Abstract
Porphyromonas gingivalis, one of the causative agents of adult periodontitis, develops biofilm microcolonies on substrata of Streptococcus gordonii but not on Streptococcus mutans. P. gingivalis genome microarrays were used to identify genes differentially regulated during accretion of P. gingivalis in heterotypic biofilms with S. gordonii. Thirty-three genes showed up- or downregulation by array analysis, and differential expression was confirmed by quantitative reverse transcription-PCR. The functions of the regulated genes were predominantly related to metabolism and energy production. In addition, many of the genes have no current known function. The roles of two upregulated genes, ftsH (PG0047) encoding an ATP-dependent zinc metallopeptidase and ptpA (PG1641) encoding a putative tyrosine phosphatase, were investigated further by mutational analysis. Strains with mutations in these genes developed more abundant biofilms with S. gordonii than the parental strain developed. ftsH and ptpA may thus participate in a regulatory network that constrains P. gingivalis accumulation in heterotypic biofilms. This study provided a global analysis of P. gingivalis transcriptional responses in an oral microbial community and also provided insight into the regulation of heterotypic biofilm development.
Periodontal diseases are a group of infections characterized by destruction of the supporting structures of the teeth. Porphyromonas gingivalis is a gram-negative anaerobe that is an important pathogen in severe manifestations of these diseases (15, 41). With regard to the disease process, the primary ecological niche of P. gingivalis is in the subgingival area, where toxic products, such as proteases, can readily access the periodontal tissues. However, initial colonization of the oral cavity by P. gingivalis involves attachment to sites remote from the subgingival area, including the supragingival tooth surface (28, 39, 43, 47, 57, 58). Indeed, introduction of P. gingivalis into the mouths of human volunteers results in localization almost exclusively on supragingival surfaces (39). The bacterial inhabitants of the supragingival tooth surface comprise a complex multispecies biofilm (42), and numerous in vitro studies have demonstrated the ability of P. gingivalis to attach to common constituents of the supragingival biofilm, including Actinomyces species and oral streptococci (12, 35). The molecular basis of P. gingivalis adhesion to Streptococcus gordonii has been investigated in some detail and has been shown to be multivalent (3, 4, 23, 24, 45). The P. gingivalis long fimbriae (FimA) bind to glyceraldehyde-3-phosphate dehydrogenase present on the streptococcal surface (27). In addition, the P. gingivalis short fimbriae (Mfa) engage the streptococcal SspA/B (antigen I/II) adhesins (33) through an approximately 80-amino-acid binding epitope of SspA/B termed BAR (11). Coadhesion mediated through these effectors is required for P. gingivalis to accumulate in a heterotypic biofilm with S. gordonii (23).
In contrast to the synergistic relationship between P. gingivalis and S. gordonii, biofilm formation does not occur with P. gingivalis and other oral streptococci, such as Streptococcus cristatus and Streptococcus mutans (23, 56). Thus, pioneer colonizers, such as S. gordonii, can influence the composition of the multispecies plaque biofilm through the specificity of adherence and signaling mechanisms. Indeed, it is becoming evident that in general, biofilm formation proceeds through a series of developmental steps that involve expression of specific sets of genes (9, 37, 46). An increase in biofilm biomass can occur in two ways: through accumulation of planktonic cells from the fluid phase and through proliferation of the cells comprising the biofilm. Our laboratory is interested in the former process, specifically the means by which P. gingivalis cells are recruited from the planktonic phase and accumulate on an S. gordonii substratum. While it has been shown that LuxS-dependent signaling is required for the development of P. gingivalis-S. gordonii biofilm communities (29), little else is known about the range of genes and genetic pathways utilized by P. gingivalis for heterotypic biofilm development.
In this study we used an array-based approach to identify genes of P. gingivalis that are regulated in the context of accumulation with S. gordonii. Transcriptional profiling revealed broadly based changes in gene expression in P. gingivalis. The roles of two of these genes, ftsH (PG0047) encoding an ATP-dependent zinc metallopeptidase and ptpA (PG1641) encoding a putative tyrosine phosphatase, were investigated through construction of deletion mutations. Mutants formed more abundant heterotypic biofilms with S. gordonii, indicating that ftsH and ptpA participate in a regulatory network that restrains biofilm accumulation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used are listed in Table 1. P. gingivalis ATCC 33277 and derivatives of this strain were grown anaerobically at 37°C in Trypticase soy broth supplemented with (per liter) 1 g of yeast extract, 5 mg of hemin, and 1 mg of menadione. When necessary, erythromycin was added to the medium at a final concentration of 10 μg/ml. Solid medium was prepared by supplementation with 5% sheep blood and 1.5% agar. S. gordonii DL1 and S. mutans KPSK2 were cultured under static conditions in Trypticase soy broth supplemented with 5 g of yeast extract per liter and with 0.5% glucose as a carbon source. Escherichia coli DH5α was grown in Luria-Bertani broth containing 100 μg/ml ampicillin when necessary.
TABLE 1.
Bacterial strains
| Strain | Source or reference |
|---|---|
| P. gingivalis ATCC 33277 | American Type Culture Collection |
| P. gingivalis 33277 ΔftsH | This study |
| P. gingivalis 33277 ΔptpA | This study |
| S. gordonii DL1 | 25 |
| S. mutans KPSK2 | Laboratory stock |
| E. coli DH5α | Invitrogen |
Genetic techniques.
General recombinant DNA and RNA techniques were performed as described by Sambrook and Russell (36), unless indicated otherwise, and in accordance with the manufacturers' recommendations for DNA isolation, restriction enzyme digestion (Promega, Madison, WI), ligation (NEB, Ipswich, MA), plasmid purification (QIAGEN, Valencia, CA), and RNA isolation (Invitrogen, Carlsbad, CA). For Southern blotting, DNA probes were labeled and hybridization was detected using the Gene Images AlkPhos direct labeling and detection system (Amersham, Piscataway, NJ). Standard PCR experiments were performed using the Taq polymerase system (Eppendorf, Westbury, NY). For cloning and sequencing experiments a high-fidelity enzyme, PfuTurbo (Stratagene, La Jolla, CA), was used. PCR products that required sequencing were cloned into pGEM-T (Promega), and nucleotide sequencing was performed by the University of Florida Sequencing Core using an ABI Prism 377XL automated DNA sequencer.
Isolation of RNA from P. gingivalis-Streptococcus consortia.
Cells of P. gingivalis, S. gordonii, and S. mutans were washed and resuspended in prereduced phosphate-buffered saline (PBS) to a final concentration of 1 × 108 CFU/ml. A total of 2 × 109 P. gingivalis cells were incubated anaerobically with an equal number of either S. gordonii or S. mutans cells for 40 min at 37°C. Cells were recovered by centrifugation and processed immediately for RNA extraction. Total RNA was isolated in triplicate independent experiments from P. gingivalis-S. gordonii or P. gingivalis-S. mutans consortia. Bacterial cells were lysed using Trizol (Invitrogen) as described by the manufacturer for gram-negative bacteria. RNA was extracted with phenol-chloroform and precipitated with isopropanol. RNA preparations were washed with 70% ethanol, dissolved in RNase-free H2O, and treated with RNase-free DNase I (Ambion, Austin, TX). The RNA was then purified on RNeasy columns (QIAGEN). Reverse transcription (RT)-PCR was performed with primers for the S. gordonii sspB gene or the S. mutans pac gene to verify that the RNA preparation did not contain mRNA derived from the streptococci. Furthermore, a conventional PCR with primers for fimA was performed to confirm the absence of DNA.
