severely curtailed in the strain lacking Fus3 (Fig. 5B and D).
FIG. 5.
The MAPK Fus3 is required for pheromone-induced degradation of Ste12. (A to C) Representative immunoblots showing the amount of Ste12-GST from extracts of STE12-GST FUS3 KSS1 (E929-6C-92), STE12-GST fus3Δ KSS1 (E929-6C-95), and STE12-GST FUS3 kss1Δ (E929-6C-97) strains at different times after the addition of pheromone to cultures. Tubulin (Tub1) serves as an internal reference for loading and blotting. (D) Plots comparing the relative amounts of Ste12-GST versus time from FUS3 KSS1 (triangles), fus3Δ KSS1 (circles), and FUS3 kss1Δ (squares) strains undergoing pheromone induction. Relative amounts of Ste12-GST at each time point are as specified in Fig. 2. Values are averages from three independent experiments. Bars show the average deviations.
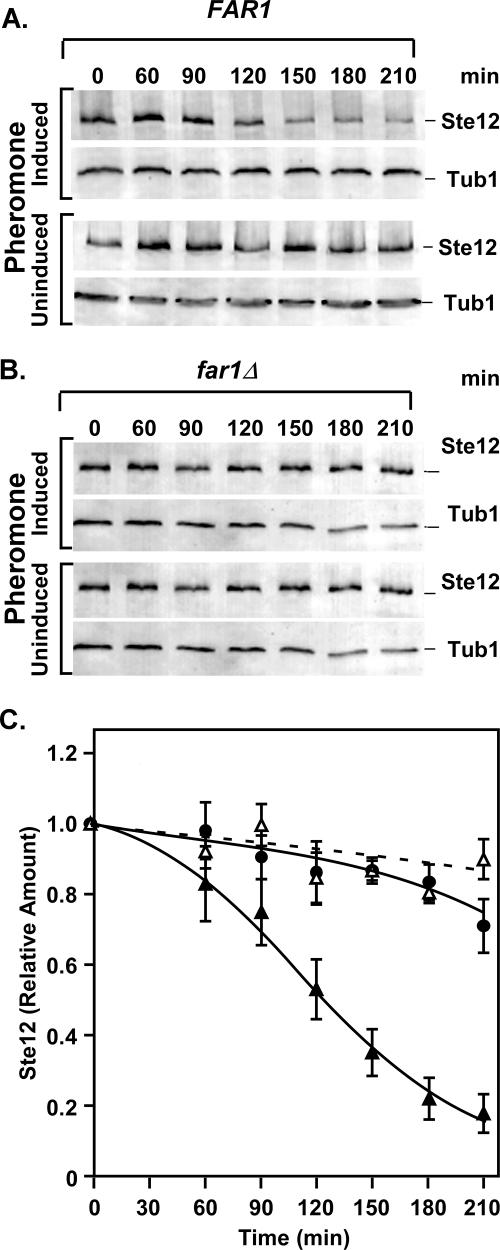
Fus3 and Kss1 differentially modify a subset of Ste12 residues (5). This situation is consistent with the possibility that those sites phosphorylated by Fus3, but not Kss1, comprise a phosphodegron signal critical for Ste12 ubiquitination. An alternative, but not mutually exclusive, possibility is that a response uniquely mediated by Fus3 leads to Ste12 degradation. Assessment of a potential Fus3-generated phosphodegron signal in Ste12 would be impractical due to the large number of Fus3-phosphorylated residues (5). For this reason we elected to pursue a hypothesis consistent with the latter possibility. Far1 is a substrate for Fus3, but not Kss1, and Far1 phosphorylation by Fus3 is essential for the inhibition of Cdc28-Cln that results in G1 arrest (11, 53). Therefore, Fus3 may promote Ste12 degradation indirectly through its regulation of Far1. To test this hypothesis, amounts of Ste12-GST from uninduced and pheromone-induced cultures were monitored as before in FAR1 and far1Δ strains (Fig. 6A and B). The comparison shows no significant depletion of Ste12 during pheromone induction in the strain lacking Far1 (Fig. 6C). This outcome is consistent with either Far1 itself or the G1 arrest imposed by Far1 having some role in Ste12 depletion during pheromone induction.
FIG. 6.
Far1 is required for pheromone-induced degradation of Ste12. (A and B) Representative immunoblots comparing amounts of Ste12-GST from extracts of STE12-GST FAR1 (E929-6C-92) and STE12-GST far1Δ (E929-6C-96) strains cultured at 30°C either under pheromone-induced or uninduced conditions. Tubulin (Tub1) serves as an internal reference for loading and blotting. (C) Plots comparing the relative amounts of Ste12-GST versus time from the FAR1 strain with (▴) and without (▵) pheromone induction and the far1Δ strain (•) with pheromone induction. Relative amounts of Ste12-GST at each time point are as specified in Fig. 2. Values are averages from three independent experiments. Bars show the average deviations.
