Abstract
The Lyme disease spirochete Borrelia burgdorferi expresses a broad array of adhesive molecules, including the decorin-binding proteins A and B (DbpA and DbpB), which are believed to play important roles in mammalian infection. The dbpBA locus was deleted; resulting mutants were able to infect both immunodeficient and immunocompetent mice, indicating that neither DbpA nor DbpB is essential for the infection of mammals, although the DbpAB deficiency may significantly attenuate infectivity potential.
The Lyme disease spirochete Borrelia burgdorferi expresses a broad array of adhesive molecules (4), including the decorin-binding proteins A and B (DbpA and DbpB) (2, 11-13), the fibronectin-binding protein BBK32 (23, 24), Bgp (Borrelia glycosaminoglycan-binding protein) (20, 21), and P66 (3, 6). The lipoproteins DbpAB and BBK32 bind to components of the mammalian extracellular matrix (ECM) decorin and fibronectin, respectively (2, 11, 23, 24); the outer membrane proteins P66 and Bgp bind to either integrin αIIbβ3 (3, 6) or other ECM components, glycosaminoglycans (20, 21). A recent study showed that BBK32 also interacts with glycosaminoglycans (9). The interactions of these proteins with host ligands have been proposed to play crucial roles in dissemination, tissue colonization, and/or persistence during B. burgdorferi infection of mammalian hosts.
DbpA and DbpB were among the first identified adhesive molecules of B. burgdorferi (11); DbpA is probably the best-characterized borrelial adhesin (2, 22). The two lipoproteins are encoded within a two-gene operon (12) which is located on the plasmid lp54 (10). DbpA and DbpB are not expressed by B. burgdorferi in the tick vector (13) but are up-regulated during mammalian infection (5, 17, 18), suggesting a potential role of the lipoproteins in the infection of mammals. However, it is unknown if these two lipoproteins are required for the infection of mammalian hosts, which is the focus of the current study.
Generation of dbpAB mutants.
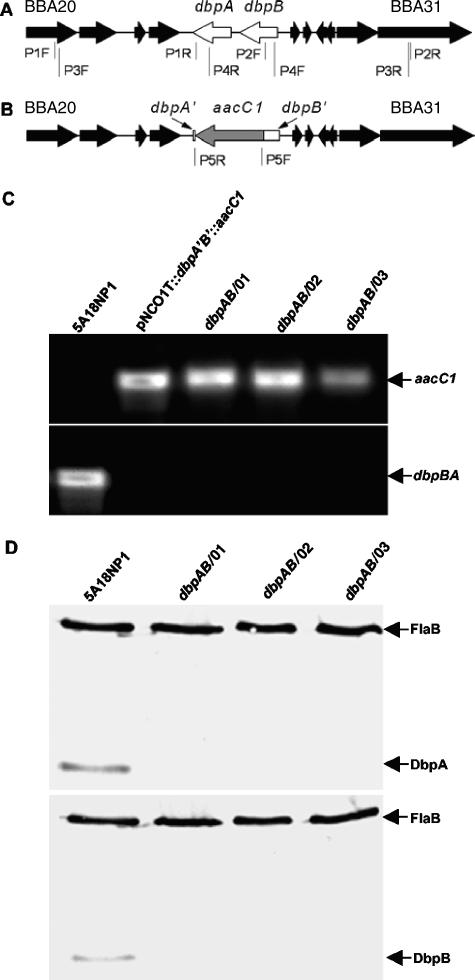
To delete the dbpBA locus, a disruption plasmid was first constructed. A 2,047-bp fragment covering a partial sequence of the open reading frame (ORF) BBA20, the entire ORFs BBA21, BBA22, and BBA23, and a partial sequence of dbpA (BBA24) was amplified by use of primers P1F and P1R (Fig. 1A; Table 1). A second, 2,145-bp fragment, including a partial sequence of dbpB (BBA25), the entire ORFs BBA26, BBA27, BBA28, and BBA29, and a partial sequence of the ORF BBA31, was amplified using primers P2F and P2R. The two PCR products were pooled, purified using a QIAquick PCR purification kit (QIAGEN Inc., Valencia, CA), digested with NheI, repurified, and ligated. The resultant product was used as a template and amplified by nested PCR using primers P3F and P3R. The PCR product was purified, digested with Acc65I, and cloned into the TA vector pNCO1T as described previously (7) to generate pNCO1T::dbpA′B′, which cannot replicate in the borrelial system. A gentamicin cassette (aacC1) was amplified by use of primers P5F and P5R (Fig. 1B; Table 1) from the shuttle vector pBSV2G (a gift from P. Rosa and P. Stewart), which confers gentamicin resistance both in Escherichia coli and in B. burgdorferi (8). The amplicon was purified, digested with XbaI, and cloned into pNCO1T::dbpA′B′ to complete the construction of the disruption plasmid pNCO1T::dbpA′B′::aacC1. The insert within the plasmid was sequenced to ensure it was as designed.