Microarray RNA labeling.
cDNA was synthesized from 8 μg of P. gingivalis RNA in a solution containing 2 μl of random hexamer primers (3 mg/ml; Invitrogen) in RNase-free water (final volume, 18.5 μl). After denaturation at 70°C for 10 min, reverse transcription was accomplished with 2 μl of SuperScript III reverse transcriptase (200 U/μl), 3 μl of 0.1 M dithiothreitol, and 0.6 μl of 50× aminoallyl-labeled nucleotides in 6 μl of 5× First Strand buffer. After incubation at 42°C for 16 h, RNA was hydrolyzed with 10 μl of 1 M NaOH and 10 μl of 0.5 M EDTA at 65°C for 15 min, and the pH was neutralized with 25 μl of 1 M Tris (pH 7.4). cDNA was purified with a QIAquick PCR purification kit (QIAGEN) and dried with a speed vac. Labeling of cDNA was performed in 4.5 μl of 0.1 M carbonate buffer (pH 9.0) with 4.5 μl of the appropriate N-hydroxysuccinimide-Cy (Cy3 or Cy5) suspended in dimethyl sulfoxide. The reaction mixture was incubated for 1 h in the dark at room temperature, and the reaction was stopped with 35 μl of 100 mM sodium acetate (pH 5.2). Labeled cDNA probes were purified with QIAGEN PCR spin columns, combined, and dried with a speed vac.
P. gingivalis microarray slide hybridization.
P. gingivalis genome microarray slides (TIGR) were washed in 1% sodium dodecyl sulfate (SDS) for 2 min, rinsed in distilled water for 10 s, and incubated at 95°C for 2 min. Prehybridization was performed in a solution containing 1% bovine serum albumin, 0.1% SDS, 5× SSC, and 10 mM EDTA at 42°C for 45 min (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). The slides were rinsed in distilled water, followed by isopropanol, and air dried. The probes were resuspended in 7.6 μl of water containing 1 μl of 0.5 M EDTA and 1 μl of salmon sperm DNA. After denaturation at 95°C for 5 min, 20 μl of formamide, 10 μl of 20× SSC, and 0.4 μl of 10% SDS were added. Labeled probe was applied to a prehybridized microarray slide, which was placed in a sealed hybridization chamber, to which 20 μl of 5× SSC was added. Hybridization was performed in the dark at 42°C in a water bath for 18 h. After removal from the hybridization chamber, slides were washed first in low-stringency buffer containing 1× SSC and 0.2% SDS at 42°C and then in high-stringency buffer containing 0.1× SSC and 2% SDS at room temperature for 4 min. Finally, the slides were washed twice at room temperature in 0.1× SSC. The slides were dipped briefly in water and air dried.
Slide scanning and data analysis.
Microarrays were scanned with a GenePix 4000B scanner operating at 532 nm and 635 nm to excite Cy3 and Cy5, respectively. Data from each fluorescence channel were collected and stored as a separate 16-bit TIFF image. The images were analyzed to calculate the relative levels of expression of each gene and to identify differentially expressed genes using TIGR-Spotfinder 1.0 and TIGR ArrayViewer (www.tigr.org). Only spots with intensities in both channels that were 2 standard deviations above the background were included in the final analysis. For statistical analyses results were expressed as averages ± standard deviations, and differences in gene expression were evaluated by a two-tailed t test.
Construction of P. gingivalis mutant strains.
Mutations in ptpA (PG1641) and ftsH (PG0047) were obtained by allelic replacement, and the mutant alleles were constructed by using a PCR fusion technique. The primers used are listed in Table 2. For ptpA, a DNA sequence containing 999 bp upstream of the ptpA ATG initiation codon was amplified from P. gingivalis ATCC 33277 chromosomal DNA using primers 1642 lower and 1642 upper. The 1,010-bp region downstream of the stop codon was amplified using primers dinF upper and dinF lower. To replace the ptpA gene, an ermF cassette was constructed using primers that exhibited 5′ homology to primers 1642 upper and dinF upper. A fusion PCR product was produced using the technique of Kuwayama et al. (22). The final fusion product was cloned into the pGEM-T vector (Promega) and sequenced through the fusion region using the ermF start and ermF stop sequencing primers. Once the construct was confirmed, the plasmid was linearized with ScaI and introduced into P. gingivalis ATCC 33277 by electroporation (33). A double-crossover recombination event was selected by plating on Trypticase soy agar supplemented with yeast extract and erythromycin (10 μg ml−1). Insertion of the replacement allele was confirmed by PCR and Southern hybridization, and the resulting mutant was designated ΔptpA (ΔPG1641). The same fusion technique was used to create the ftsH mutant. Primers PG47A and PG47B and primers PG47C and PG47D were used to amplify the 1,072-bp region upstream of the ftsH open reading frame and the 896-bp region downstream of the ftsH open reading frame. Primers ermF-PG47B and ermF-PG47C contained 5′ ends homologous to primers PG47B and PG47C, respectively, and 3′ ends homologous to the ermF coding region. The resulting mutant was designated ΔftsH (ΔPG47).
TABLE 2.
Oligonucleotides used for mutant construction and sequencing
| Primer | Sequence (5′-3′)a |
|---|---|
| 1642 upper | AAGCGGATTATCGAAGCAAAGGT |
| 1642 lower | TACGCAATGCCGAAGCACTGGAG |
| dinF upper | ATCCGCATGCGACTGACCGCGGGGC |
| dinF lower | GGGAACAGCAGATAGAGCAAGGTG |
| ermF-1642 | CCTTTGCTTCGATAATCCGCTTATGACAAAAAAGAAATTGCCCG |
| ermF-dinF | GCCCCGCGGTCAGTCGCATGCGGATCTACGAAGGATGAAATTTTTCAGG |
| ermF start | ATGAACAGTAAGAAACCCCT |
| ermF stop | CTGTCAAATCAGCCCTGTTA |
| PG47A | GCGCCAGCAAGGGCGAAGGGTTAGGGAATA |
| PG47B | ATTGTTTGCGCAGTTTTATTTTTTCTACTT |
| PG47C | AATATAAAGAGACTATCGAGCTTACGTTAT |
| PG47D | CCGGGAAAAATGCTTACAATCCGAAGATCA |
| ermF-PG47B | AAGTAGAAAAAATAAAACTGCGCAAACAATATGACAAAAAAGAAATTGCCCG |
| ermF-PG47C | ATAACGTAAGCTCGATAGTCTCTTTATATTCTAGGAAGGATGAAATTTTTCAGG |
Underlined sequences hybridize to the ermF gene.
Quantitative RT-PCR.
Real time RT-PCR was used to confirm differential gene expression observed in the microarrays and to verify that the mutational strategy did not disrupt expression of the genes downstream of ptpA or ftsH. The primers were designed using the Beacon Designer 2.0 software (Premier Biosoft, Palo Alto, CA) and are listed in Table 3. cDNA was synthesized from RNA using Superscript III and the reverse primer for each gene. In experiments to validate microarray data, PG0178, a gene that was not regulated by array analysis, was used as a control. Specific DNA standards for each gene under investigation were synthesized from chromosomal DNA using standard PCR methods and were visualized by gel electrophoresis to verify that a single specific product had been generated. Each product was purified using a QIAquick PCR purification kit and was quantified using an Eppendorf BioPhotometer. The DNA product copy number was calculated using the formula of Yin et al. (60): starting quantity (number of copies μl−1) = (6.023 × 1023 × [DNA g/ml]/molecular weight of product [bp × 6.58 × 102 g])/1,000.