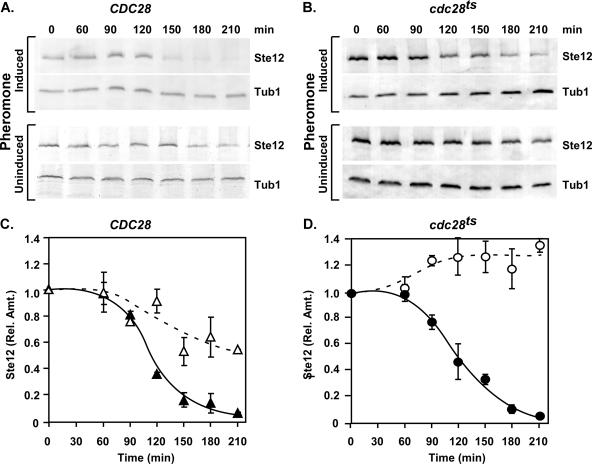
If the sole requirement for Fus3 and Far1 in Ste12 degradation involves their role in imposing G1 arrest, establishment of G1 arrest by another mechanism might be sufficient to cause a decrease in the amounts of Ste12. To test this possibility, we exploited a temperature-sensitive allele of CDC28 (cdc28-13) that causes G1 arrest at the nonpermissive temperature. Cultures of cdc28-13 and CDC28 reference strains were shifted to the nonpermissive temperature (37°C) and maintained at this temperature in either the presence or absence of pheromone. Ste12-GST levels were monitored as before at different times after the temperature shift. The amount of Ste12-GST decreased to the same extent during the pheromone induction time course in the cdc28-13 strain as in the reference CDC28 strain (Fig. 7). This result shows that Cdc28 function is not essential for pheromone-induced Ste12 degradation. In the uninduced cultures of the CDC28 reference strain, which do not arrest in G1, the amounts of Ste12-GST gradually decreased at 37°C (Fig. 7A and C). By contrast, Ste12-GST overaccumulates in the cdc28-13 strain, which arrests in G1 independently of pheromone induction (Fig. 7B and D). These results reveal the existence of a temperature- and Cdc28-dependent mechanism that destabilizes Ste12 during vegetative growth and show that G1 arrest per se is not sufficient to promote Ste12 degradation.
FIG. 7.
G1 arrest is not sufficient for promoting degradation of Ste12. (A and B) Representative immunoblots comparing amounts of Ste12-GST from extracts of STE12-GST CDC28 (BB151) and STE12-GST cdc28-13 (BB153) strains cultured at 37°C either under pheromone-induced or uninduced conditions. Tubulin (Tub1) serves as an internal reference for loading and blotting. (C and D) Plots comparing the relative amounts of Ste12-GST versus time from the CDC28 strain with (▴) or without (▵) pheromone induction and from the cdc28 temperature-sensitive (cdc28ts) strain with (•) or without (○) pheromone induction. Relative amounts of Ste12-GST at each time point are as specified in Fig. 2. Values are averages from three independent experiments. Bars show the average deviations.
Pheromone-induced depletion of Ste12 contributes to attenuation of mating-specific gene transcription.
The responsiveness of cells to persistent pheromone induction diminishes with time. This phenomenon occurs through several desensitization or adaptation mechanisms that impinge on different components of the signaling pathway (see the introduction.) The observed degradation of Ste12 could be an additional mechanism that contributes to desensitization. If this is the case, attenuation of Ste12-dependent mating gene transcription should be compromised in the fus3Δ and far1Δ genetic backgrounds, where amounts of Ste12 are relatively constant throughout pheromone induction.
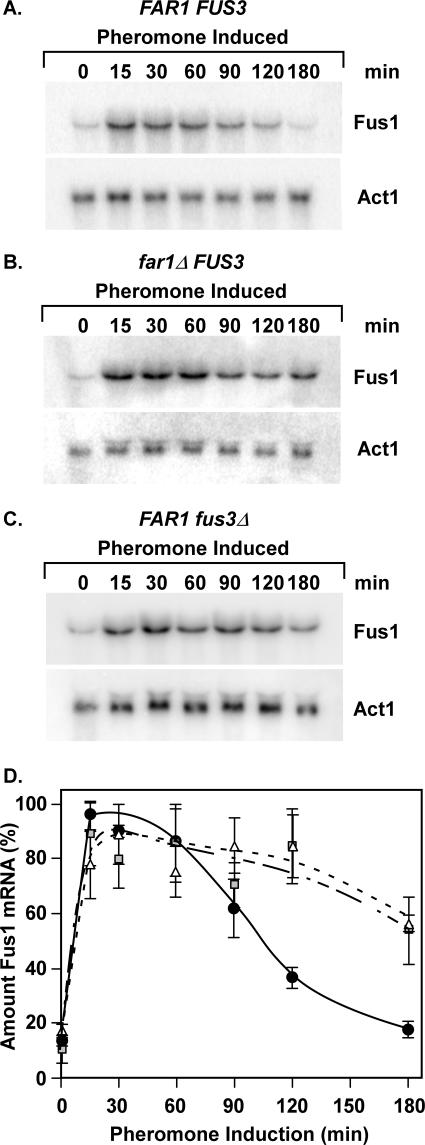
To test this prediction we compared the amounts of FUS1 mRNA in wild-type, fus3Δ, and far1Δ strains at different times during pheromone induction. FUS1 is a prototypical mating-specific gene with a promoter configuration that is dependent on Ste12 homomultimers for activation (21, 47, 66). Northern blots of total RNA isolated from culture samples were first hybridized to a probe for detection of FUS1 and then stripped and hybridized to a probe for detection of ACT1 mRNA as an internal reference (Fig. 8A to C). The amount of FUS1 mRNA at each time point was determined as the ratio of the FUS1 to ACT1 signal intensity. The relative amounts are expressed as a percentage of the maximum value for each trial. The averages for three independent experiments are shown (Fig. 8D).
FIG. 8.
