Abstract
Evasion of host immune defenses is critical for the progression of invasive infections caused by the leading neonatal pathogen, group B streptococcus (GBS). Upon characterizing the factors required for virulence in a neonatal rat sepsis model, we found that a surface-associated penicillin-binding protein (PBP1a), encoded by ponA, played an essential role in resistance of GBS to phagocytic clearance. In order to elucidate how PBP1a promotes resistance to innate immunity, we compared the susceptibility of wild-type GBS and an isogenic ponA mutant to the bactericidal components of human neutrophils. The isogenic strains were found to be equally capable of blocking complement activation on the bacterial surface and equally associated with phagocytes and susceptible to oxidative killing. In contrast, the ponA mutant was significantly more susceptible to killing by cationic antimicrobial peptides (AMPs) of the cathelicidin and defensin families, which are now recognized as integral components of innate host defense against invasive bacterial infection. These observations may help explain the sensitivity to phagocytic killing and attenuated virulence of the ponA mutant. This novel function for PBP1a in promoting resistance of GBS to AMP did not involve an alteration in bacterial surface charge or peptidoglycan cross-linking. While the peptidoglycan polymerization and cross-linking activity of PBPs are essential for bacterial survival, our study is the first to identify a role for a PBP in resistance to host AMPs.
Streptococcus agalactiae (group B streptococcus [GBS]) is a major cause of neonatal pneumonia, sepsis, and meningitis in the United States (59). While mortality due to GBS infection has declined recently due to advances in care and improved disease recognition, this organism remains a leading cause of invasive infections in neonates (4). Neonatal GBS infections manifest as either early-onset disease, which occurs in the first 7 days of life, or as the less common late-onset disease that develops after the first 7 days. Bacteremia is a common manifestation of both forms (4); thus, the ability of GBS to survive in the bloodstream is thought to be an important aspect of invasive disease. To increase our understanding of invasive GBS disease, we previously performed a genome-wide screen to identify genes that are required for survival in the bloodstream and the development of sepsis in a neonatal rat infection model. In this screen, penicillin-binding protein 1a (PBP1a), encoded by ponA, was found to be critical for the virulence of GBS (28). PBP1a was subsequently found to promote resistance of GBS to innate immunity by protecting the organism from phagocytic killing by neutrophils (29).
PBPs are traditionally known for their role in the biosynthesis of cell wall peptidoglycan (PG). Bacterial species express between 2 and 16 PBPs, some of which have redundant functions (10). These proteins are divided into three classes, low-molecular-weight (with a mass of ∼40 kDa) PBPs and class A and B high-molecular-weight (HMW; with a mass of ∼50 to 100 kDa) PBPs. Low-molecular-weight PBPs are monofunctional carboxypeptidases that are involved in regulating the number of peptide cross-links. The HMW class B PBPs are monofunctional transpeptidases that are responsible for cross-linking of the inter peptide bridges. The HMW class A PBPs are bifunctional proteins with both transpeptidase (TP) activity and glycosyltransferase (GT) activity, which is required for the polymerization of the glycan chains (for a review, see reference 18). GBS PBP1a is representative of a bifunctional class A HMW PBP (29). In addition to our studies linking PBP1a with virulence in GBS, PBPs have been implicated in the virulence of a number of other pathogens including mycobacterial, gram-positive, and gram-negative bacterial species (2, 21, 27, 33, 35, 40, 49, 58, 69). Despite their frequent identification in screens for virulence factors, the mechanism by which PBPs contribute to virulence has not been determined.
The innate immune system represents the first line of defense against invading bacteria and is particularly important in neonates with suboptimal levels of protective antibody directed against GBS (3, 5, 25). Key factors in innate immune defense are the components of complement and neutrophils. GBS uses a number of strategies to avoid killing by these components of host defense including the elaboration of capsular polysaccharide that inhibits opsonophagocytosis (37, 56, 57, 64). Sialylated capsular polysaccharide prevents deposition of opsonically active C3 on the bacterial surface and promotes increased conversion of C3b to iC3b, thereby rendering GBS resistant to uptake and killing by phagocytes (36) and promoting virulence in a variety of animal models of infection (37, 56, 57, 64). Bacteria that are phagocytosed by neutrophils are exposed to microbicidal products including reactive oxygen intermediates (ROI) and antimicrobial peptides (AMPs). For defense against ROI generated during the respiratory burst, GBS relies on expression of a superoxide dismutase (53), a carotenoid pigment that is linked to hemolysin (34) and high intracellular levels of the oxygen-metabolite scavenger glutathione (65).
AMPs are integral components of innate immunity that have bactericidal activity against a wide range of bacterial species. AMPs are characteristically highly cationic and have the capacity to kill a broad spectrum of microorganisms by creating pores or otherwise affecting the integrity of membranes (68). The AMPs found in human neutrophils include members of the defensin and cathelicidin classes. The human neutrophil peptides 1 to 4 (HNP-1 to HNP-4) are members of the defensin family that are stored in cytoplasmic granules of the neutrophil, where they make up about 30% of the total granule protein. These AMPs are released into phagolysosomes where they contribute to the killing of engulfed microorganisms. Cathelicidins are a second class of AMP found in neutrophils that are structurally distinct from defensins. The best-characterized cathelicidin is LL-37, a 37-amino-acid peptide found in human neutrophils. The LL-37 homolog found in mice is known as cathelin-related antimicrobial peptide (CRAMP). In addition to being a major component of the bactericidal granule content of neutrophils, defensins and cathelicidins are also widely expressed by epithelial cells (for review, see reference 17).
Bacteria possess numerous mechanisms for resisting the activity of AMPs (for a review, see reference 45). These include modification of the surface charge, which results in decreased binding of the cationic AMPs, and the expression of proteins that inactivate or degrade AMPs. To date, the only known mechanism of resistance to AMPs that has been reported in GBS is electrostatic repulsion of AMPs by incorporation of d-alanine substitutions into teichoic acids on the bacterial cell surface (54). Here we demonstrate that PBP1a protects GBS from phagocytic killing by promoting resistance of the organism to AMPs through a novel mechanism independent of cell surface charge.
MATERIALS AND METHODS
Bacterial strains and growth medium.
The bacterial strains used in this study are listed in Table 1. GBS strains were grown in Todd Hewitt broth (THB; Difco Laboratories) at 37°C in 5% CO2 except as noted. Staphylococcus aureus strains were grown in THB at 37°C with aeration.
TABLE 1.
Bacterial strains used in this study
Opsonization and hydroxylamine release of bound C3.
Purified C3 protein (a kind gift from M. Hostetter) was digested to completion with 16 μg/ml trypsin (sequencing grade; Sigma) in 1 mM HCl during a 1-h incubation at 30°C. Complete digestion was confirmed by analyzing C3 fragments by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (31), staining with Coomassie blue, and comparing the fragments that we observed to published tryptic cleavage patterns (41). Western blotting was performed using an anti-C3 antibody (MP Biomedicals) diluted 1:10,000 in blocking buffer (5% nonfat milk in phosphate-buffered saline [PBS], 0.05% Tween 20), and an anti-goat immunoglobulin G-peroxidase-conjugated secondary antibody (Sigma) diluted 1:5,000 in blocking buffer.