TABLE 3.
Real-time PCR primers used for validation of array data and quantitation of operon gene expression
| Open reading framea | Directionb | Primer sequence (5′-3′) | Amplicon size (bp) |
|---|---|---|---|
| PG0026 | F | TCGCATCGCAGTGGACAAGAC | 158 |
| R | GCATGATGGCAGGAGCATAACG | ||
| PG0046 | F | ACAGTGAACAGCAGCCCTCCGATAAAGC | 186 |
| R | GGAGTTGGCAGTAGGAGAAGTGGCAGAC | ||
| PG0047 | F | CGTCGCAGCATCGCCATCC | 162 |
| R | CAGAGCCTCCGTTGTCGTGATC | ||
| PG0064 | F | GCCCATTTCGCACCGCATC | 168 |
| R | TCCACCTGCTGCTCCACATTC | ||
| PG0082 | F | GGTCGTTGCGTGCGGATG | 153 |
| R | GCGTACTCTGTGCCCGAAG | ||
| PG0083 | F | ACTGAATCCGATGGTGCAACTG | 153 |
| R | TAAGGCGAGTCCGACACGATAG | ||
| PG0104 | F | GCAGACGGACGATGATGGAGAG | 150 |
| R | ATGGCACGCAGGAGTGTAGC | ||
| PG0178 | F | CTCAATGACATCGCAGGCAAGG | 150 |
| R | AGCCGCCGTAGCAGGATTTG | ||
| PG0258 | F | GCGAGAGAAGCGTGCCATTGTG | 195 |
| R | TTTACCCCGCCTGCTACGATGC | ||
| PG0273 | F | CCCTCACGATACTCCGCAAATC | 167 |
| R | GCAGAGCACGATCCGATAGC | ||
| PG0275 | F | GGCTTCTCTGCTCCTCCTCTC | 184 |
| R | CCATTCTTGGGGATTGGCTGTG | ||
| PG0296 | F | TGTAGTAGGCGGTCTGGAGGTG | 173 |
| R | CTTGGGTCGGCTTGGCTATCG | ||
| PG0393 | F | GCTGCTCCTGGTGCTGCC | 169 |
| R | CGGTAGAAGGTGCTGCGTCC | ||
| PG0432 | F | GCAGGCAGCAGCGTATCC | 159 |
| R | TGAGAACATCCAAAGCCACAGG | ||
| PG0434 | F | GAACTGTCAGCCGTCCGTATTG | 168 |
| R | CCCTCTCTGCCACTGCTTCTG | ||
| PG0514 | F | GATACCGCCGTCTACCAAAACC | 192 |
| R | GCCGTTACCAGTCAGAGCATTG | ||
| PG0548 | F | TCGCAGCCAATGTGAAGAACTC | 179 |
| R | TGTAGAAGGAGCCGAAGCAGAG | ||
| PG0575 | F | CAGGGCAGGTAAAGGCTATCAC | 196 |
| R | CCTACGGAGAAGAGACCATTGC | ||
| PG0620 | F | GTGCTGGAGATGCCCGATGG | 178 |
| R | CCTGTATGGTGGAGACGAGTGC | ||
| PG0665 | F | CAGAAGAAGCCAAGCACAGACC | 151 |
| R | TCCACGCACACCTCTACGAAG | ||
| PG0686 | F | CTGGATGTAGCCGAAGGAAAGC | 186 |
| R | CTTGGGCGGATGGCAGTTG | ||
| PG0699 | F | TCGGCATCGGTGGCATCC | 171 |
| R | AGAGCGAGGAGGAACGAACAG | ||
| PG0707 | F | GTAGGGGTTGTAGGCGGTGAG | 181 |
| R | GCTGCTCCTCGTTGGTGAATAC | ||
| PG0759 | F | TCGTTACCGCCGCAGCATC | 150 |
| R | GCAGCCTCTTCGTCCGTCAG | ||
| PG0788 | F | CACGAGTATTGGCACGACGAC | 200 |
| R | TCGCACAGTATCGGCAATGAAG | ||
| PG0889 | F | CAGAGTAGAGGAGGCGTATTGC | 194 |
| R | CCGTGGATGGCAGCAGTC | ||
| PG1010 | F | GCTCTTGCTGATGGACGAACC | 198 |
| R | TCGGCTACGGACCTGTTGC | ||
| PG1017 | F | GACCAACGACCTTACGCAGATG | 199 |
| R | ACCGCAGATACCGCACTTCGA | ||
| PG1036 | F | CCCGATGTATTTGCTCCGTGTG | 192 |
| R | ATAGCCCAAACCGACCTCCTG | ||
| PG1095 | F | AGACATCCTGACCAACGACTTC | 198 |
| R | CGGTATCTGCCATCTGCCATC | ||
| PG1116 | F | ACCTCCTCGCTCATTCGCTAC | 182 |
| R | TGAAATCCGTCCACATCCTTGC | ||
| PG1129 | F | GAACTCCGCCCTCACATCCAC | 177 |
| R | GCTCCCGTCACCTTGCCTTC | ||
| PG1321 | F | ACGGAAATCAAGGCACCCAATG | 168 |
| R | CCCGATACAATGCTCACGAACG | ||
| PG1545 | F | TTGGTTCGGGCTGGGTATGG | 162 |
| R | CGGCACGACGATTCTGGTAAG | ||
| PG1551 | F | AGAGAAATACTGCCACGTTTCG | 156 |
| R | ATCATTTTCTCCGCACTCTGTG | ||
| PG1640 | F | CTCTCGTCCGATGGTCTCCTG | 200 |
| R | TCTTCGCCTTCCTCCGTATGG | ||
| PG1641 | F | TTCAGCAGTAGCGGTATTCACG | 150 |
| R | TGCGGATAGGGAGGAGTTGTC | ||
| PG1824 | F | TTCCTTCGCCGTGGTATCGC | 199 |
| R | TCTGTCCCGAGTTGGTTGCTAC | ||
| PG1841 | F | GCTATTGTGGAGGGCTGTCTG | 186 |
| R | ACATCGGTGAATATCGGGAGTC | ||
| PG2205 | F | CGGCAGGAGGTCTACGAGTG | 194 |
| R | GTTGAGCAAGGGCAGGATCTTC | ||
| sspB | F | CCACCAGTTGTACCGACAGTTC | 213 |
| R | GCCAGTTGGAAGCGGATCTAC | ||
| pac | F | TGGAGCCAGCACCTGTTGAG | 243 |
| R | CAGTCGGCGGTGTTGGAATAAC |
Locus number from TIGR (www.tigr.org).
F, forward; R, reverse.