Induction profiles for FUS1 mRNA correlate with Ste12 abundance. (A to C) Representative Northern blots comparing the amounts of FUS1 mRNA from FAR1 FUS3 (E929-6C-92), far1Δ FUS3 (E929-6C-96), and FAR1 fus3Δ (E929-6C-95) strains at different times after the addition of pheromone (3 μM) to cultures. Actin mRNA (ACT1) serves as an internal reference for loading and blotting. (D) Plots showing the relative amounts of FUS1 mRNA versus time for FAR1 FUS3 (circles), far1Δ FUS3 (squares), and FAR1 fus3Δ (triangles) backgrounds. The amount of FUS1 mRNA at each time point is the FUS1 signal intensity divided by the signal intensity for the ACT1 internal reference. Relative amounts are normalized to the maximum amount for each separate time course. Values are averages from three or more independent experiments. (Note that the averages may not show a 100% value, because the maximum in some trials occurred at 15 min but in others at 30 min.) Bars show the average deviation for each point.
Pheromone rapidly causes a 5- to 10-fold induction of FUS1 mRNA. The abundance of the mRNA then declines 30 to 60 min after pheromone addition in the wild-type background but persists for a longer time in the fus3Δ and far1Δ backgrounds (Fig. 8A to D). These results strictly show that FUS3 and FAR1 are required for the transient transcription profiles typical of FUS1 mRNA. However, the correlation between FUS1 mRNA levels and Ste12 protein abundance in these strains is consistent with the interpretation that depletion of Ste12 contributes to the attenuation of the mating-specific transcriptional response (Fig. 5, 6, and 8).
Pheromone-induced haploid invasion.
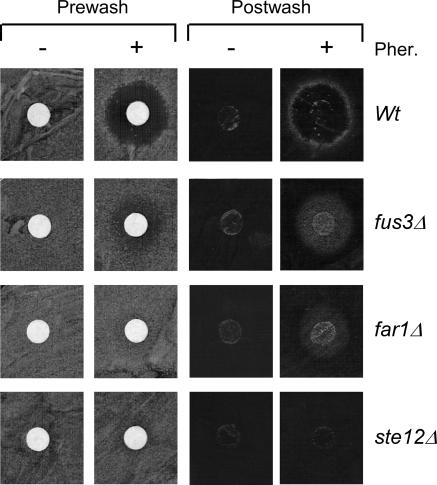
Nonsaturating doses of pheromone have been reported to cause haploid cells to invade an agar substrate, to elongate, and to switch from an axial to bipolar budding pattern (22, 25, 57). These characteristics are associated with filamentous growth but not mating differentiation. Agar invasion is readily observed in standard halo assays for pheromone-induced growth arrest. In wild-type cells, growth inhibition occurs in a zone (halo) where pheromone diffusion from a point source generates concentrations that are sufficient to induce G1 arrest. A ring of cells at the periphery of this halo invade the agar substrate and can be visualized after a gentle wash of surface cells from the plate (Fig. 9) (57). Pheromone-induced invasion is dependent on Ste12 and other components of the pheromone-induced pathway, such as Ste2 (57). Roberts et al. (57) also reported that a far1Δ strain, which fails to arrest or only transiently arrests in G1, forms a uniform halo of cells that are invasive. We see the same phenomenon for a fus3Δ strain (Fig. 9).
FIG. 9.
Pheromone-induced haploid invasion assays. Lawns of 105 cells from wild-type (Wt) (E929-6C-92), fus3Δ (E929-6C-95), far1Δ (E929-6C-96), and ste12Δ (E929-6C-6) strains were spread onto YPD plates either in the presence (+) or absence (−) of mating pheromone (20 μg α-factor) applied to a disc at the center of the lawn. Plates were incubated for 48 h at 30°C. Photographs show plates before and after washing of cells from the agar surface. Prewash plates show the extent of pheromone-induced G1 arrest apparent as a zone of growth inhibition (halo). Postwash plates show cells that invaded the agar substrate.
The zone of invasion in the far1Δ and fus3Δ lawns is apparent throughout the pheromone diffusion gradient. Thus, the invasive response occurs even in the presence of saturating amounts of pheromone. The ability of these deletion strains to invade at saturating pheromone levels can be attributed to their ability to continue through the successive rounds of cell division necessary for filamentous growth. Because pheromone-induced agar invasion requires Ste12, we predict that its stabilization contributes to the ability of these strains to follow this developmental program at saturating pheromone concentrations.
Pheromone-induced haploid filamentation correlates with Ste12 stabilization.