FIG. 1.
Generation of dbpAB mutants. (A) Diagram of the dbpBA locus and adjacent ORFs. The two-gene operon is located within lp54, which carries 76 ORFs, BBA01 to BBA76. The two open arrows represent dbpA and dbpB; the filled arrows denote BBA20 to BBA23 and BBA26 to BBA31. The binding sites of eight primers, i.e., P1F to P4F and P1R to P4R, are also indicated. (B) Diagram of the disrupted dbpBA locus, showing major portions of the dbpA and dbpB genes replaced with a gentamicin cassette (aacC1). The gray arrow represents the aacC1 cassette flanked by small residual sequences (dbpA′ and dbpB′) of the genes dbpA and dbpB (open bars). The binding sites of primers P5F and P5R are also indicated. (C) PCR analysis of dbpAB mutants. 5A18NP1 spirochetes, the disruption plasmid pNCO1T::dbpA′B′::aacC1, and three dbpAB mutants were used as DNA sources and subjected to PCR amplification using primers P5F and P5R (top) or primers P4F and P4R (bottom). (D) Immunoblot analysis of dbpAB mutants. 5A18NP1 and three dbpAB mutants were analyzed by immunoblotting probed with a mixture of FlaB MAb and mouse antisera raised against recombinant DbpA (top) or DbpB (bottom).
TABLE 1.
Primers used in the study
| Primer name | Primer sequence (5′ to 3′)a |
|---|---|
| P1F | AAATGCCCTTGTATGTAGCAATTACA |
| P1R | CAGCTAGCAAAAATAACTAATAAACAACAT |
| P2F | TTGCTAGCTTCTCTTAAGGCTAGTCCA |
| P2R | AAGCTTAAGACCTGCTATCTTACAA |
| P3F | AAGGTACCAATAACAGCAGAGAAAT |
| P3R | TAGGTACCTGCTATCTTACAAAAAGACT |
| P4F | CGCATGTAGTATTGGATTAG |
| P4R | CATTGCTGAAAATTCACCAC |
| P5F | TATCTAGAGCTTCAAGGAAGATTTCC |
| P5R | AAATCTAGACGCTCAGTGGAACGAA |
P1R and P2F, P3F and P3R, and P5F and P5R contain restriction enzyme sites NheI, Acc65I, and XbaI (underlined), respectively.