An eightfold dilution series of each DNA standard was prepared for starting quantities of 108 to 101 copies μl−1. The resulting preparations were used in duplicate in each real-time PCR assay to allow the real-time PCR software to estimate the starting quantities of the gene in cDNA samples. The standard DNA dilution series (starting quantities, 108 to 101 copies μl−1) or cDNA templates (2 μl) were added in duplicate to an iCycler iQ 96-well PCR plate (Bio-Rad, Hercules, CA). RNA extracts were prepared in triplicate from independent experiments, and cDNA samples were loaded in duplicate. To each well, the following were added: 1 μl of 5′-specific primer and 1 μl of 3′-specific primer (50 pmol each); 12.5 μl of iQSYBRGreen Supermix (Bio-Rad) containing 100 mM KCl, 40 mM Tris-HCl (pH 8.4), 0.4 mM dATP, 0.4 mM dCTP, 0.4 mM dGTP, 0.4 mM dTTP, iTaq DNA polymerase (50 U ml−1), 6 mM MgCl2, 20 nM fluorescein, and stabilizers; and 8.5 μl of distilled H2O, which resulted in a final volume of 25 μl per well. The 96-well plate was sealed with optical tape, and samples were quantified with the iCycler (Bio-Rad) using a standard thermal cycling program. Real-time results were analyzed using the iCycler iQ optical system software, version 3.0a (Bio-Rad). The melting curve profile was analyzed to verify that there was a single peak for each sample, indicating primer specificity.
P. gingivalis-S. gordonii biofilms.
Heterotypic P. gingivalis-S. gordonii biofilms were generated as described previously (21), and quantitative and structural analyses of these communities were performed by confocal scanning laser microscopy and subsequent image analysis. S. gordonii was stained with hexidium iodide (15 μg ml−1; Molecular Probes, Carlsbad, CA) and then cultured anaerobically at 37°C for 16 h in individual chambers of a Culture Well chambered coverglass system (Grace Bio-Labs, Bend, OR). P. gingivalis was stained with 5-(and 6)-carboxyfluorescein, succinimidyl ester (4 μg ml−1; Molecular Probes), and 2 × 106 cells in prereduced PBS were allowed to react with the S. gordonii biofilm for 24 h anaerobically at 37°C in the dark on a rotator. After washing, the heterotypic biofilms that developed on the coverglass were observed with a Bio-Rad MRC1024 confocal laser scanning microscope (Kr/Ar) system with an Olympus IMT-2 inverted light microscope and an MS plan 40 × 0.85 NA objective using reflected laser light at wavelengths of 488, 546, and 647 nm. A series of fluorescent optical x-y sections in the z plane to the maximum vertical extent of the biofilm was collected with the Laser Sharp software. Images were digitally reconstructed (x-z section and z projection of x-y sections) with Image J V1.33u (National Institutes of Health). P. gingivalis-specific fluorescence and volume were then quantified using the Segmentation/Analysis functions of the daime software (10).
RESULTS
Transcriptome analysis of P. gingivalis from mixed communities.
A microarray approach was used to investigate genes of P. gingivalis regulated by specific interactions with its biofilm partner S. gordonii. As a control for nonspecific gene regulation in the model system and for non-species-specific bacterium-bacterium interactions, the transcriptome of P. gingivalis in association with S. mutans was also investigated. In order to focus on the early developmental stages of heterotypic P. gingivalis-S. gordonii biofilm formation (8, 21, 23), RNA was extracted from P. gingivalis after 40 min of reaction with the streptococci under conditions that preceded biofilm formation. Genes differentially regulated by P. gingivalis in association with S. gordonii with a >1.5-fold change at a P value of <0.05 are listed in Table 4. Thirty genes were upregulated, and three genes were downregulated. Quantitative RT-PCR confirmed that there was differential expression of these genes. The ratios were in the range from 1.5 to 3.1, as determined by both techniques. Such changes in mRNA levels are generally considered to have the potential to be biologically relevant (16, 40). Furthermore, small changes in the level of expression of one gene can be amplified through regulatory networks and result in significant phenotypic alteration (49). Functional classes containing regulated genes spanned a number of categories, indicating that adaptation of P. gingivalis to a community lifestyle requires broadly based transcriptional modulation.
TABLE 4.
P. gingivalis genes differentially expressed during development of a community with S. gordonii as opposed to S. mutans
| Open reading framea | Genea | Protein or functiona | Expression ratiob
|
|
|---|---|---|---|---|
| Microarray | Quantitative RT-PCR | |||
| PG0026 | Conserved hypothetical | 2.22 | 1.8 | |
| PG0047 | ftsH | Cell division, ATP-dependent zinc metallopeptidase | 1.57 | 2.0 |
| PG0064 | czcA | Cation efflux pump | 1.87 | 1.88 |
| PG0082 | Conserved hypothetical | 1.88 | 1.88 | |
| PG0083 | Hypothetical | 1.73 | 1.7 | |
| PG0104 | topB | DNA topoisomerase III | 1.56 | 1.68 |
| PG0258 | abcX | ABC transporter, ATP-binding protein | 1.96 | 1.87 |
| PG0273 | Hypothetical | 1.88 | 1.57 | |
| PG0275 | trxA | Thioredoxin | 1.90 | 2.05 |
| PG0432 | nol1 | Proliferating cell nucleolar protein/rRNA methylase | 2.04 | 2.79 |
| PG0434 | Hypothetical | 1.71 | 2.03 | |
| PG0491 | Conserved hypothetical | 1.82 | 1.74 | |
| PG0514 | secA | Preprotein translocase subunit A protein | 1.62 | 1.53 |
| PG0548 | nifJ | Pyruvate ferredoxin/flavodoxin oxidoreductase | 1.99 | 1.85 |
| PG0620 | lon | ATP-dependent protease LA | 1.86 | 1.59 |
| PG0665 | lacZ | Beta-galactosidase | 1.74 | 1.53 |
| PG0686 | Conserved hypothetical | 1.79 | 2.5 | |
| PG0699 | malP | Maltodextrin phosphorylase | 2.44 | 1.71 |
| PG0707 | TonB-dependent receptor, putative | 3.12 | 3.13 | |
| PG0788 | Hypothetical | 1.90 | 1.65 | |
| PG0889 | Peptidase, M24 family | 1.62 | 1.8 | |
| PG1017 | ppdk | Pyruvate phosphate dikinase | 1.95 | 1.7 |
| PG1036 | uvrA | Excinuclease ABC, A subunit | 2.39 | 2.52 |
| PG1116 | folD | Methylenetetrahydrofolate dehydrogenase | 1.67 | 1.77 |
| PG1129 | nrd | Ribonucleotide reductase alpha subunit | 2.10 | 2.46 |
| PG1321 | fhs | Formate-tetrahydrofolate ligase | 2.16 | 2.46 |
| PG1545 | sod | Superoxide dismutase, Fe-Mn | 1.81 | 1.5 |
| PG1641 | ptpA | Phosphotyrosine phosphatase | 1.69 | 2.06 |
| PG1841 | Conserved hypothetical | 1.62 | 1.86 | |
| PG2205 | panE | 2-Dehydropantoate 2-reductase | 2.04 | 2.21 |
| PG1010 | yvfR | ABC transporter, ATP-binding protein | −1.69 | −1.77 |
| PG1551 | hmuY | Iron transport | −1.65 | −1.59 |
| PG1824 | eno | Enolase | −1.54 | −1.82 |
Information from TIGR (www.tigr.org).
Fold expression after interaction with S. gordonii or S. mutans. All P values were <0.05 for the microarray analysis and <0.001 for the quantitative RT-PCR analysis, as determined by a t test.
Genetic organization of the PG0047 and PG1641 loci.