Serum for C3 deposition assays was isolated from human blood obtained from volunteers following informed consent, as required by Seattle Children's Hospital and Regional Medical Center Institutional Review Board. GBS-specific antibody was removed by preadsorbing the serum against the appropriate isogenic strains (7) or using an ImmunoPure Immobilized Protein A column (Pierce) according to the manufacturer's directions. Analysis of C3 fragments on the surface of GBS strains following opsonization was performed using a modification of the methods of Gordon et al. (20). Briefly, isogenic strains were grown to an optical density at 600 nm (OD600) of 0.8, incubated in 50% pooled normal human serum (NHS) for 30 min at 37°C, and then washed in PBS-1% SDS to release noncovalently bound C3. Covalently attached C3 was released from the bacterial surface by hydroxylamine treatment (1 M hydroxylamine-1% SDS, pH 9.0, in 0.1 M carbonate buffer). The supernatant containing C3 was reduced by incubation with PBS containing 1% SDS and 10 mM dithiothreitol (10%, wt/vol) for 60 min at 37°C and then N-acetylated again by incubation in 22 mM iodoacetamide (10%, wt/vol) in TE buffer (10 mM Tris, 1 mM EDTA), pH 8, for 60 min at 37°C. C3 fragments released from the bacterial surface were resolved on 10% SDS-polyacrylamide gels and transferred to polyvinylidene difluoride membrane (Millipore) and detected by Western blotting using anti-C3 antibody and an anti-goat immunoglobulin G-peroxidase-conjugated secondary antibody as described above.
Neutrophil isolation and phagocytosis assay.
Dextran sedimentation was used to isolate neutrophils from heparinized human blood obtained from volunteers (6). Contaminating red blood cells were lysed by the addition of ice-cold 0.2% NaCl, and the neutrophils were resuspended in RPMI 1640 medium (Mediatech) and counted, and the viability was confirmed using trypan blue dye exclusion. Phagocytosis assays were performed as previously described (7) with modifications. The isogenic strains were incubated with neutrophils and 10% NHS at a ratio of 9 bacteria to 1 neutrophil (9 × 106 bacteria to 1 × 106 neutrophils) for 10 min at 37°C with constant mixing. The neutrophils were pelleted by centrifugation (150 × g at 4°C), washed twice to remove nonadherent bacteria, and lysed by vortexing vigorously in ice-cold distilled H2O. The number of neutrophil-associated bacteria was determined by plating serial dilutions of the lysates on Todd Hewitt agar (THA). Each assay was performed in triplicate.
Sensitivity to oxidative killing.
The sensitivity of the isogenic GBS strains to killing by oxidants was determined using a modification of the method of Liu et al. (34). Briefly, the isogenic strains were grown to an OD600 of 0.5, washed, and resuspended in the original volume of PBS. Paraquat, sodium hypochlorite, or hydrogen peroxide (30 mM, 0.002%, or 0.003% final concentration, respectively; Sigma) was added to the bacteria, and the samples were incubated for 2 h at 37°C. The number of surviving bacteria was determined by plating serial dilutions of the samples on THA.
MIC assays and AMP killing kinetics.
Gramicidin D was purchased from Sigma. The MIC required to inhibit growth of the isogenic strains was determined in Tryptic soy broth (TSB) after an overnight incubation at 37°C using the standard broth dilution method (42). HNP-1 (Peptides International) was resuspended in 0.01% acetic acid as recommended by the manufacturer and served as a representative of the neutrophil defensins. The cathelicidins LL-37 and CRAMP (murine cathelicidin) were purified as previously described (67). The MICs of HNP-1, LL-37, and CRAMP required to inhibit growth of the isogenic strains and the kinetics of killing were determined using a modification of a previously described method (44). For MIC assays the isogenic strains were grown to logarithmic phase (OD600 of 0.4) in THB, washed, diluted in 10 mM sodium phosphate, pH 7.4, containing 0.2 × TSB, and added to 96-well plates containing serially diluted peptide. The MIC was defined as the lowest concentration of peptide that inhibited growth after overnight incubation at 37°C. For killing kinetics assays, ∼1 × 104 bacteria of the logarithmic phase bacteria were incubated at 37°C in 10 mM sodium phosphate, pH 7.4, and 0.2 × TSB containing HNP-1, LL-37, CRAMP, or acetic acid as a control. The number of surviving bacteria was determined over a 2-h time period by plating serial dilutions of the samples on THA. The percent survival was calculated relative to the bacterial survival in the acetic acid control samples.
Cytochrome c surface charge assay.
The binding of cytochrome c to the surface of the isogenic GBS strains was determined as previously described (47). A wild-type (WT) S. aureus strain (SA113) and an isogenic dltA mutant (AG1) were included as controls. Strains were grown overnight, and the bacteria were collected by centrifugation, washed twice in 20 mM morpholinepropanesulfonic acid, pH 7.0, and concentrated to a final OD600 of 7.0. An aliquot of each strain was incubated with 0.5 mg/ml cytochrome c (Sigma) for 10 min at room temperature. The cells were pelleted, absorbance of the supernatants was measured at 530 nm, and the amount of cytochrome c captured was determined by comparison to a standard curve.
PG purification and analysis.
PG was isolated from cultures of the WT strain and isogenic ponA mutant grown to an OD600 of 0.5 using the previously described method of Meador-Parton and Popham (39) except that group antigen and capsular polysaccharides were first removed as described by Wessels et al. (63). Briefly, bacterial cells were incubated in 0.25 N NaOH for 28 h at 37°C to remove polysaccharides. The insoluble PG was treated with trypsin, washed, and then digested with mutanolysin (Sigma). The muropeptides were reduced with borohydride and analyzed by reverse-phase high-pressure liquid chromatography (HPLC) using the previously described method of Hakenbeck et al. (23). The HPLC system consisted of a Waters 600E controller and pump, a 717 Autosampler, and a 486 UV detector. A Powerchrom hardware and software system (AD Instruments Inc.) on an Apple PowerMacintosh 5400 computer was used for signal integration. Muropeptides were eluted at 0.5 ml/min from a Hypersil ODS (octyldecylsilane) column (3-μm particle size; 250 × 4.6 mm; Keystone Science) for 10 min with 0.05% (vol/vol) trifluoroacetic acid in water and subsequently with a 90-min linear gradient to 20% acetonitrile in 0.035% trifluoroacetic acid. The eluted compounds were detected by absorption at 210 nm. Bacterial cell pellets were acid hydrolyzed and subjected to amino acid/amino sugar analysis as previously described (39). The relative amounts of amino acids and glucosamine (produced from N-acetyl-glucosamine during acid hydrolysis) were normalized to the amount of isoglutamine (iGln). For calculating the PG per ml of culture, the muramic acid content was used as a measure of PG.