The B. burgdorferi B31 BBE02 disruptant 5A18NP1 (a gift from H. Kawabata and S. Norris) was grown in 50 ml of Barbour-Stoenner-Kelly H (BSK-H) complete medium (Sigma Chemical Co., St. Louis, MO) at densities of 5 × 107 to 1 × 108 cells/ml (mid- to late exponential phase), harvested, washed, and transformed with 8.0 μg of the disruption plasmid DNA under standard electroporation conditions (26, 28). 5A18NP1 was utilized for its high transformability resulting from the disruption of the gene BBE02, a putative restriction-modification gene (14). The cells were allowed to recover in 20 ml of BSK-H complete medium at 33°C for 18 h. After a gentamicin concentration of 50 μg/ml was added, the suspension was transferred into 96 PCR tubes (200 μl/tube). Aliquots were incubated at 33°C for 10 days; live spirochetes were examined under a dark-field microscope and found in 10 of the 96 tubes. Approximately 30 μl of gentamicin resistance culture was transferred to 1.4 ml of BSK-H medium in a 1.5-ml microcentrifuge tube and grown to near-stationary phase at 33°C. Spirochetes were harvested from 500 μl of culture by centrifugation at 13,000 × g for 10 min at room temperature, washed twice with excess volumes of phosphate-buffered saline (pH = 7.3) to remove the residual disruption plasmid DNA, and resuspended in 500 μl of deionized H2O. One microliter of suspension was used as a DNA source for the examination of the aacC1 cassette by PCR using primers P5F and P5R (Fig. 1B; Table 1). The cassette was found in 7 of the 10 clones. Spontaneous mutations most likely contributed to the three clones lacking the aacC1 cassette but developing gentamicin resistance; these clones were discarded. The presence of lp28-1, the plasmid essential for infection in immunocompetent hosts (15, 25), was examined by PCR as described previously (29); three of the clones maintaining lp28-1 were chosen for the study and were designated dbpAB/01, dbpAB/02, and dbpAB/03.
The plasmid contents of the three selected clones were surveyed by PCR and further confirmed by microarray hybridization as described previously (29) and are presented in Table 2. The insertion of the aacC1 cassette was reconfirmed by PCR using primers P5F and P5R (Fig. 1C, top); the deletion of the dbpA and dbpB genes was shown by PCR using primers P4F and P4R (Fig. 1C, bottom). The lack of DbpA and DbpB expression was confirmed by immunoblotting probed with a mixture of FlaB monoclonal antibody (MAb) and antisera raised against either recombinant DbpA or DbpB (Fig. 1D). The FlaB MAb was developed by Barbour et al. (1); the DbpA and DbpB antisera were produced by immunizing mice with recombinant DbpA or DbpB emulsified with Freund's complete (first injection) or incomplete (remaining injections) adjuvant.
TABLE 2.
The dbpBA locus is not required for infection of immunodeficient micea
| Inoculum | Plasmids missing | No. of cultures positive/total no. of specimens examined
|
|||
|---|---|---|---|---|---|
| Heart | Joint | Skin | All sites | ||
| 5A18NP1 | lp28-4, lp56, lp5 | 5/5 | 5/5 | 5/5 | 15/15 |
| dbpAB/01 | cp9, lp28-4, lp56, lp5 | 2/2 | 2/2 | 2/2 | 6/6 |
| dbpAB/02 | cp9, lp28-4, lp56, lp5 | 2/2 | 2/2 | 2/2 | 6/6 |
| dbpAB/03 | cp9, lp28-4, lp56, lp21, lp5 | 2/2 | 2/2 | 2/2 | 6/6 |
Each of the three dbpAB clones was inoculated into two SCID mice, and the parental clone 5A18NP1 was inoculated into five mice as a control. All animals were sacrificed 1 month after inoculation. Heart, tibiotarsal joint, and skin specimens were harvested and cultured for spirochetes in BSK-H complete medium.
The dbpBA locus is not essential for infection of either SCID or wild-type mice.
Mice with severe combined immunodeficiency (SCID) were first used to examine whether the dbpBA locus is required for infection. For each of the three dbpAB mutants, two SCID mice on a BALB/c background (provided by the Division of Laboratory Animal Medicine at Louisiana State University, Baton Rouge, LA) were inoculated via one single intradermal/subcutaneous injection of 105 organisms. An additional five mice were challenged with the parental clone 5A18NP1 as a control. All mice were euthanized 1 month after inoculation; heart, tibiotarsal joint, and skin specimens (not from inoculation sites) were aseptically collected for spirochete culture as previously described (29). Spirochetes were recovered successfully from each of the heart, joint, and skin specimens from all of the 11 mice, regardless of whether they received the parental clone or mutants (Table 2), indicating that neither DbpA nor DbpB is required for the infection of immunodeficient mice.