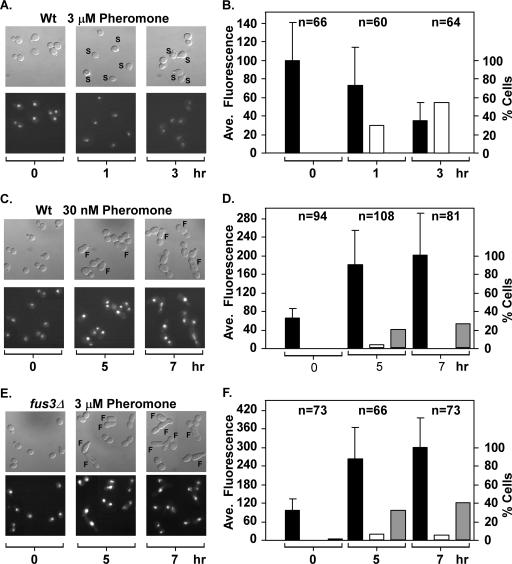
To compare Ste12 stability in individual living cells undergoing pheromone-induced mating differentiation versus filamentous growth, we used imaging analysis with cells expressing a Ste12 fusion to GFP (Ste12-GFP). We first examined Ste12-GFP levels in cells undergoing pheromone-induced mating differentiation. Exponentially growing wild-type cells that express Ste12-GFP from the endogenous locus were spread on a slide with agar containing a uniform and saturating concentration of mating pheromone (3 μM α-factor). DIC and fluorescence images of the cells were taken immediately (time zero), at 1 h, and at 3 h after preparation of the slides (Fig. 10A). Cells with incipient mating projections, or shmoos, were apparent after 1 h in the presence of pheromone-containing medium. By 3 h, the shmoos of mating-differentiated cells had matured. The percentage of cells with incipient or mature shmoos is shown in the bar graph (Fig. 10B). Notably, no cells with a filamentous morphology developed in the presence of saturating pheromone, even after 5 h on pheromone-containing medium (data not shown). The signal intensity for Ste12-GFP in individual cells was measured from the fluorescence images of the corresponding fields and is reported as the average for the population scored at each time interval (Fig. 10B). Ste12-GFP gives a strong nuclear signal in all cells of the initial image (time zero), but the signal declines to ∼30% of that amount by 3 h on pheromone-containing medium. The percentage decrease in signal intensity for Ste12-GFP is comparable to the estimates obtained by Western blot analysis for amounts of Ste12-GST protein. The imaging analysis further shows that the timing of the decline in Ste12 abundance occurs close to the time when morphological evidence of differentiation is already apparent. Therefore, this analysis confirms that Ste12 protein levels decline late in the mating differentiation response.
FIG. 10.
Ste12 depletion or persistence correlates with different pheromone-induced developmental fates. (A, C, and E) DIC and corresponding fluorescence images of STE12-GFP FUS3 (Wt) (E929-6C-94) or STE12-GFP fus3Δ (E929-6C-106) cells on medium containing the indicated amounts of mating pheromone α-factor. The micrographs are a collage of cells from one or more fields that have been arranged to eliminate empty regions and minimize the size of the figure. Images are from fields captured immediately upon placing cells on α-factor medium (0 h) and from different fields at the indicated times after slide preparation. Cells showing shmoo (S) or filamentous (F) morphologies are indicated. (B, D, and F) Bars show the percentages of cells scored as having shmoo (open bares) or filamentous (gray bars) morphologies and the average Ste12-GFP fluorescence signal (black bars) at the indicated times. The number of cells scored for morphology and quantified for Ste12-GFP fluorescence at each time interval is specified above the bars.
In parallel experiments, wild-type cells were spread on a slide with agar containing a nonsaturating concentration of mating pheromone (30 nM α-factor) and fus3Δ cells were spread on agar containing saturating amounts (3 μM α-factor). DIC and fluorescence images of different fields were taken immediately (time zero), at 5 h, and at 7 h after preparation of the slides (Fig. 10C and E). Wild-type and fus3Δ cells incubated at the respective concentrations of pheromone failed to form shmoos by 3 h (data not shown). Only a small percentage of cells from either background had a shmoo-like morphology even at later times (Fig. 10C to F). By 5 h, cells in both populations began to exhibit a filamentous morphology, which we define as cells with a length-to-width ratio of >1.6. After 7 h, the percentage of cells meeting this criterion increased, with many filamentous cells showing greater elongation than was evident at earlier times (Fig. 10C to F). By 7 h, some wild-type cells with the filamentous morphology showed evidence of bipolar budding (Fig. 10C, 7 h) and fus3Δ cells with filamentous morphology showed evidence of both bipolar and unipolar budding (Fig. 10E, 7 h). Thus, similar to what was reported by Erdman and Snyder (25), we also observed that the budding pattern of the elongated cells departs from the axial pattern characteristic of haploid vegetative yeast.
In contrast to wild-type cells exposed to saturating concentrations of pheromone, the Ste12-GFP signal intensity for wild-type cells at low pheromone concentrations increases threefold rather than declining over the imaging time course (Fig. 10C and D). Similarly, the Ste12-GFP signal intensity for fus3Δ cells at saturating concentrations of pheromone increases threefold over the imaging time course (Fig. 10E and F). Results similar to those shown for fus3Δ cells were obtained with far1Δ cells at a saturating concentration of pheromone (data not shown.) These comparisons confirm that the pheromone-inducing conditions under which Ste12 protein persists correlate with conditions that promote cells in the population to take on a filamentous morphology rather than to differentiate into a mating-competent form. These correlations are consistent with the model that pheromone-induced degradation of Ste12 normally contributes to the suppression of an inappropriate filamentous growth response to pheromone.
DISCUSSION
Ubiquitin-mediated protein turnover defines the pheromone-induced signal profile.
Mounting evidence shows that ubiquitin-mediated protein turnover plays a pivotal role in determining the amplitude and duration of MAPK activation in the mating response pathway. Pheromone induction promotes monoubiquitination of the Ste2 and Ste3 receptors. This modification leads to receptor endocytosis and sorting to the vacuole for degradation. Destruction of the receptors plays a determining role in attenuation at the level of signal perception (33, 59). Amounts of Gpa1-Gα and Sst2, its GTPase regulator, are influenced by polyubiquitination that targets these components to the N-end rule degradation pathway (31, 43, 72). More recently it has been learned that Gpa1-Gα also undergoes monoubiquitination that targets it to the vacuole (31, 43, 72). Destruction of Sst2 and Gpa1 frees the Gβγ dimer to stimulate its downstream effectors and consequently makes cells more responsive to pheromone. A ubiquitin-dependent mechanism underlies pheromone-induced accelerated turnover of the MAPKKK Ste11 (27). The MAPKK Ste7 undergoes SCF-dependent polyubiquitination and turnover in response to pheromone (69, 71). Destruction of the Ste11 and Ste7 kinases counteracts the effects of destruction of Gα and Sst2 and contributes to the attenuation of signal output (27, 69).