Analysis of cell wall integrity and permeability.
Susceptibility of the isogenic strains to lysis following incubation with a cell wall hydrolase, mutanolysin, was used to assess the integrity of the cell wall. Bacterial cells were incubated with mutanolysin (10 to 50 units/ml) in 50 mM NaH2PO4 buffer (pH 6.8) at 37°C, and lysis was monitored by following the decrease in the OD550 of the sample over time as previously described (48). The concentration of the aminoglycoside antibiotics kanamycin and gentamicin required to inhibit growth of the isogenic strains was used as a measure of cell wall permeability. MICs were determined with Etest strips (AB Biodisk), used according to the manufacturer's directions, and confirmed using the broth dilution method (42).
RESULTS
PBP1a does not promote processing of C3 on the bacterial surface.
Capsule is well known for its ability to protect GBS from phagocytic killing by preventing opsonization by C3 and by promoting conversion of active C3b to enzymatically inactive iC3b on the bacterial surface (36). While capsule expression in the ponA mutant is unchanged compared to WT (29), we investigated whether PBP1a also had antiopsonic activity in GBS. In previous studies, the absence of PBP1a was not found to impact the relative amount of C3 deposited on the bacterial surface (29). Since conversion of C3b to iC3b is also another strategy for innate immune invasion, in this study we examined whether PBP1a expression affected the processing of C3 on the bacterial surface.
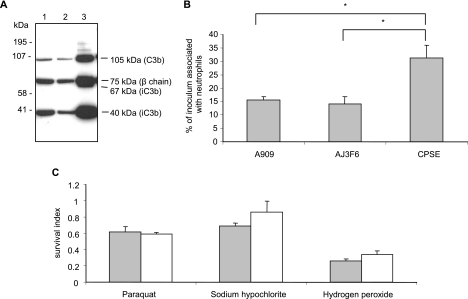
We first confirmed that the antibody that we used could detect the relevant C3 fragments since commercially available anti-C3 antibodies vary in their ability to detect C3 fragments. Comparison of the products that we obtained to published tryptic cleavage profiles (41) revealed that all of the expected processed forms of C3 were generated in assays and were detectable using this antibody (data not shown). As seen in Fig. 1A, the C3 fragments released from equivalent numbers of WT and ponA mutant GBS following opsonization in 50% NHS included the 105-kDa fragment of C3b, the 40-kDa fragment of iC3b and the 75-kDa β-chain that is common to all forms of C3 (Fig. 1, lanes 1 and 2, respectively). The same C3 fragments were released from the opsonized isogenic acapsular mutant (ΔcpsE) (Fig. 1, lane 3) with the addition of the 67-kDa fragment of iC3b. As expected, a significantly greater amount of C3 was detected on the surface of the acapsular mutant. These data indicated that PBP1a does not protect GBS from phagocytic killing by promoting the processing of C3 on the surface of GBS.
FIG. 1.
The mutation in ponA does not affect opsonization, association with neutrophils, or resistance to phagocyte oxidants. (A) Western blot analysis of C3 fragments. Lanes 1 to 3, C3 fragments released by hydroxylamine treatment of opsonized WT bacteria, ponA mutant, and ΔcpsE acapsular mutant, respectively. The identity of the C3 fragments that were released is indicated. (B) Association of GBS strains with human neutrophils. Isogenic strains were incubated with neutrophils at a bacteria-to-neutrophil ratio of ∼9:1. The percentage of the inoculum associated with the neutrophils was determined after 10 min. Data represent the means ± standard deviations of triplicate values and are representative of three independent experiments. *, P < 0.001 (two-tailed t test). (C) Sensitivity of the isogenic strains to ROI. The isogenic strains were incubated with the indicated oxidant, and the survival index was calculated as CFU recovered at the end of the assay/input CFU. Gray bars, WT strain; white bars, ponA mutant. Data represent the means ± standard deviations of triplicate values and are representative of three independent experiments.
PBP1a does not affect association of GBS with human neutrophils.
We investigated whether the absence of PBP1a affected uptake of GBS by neutrophils using a phagocytosis assay. An incubation time of 10 min was chosen to minimize the impact of the differential survival of the isogenic strains in neutrophils that we had previously observed (29). As shown in Fig. 1B, we detected equivalent numbers of WT and ponA bacteria associated with neutrophils following incubation, indicating that PBP1a does not affect the uptake or association of GBS with neutrophils. As a positive control for the phagocytosis assay, we also determined the number of the isogenic acapsular mutant bacteria that were associated with the neutrophils after the same time period. As expected, significantly more ΔcpsE bacteria were associated with the neutrophils compared to WT and ponA strains.
PBP1a is not required for resistance of GBS to killing by oxidants.
To investigate whether the mutation in ponA affected resistance of GBS to the products of the oxidative burst, we compared the survival of the isogenic strains in the presence of the principal oxidants produced by phagocytes: superoxide, hypochlorite, and hydrogen peroxide. As shown in Fig. 1C, there was no difference in susceptibility between the WT strain and the ponA mutant to any of the oxidants that we tested under these conditions.
The absence of PBP1a renders GBS sensitive to killing by neutrophil AMPs.
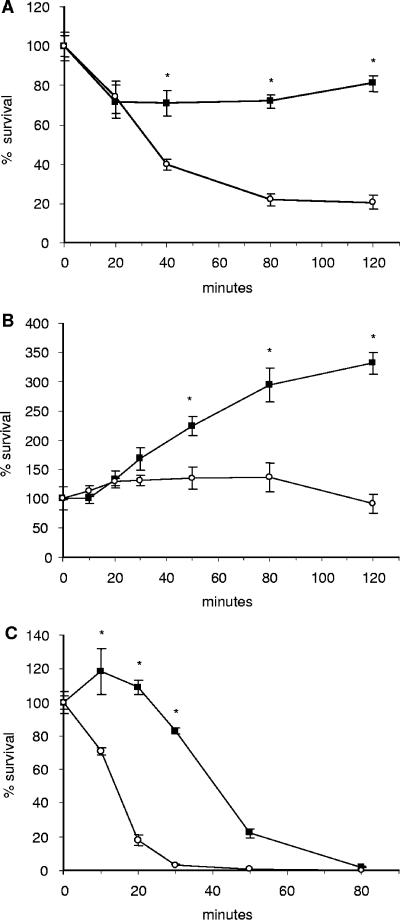
AMPs are a major component of the nonoxidative killing capacity of neutrophils and are present in high concentrations in the lysosomal granules. The AMPs found in human neutrophils include members of the defensin and cathelicidin classes. We compared the sensitivity of the isogenic strains to the defensin, HNP-1, and the cathelicidins, LL-37 and CRAMP, by performing killing kinetic and MIC assays. HNP-1, LL-37, and CRAMP are all cationic with charges ranging from +2 to +7. As seen in Fig. 2, while the kinetics varied depending on the specific AMP and concentration used, in all cases the ponA mutant was significantly more susceptible to the activity of AMPs than the WT strain. Additionally, in MIC assays growth of the ponA mutant was inhibited at lower AMP concentrations compared to the WT strain. In general, the concentration of peptide required to inhibit growth of the ponA mutant was twofold lower than for the WT parent strain (Table 2).