Changes in gene expression provide a useful means to identify potentially relevant participants in a biological process. In order to begin to assess the functionality of the differentially regulated genes in more detail, two genes were selected for further study. We are particularly interested in genes that may comprise part of master regulatory pathways for biofilm formation and have the potential to impact the activity of several biofilm effector molecules. PG0047 encodes a predicted FtsH protein, an outer-membrane-associated ATP-dependent zinc metalloprotease (17). In other organisms, FtsH is involved in membrane stability and can degrade a set of short-lived proteins, enabling cellular regulation at the level of protein stability and turnover (17). PG1641 encodes a predicted low-molecular-weight phosphotyrosine phosphatase, an enzyme that could play a role in signal transduction or enzyme modification within P. gingivalis. We hypothesized that both these gene products could play important regulatory roles in P. gingivalis community interactions with S. gordonii.
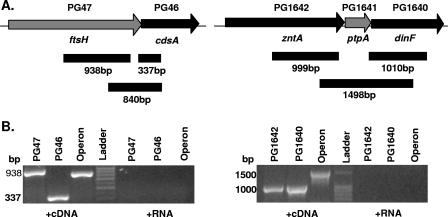
Both ftsH and ptpA are located in putative operons (Fig. 1A), based on the close proximity of their contiguous open reading frames in the sequenced strain W83. ftsH is the first gene in a potential two-gene operon and is followed by a gene encoding phosphatidate cytidylyltransferase (cdsA), a class of enzyme which is membrane bound and involved in phospholipid metabolism. In the PtpA potential operon, ptpA is preceded by zntA, encoding a predicted cation ATPase belonging to the E1-E2 family, and is followed by dinF, encoding an integral membrane protein with 10 predicted membrane-spanning regions. The E. coli homolog of dinF is a MATE family efflux pump, and expression is induced by DNA damage. To confirm that these loci were cotranscribed in operons, we performed PCRs with cDNA produced from wild-type P. gingivalis ATCC 33277 RNA. PCRs were performed individually for the first gene and last gene of each operon and for a region spanning all genes of the operon. Polycistronic RT-PCR products were detected in both regions (Fig. 1B), confirming that the genetic loci of interest are both cotranscribed in an operon. The results also show that the genetic organization of these loci in strain ATCC 33277 is similar to the genetic organization published in the W83 sequence databases (www.tigr.org and www.oralgen.lanl.gov).
FIG. 1.
Gene organization of PG47 and PG1641 regions of the P. gingivalis chromosome. (A) Schematic diagram of the genomic regions surrounding the genes of interest. In W83, PG46 and PG47 are separated by 30 bp and PG1641 is separated from PG1642 and PG1640 by 9 bp and 6 bp, respectively. PCR primers were designed to determine the presence of mRNA spanning PG47 and PG46 or PG1642, PG1641, and PG1640. The solid bars indicate the regions amplified from cDNA. (B) RT-PCR products from P. gingivalis ATCC 33277, showing cotranscription of PG47 and PG46 (left panel) or cotranscription of PG1642, PG1641, and PG1640 (right panel). cDNA was synthesized using gene-specific primers and then amplified to detect potential operon products. Individual gene products and operon products were amplified with unique primer sets. Negative controls were amplified from RNA that was not reverse transcribed.
Construction of nonpolar PG0047 and PG1641 mutants.
As ftsH and ptpA are located in operons containing other genes likely to be involved in outer membrane structure or function, we used a mutagenesis technique that allowed us to specifically replace one gene in the operon with minimal polar effects. Mutations were constructed by allelic replacement of the open reading frame of interest with the ermF open reading frame. Essentially, this deleted the P. gingivalis gene from the start codon to the stop codon and replaced it with the ermF open reading frame that did not contain termination sequences. This allowed the erythromycin cassette to be expressed as part of the operon, thus avoiding disruption of downstream genes. To confirm that this mutagenic technique preserved the expression of the terminal genes of the two operons, we used quantitative RT-PCR to determine the levels of transcription of either dinF or cdsA in parental and mutant strains. As shown in Table 5, the levels of expression of both genes were comparable for the wild type and the isogenic mutants. Thus, the mutant strains allowed the effect of the loss of the gene of interest to be investigated without pleiotropic effects on cotranscribed genes.
TABLE 5.
Levels of transcription downstream of mutated genes
| Strain | PG46 (cdsA) copy no. (103) | PG1640 (dinF) copy no. (106) |
|---|---|---|
| Wild type | 75.3 | 18.7 |
| ΔPG47 | 89.4a | ND |
| ΔPG1641 | NDb | 20.1a |
No statistical difference compared to the wild-type level (P > 0.3).
ND, not determined.
Effect of ftsH or ptpA mutation on heterotypic biofilm architecture.
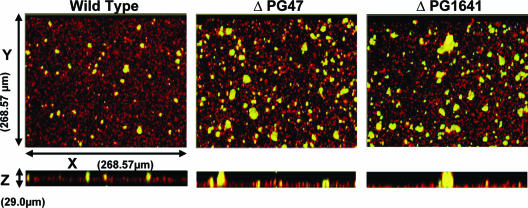
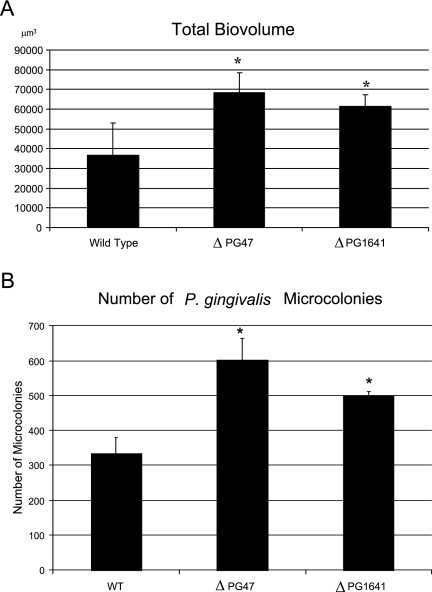
We hypothesized that genes differentially regulated during initial contact between P. gingivalis and S. gordonii would have relevance for the subsequent development of mixed P. gingivalis-S. gordonii communities. The role of ftsH or ptpA in the development of mixed P. gingivalis-S. gordonii biofilms was investigated by confocal laser scanning microscopy. S. gordonii cells were first cultured for 16 h on a glass surface, before exposure to P. gingivalis cells in PBS. As P. gingivalis cells were suspended in buffer, the number of cells did not increase due to cell division during the course of the assay. Hence, this assay modeled one of the early stages of biofilm development, namely, recruitment, coadhesion, and accumulation of planktonic P. gingivalis cells in a biofilm, prior to a further increase in the number of cells through growth and division. x-y and x-z images of the resulting heterotypic S. gordonii-P. gingivalis biofilms are shown in Fig. 2. S. gordonii cells developed accumulations that extensively covered the glass surface, and cells of P. gingivalis formed discrete accumulations clearly separated from each other. This morphology is consistent with previous studies of mixed S. gordonii-P. gingivalis biofilms performed with both saliva-coated and uncoated abiotic surfaces (8, 21, 23). Interestingly, in the absence of either ftsH or ptpA, P. gingivalis formed more extensive biofilm accumulations containing higher numbers of microcolonies (Fig. 2). The P. gingivalis-specific fluorescence and volume were determined using the daime software. The parameters measured were total P. gingivalis biovolume, number of microcolonies, microcolony volume, and microcolony surface/volume ratios. As shown in Fig. 3A, the total biovolumes of the ftsH and ptpA mutants were 86% and 67% greater, respectively, than the total biovolume of the parental strain. In addition, the biofilm containing the ftsH mutant had 76% more microcolonies and the biofilm containing the ptpA mutant had 47% more microcolonies than the wild-type biofilm (Fig. 3B). The increases in the size of the microcolonies formed by the mutant strains were reflected in changes in the surface-to-volume ratios. The median surface-to-volume ratio for the wild-type strain was 3.3. The ftsH mutant had a median ratio of 2.7, while the median ratio for the ptpA mutant was 3.0. Higher surface-to-volume ratios can be assumed to improve nutrient diffusion into the microcolony structures. Thus, these data indicate that the ftsH and ptpA gene products are required for normal development of heterotypic P. gingivalis-S. gordonii biofilms, and in their absence P. gingivalis forms more abundant biofilm accumulations.