MAPK targets required for responses to pheromone are also regulated by ubiquitin-mediated protein turnover. Early studies revealed that Far1 destruction by the G1-S ubiquitination system, which involves SCF-catalyzed polyubiquitination of target proteins and their proteasomal degradation, contributes to recovery from pheromone-induced cell cycle arrest (32). Our work shows that the Ste12 transcription factor also is depleted in the late stages of pheromone induction by ubiquitin-mediated proteasomal degradation (Fig. 2 to 4).
Pheromone-induced Ste12 depletion is dependent on Fus3 and Far1 (Fig. 5 and 6). Ubiquitination of the MAPKK Ste7 is similarly dependent on Fus3 and Far1 (71). Stabilization of both proteins in fus3Δ or far1Δ backgrounds would likely be cooperative for prolonging Ste12 activation and, therefore, the duration of target gene transcription. The direct correlation that we find between Ste12 stability and the duration of target gene expression is consistent with this interpretation and provides evidence that Ste12 destruction contributes to attenuation of the pheromone-induced transcriptional response (Fig. 5, 6, and 8).
Ste12 regulation of transcription has an impact not only on terminal responses to pheromone but also on signal transmission leading to MAPK activation. Transcription of mating pathway components such as the receptors, Gα subunit, Sst2, and Far1 is pheromone induced and dependent on Ste12 (see reference 73 for a review). Because of this circuitry, destruction of Ste12 reinforces the effects that ubiquitin-mediated turnover has on these other components of the pathway. This situation makes it clear that the amplitude of the signal output at different times during pheromone induction represents the dynamic balance of rates of synthesis, activation, and destruction of multiple components in this pathway.
Mechanisms imparting specificity to signal output.
Pheromone-induced Ste12 depletion is apparent during the later stages of the pheromone response when the differentiation program is already established (Fig. 2 and 10A and B). As discussed above, this destruction of Ste12 contributes critically to the attenuation of pheromone-induced transcription. The physiological contribution that this regulation imparts to signal identity is best supported by observations made under conditions where Ste12 is found to persist during pheromone induction. One condition is at low concentrations of pheromone, and the other is at high concentrations of pheromone but in genetic backgrounds (far1Δ and fus3Δ) that do not arrest or only transiently arrest in the G1 phase of the cell cycle. In both circumstances, cells fail to differentiate into a mating-competent form but instead initiate a haploid filamentation program. It has been suggested by others that this program could be physiologically important for disperse cell populations where pheromone concentrations are diffuse. Their reasoning is that the cell elongation, continued division, and modified budding patterns characteristic of this developmental fate would provide a mechanism for nonmotile cells to eventually encounter a mating partner (25, 57).
The difference we observe in Ste12 stability under saturating versus low pheromone concentrations is compatible with our understanding of the requirements for the developmental programs supporting mating differentiation compared with that for filamentous growth. At a saturating pheromone concentration, Ste12 activation is maximal, but its abundance diminishes substantially in the later stages of the response. This profile is compatible with mating differentiation, which is already established by the time Ste12 depletion becomes evident and the response attenuates. At a low pheromone concentration or in deletion backgrounds that abrogate Ste12 depletion, cells are able to sustain more a prolonged Ste12-dependent program that is required to support filamentous growth through successive rounds of cell division.
The idea that the magnitude and duration of signaling can specify distinct responses has been attractive for some time. This model originated with the observation that rat PC12 cells proliferate in response to epidermal growth factor but differentiate in response to nerve growth factor. Both stimuli activate the MAPK Erk2. However, epidermal growth factor promotes transient activation whereas nerve growth factor promotes sustained activation (45). The correlation between Ste12 abundance upon signal activation and the duration of transcription that we describe is consistent with this model as it might apply to specifying the transcription programs required for mating differentiation versus filamentous growth.
Acknowledgments
We thank M. Hochsrasser (Yale University) for plasmid YEp120, R. A. Young (Whitehead Institute for Biomedical Research) for strain Z1315, and H. Dohlman and T. Elston (University of North Carolina) for valuable discussions and comments on the manuscript.
This work was supported by NIH grants GM39852 and GM067809 (B.E.) and by American Heart Association Scientist Development grant 0635271N (Y.W.).
Footnotes
Published ahead of print on 13 October 2006.
REFERENCES
- 1.Bao, M. Z., M. A. Schwartz, G. T. Cantin, J. R. Yates III, and H. D. Madhani. 2004. Pheromone-dependent destruction of the Tec1 transcription factor is required for MAP kinase signaling specificity in yeast. Cell 119:991-1000. [DOI] [PubMed] [Google Scholar]
- 2.Bardwell, L., J. G. Cook, V. Depak, D. M. Baggott, A. R. Martinez, and J. Thorner. 1998. Repression of yeast Ste12 transcription factor by direct binding of unphosphorylated Kss1 MAPK and its regulation by the Ste7 MEK. Genes Dev. 12:2887-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardwell, L., J. G. Cook, J. X. Zhu-Shimoni, D. Voora, and J. Thorner. 1998. Differential regulation of transcription: repression by unactivated mitogen-activated protein kinase Kss1 requires the Dig1 and Dig2 proteins. Proc. Natl. Acad. Sci. USA 95:15400-15405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baur, M., R. K. Esch, and B. Errede. 1997. Cooperative binding interactions required for function of the Ty1 sterile responsive element. Mol. Cell. Biol. 17:4330-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breitkreutz, A. 2002. Signal specificity in the budding yeast mating/filamentous MAPK pathway. University of Toronto, Toronto, Canada.