FIG. 2.
Analysis of the activity of AMPs against the isogenic strains using killing kinetics and MIC assays. For kinetic assays, the WT strain (▪) and ponA mutant (○) were incubated with 3 μM HNP-1 (A), 32 μM LL-37 (B), or 16 μM CRAMP (C). The number of viable bacteria was determined over time by plating aliquots of each sample. The MIC of each peptide is shown for the WT strain and ponA mutant. Data represent the means ± standard deviations of triplicate values and are representative of three independent experiments. *, P < 0.05 (two-tailed t test).
TABLE 2.
Activity of AMPs against GBS strainsa
| AMP | Net charge | MIC (μM)
|
|
|---|---|---|---|
| A909 | AJ3F6 | ||
| Gramicidin | 0 | 0.083b | 0.016b |
| HNP-1 | +2 to +3 | 0.8 | 0.4 |
| LL-37 | +6 | >32 | 24 |
| CRAMP | +7 | 8 | 4 |
A909 is wild type; AJ3F6 is a ponA mutant strain.
Gramicidin MICs are in nanomolars.
The absence of PBP1a does not alter surface charge.
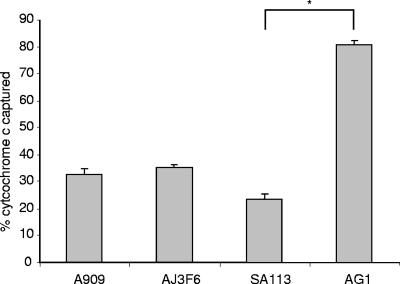
We used a cytochrome c capture assay to compare the effective surface charge of the WT and ponA mutant strains. Cytochrome c is a highly positively charged molecule; thus, binding to bacterial strains is dependent on the negative charge on the surface. This assay is commonly used to model the electrostatic interaction between AMPs and the bacterial surface (30, 47). As shown in Fig. 3, there was no difference in the amount of cytochrome c captured by the isogenic strains, indicating that the mutation in ponA does not alter the effective surface charge of the bacterium. As a control we used an S. aureus strain with a known alteration in surface charge since the corresponding GBS mutant strain was not available. S. aureus AG1 has a mutation in dltA that causes a reduction in the d-alanylation of the lipoteichoic acid and an increased net negative surface charge (47). As expected, S. aureus AG1 captured significantly more cytochrome c than the WT parental strain, SA113. To further investigate whether the electrostatic interaction with AMPs had been disrupted in the ponA mutant, we assessed the sensitivity of the isogenic strains to a neutral peptide antibiotic, gramicidin. While uncharged, gramicidin exerts its bactericidal activity by permeabilizing membranes in a mechanism similar to cationic AMPs (24). The ponA mutant was also more sensitive to gramicidin in our MIC assays than the WT strain (Table 2). These data indicate that the mutation in ponA does not affect the charge on the bacterial surface. Our observations further suggest that unlike previously described mechanisms of AMP resistance in GBS, PBP1a does not promote resistance to AMPs by disrupting the electrostatic interaction between the bacterial surface and the peptides.
FIG. 3.
Binding of the cationic protein cytochrome c to the bacterial strains. Bacterial strains were incubated at a neutral pH with cytochrome c. The amount of cytochrome c remaining in the supernatant following pelleting of the bacterial cells was determined by measuring the absorbance at 530 nm and comparing these values to a standard curve. The data represent the means ± standard deviations of triplicate values and are representative of three independent experiments *, P < 0.05 (two-tailed t test). A909, WT GBS; AJ3F6, ponA GBS mutant; SA113, WT S. aureus; AG1, S. aureus dltA mutant.
The absence of PBP1a does not produce major changes in PG.
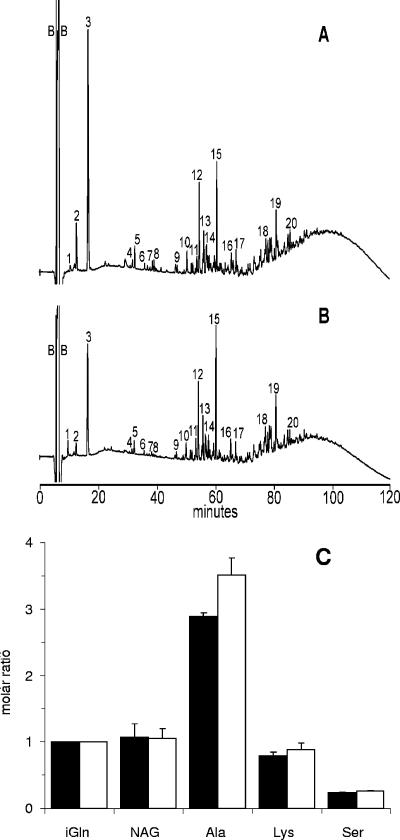
GBS PBP1a is predicted to have both GT and TP activities that contribute to cell wall biosynthesis (29). To determine if there were any changes in the PG that could affect susceptibility to AMPs, we compared the composition of the PG produced by the isogenic strains. Chromatograms of HPLC elution profiles of the muropeptides are shown in Fig. 4A and B. While these analyses did not identify individual muropeptides, peaks 1 to 8 generally represent disaccharides with attached peptides that are not cross-linked. We observed minor decreases in the abundance of some of the smaller muropeptides (Fig. 4B, peaks 1 to 3) from the ponA mutant, relative to the WT strain. The amount of material represented by these peaks is proportionally small relative to that in the later regions of the chromatogram. Thus, any effect of the change in these muropeptides on the overall PG structure would be expected to be minor in comparison to a change in the amount of the highly cross-linked multimers. In the chromatograms, peaks 12 to 15 represent dimers of cross-linked disaccharides, and peaks 16 through 20 and the unresolved mound that elutes at ∼90 min represent highly cross-linked multimers of muropeptide. The reduced size of the mound on the chromatogram for the muropeptides from the ponA mutant suggests that there is a slight reduction in the degree of cross-linking in the PG in this strain.
FIG. 4.
Analysis of PG produced by the isogenic strains. PG was purified and analyzed as described in Materials and Methods. HPLC elution profiles of muropeptides from the WT strain A909 (A) and ponA mutant AJ3F6 (B) are shown. Peaks labeled as 1 to 8 represent disaccharides, peaks 12 to 15 represent dimers of cross-linked disaccharides, and peaks 16 to 20 and the unresolved mound represent highly cross-linked oligomeric components. (C) Amino acid/sugar analysis of PG produced by WT (black bars) and ponA mutant (white bars) GBS. The relative amounts of amino acids and glucosamine (produced from N-acetyl-glucosamine [NAG] during acid hydrolysis) were normalized to the amount of iGln. Analyses were performed on two separate PG preparations, and the data represent means ± standard deviations.