FIG. 2.
Mixed biofilm analyzed by confocal laser scanning microscopy. S. gordonii, stained with hexidium iodide (red), was cultured in individual chambers of a coverglass system. P. gingivalis parent and mutant strains were stained with fluorescein (green) and reacted with the S. gordonii biofilms for 24 h. After washing, the mixed-species biofilms that developed on the coverglass were observed with a confocal laser scanning microscope (Kr/Ar) system with a 40× objective. A series of fluorescent optical x-y sections (268.57 μm by 268.57 μm) were collected to create digitally reconstructed images (x-z section and z projection of x-y sections) of the two species with Image J V1.33u. Colocalized bacteria are yellow.
FIG. 3.
Parent and mutant P. gingivalis volume and microcolony number in mixed P. gingivalis-S. gordonii biofilms. (A) Total accumulation of P. gingivalis cells. The bars indicate averages and the error bars indicate standard deviations for three images (268.57 μm by 268.57 μm). (B) Numbers of microcolonies formed by P. gingivalis strains. Microcolonies were defined as clusters of colocalized cells larger than 40 μm3 in an area that was 268.57 μm by 268.57 μm in a representative biofilm. The bars indicate averages, and the error bars indicate standard deviations. WT, wild type. *, P < 0.05 compared to control.
DISCUSSION
Bacteria on host surfaces exist primarily as single-species or multispecies biofilms (44, 46). Patterns of gene expression that characterize distinct developmental stages of biofilm formation have been studied in some detail for single-species biofilms (9, 37, 44, 46, 53). However, less is known about gene expression that is regulated by interspecies interactions in mixed-species biofilms. Oral biofilms are complex multispecies communities that develop through a variety of coadhesive, nutritional, metabolic, and signaling interactions (19, 35). In this study P. gingivalis microarrays were utilized to analyze gene expression in P. gingivalis that is dependent on interactions with S. gordonii. The transcriptome data revealed that approximately 1 to 2% of the P. gingivalis genome was regulated during the initial stages of development of a community with S. gordonii. This degree of adaptation is similar to that reported for single-species biofilms of Pseudomonas aeruginosa and E. coli (38, 53). Higher numbers of biofilm-regulated genes have also been reported (51), and it is likely that factors such as the strain used, the incubation time and conditions, and the array technology used influence the results. It should be noted that our interbacterial community model represents a prelude to biofilm formation and that other transcriptional changes in P. gingivalis are likely to occur during different stages of the development of mature heterotypic biofilms. In addition, P. gingivalis sequenced strain W83 is deficient in biofilm formation, possibly due to impaired production of both the long and short fimbriae (32, 33, 52). Hence, for this study we employed strain ATCC 33277, which produces both fimbrial types, has been used as a model biofilm organism (21, 23, 55), and is pathogenic in the rat model of periodontitis (20, 34). Genomic differences between ATCC 33277 and W83 could limit the ability of W83-based arrays to detect ATCC 33277 gene expression, although array experiments with ATCC 33277 have been performed successfully and the genomes have been reported to be 93% similar (6). Approximately 30% of the P. gingivalis regulated genes are classified as genes that are involved in metabolic pathways, and ∼12% encode transport and binding proteins. These results suggest that the initial adaptation to a heterotypic biofilm involves a shift in metabolic and physiologic status. Both superoxide dismutase and excinuclease were upregulated, indicating that the P. gingivalis cells experience stress as they transition to the biofilm mode. Interestingly, ∼30% of differentially regulated genes were classified as hypothetical genes. These genes, therefore, may have unique biofilm-associated properties.
Beyond the housekeeping and hypothetical genes, two of the genes that have functions consistent with a role in regulatory networks for biofilm control were ftsH and ptpA. FtsH is an AAA+ (ATPases associated with diverse cellular activities plus) ATP-dependent integral membrane protease that is universally conserved in bacteria (17). In addition to its role in the degradation of specific proteins, the ftsH gene of E. coli has been shown to be involved in the processing of inner membrane proteins (1, 2) and in RNA stability (13). Furthermore, FtsH can degrade the transcription factors σ32 and SoxS in E. coli (17). Thus, FtsH can play a role in regulating both the transcriptome and the proteome of bacterial cells. Indeed, in some species FtsH is required for bacterial growth (18), and while the P. gingivalis ftsH mutant initially grew at a rate equivalent to the rate of the parent in batch culture, after multiple passages on agar plates this mutant ceased to grow. For our studies, we used the ftsH mutant freshly streaked from frozen stocks. PtpA is a predicted eukaryote-like low-molecular-weight phosphotyrosine protein phosphatase. The biological role of this class of enzymes has yet to be fully defined; however, in bacteria these enzymes participate in pigment production in Streptomyces coelicolor (48) and in the control of biosynthesis and/or transport of exopolysaccharides in E. coli (54) and Streptococcus pneumoniae (31). Moreover, the phosphoproteome of bacteria is implicated in a wide variety of cellular processes (26). Thus, both FstH and PtpA could participate in regulatory networks that reverberate throughout the transcriptome and expressed proteome and control heterotypic biofilm formation. In order to test this concept, mutant strains deficient in FtsH and PtpA were examined for formation of a heterotypic biofilm with S. gordonii. Under the environmental conditions of a surface-attached mixed biofilm, both mutant strains resulted in more abundant P. gingivalis accumulation than the wild type. Thus, one role of both FtsH and PtpA is in constraining P. gingivalis biofilm development. Regulation of biofilm development involves mechanisms that both stimulate an increase in biomass and limit or stabilize the accumulation according to environmental conditions. For example, in P. aeruginosa, the transcription factor RpoS limits biofilm depth (14, 53), and RpoS mutants of P. aeruginosa form deeper biofilms under flowing conditions (53). RpoS production is regulated at multiple levels, including transcription, translation, and proteolysis, in response to different stress conditions, such as nutrient limitation (50). In S. mutans, Staphylococcus epidermidis, and Helicobacter pylori, the action of the AI-2 synthase LuxS represses biofilm formation (7, 30, 59). There are also examples of biofilm restraint mechanisms in P. gingivalis. The internalin family protein InlJ limits mixed-biofilm accumulation with S. gordonii, and an InlJ mutant strain forms more expansive heterotypic biofilms (5). We speculate that for P. gingivalis in the oral cavity exposure to oxygen or nutrient diffusion could limit the optimal size of biofilm microcolonies; however, this hypothesis requires further investigation.
Acknowledgments
This study was supported by NIDCR grants DE12505 and FAPESP and by the University of Florida Undergraduate Scholars Program.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 21 August 2006.