- 6.Breitkreutz, A., L. Boucher, B. J. Breitkreutz, M. Sultan, I. Jurisica, and M. Tyers. 2003. Phenotypic and transcriptional plasticity directed by a yeast mitogen-activated protein kinase network. Genetics 165:997-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breitkreutz, A., L. Boucher, and M. Tyers. 2001. MAPK specificity in the yeast pheromone response independent of transcriptional activation. Curr. Biol. 11:1266-1271. [DOI] [PubMed] [Google Scholar]
- 8.Broach, J. R., J. F. Atkins, C. McGill, and L. Chow. 1979. Identification and mapping of the transcriptional and translational products of the yeast plasmid, 2mu circle. Cell 16:827-839. [DOI] [PubMed] [Google Scholar]
- 9.Bruckner, S., T. Kohler, G. H. Braus, B. Heise, M. Bolte, and H. U. Mosch. 2004. Differential regulation of Tec1 by Fus3 and Kss1 confers signaling specificity in yeast development. Curr. Genet. 46:331-342. [DOI] [PubMed] [Google Scholar]
- 10.Cameron, J. R., E. Y. Loh, and R. W. Davis. 1979. Evidence for transposition of dispersed repetitive DNA families in yeast. Cell 16:739-751. [DOI] [PubMed] [Google Scholar]
- 11.Chang, F., and I. Herskowitz. 1992. Phosphorylation of FAR1 in response to alpha-factor: a possible requirement for cell-cycle arrest. Mol. Biol. Cell 3:445-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou, S., L. Huang, and H. Liu. 2004. Fus3-regulated Tec1 degradation through SCFCdc4 determines MAPK signaling specificity during mating in yeast. Cell 119:981-990. [DOI] [PubMed] [Google Scholar]
- 13.Chou, S., S. Lane, and H. Liu. 2006. Regulation of mating and filamentation genes by two distinct Ste12 complexes in Saccharomyces cerevisiae. Mol. Cell. Biol. 26:4794-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Company, M., C. Adler, and B. Errede. 1988. Identification of a Ty1 regulatory sequence responsive to STE7 and STE12. Mol. Cell. Biol. 8:2545-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cook, J. G., L. Bardwell, S. J. Kron, and J. Thorner. 1996. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes Dev. 10:2831-2848. [DOI] [PubMed] [Google Scholar]
- 16.Cook, J. G., L. Bardwell, and J. Thorner. 1997. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature 390:85-88. [DOI] [PubMed] [Google Scholar]
- 17.Cullen, P. J., W. Sabbagh, Jr., E. Graham, M. M. Irick, E. K. van Olden, C. Neal, J. Delrow, L. Bardwell, and G. F. Sprague, Jr. 2004. A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes Dev. 18:1695-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davenport, K. D., K. E. Williams, B. D. Ullmann, and M. C. Gustin. 1999. Activation of the Saccharomyces cerevisiae filamentation/invasion pathway by osmotic stress in high-osmolarity glycogen pathway mutants. Genetics 153:1091-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dohlman, H. G., and J. W. Thorner. 2001. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu. Rev. Biochem. 70:703-754. [DOI] [PubMed] [Google Scholar]
- 20.Doi, K., A. Gartner, G. Ammerer, B. Errede, H. Shinkawa, K. Sugimoto, and K. Matsumoto. 1994. MSG5, a novel protein phosphatase, promotes adaptation to pheromone response in S. cerevisiae. EMBO J. 13:61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dolan, J. W., C. Kirkman, and S. Fields. 1989. The yeast STE12 protein binds to the DNA sequence mediating pheromone induction. Proc. Natl. Acad. Sci. USA 86:5703-5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dorer, R., P. M. Pryciak, and L. H. Hartwell. 1995. Saccharomyces cerevisiae cells execute a default pathway to select a mate in the absence of pheromone gradients. J. Cell Biol. 131:845-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ecker, D. J., M. I. Khan, J. Marsh, T. R. Butt, and S. T. Crooke. 1987. Chemical synthesis and expression of a cassette adapted ubiquitin gene. J. Biol. Chem. 262:3524-3527. [PubMed] [Google Scholar]
- 24.Elion, E. A., J. A. Brill, and G. R. Fink. 1991. FUS3 represses CLN1 and CLN2 and in concert with KSS1 promotes signal transduction. Proc. Natl. Acad. Sci. USA 88:9392-9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erdman, S., and M. Snyder. 2001. A filamentous growth response mediated by the yeast mating pathway. Genetics 159:919-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Errede, B., and G. Ammerer. 1989. STE12, a protein involved in cell-type-specific transcription and signal transduction in yeast, is part of protein-DNA complexes. Genes Dev. 3:1349-1361. [DOI] [PubMed] [Google Scholar]
- 27.Esch, R. K., and B. Errede. 2002. Pheromone induction promotes Ste11 degradation through a MAPK feedback and ubiquitin-dependent mechanism. Proc. Natl. Acad. Sci. USA 99:9160-9165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evan, G. I., G. K. Lewis, G. Ramsay, and J. M. Bishop. 1985. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol. 5:3610-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farley, F. W., B. Satterberg, E. J. Goldsmith, and E. A. Elion. 1999. Relative dependence of different outputs of the Saccharomyces cerevisiae pheromone response pathway on the MAP kinase Fus3p. Genetics 151:1425-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garrison, T. R., Y. Zhang, M. Pausch, D. Apanovitch, R. Aebersold, and H. G. Dohlman. 1999. Feedback phosphorylation of an RGS protein by MAP kinase in yeast. J. Biol. Chem. 274:36387-36391. [DOI] [PubMed] [Google Scholar]