Consistent with previous reports, we identified the amino acids alanine, serine, iGln, and lysine in the PG from both strains (9, 55) using amino acid analysis. However, as seen in Fig. 4C, there was a 22% increase in the amount of alanine in PG from the ponA mutant compared to the WT strain. Similar to many other gram-positive bacteria, the GBS PG stem peptide is composed of the pentapeptide l-Ala-d-iGln-l-Lys-d-Ala-d-Ala (9). Taken together with the muropeptide profiles, our observations suggest that the minor reduction in cross-linking of PG from the ponA mutant results in an increase in stem peptides that retain the d-Ala in position four of the peptide side chains. We also quantitated the muramic acid produced by the isogenic strains as a measure of PG. The two strains produced equivalent amounts of PG (WT GBS produced 29.5 ± 2.1 nmol, and the isogenic ponA mutant produced 28.5 ± 0.7 nmol of muramic acid per ml of culture). Collectively, these data indicate that the absence of PBP1a does not affect the composition or the total amount of PG produced and has only a minor impact on the cross-linking.
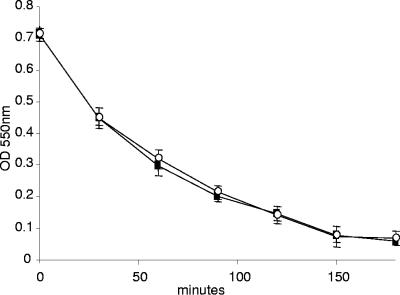
The absence of PBP1a does not affect cell wall integrity or permeability.
To investigate the possibility that the ponA mutant was more sensitive to killing by AMPs due to a change in the integrity of the cell wall, we compared the sensitivity of the isogenic strains to lysis by mutanolysin. Mutanolysin cleaves the β1-4 glycosidic bond between N-acetylmuramic acid and N-acetylglucosamine of the glycan chain (66) and is commonly used to digest the cell wall of gram-positive bacteria, including GBS (9). The isogenic strains were incubated in mutanolysin at a final concentration of 50, 25, or 10 U/ml, and lysis was monitored by measuring the decrease in OD550 over time. Both strains were equally susceptible to lysis during incubation in all concentrations of mutanolysin that we tested. A representative assay using 10 U/ml mutanolysin is shown in Fig. 5. No lysis was observed when the strains were incubated for the same length of time in buffer that did not contain mutanolysin (data not shown). Sensitivity of bacterial strains to aminoglycoside antibiotics is known to be affected by the permeability of the bacterial cell (3); thus, we also compared the sensitivity of the isogenic strains to the bactericidal effects of kanamycin and gentamicin. The MIC of kanamycin was 400 μg/ml for both the WT and ponA mutant strains, and the MIC of gentamicin was 48 μg/ml for both strains. Collectively, these data indicate that the minor decrease in cross-linking that was observed in PG produced by the isogenic ponA mutant does not affect the integrity or permeability of the cell wall.
FIG. 5.
Susceptibility of the isogenic strains to lysis during incubation with mutanolysin. The isogenic strains A909 (▪) and AJ3F6 (○) were incubated in mutanolysin (10 U/ml) at 37°C, and lysis was monitored by following the decrease in OD550 over time. Incubations were performed in triplicate, and the data shown are a representative of triplicate assays.
DISCUSSION
PBPs are traditionally viewed as cell wall metabolic enzymes that play an important role in maintaining cellular integrity and shape. More recently, PBPs have been linked with the virulence of a number of bacterial species (2, 21, 27, 33, 35, 40, 49, 58, 69). In GBS, PBP1a encoded by ponA, was shown to be required for the development of sepsis in a neonatal rat model of infection (28). The attenuated virulence of the ponA mutant correlated with an increased susceptibility to phagocytic killing by neutrophils, suggesting that PBP1a promotes resistance of GBS to innate immune clearance (29). In this study, we examined the sensitivity of our isogenic strains to several components of host innate immune defense in order to determine the role of PBP1a in GBS virulence.
Similar to other bacterial pathogens, effective phagocytosis of GBS by neutrophils and macrophages requires opsonization by specific antibodies or complement (1, 14, 60). In neonates that lack specific antibodies, opsonization with C3 via the alternative complement pathway is essential for effective phagocytic uptake and killing. While the expression of capsule is known to function in immune evasion by inhibiting opsonization with C3 and by promoting the conversion of C3b to enzymatically inactive iC3b (8, 36), PBP1a does not appear to play a similar role. We previously demonstrated that the mutation in ponA did not affect the quantity of C3 deposited on the surface of the ponA GBS mutant (29), and in further studies presented here, we also did not detect any difference in the type of C3 fragments on the surface of the mutant following opsonization in NHS. The absence of PBP1a did not appear to affect the uptake by or association of GBS with neutrophils.
Once bacteria are taken up by neutrophils, they are exposed to their microbicidal components. The major microbicidal products of the neutrophil include the ROI generated during the respiratory burst and the elaboration of AMPs. We were unable to detect any difference in susceptibility of the ponA mutant to killing by ROI. However, the ponA mutant was more sensitive to killing by human neutrophil defensins and the human cathelicidin LL-37, suggesting that PBP1a promotes resistance of GBS to AMPs. The ponA mutant was also more sensitive to killing by cathelicidin purified from mouse neutrophils (CRAMP). Rat neutrophils also express cathelicidin (62) and homologs of HNP-1 to HNP-4 (15); thus, the increased sensitivity of the ponA mutant to killing by AMPs may contribute to the attenuated virulence that we previously observed in the neonatal rat sepsis model. We observed two patterns in our kinetic killing assays depending on the AMP and concentration used. During exposure to HNP-1 and CRAMP, the ponA mutant was killed far more rapidly than the WT GBS strain. When exposed to LL-37 at the concentration that we used, the ponA mutant remained static while the WT strain was able to replicate. In all assays, the ponA mutant was far more sensitive to the inhibitory or bactericidal effects of the AMP. In MIC assays, the concentration of a single AMP required to inhibit growth of the ponA mutant was two- to fourfold lower than for the WT strain, which is consistent with what has been observed for AMP-sensitive mutants from other bacterial species (13, 22). It is important to note that defensins and cathelicidins act synergistically (11) and that a seemingly small difference in MIC in vitro can manifest as a significant change in virulence in vivo (16, 43, 44). Our data support these observations as we have previously shown that the 50% lethal dose for the ponA mutant in the neonatal rat sepsis model is 1 to 2 log units greater than that of the WT strain (29).