REFERENCES
- 1.Akiyama, Y., Y. Shirai, and K. Ito. 1994. Involvement of FtsH in protein assembly into and through the membrane. II. Dominant mutations affecting FtsH functions. J. Biol. Chem. 269:5225-5229. [PubMed] [Google Scholar]
- 2.Akiyama, Y., T. Yoshihisa, and K. Ito. 1995. FtsH, a membrane-bound ATPase, forms a complex in the cytoplasmic membrane of Escherichia coli. J. Biol. Chem. 270:23485-23490. [DOI] [PubMed] [Google Scholar]
- 3.Amano, A., T. Fujiwara, H. Nagata, M. Kuboniwa, A. Sharma, H. T. Sojar, R. J. Genco, S. Hamada, and S. Shizukuishi. 1997. Porphyromonas gingivalis fimbriae mediate coaggregation with Streptococcus oralis through specific domains. J. Dent. Res. 76:852-857. [DOI] [PubMed] [Google Scholar]
- 4.Brooks, W., D. R. Demuth, S. Gil, and R. J. Lamont. 1997. Identification of a Streptococcus gordonii SspB domain that mediates adhesion to Porphyromonas gingivalis. Infect. Immun. 65:3753-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Capestany, C. A., M. Kuboniwa, I.-Y. Jung, Y. Park, G. D. Tribble, and R. J. Lamont. 2006. Role of Porphyromonas gingivalis InlJ protein in homotypic and heterotypic biofilm development. Infect. Immun. 74:3002-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, T., Y. Hosogi, K. Nishikawa, K. Abbey, R. D. Fleischmann, J. Walling, and M. J. Duncan. 2004. Comparative whole-genome analysis of virulent and avirulent strains of Porphyromonas gingivalis. J. Bacteriol. 186:5473-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole, S. P., J. Harwood, R. Lee, R. She, and D. G. Guiney. 2004. Characterization of monospecies biofilm formation by Helicobacter pylori. J. Bacteriol. 186:3124-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cook, G. S., J. W. Costerton, and R. J. Lamont. 1998. Biofilm formation by Porphyromonas gingivalis and Streptococcus gordonii. J. Periodontal Res. 33:323-327. [DOI] [PubMed] [Google Scholar]
- 9.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 10.Daims, H., S. Lucker, and M. Wagner. 2006. daime, a novel image analysis program for microbial ecology and biofilm research. Environ. Microbiol. 8:200-213. [DOI] [PubMed] [Google Scholar]
- 11.Demuth, D. R., D. C. Irvine, J. W. Costerton, G. S. Cook, and R. J. Lamont. 2001. Discrete protein determinant directs the species-specific adherence of Porphyromonas gingivalis to oral streptococci. Infect. Immun. 69:5736-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellen, R. P., G. Lepine, and P. M. Nghiem. 1997. In vitro models that support adhesion specificity in biofilms of oral bacteria. Adv. Dent. Res. 11:33-42. [DOI] [PubMed] [Google Scholar]
- 13.Granger, L. L., E. B. O'Hara, R. F. Wang, F. V. Meffen, K. Armstrong, S. D. Yancey, P. Babitzke, and S. R. Kushner. 1998. The Escherichia coli mrsC gene is required for cell growth and mRNA decay. J. Bacteriol. 180:1920-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heydorn, A., B. Ersboll, J. Kato, M. Hentzer, M. R. Parsek, T. Tolker-Nielsen, M. Givskov, and S. Molin. 2002. Statistical analysis of Pseudomonas aeruginosa biofilm development: impact of mutations in genes involved in twitching motility, cell-to-cell signaling, and stationary-phase sigma factor expression. Appl. Environ. Microbiol. 68:2008-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holt, S. C., and J. L. Ebersole. 2005. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex,” a prototype polybacterial pathogenic consortium in periodontitis. Periodontol. 2000 38:72-122. [DOI] [PubMed] [Google Scholar]
- 16.Hughes, T. R., M. J. Marton, A. R. Jones, C. J. Roberts, R. Stoughton, C. D. Armour, H. A. Bennett, E. Coffey, H. Dai, Y. D. He, M. J. Kidd, A. M. King, M. R. Meyer, D. Slade, P. Y. Lum, S. B. Stepaniants, D. D. Shoemaker, D. Gachotte, K. Chakraburtty, J. Simon, M. Bard, and S. H. Friend. 2000. Functional discovery via a compendium of expression profiles. Cell 102:109-126. [DOI] [PubMed] [Google Scholar]
- 17.Ito, K., and Y. Akiyama. 2005. Cellular functions, mechanism of action, and regulation of FtsH protease. Annu. Rev. Microbiol. 59:211-231. [DOI] [PubMed] [Google Scholar]
- 18.Jayasekera, M. M., S. K. Foltin, E. R. Olson, and T. P. Holler. 2000. Escherichia coli requires the protease activity of FtsH for growth. Arch. Biochem. Biophys. 380:103-107. [DOI] [PubMed] [Google Scholar]
- 19.Jenkinson, H. F., and R. J. Lamont. 2005. Oral microbial communities in sickness and in health. Trends Microbiol. 13:589-595. [DOI] [PubMed] [Google Scholar]
- 20.Katz, J., D. C. Ward, and S. M. Michalek. 1996. Effect of host responses on the pathogenicity of strains of Porphyromonas gingivalis. Oral Microbiol. Immunol. 11:309-318. [DOI] [PubMed] [Google Scholar]
- 21.Kuboniwa, M., G. D. Tribble, C. E. James, A. O. Kilic, L. Tao, M. C. Herzberg, S. Shizukuishi, and R. J. Lamont. 2006. Streptococcus gordonii utilizes several distinct gene functions to recruit Porphyromonas gingivalis into a mixed community. Mol. Microbiol. 60:121-139. [DOI] [PubMed] [Google Scholar]
- 22.Kuwayama, H., S. Obara, T. Morio, M. Katoh, H. Urushihara, and Y. Tanaka. 2002. PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res. 30:E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamont, R. J., A. El-Sabaeny, Y. Park, G. S. Cook, J. W. Costerton, and D. R. Demuth. 2002. Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiol. 148:1627-1636. [DOI] [PubMed] [Google Scholar]
- 24.Lamont, R. J., S. G. Hersey, and B. Rosan. 1992. Characterization of the adherence of Porphyromonas gingivalis to oral streptococci. Oral Microbiol. Immunol. 7:193-197. [DOI] [PubMed] [Google Scholar]
- 25.LeBlanc, D. J., and F. P. Hassell. 1976. Transformation of Streptococcus sanguis Challis by plasmid deoxyribonucleic acid from Streptococcus faecalis. J. Bacteriol. 128:347-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levine, A., F. Vannier, C. Absalon, L. Kuhn, P. Jackson, E. Scrivener, V. Labas, J. Vinh, P. Courtney, J. Garin, and S. J. Seror. 2006. Analysis of the dynamic Bacillus subtilis Ser/Thr/Tyr phosphoproteome implicated in a wide variety of cellular processes. Proteomics 6:2157-2173. [DOI] [PubMed] [Google Scholar]
- 27.Maeda, K., H. Nagata, Y. Yamamoto, M. Tanaka, J. Tanaka, N. Minamino, and S. Shizukuishi. 2004. Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus oralis functions as a coadhesin for Porphyromonas gingivalis major fimbriae. Infect. Immun. 72:1341-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayanagi, G., T. Sato, H. Shimauchi, and N. Takahashi. 2004. Detection frequency of periodontitis-associated bacteria by polymerase chain reaction in subgingival and supragingival plaque of periodontitis and healthy subjects. Oral Microbiol. Immunol. 19:379-385. [DOI] [PubMed] [Google Scholar]