- 31.Hao, N., N. Yildirim, Y. Wang, T. C. Elston, and H. G. Dohlman. 2003. Regulators of G protein signaling and transient activation of signaling: experimental and computational analysis reveals negative and positive feedback controls on G protein activity. J. Biol. Chem. 278:46506-46515. [DOI] [PubMed] [Google Scholar]
- 32.Henchoz, S., Y. Chi, B. Catarin, I. Herskowitz, R. J. Deshaies, and M. Peter. 1997. Phosphorylation- and ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Far1p in budding yeast. Genes Dev. 11:3046-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hicke, L., and H. Riezman. 1996. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell 84:277-287. [DOI] [PubMed] [Google Scholar]
- 34.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kranz, J. E., B. Satterberg, and E. A. Elion. 1994. The MAP kinase Fus3 associates with and phosphorylates the upstream signaling component Ste5. Genes Dev. 8:313-327. [DOI] [PubMed] [Google Scholar]
- 36.Lee, D. H., and A. L. Goldberg. 1996. Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J. Biol. Chem. 271:27280-27284. [DOI] [PubMed] [Google Scholar]
- 37.Lo, W. S., and A. M. Dranginis. 1996. FLO11, a yeast gene related to the STA genes, encodes a novel cell surface flocculin. J. Bacteriol. 178:7144-7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 39.MacKay, V. L., X. Li, M. R. Flory, E. Turcott, G. L. Law, K. A. Serikawa, X. L. Xu, H. Lee, D. R. Goodlett, R. Aebersold, L. P. Zhao, and D. R. Morris. 2004. Gene expression analyzed by high-resolution state array analysis and quantitative proteomics: response of yeast to mating pheromone. Mol. Cell. Proteomics 3:478-489. [DOI] [PubMed] [Google Scholar]
- 40.Madhani, H. D., and G. R. Fink. 1997. Combinatorial control required for the specificity of yeast MAPK signaling. Science 275:1314-1317. [DOI] [PubMed] [Google Scholar]
- 41.Madhani, H. D., T. Galitski, E. S. Lander, and G. R. Fink. 1999. Effectors of a developmental mitogen-activated protein kinase cascade revealed by expression signatures of signaling mutants. Proc. Natl. Acad. Sci. USA 96:12530-12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madhani, H. D., C. A. Styles, and G. R. Fink. 1997. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell 91:673-684. [DOI] [PubMed] [Google Scholar]
- 43.Madura, K., and A. Varshavsky. 1994. Degradation of G alpha by the N-end rule pathway. Science 265:1454-1458. [DOI] [PubMed] [Google Scholar]
- 44.Marini, N. J., E. Meldrum, B. Buehrer, A. V. Hubberstey, D. E. Stone, A. Traynor-Kaplan, and S. I. Reed. 1996. A pathway in the yeast cell division cycle linking protein kinase C (Pkc1) to activation of Cdc28 at START. EMBO J. 15:3040-3052. [PMC free article] [PubMed] [Google Scholar]
- 45.Marshall, C. J. 1995. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell 80:179-185. [DOI] [PubMed] [Google Scholar]
- 46.Mattison, C. P., S. S. Spencer, K. A. Kresge, J. Lee, and I. M. Ota. 1999. Differential regulation of the cell wall integrity mitogen-activated protein kinase pathway in budding yeast by the protein tyrosine phosphatases Ptp2 and Ptp3. Mol. Cell. Biol. 19:7651-7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCaffrey, G., F. J. Clay, K. Kelsay, and G. F. Sprague, Jr. 1987. Identification and regulation of a gene required for cell fusion during mating of the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 7:2680-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muratani, M., C. Kung, K. M. Shokat, and W. P. Tansey. 2005. The F box protein Dsg1/Mdm30 is a transcriptional coactivator that stimulates Gal4 turnover and cotranscriptional mRNA processing. Cell 120:887-899. [DOI] [PubMed] [Google Scholar]
- 49.Ng, R., and J. Abelson. 1980. Isolation and sequence of the gene for actin in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 77:3912-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Olson, K. A., C. Nelson, G. Tai, W. Hung, C. Yong, C. Astell, and I. Sadowski. 2000. Two regulators of Ste12p inhibit pheromone-responsive transcription by separate mechanisms. Mol. Cell. Biol. 20:4199-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O'Rourke, S. M., and I. Herskowitz. 1998. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 12:2874-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Palecek, S. P., A. S. Parikh, and S. J. Kron. 2002. Sensing, signalling and integrating physical processes during Saccharomyces cerevisiae invasive and filamentous growth. Microbiology 148:893-907. [DOI] [PubMed] [Google Scholar]