GBS PBP1a is class A HMW bifunctional PBP that is predicted to possess both GT activity, which is needed for PG strand polymerization, and TP activity, which is required for cross-linking of peptide side chains on PG strands. As cell wall integrity is critical for bacterial survival, PBPs are thought to be functionally redundant, and inactivation of a single one can often be compensated for by other PBPs (10, 12, 19, 38, 50, 51). In some bacterial species, mutations in class A PBPs can result in minor changes in PG structure. A mutation in Bacillus subtilis ponA resulted in a minor reduction in cell diameter (51), while a mutation in Streptococcus pneumoniae ponA did not produce any detectable changes in morphology (26). Previous examination of the GBS ponA mutant using electron microscopy did not reveal any gross changes in cellular morphology (29). In this study, we used the highly sensitive method of HPLC to analyze the PG produced by the ponA mutant in comparison to the WT strain. While the total amount of PG produced was unaffected, we did detect a minor decrease in the amount of cross-linking of the PG produced by the ponA mutant. While substantial decreases in PG cross-linking are known to increase the sensitivity of bacteria to lysis following incubation with cell wall hydrolases such as mutanolysin (61), the minor decrease in cross-linking that we observed in the PG produced by the GBS ponA mutant did not affect the susceptibility to lysis by mutanolysin. Additionally, we were unable to detect any change in the permeability of the cell wall of the mutant as measured by sensitivity to aminoglycosides antibiotics. Thus, we concluded that the mutation in ponA had not affected the permeability or structural integrity of the cell wall.
Resistance to AMPs is being increasingly recognized as a virulence trait in bacterial pathogens (see reference 45 for a review). To date, the only known mechanism of resistance to AMPs that has been reported in GBS is modification of the surface charge to repel binding of AMPs (54). The surface of bacteria tends to be negatively charged; thus, cationic AMPs are drawn electrostatically to the bacteria. To reduce the net negative charge on the surface, bacteria incorporate d-alanine into the teichoic acid. The genes responsible for d-alanylation of teichoic acid in GBS have been identified as dltABCD (52). The charge on the surface of a dltA mutant is more negative than the WT strain, and the mutant is more susceptible to killing by AMP (54). In this study, we were unable to detect any difference in the surface charge of the ponA mutant using cytochrome c capture assays. Additionally, the ponA mutant was more susceptible than the WT strain to the neutral peptide antibiotic gramicidin. In contrast, mutant bacterial strains that are sensitive to AMPs due to an alteration in surface charge are as sensitive to gramicidin as their wild-type parent (46, 54). Collectively, these data indicate that PBP1a promotes resistance to AMPs by a novel mechanism that does not involve modification of the surface charge. While PBPs are traditionally known for their role in the biosynthesis of PG, our study represents the first demonstration of a role for PBPs in resistance to AMPs of the innate immune system. Studies are under way to characterize the precise mechanism by which PBP1a promotes the resistance of GBS to AMPs.
In summary, these data represent the first description of a mechanism of resistance to AMPs in GBS that is not related to surface charge modification. While PBPs are traditionally known for their PG biosynthetic activity, our study represents the first demonstration of a role for these surface proteins in resistance to AMPs of the innate immune system. PBPs have been reported to impact the virulence of a number of bacterial species; but to date, their role in virulence has not been determined, and no functions other than cell wall metabolic activity have been proposed for PBPs. In vivo virulence screens identified ponA homologs as being required for virulence of S. pneumoniae, S. aureus, Listeria monocytogenes, Mycobacterium tuberculosis, and Salmonella enterica (2, 33, 35, 49, 69). Additionally, PBP1a appears to be upregulated during growth of group A streptococcus (GAS) in vivo during a murine infection, and antibodies directed against PBP1a were detected using in vivo induced antigen technology in convalescent-phase sera from patients with invasive GAS disease (58). Resistance to AMPs is being increasingly recognized as a virulence trait in bacterial pathogens, and it is tempting to speculate that PBP-mediated resistance to AMPs represents a widespread mechanism for evading innate immunity.
Acknowledgments
This work was supported by grant R01AI52299 from the National Institutes of Health awarded to A.L.J., NIGMS grant R01GM56695 awarded to D.L.P., and an American Heart Association Grant awarded to V.N.
The authors thank Marissa Braff and Christy Ventura for helpful discussions and critical review of the manuscript.
Editor: J. N. Weiser
REFERENCES
- 1.Anderson, D. C., B. J. Hughes, M. S. Edwards, G. J. Buffone, and C. J. Baker. 1983. Impaired chemotaxigenesis by type III group B streptococci in neonatal sera: relationship to diminished concentration of specific anticapsular antibody and abnormalities of serum complement. Pediatr. Res. 17:496-502. [DOI] [PubMed] [Google Scholar]
- 2.Autret, N., I. Dubail, P. Trieu-Cuot, P. Berche, and A. Charbit. 2001. Identification of new genes involved in the virulence of Listeria monocytogenes by signature-tagged transposon mutagenesis. Infect. Immun. 69:2054-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, C. J., M. S. Edwards, and D. L. Kasper. 1981. Role of antibody to native type III polysaccharide of group B Streptococcus in infant infection. Pediatrics 68:544-549. [PubMed] [Google Scholar]
- 4.Baker, C. J., and M. W. Edwards. 2005. Group B streptococcal infections, p. 1091-1156. In J. S. Remington, J. O. Klein, C. B. Wilson, and C. J. Baker (ed.), Infectious diseases of the fetus and newborn infant, 6 ed. Elsevier/Saunders, Philadelphia, Pa.