Next, six BALB/c mice (provided by the Division of Laboratory Animal Medicine at Louisiana State University, Baton Rouge, LA) were challenged with the three mutants via one single intradermal/subcutaneous injection of 105 organisms; an additional five mice received 5A18NP1 as a control. In a separate experiment, 10 animals were inoculated with the three mutants; an additional five mice were given the parental clone. All mice were euthanized 1 month later; 5A18NP1 spirochetes were recovered from each of the heart, joint, and skin specimens of all 10 mice from the two experiments (Table 3). Fourteen of the 16 mice inoculated with dbpAB mutants had at least one positive specimen, indicating that neither DbpA nor DbpB is required for the infection of immunocompetent mice. The mutants were not recovered from two inoculated mice probably because of attenuated infectivity resulting from the disruption of the dbpBA locus. Alternatively, genetic manipulating processes might have introduced unnoted defects, which, in turn, could be responsible for the negative culture results in 13 heart, 5 joint, and 2 skin specimens; a complementation study, which we have been unable to accomplish, may help clarify the issue. Another unaddressed issue is whether the DbpAB deficiency affects infectivity reflected by the 50% infective dose (ID50). In the current study, up to 105 dbpAB bacteria, a dose that is 1,000-fold higher than the ID50 value of the parental clone 5A18NP1 (14), were inoculated into a mouse. Nevertheless, our study clearly shows that the dbpBA locus is not essential for the infection of either immunodeficient or immunocompetent mice, although the DbpAB deficiency may severely attenuate infectivity potential.
TABLE 3.
The dbpBA locus is not required for infection of immunocompetent micea
| Expt no. | Inoculum | No. of cultures positive/total no. of specimens examined
|
No. of mice infected/total no. of mice inoculated | |||
|---|---|---|---|---|---|---|
| Heart | Joint | Skin | All sites | |||
| I | 5A18NP1 | 5/5 | 5/5 | 5/5 | 15/15 | 5/5 |
| dbpAB/01 | 1/2 | 1/2 | 2/2 | 4/6 | 2/2 | |
| dbpAB/02 | 1/3 | 3/3 | 3/3 | 7/9 | 3/3 | |
| dbpAB/03 | 0/1 | 1/1 | 1/1 | 2/3 | 1/1 | |
| II | 5A18NP1 | 5/5 | 5/5 | 5/5 | 15/15 | 5/5 |
| dbpAB/01 | 0/3 | 2/3 | 3/3 | 5/9 | 3/3 | |
| dbpAB/02 | 1/3 | 2/3 | 2/3 | 5 /9 | 2/3 | |
| dbpAB/03 | 0/4 | 2/4 | 3/4 | 5/12 | 3/4 | |
Each of the three dbpAB clones was inoculated into one to three (in experiment I) or three or four (in experiment II) BALB/c mice, and 5A18NP1 was used as a control to infect five mice in each experiment. All animals were sacrificed 1 month after inoculation. Heart, tibiotarsal joint, and skin specimens (not from the inoculation site) were harvested and cultured for spirochetes in BSK-H complete medium.
B. burgdorferi expresses multiple adhesins that potentially mediate interactions of spirochetes with ECM components and other host ligands, including DbpA, DbpB, BBK32, Bgp, and P66. Seshu et al. showed that a BBK32 deficiency increases the ID50 value in immunocompetent mice (27), consistent with a subsequent study by Li et al. indicating that the BBK32 gene is essential for the life cycle of B. burgdorferi neither in the tick vector nor in the murine host (16). Parveen et al. reported that Bgp is not required for the infection of immunodeficient mice, although it remains to be examined whether the adhesin is essential for the infection of immunocompetent mice (19). The current study shows that neither DbpA nor DbpB is required for the infection of immunodeficient or immunocompetent mice. The data presented in conjunction with studies by others (16, 19, 27) firmly demonstrate that the lack of one or two individual adhesins does not completely diminish the ability of B. burgdorferi to infect mammals, although these adhesive molecules as a whole may play an essential role in dissemination, tissue colonization, and/or persistence during mammalian infection.