- 53.Peter, M., A. Gartner, J. Horecka, G. Ammerer, and I. Herskowitz. 1993. FAR1 links the signal transduction pathway to the cell cycle machinery in yeast. Cell 73:747-760. [DOI] [PubMed] [Google Scholar]
- 54.Primig, M., H. Winkler, and G. Ammerer. 1991. The DNA binding and oligomerization domain of MCM1 is sufficient for its interaction with other regulatory proteins. EMBO J. 10:4209-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reed, S. I., J. A. Hadwiger, and A. T. Lorincz. 1985. Protein kinase activity associated with the product of the yeast cell division cycle gene CDC28. Proc. Natl. Acad. Sci. USA 82:4055-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ren, B., F. Robert, J. J. Wyrick, O. Aparicio, E. G. Jennings, I. Simon, J. Zeitlinger, J. Schreiber, N. Hannett, E. Kanin, T. L. Volkert, C. J. Wilson, S. P. Bell, and R. A. Young. 2000. Genome-wide location and function of DNA binding proteins. Science 290:2306-2309. [DOI] [PubMed] [Google Scholar]
- 57.Roberts, C. J., B. Nelson, M. J. Marton, R. Stoughton, M. R. Meyer, H. A. Bennett, Y. D. He, H. Dai, W. L. Walker, T. R. Hughes, M. Tyers, C. Boone, and S. H. Friend. 2000. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287:873-880. [DOI] [PubMed] [Google Scholar]
- 58.Roberts, R. L., and G. R. Fink. 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 8:2974-2985. [DOI] [PubMed] [Google Scholar]
- 59.Roth, A. F., and N. G. Davis. 1996. Ubiquitination of the yeast a-factor receptor. J. Cell Biol. 134:661-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rothstein, R. J. 1983. One-step gene disruption in yeast. Methods Enzymol. 101:202-211. [DOI] [PubMed] [Google Scholar]
- 61.Sabbagh, W., Jr., L. J. Flatauer, A. J. Bardwell, and L. Bardwell. 2001. Specificity of MAP kinase signaling in yeast differentiation involves transient versus sustained MAPK activation. Mol. Cell 8:683-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 63.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 64.Song, D., J. W. Dolan, Y. L. Yuan, and S. Fields. 1991. Pheromone-dependent phosphorylation of the yeast STE12 protein correlates with transcriptional activation. Genes Dev. 5:741-750. [DOI] [PubMed] [Google Scholar]
- 65.Tedford, K., S. Kim, D. Sa, K. Stevens, and M. Tyers. 1997. Regulation of the mating pheromone and invasive growth responses in yeast by two MAP kinase substrates. Curr. Biol. 7:228-238. [DOI] [PubMed] [Google Scholar]
- 66.Trueheart, J., J. D. Boeke, and G. R. Fink. 1987. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol. Cell. Biol. 7:2316-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Drogen, F., S. M. O'Rourke, V. M. Stucke, M. Jaquenoud, A. M. Neiman, and M. Peter. 2000. Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr. Biol. 10:630-639. [DOI] [PubMed] [Google Scholar]
- 68.Wang, H., X. Wang, and Y. Jiang. 2003. Interaction with Tap42 is required for the essential function of Sit4 and type 2A phosphatases. Mol. Biol. Cell 14:4342-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang, Y., and H. G. Dohlman. 2002. Pheromone-dependent ubiquitination of the mitogen-activated protein kinase kinase Ste7. J. Biol. Chem. 277:15766-15772. [DOI] [PubMed] [Google Scholar]
- 70.Wang, Y., and H. G. Dohlman. 2006. Pheromone-regulated sumoylation of transcription factors that mediate the invasive to mating developmental switch in yeast. J. Biol. Chem. 281:1964-1969. [DOI] [PubMed] [Google Scholar]
- 71.Wang, Y., Q. Ge, D. Houston, J. Thorner, B. Errede, and H. G. Dohlman. 2003. Regulation of Ste7 ubiquitination by Ste11 phosphorylation and the SCF (Skp1/Cullin/F-box) complex. J. Biol. Chem. 31:31. [DOI] [PubMed] [Google Scholar]
- 72.Wang, Y., L. A. Marotti, Jr., M. J. Lee, and H. G. Dohlman. 2005. Differential regulation of G protein alpha subunit trafficking by mono- and polyubiquitination. J. Biol. Chem. 280:284-291. [DOI] [PubMed] [Google Scholar]
- 73.Whiteway, M., and B. Errede. 1993. Pheromone response in Saccharomyces cerevisiae, p. 189-237. In R. P. Dottin, J. Kurjan, and B. L. Taylor (ed.), Molecular mechanisms of signal transduction in genetically tractable organisms. Academic Press, Inc., Orlando, Fla.
- 74.Zeitlinger, J., I. Simon, C. T. Harbison, N. M. Hannett, T. L. Volkert, G. R. Fink, and R. A. Young. 2003. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell 113:395-404. [DOI] [PubMed] [Google Scholar]
- 75.Zhan, X. L., R. J. Deschenes, and K. L. Guan. 1997. Differential regulation of FUS3 MAP kinase by tyrosine-specific phosphatases PTP2/PTP3 and dual-specificity phosphatase MSG5 in Saccharomyces cerevisiae. Genes Dev. 11:1690-1702. [DOI] [PubMed] [Google Scholar]
- 76.Zhou, Z., A. Gartner, R. Cade, G. Ammerer, and B. Errede. 1993. Pheromone-induced signal transduction in Saccharomyces cerevisiae requires the sequential function of three protein kinases. Mol. Cell. Biol. 13:2069-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]