- 5.Baker, C. J., and D. L. Kasper. 1976. Correlation of maternal antibody deficiency with susceptibility to neonatal group B streptococcal infection. N. Engl. J. Med. 294:753-756. [DOI] [PubMed] [Google Scholar]
- 6.Baltimore, R. S., D. L. Kasper, C. J. Baker, and D. K. Goroff. 1977. Antigenic specificity of opsonophagocytic antibodies in rabbit anti-sera to group B streptococci. J. Immunol. 118:673-678. [PubMed] [Google Scholar]
- 7.Butko, P., A. Nicholson-Weller, and M. R. Wessels. 1999. Role of complement component C1q in the IgG-independent opsonophagocytosis of group B streptococcus. J. Immunol. 163:2761-2768. [PubMed] [Google Scholar]
- 8.Campbell, J. R., C. J. Baker, and M. S. Edwards. 1991. Deposition and degradation of C3 on type III group B streptococci. Infect. Immun. 59:1978-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Cueninck, B. J., G. D. Shockman, and R. M. Swenson. 1982. Group B, type III streptococcal cell wall: composition and structural aspects revealed through endo-N-acetylmuramidase-catalyzed hydrolysis. Infect. Immun. 35:572-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denome, S. A., P. K. Elf, T. A. Henderson, D. E. Nelson, and K. D. Young. 1999. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J. Bacteriol. 181:3981-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorschner, R. A., K. H. Lin, M. Murakami, and R. L. Gallo. 2003. Neonatal skin in mice and humans expresses increased levels of antimicrobial peptides: innate immunity during development of the adaptive response. Pediatr. Res. 53:566-572. [DOI] [PubMed] [Google Scholar]
- 12.Duez, C., S. Hallut, N. Rhazi, S. Hubert, A. Amoroso, F. Bouillenne, A. Piette, and J. Coyette. 2004. The ponA gene of Enterococcus faecalis JH2-2 codes for a low-affinity class A penicillin-binding protein. J. Bacteriol. 186:4412-4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edelstein, P. H., B. Hu, F. Higa, and M. A. Edelstein. 2003. lvgA, a novel Legionella pneumophila virulence factor. Infect. Immun. 71:2394-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards, M. S., A. Nicholson-Weller, C. J. Baker, and D. L. Kasper. 1980. The role of specific antibody in alternative complement pathway-mediated opsonophagocytosis of type III, group B Streptococcus. J. Exp. Med. 151:1275-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisenhauer, P. B., S. S. Harwig, and R. I. Lehrer. 1992. Cryptdins: antimicrobial defensins of the murine small intestine. Infect. Immun. 60:3556-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fields, P. I., R. V. Swanson, C. G. Haidaris, and F. Heffron. 1986. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. USA 83:5189-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ganz, T. 2003. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3:710-720. [DOI] [PubMed] [Google Scholar]
- 18.Goffin, C., and J. M. Ghuysen. 2002. Biochemistry and comparative genomics of SxxK superfamily acyltransferases offer a clue to the mycobacterial paradox: presence of penicillin-susceptible target proteins versus lack of efficiency of penicillin as therapeutic agent. Microbiol. Mol. Biol. Rev. 66:702-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goffin, C., and J. M. Ghuysen. 1998. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 62:1079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gordon, D. L., J. Rice, J. J. Finlay-Jones, P. J. McDonald, and M. K. Hostetter. 1988. Analysis of C3 deposition and degradation on bacterial surfaces after opsonization. J. Infect. Dis. 157:697-704. [DOI] [PubMed] [Google Scholar]
- 21.Graham, J. E., and J. E. Clark-Curtiss. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. USA 96:11554-11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groisman, E. A., C. Parra-Lopez, M. Salcedo, C. J. Lipps, and F. Heffron. 1992. Resistance to host antimicrobial peptides is necessary for Salmonella virulence. Proc. Natl. Acad. Sci. USA 89:11939-11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hakenbeck, R., A. Konig, I. Kern, M. van der Linden, W. Keck, D. Billot-Klein, R. Legrand, B. Schoot, and L. Gutmann. 1998. Acquisition of five high-Mr penicillin-binding protein variants during transfer of high-level β-lactam resistance from Streptococcus mitis to Streptococcus pneumoniae. J. Bacteriol. 180:1831-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hancock, R. E. 1999. Host defence (cationic) peptides: what is their future clinical potential? Drugs 57:469-473. [DOI] [PubMed] [Google Scholar]
- 25.Hemming, V. G., R. T. Hall, P. G. Rhodes, A. O. Shigeoka, and H. R. Hill. 1976. Assessment of group B streptococcal opsonins in human and rabbit serum by neutrophil chemiluminescence. J. Clin. Investig. 58:1379-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoskins, J., P. Matsushima, D. L. Mullen, J. Tang, G. Zhao, T. I. Meier, T. I. Nicas, and S. R. Jaskunas. 1999. Gene disruption studies of penicillin-binding proteins 1a, 1b, and 2a in Streptococcus pneumoniae. J. Bacteriol. 181:6552-6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsieh, W. J., and M. J. Pan. 2004. Identification Leptospira santarosai serovar shermani specific sequences by suppression subtractive hybridization. FEMS Microbiol. Lett. 235:117-124. [DOI] [PubMed] [Google Scholar]
- 28.Jones, A. L., K. M. Knoll, and C. E. Rubens. 2000. Identification of Streptococcus agalactiae virulence genes in the neonatal rat sepsis model using signature-tagged mutagenesis. Mol. Microbiol. 37:1444-1455. [DOI] [PubMed] [Google Scholar]
- 29.Jones, A. L., R. H. Needham, A. Clancy, K. M. Knoll, and C. E. Rubens. 2003. Penicillin-binding proteins in Streptococcus agalactiae: a novel mechanism for evasion of immune clearance. Mol. Microbiol. 47:247-256. [DOI] [PubMed] [Google Scholar]
- 30.Kristian, S. A., V. Datta, C. Weidenmaier, R. Kansal, I. Fedtke, A. Peschel, R. L. Gallo, and V. Nizet. 2005. d-Alanylation of teichoic acids promotes group a streptococcus antimicrobial peptide resistance, neutrophil survival, and epithelial cell invasion. J. Bacteriol. 187:6719-6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 32.Lancefield, R. C., M. McCarty, and W. N. Everly. 1975. Multiple mouse-protective antibodies directed against group B streptococci. Special reference to antibodies effective against protein antigens. J. Exp. Med. 142:165-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lau, G. W., S. Haataja, M. Lonetto, S. E. Kensit, A. Marra, A. P. Bryant, D. McDevitt, D. A. Morrison, and D. W. Holden. 2001. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 40:555-571. [DOI] [PubMed] [Google Scholar]
- 34.Liu, G. Y., K. S. Doran, T. Lawrence, N. Turkson, M. Puliti, L. Tissi, and V. Nizet. 2004. Sword and shield: linked group B streptococcal beta-hemolysin/cytolysin and carotenoid pigment function to subvert host phagocyte defense. Proc. Natl. Acad. Sci. USA 101:14491-14496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lowe, A. M., D. T. Beattie, and R. L. Deresiewicz. 1998. Identification of novel staphylococcal virulence genes by in vivo expression technology. Mol. Microbiol. 27:967-976. [DOI] [PubMed] [Google Scholar]
- 36.Marques, M. B., D. L. Kasper, M. K. Pangburn, and M. R. Wessels. 1992. Prevention of C3 deposition by capsular polysaccharide is a virulence mechanism of type III group B streptococci. Infect. Immun. 60:3986-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin, T. R., J. T. Ruzinski, C. E. Rubens, E. Y. Chi, and C. B. Wilson. 1992. The effect of type-specific polysaccharide capsule on the clearance of group B streptococci from the lungs of infant and adult rats. J. Infect. Dis. 165:306-314. [DOI] [PubMed] [Google Scholar]