Acknowledgments
We thank H. Kawabata and S. Norris for providing the B. burgdorferi B31 disruptant 5A18NP1, P. Stewart and P. Rosa for providing the shuttle vector pBSV2G, and K. DePonte and N. Marcantonio for providing the FlaB hybridoma cell line.
This work was supported in part by an NIH/NIAMS career development award and an Arthritis Foundation Investigators award.
Editor: J. T. Barbieri
Footnotes
Published ahead of print on 5 September 2006.
REFERENCES
- 1.Barbour, A. G., S. F. Hayes, R. A. Heiland, M. E. Schrumpf, and S. L. Tessier. 1986. A Borrelia-specific monoclonal antibody binds to a flagellar epitope. Infect. Immun. 52:549-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, E. L., B. P. Guo, P. O'Neal, and M. Höök. 1999. Adherence of Borrelia burgdorferi. Identification of critical lysine residues in DbpA required for decorin binding. J. Biol. Chem. 274:26272-26278. [DOI] [PubMed] [Google Scholar]
- 3.Coburn, J., W. Chege, L. Magoun, S. C. Bodary, and J. M. Leong. 1999. Characterization of a candidate Borrelia burgdorferi αIIbβ3 chain integrin ligand identified using a phage display library. Mol. Microbiol. 34:926-940. [DOI] [PubMed] [Google Scholar]
- 4.Coburn, J., J. R. Fischer, and J. M. Leong. 2005. Solving a sticky problem: new genetic approaches to host cell adhesion by the Lyme disease spirochete. Mol. Microbiol. 57:1182-1195. [DOI] [PubMed] [Google Scholar]
- 5.Crother, T. R., C. I. Champion, X. Y. Wu, D. R. Blanco, J. N. Miller, and M. A. Lovett. 2003. Antigenic composition of Borrelia burgdorferi during infection of SCID mice. Infect. Immun. 71:3419-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Defoe, G., and J. Coburn. 2001. Delineation of Borrelia burgdorferi P66 sequences required for integrin αIIbβ3 recognition. Infect. Immun. 69:3455-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Downie, A. B., L. M. Dirk, Q. Xu, J. Drake, D. Zhang, M. Dutt, A. Butterfield, R. R. Geneve, J. W. Corum III, K. G. Lindstrom, and J. C. Snyder. 2004. A physical, enzymatic, and genetic characterization of perturbations in the seeds of the brown tomato mutants. J. Exp. Bot. 55:961-973. [DOI] [PubMed] [Google Scholar]
- 8.Elias, A. F., J. L. Bono, J. J. Kupko III, P. E. Stewart, J. G. Krum, and P. A. Rosa. 2003. New antibiotic resistance cassettes suitable for genetic studies in Borrelia burgdorferi. J. Mol. Microbiol. Biotechnol. 6:29-40. [DOI] [PubMed] [Google Scholar]
- 9.Fischer, J. R., K. T. LeBlanc, and J. M. Leong. 2006. Fibronectin binding protein BBK32 of the Lyme disease spirochete promotes bacterial attachment to glycosaminoglycans. Infect. Immun. 74:435-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidman, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fuji, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 11.Guo, B. P., E. L. Brown, D. W. Dorward, L. C. Rosenberg, and M. Höök. 1998. Decorin-binding adhesins from Borrelia burgdorferi. Mol. Microbiol. 30:711-723. [DOI] [PubMed] [Google Scholar]
- 12.Hagman, K. E., P. Lahdenne, T. G. Popova, S. F. Porcella, D. R. Akins, J. D. Radolf, and M. V. Norgard. 1998. Decorin-binding protein of Borrelia burgdorferi is encoded within a two-gene operon and is protective in the murine model of Lyme borreliosis. Infect. Immun. 66:2674-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagman, K. E., X. Yang, S. K. Wikel, G. B. Schoeler, M. J. Caimano, J. D. Radolf, and M. V. Norgard. 2000. Decorin-binding protein A (DbpA) of Borrelia burgdorferi is not protective when immunized mice are challenged via tick infestation and correlates with the lack of DbpA expression by B. burgdorferi in ticks. Infect. Immun. 68:4759-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawabata, H., S. J. Norris, and H. Watanabe. 