- 29.McNab, R., and R. J. Lamont. 2003. Microbial dinner-party conversations: the role of LuxS in interspecies communication. J. Med. Microbiol. 52:541-545. [DOI] [PubMed] [Google Scholar]
- 30.Merritt, J., F. Qi, S. D. Goodman, M. H. Anderson, and W. Shi. 2003. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect. Immun. 71:1972-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morona, J. K., J. C. Paton, D. C. Miller, and R. Morona. 2000. Tyrosine phosphorylation of CpsD negatively regulates capsular polysaccharide biosynthesis in Streptococcus pneumoniae. Mol. Microbiol. 35:1431-1442. [DOI] [PubMed] [Google Scholar]
- 32.Noiri, Y., L. Li, F. Yoshimura, and S. Ebisu. 2004. Localization of Porphyromonas gingivalis-carrying fimbriae in situ in human periodontal pockets. J. Dent. Res. 83:941-945. [DOI] [PubMed] [Google Scholar]
- 33.Park, Y., M. R. Simionato, K. Sekiya, Y. Murakami, D. James, W. Chen, M. Hackett, F. Yoshimura, D. R. Demuth, and R. J. Lamont. 2005. Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii. Infect. Immun. 73:3983-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rajapakse, P. S., N. M. O'Brien-Simpson, N. Slakeski, B. Hoffmann, and E. C. Reynolds. 2002. Immunization with the RgpA-Kgp proteinase-adhesin complexes of Porphyromonas gingivalis protects against periodontal bone loss in the rat periodontitis model. Infect. Immun. 70:2480-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosan, B., and R. J. Lamont. 2000. Dental plaque formation. Microbes Infect. 2:1599-1607. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schembri, M. A., K. Kjaergaard, and P. Klemm. 2003. Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48:253-267. [DOI] [PubMed] [Google Scholar]
- 39.Slots, J., and R. J. Gibbons. 1978. Attachment of Bacteroides melaninogenicus subsp. asaccharolyticus to oral surfaces and its possible role in colonization of the mouth and of periodontal pockets. Infect. Immun. 19:254-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smoot, L. M., J. C. Smoot, M. R. Graham, G. A. Somerville, D. E. Sturdevant, C. A. Migliaccio, G. L. Sylva, and J. M. Musser. 2001. Global differential gene expression in response to growth temperature alteration in group A streptococcus. Proc. Natl. Acad. Sci. USA 98:10416-10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Socransky, S. S., and A. D. Haffajee. 1992. The bacterial etiology of destructive periodontal disease: current concepts. J. Periodontol. 63:322-331. [DOI] [PubMed] [Google Scholar]
- 42.Socransky, S. S., and A. D. Haffajee. 2005. Periodontal microbial ecology. Periodontol. 2000 38:135-187. [DOI] [PubMed] [Google Scholar]
- 43.Socransky, S. S., A. D. Haffajee, L. A. Ximenez-Fyvie, M. Feres, and D. Mager. 1999. Ecological considerations in the treatment of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis periodontal infections. Periodontol. 2000 20:341-362. [DOI] [PubMed] [Google Scholar]
- 44.Stanley, N. R., and B. A. Lazazzera. 2004. Environmental signals and regulatory pathways that influence biofilm formation. Mol. Microbiol. 52:917-924. [DOI] [PubMed] [Google Scholar]
- 45.Stinson, M. W., K. Safulko, and M. J. Levine. 1991. Adherence of Porphyromonas (Bacteroides) gingivalis to Streptococcus sanguis in vitro. Infect. Immun. 59:102-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stoodley, P., K. Sauer, D. G. Davies, and J. W. Costerton. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187-209. [DOI] [PubMed] [Google Scholar]
- 47.Tanaka, S., Y. Murakami, K. Seto, K. Takamori, M. Yosida, K. Ochiai, S. Watanabe, and S. Fujisawa. 2003. The detection of Porphyromonas gingivalis, Prevotella intermedia, and Actinobacillus actinomycetemcomitans in the supragingival plaque of children with and without caries. Pediatr. Dent. 25:143-148. [PubMed] [Google Scholar]
- 48.Umeyama, T., Y. Tanabe, B. D. Aigle, and S. Horinouchi. 1996. Expression of the Streptomyces coelicolor A3(2) ptpA gene encoding a phosphotyrosine protein phosphatase leads to overproduction of secondary metabolites in S. lividans. FEMS Microbiol. Lett. 144:177-184. [DOI] [PubMed] [Google Scholar]
- 49.VanBogelen, R. A., K. D. Greis, R. M. Blumenthal, T. H. Tani, and R. G. Matthews. 1999. Mapping regulatory networks in microbial cells. Trends Microbiol. 7:320-328. [DOI] [PubMed] [Google Scholar]
- 50.Venturi, V. 2003. Control of rpoS transcription in Escherichia coli and Pseudomonas: why so different? Mol. Microbiol. 49:1-9. [DOI] [PubMed] [Google Scholar]
- 51.Waite, R. D., A. Papakonstantinopoulou, E. Littler, and M. A. Curtis. 2005. Transcriptome analysis of Pseudomonas aeruginosa growth: comparison of gene expression in planktonic cultures and developing and mature biofilms. J. Bacteriol. 187:6571-6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watanabe, K., Y. Yamaji, and T. Umemoto. 1992. Correlation between cell-adherent activity and surface structure in Porphyromonas gingivalis. Oral Microbiol. Immunol. 7:357-363. [DOI] [PubMed] [Google Scholar]
- 53.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]
- 54.Whitfield, C., and I. S. Roberts. 1999. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 31:1307-1319. [DOI] [PubMed] [Google Scholar]
- 55.Wright, T. L., R. P. Ellen, J. M. Lacroix, S. Sinnadurai, and M. W. Mittelman. 1997. Effects of metronidazole on Porphyromonas gingivalis biofilms. J. Periodontal Res. 32:473-477. [DOI] [PubMed] [Google Scholar]
- 56.Xie, H., G. S. Cook, J. W. Costerton, G. Bruce, T. M. Rose, and R. J. Lamont. 2000. Intergeneric communication in dental plaque biofilms. J. Bacteriol. 182:7067-7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ximenez-Fyvie, L. A., A. D. Haffajee, and S. S. Socransky. 2000. Comparison of the microbiota of supra- and subgingival plaque in health and periodontitis. J. Clin. Periodontol. 27:648-657. [DOI] [PubMed] [Google Scholar]
- 58.Ximenez-Fyvie, L. A., A. D. Haffajee, and S. S. Socransky. 2000. Microbial composition of supra- and subgingival plaque in subjects with adult periodontitis. J. Clin. Periodontol. 27:722-732. [DOI] [PubMed] [Google Scholar]
- 59.Xu, L., H. Li, C. Vuong, V. Vadyvaloo, J. Wang, Y. Yao, M. Otto, and Q. Gao. 2006. Role of the luxS quorum-sensing system in biofilm formation and virulence of Staphylococcus epidermidis. Infect. Immun. 74:488-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yin, J. L., N. A. Shackel, A. Zekry, P. H. McGuinness, C. Richards, K. V. Putten, G. W. McCaughan, J. M. Eris, and G. A. Bishop. 2001. Real-time reverse transcriptase-polymerase chain reaction (RT-PCR) for measurement of cytokine and growth factor mRNA expression with fluorogenic probes or SYBR Green I. Immunol. Cell Biol. 79:213-221. [DOI] [PubMed] [Google Scholar]