- 38.McPherson, D. C., A. Driks, and D. L. Popham. 2001. Two class A high-molecular-weight penicillin-binding proteins of Bacillus subtilis play redundant roles in sporulation. J. Bacteriol. 183:6046-6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meador-Parton, J., and D. L. Popham. 2000. Structural analysis of Bacillus subtilis spore peptidoglycan during sporulation. J. Bacteriol. 182:4491-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mei, J. M., F. Nourbakhsh, C. W. Ford, and D. W. Holden. 1997. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol. Microbiol. 26:399-407. [DOI] [PubMed] [Google Scholar]
- 41.Minta, J. O., D. Man, and H. Z. Movat. 1977. Kinetic studies on the fragmentation of the third component of complement (C3) by trypsin. J. Immunol. 118:2192-2198. [PubMed] [Google Scholar]
- 42.National Committee for Clinical Laboratory Standards. 2000. Performance standard for antimicrobial susceptibility testing: tenth informational supplement (aerobic dilution). Approved standard M100-S10. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 43.Nizet, V., and R. L. Gallo. 2003. Cathelicidins and innate defense against invasive bacterial infection. Scand. J. Infect. Dis. 35:670-676. [DOI] [PubMed] [Google Scholar]
- 44.Nizet, V., T. Ohtake, X. Lauth, J. Trowbridge, J. Rudisill, R. A. Dorschner, V. Pestonjamasp, J. Piraino, K. Huttner, and R. L. Gallo. 2001. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature 414:454-457. [DOI] [PubMed] [Google Scholar]
- 45.Peschel, A. 2002. How do bacteria resist human antimicrobial peptides? Trends Microbiol. 10:179-186. [DOI] [PubMed] [Google Scholar]
- 46.Peschel, A., R. W. Jack, M. Otto, L. V. Collins, P. Staubitz, G. Nicholson, H. Kalbacher, W. F. Nieuwenhuizen, G. Jung, A. Tarkowski, K. P. van Kessel, and J. A. van Strijp. 2001. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J. Exp. Med. 193:1067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peschel, A., M. Otto, R. W. Jack, H. Kalbacher, G. Jung, and F. Gotz. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274:8405-8410. [DOI] [PubMed] [Google Scholar]
- 48.Piuri, M., C. Sanchez-Rivas, and S. M. Ruzal. 2005. Cell wall modifications during osmotic stress in Lactobacillus casei. J. Appl. Microbiol. 98:84-95. [DOI] [PubMed] [Google Scholar]
- 49.Polissi, A., A. Pontiggia, G. Feger, M. Altieri, H. Mottl, L. Ferrari, and D. Simon. 1998. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 66:5620-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Popham, D. L., and P. Setlow. 1995. Cloning, nucleotide sequence, and mutagenesis of the Bacillus subtilis ponA operon, which codes for penicillin-binding protein (PBP) 1 and a PBP-related factor. J. Bacteriol. 177:326-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Popham, D. L., and P. Setlow. 1996. Phenotypes of Bacillus subtilis mutants lacking multiple class A high-molecular-weight penicillin-binding proteins. J. Bacteriol. 178:2079-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poyart, C., M. C. Lamy, C. Boumaila, F. Fiedler, and P. Trieu-Cuot. 2001. Regulation of d-alanyl-lipoteichoic acid biosynthesis in Streptococcus agalactiae involves a novel two-component regulatory system. J. Bacteriol. 183:6324-6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Poyart, C., E. Pellegrini, O. Gaillot, C. Boumaila, M. Baptista, and P. Trieu-Cuot. 2001. Contribution of Mn-cofactored superoxide dismutase (SodA) to the virulence of Streptococcus agalactiae. Infect. Immun. 69:5098-5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poyart, C., E. Pellegrini, M. Marceau, M. Baptista, F. Jaubert, M. C. Lamy, and P. Trieu-Cuot. 2003. Attenuated virulence of Streptococcus agalactiae deficient in d-alanyl-lipoteichoic acid is due to an increased susceptibility to defensins and phagocytic cells. Mol. Microbiol. 49:1615-1625. [DOI] [PubMed] [Google Scholar]
- 55.Reinscheid, D. J., C. Stosser, K. Ehlert, R. W. Jack, K. Moller, B. J. Eikmanns, and G. S. Chhatwal. 2002. Influence of proteins Bsp and FemH on cell shape and peptidoglycan composition in group B streptococcus. Microbiology 148:3245-3254. [DOI] [PubMed] [Google Scholar]
- 56.Rubens, C. E., M. R. Wessels, L. M. Heggen, and D. L. Kasper. 1987. Transposon mutagenesis of type III group B Streptococcus: correlation of capsule expression with virulence. Proc. Natl. Acad. Sci. USA 84:7208-7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rubens, C. E., M. R. Wessels, J. M. Kuypers, D. L. Kasper, and J. N. Weiser. 1990. Molecular analysis of two group B streptococcal virulence factors. Semin. Perinatol. 14:22-29. [PubMed] [Google Scholar]
- 58.Salim, K. Y., D. G. Cvitkovitch, P. Chang, D. J. Bast, M. Handfield, J. D. Hillman, and J. C. de Azavedo. 2005. Identification of group A Streptococcus antigenic determinants upregulated in vivo. Infect. Immun. 73:6026-6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schuchat, A. 2001. Group B streptococcal disease: from trials and tribulations to triumph and trepidation. Clin. Infect. Dis. 33:751-756. [DOI] [PubMed] [Google Scholar]
- 60.Shigeoka, A. O., R. T. Hall, V. G. Hemming, C. D. Allred, and H. R. Hill. 1978. Role of antibody and complement in opsonization of group B streptococci. Infect. Immun. 21:34-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Strominger, J. L., and J. M. Ghuysen. 1967. Mechanisms of enzymatic bacteriaolysis. Cell walls of bacteria are solubilized by action of either specific carbohydrases or specific peptidases. Science 156:213-221. [DOI] [PubMed] [Google Scholar]
- 62.Termen, S., M. Tollin, B. Olsson, T. Svenberg, B. Agerberth, and G. H. Gudmundsson. 2003. Phylogeny, processing and expression of the rat cathelicidin rCRAMP: a model for innate antimicrobial peptides. Cell Mol. Life Sci. 60:536-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wessels, M. R., W. J. Benedi, H. J. Jennings, F. Michon, J. L. DiFabio, and D. L. Kasper. 1989. Isolation and characterization of type IV group B Streptococcus capsular polysaccharide. Infect. Immun. 57:1089-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wessels, M. R., C. E. Rubens, V. J. Benedi, and D. L. Kasper. 1989. Definition of a bacterial virulence factor: sialylation of the group B streptococcal capsule. Proc. Natl. Acad. Sci. USA 86:8983-8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilson, C. B., and W. M. Weaver. 1985. Comparative susceptibility of group B streptococci and Staphylococcus aureus to killing by oxygen metabolites. J. Infect. Dis. 152:323-329. [DOI] [PubMed] [Google Scholar]
- 66.Yokogawa, K., S. Kawata, S. Nishimura, Y. Ikeda, and Y. Yoshimura. 1974. Mutanolysin, bacteriolytic agent for cariogenic streptococci: partial purification and properties. Antimicrob. Agents Chemother. 6:156-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zaiou, M., V. Nizet, and R. L. Gallo. 2003. Antimicrobial and protease inhibitory functions of the human cathelicidin (hCAP18/LL-37) prosequence. J. Investig. Dermatol. 120:810-816. [DOI] [PubMed] [Google Scholar]
- 68.Zasloff, M. 2002. Innate immunity, antimicrobial peptides, and protection of the oral cavity. Lancet 360:1116-1117. [DOI] [PubMed] [Google Scholar]
- 69.Zhao, Y., R. Jansen, W. Gaastra, G. Arkesteijn, B. A. van der Zeijst, and J. P. van Putten. 2002. Identification of genes affecting Salmonella enterica serovar Enteritidis infection of chicken macrophages. Infect. Immun. 70:5319-5321. [DOI] [PMC free article] [PubMed] [Google Scholar]