2004. BBE02 disruption mutants of Borrelia burgdorferi B31 have a highly transformable, infectious phenotype. Infect. Immun. 72:7147-7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Labandeira-Rey, M., J. Seshu, and J. T. Skare. 2003. The absence of linear plasmid 25 or 28-1 of Borrelia burgdorferi dramatically alters the kinetics of experimental infection via distinct mechanisms. Infect. Immun. 71:4608-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, X., X. Liu, D. S. Beck, F. S. Kantor, and E. Fikrig. 2006. Borrelia burgdorferi lacking BBK32, a fibronectin-binding protein, retains full pathogenicity. Infect. Immun. 74:3305-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang, F. T., F. K. Nelson, and E. Fikrig. 2002. Molecular adaptation of Borrelia burgdorferi in the murine host. J. Exp. Med. 196:275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang, F. T., J. Yan, M. L. Mbow, S. L. Sviat, R. D. Gilmore, M. Mamula, and E. Fikrig. 2004. Borrelia burgdorferi changes its surface antigenic expression in response to host immune responses. Infect. Immun. 72:5759-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parveen, N., K. A. Cornell, J. L. Bono, C. Chamberland, P. Rosa, and J. M. Leong. 2006. Bgp, a secreted glycosaminoglycan-binding protein of Borrelia burgdorferi strain N40, displays nucleosidase activity and is not essential for infection of immunodeficient mice. Infect. Immun. 74:3016-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parveen, N., and J. M. Leong. 2000. Identification of a candidate glycosaminoglycan-binding adhesin of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:1220-1234. [DOI] [PubMed] [Google Scholar]
- 21.Parveen, N., D. Robbins, and J. M. Leong. 1999. Strain variation in glycosaminoglycan recognition influences cell-type-specific binding by Lyme disease spirochetes. Infect. Immun. 67:1743-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pikas, D. S., E. L. Brown, S. Gurusiddappa, L. Y. Lee, Y. Xu, and M. Höök. 2003. Decorin-binding sites in the adhesin DbpA from Borrelia burgdorferi: a synthetic peptide approach. J. Biol. Chem. 278:30920-30926. [DOI] [PubMed] [Google Scholar]
- 23.Probert, W. S., and B. J. Johnson. 1998. Identification of a 47 kDa fibronectin-binding protein expressed by Borrelia burgdorferi isolate B31. Mol. Microbiol. 30:1003-1015. [DOI] [PubMed] [Google Scholar]
- 24.Probert, W. S., J. H. Kim, M. Höök, and B. J. Johnson. 2001. Mapping the ligand-binding region of Borrelia burgdorferi fibronectin-binding protein BBK32. Infect. Immun. 69:4129-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purser, J. E., and S. J. Norris. 2000. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc. Natl. Acad. Sci. USA 97:13865-13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samuels, D. S. 1995. Electrotransformation of the spirochete Borrelia burgdorferi. Methods Mol. Biol. 47:253-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seshu, J., M. D. Esteve-Gassent, M. Labandeira-Rey, J. H. Kim, J. P. Trzeciakowski, M. Höök, and J. T. Skare. 2006. Inactivation of the fibronectin-binding adhesin gene bbk32 significantly attenuates the infectivity potential of Borrelia burgdorferi. Mol. Microbiol. 59:1591-1601. [DOI] [PubMed] [Google Scholar]
- 28.Stewart, P. E., R. Thalken, J. L. Bono, and P. Rosa. 2001. Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Mol. Microbiol. 39:714-721. [DOI] [PubMed] [Google Scholar]
- 29.Xu, Q., S. V. Seemanapalli, L. Lomax, K. McShan, X. Li, E. Fikrig, and F. T. Liang. 2005. Association of linear plasmid 28-1 with an arthritic phenotype of Borrelia burgdorferi. Infect. Immun. 73:7208-7215. [DOI] [PMC free article] [PubMed] [Google Scholar]