Abstract
Porphyromonas gingivalis is a crucial component of complex plaque biofilms that form in the oral cavity, resulting in the progression of periodontal disease. To elucidate the mechanism of periodontal biofilm formation, we analyzed the involvement of several genes related to the synthesis of polysaccharides in P. gingivalis. Gene knockout P. gingivalis mutants were constructed by insertion of an ermF-ermAM cassette; among these mutants, the galE mutant showed some characteristic phenotypes involved in the loss of GalE activity. As expected, the galE mutant accumulated intracellular carbohydrates in the presence of 0.1% galactose and did not grow in the presence of galactose at a concentration greater than 1%, in contrast to the parental strain. Lipopolysaccharide (LPS) analysis indicated that the length of the O-antigen chain of the galE mutant was shorter than that of the wild type. It was also demonstrated that biofilms generated by the galE mutant had an intensity 4.5-fold greater than those of the wild type. Further, the galE mutant was found to be significantly susceptible to some antibiotics in comparison with the wild type. In addition, complementation of the galE mutation led to a partial recovery of the parental phenotypes. We concluded that the galE gene plays a pivotal role in the modification of LPS O antigen and biofilm formation in P. gingivalis and considered that our findings of a relationship between the function of the P. gingivalis galE gene and virulence phenotypes such as biofilm formation may provide clues for understanding the mechanism of pathogenicity in periodontal disease.
Porphyromonas gingivalis has been implicated in the causation and pathogenesis of periodontal disease. Although this gram-negative anaerobe possesses a diverse repertoire of virulence factors, including fimbriae, gingipains, hemagglutinins, lipopolysaccharide (LPS), and others, such as outer membrane vesicles (6, 9, 13, 14, 18), the mechanism by which it initiates periodontal disease is not fully understood. On the other hand, it has been suggested that P. gingivalis is usually a late colonizer in the oral cavity and plays a central role in subgingival pockets (18). Thus, subgingival plaque biofilms containing P. gingivalis appear to be important for the progression of periodontal disease. In P. gingivalis strain W50, the bacterial surface polysaccharide thought to contribute to biofilm formation was found to contain mannose, galactose, rhamnose, glucose, and 2-acetamido-2-deoxy-d-glucose in a relative molar ratio of 13.5:2.0:1.4:1.0:1.0 (7).
Among the virulence factors of P. gingivalis, LPS, the major integral component of the outer membrane, which exhibits immunostimulatory and inflammatory activities, has three general components: O-antigen polysaccharide, core oligosaccharide, and lipid A. Most of the biological effects of LPS result from the lipid A part; however, there is an increasing body of evidence indicating that O antigen plays an important role in its effective colonization of host tissue (5, 11, 26-28) as well as in resistance to some bactericidal effects (2, 29, 37). The location of O antigen places it at the interface between the bacterium and its environment. Hence, a critical density of the long O-antigen chain not only prevents access of detrimental molecules into the outer membrane but also plays an important role in the initial attachment of the bacterium to other organisms, such as other bacteria and mammals, or inorganic surfaces. However, it is not clear whether changes in O antigen of P. gingivalis have effects on the properties of attachment and biofilm formation.
To elucidate the virulence mechanisms associated with periodontal biofilms, we analyzed several genes of P. gingivalis related to the synthesis of polysaccharides, which are contained as building components of the bacterial surface and biofilms. We selected three genes involved in the synthesis of LPS O antigen: wecA, encoding GlcNAc-1-phosphate transferase; wbaP, encoding galactose-1-phosphate transferase; and wzt, encoding an ATP-binding protein subunit. In addition, we analyzed the galE gene, which is involved in the synthesis of sugar nucleotides in the Leloir pathway. The GalE enzyme (UDP-galactose 4-epimerase) catalyzes the interconversion between UDP-glucose and UDP-galactose and is universally conserved among all organisms from bacteria to Homo sapiens. Further, galE mutants of Salmonella enterica serovar Typhimurium, Neisseria gonorrhoeae, and Haemophilus influenzae have been shown to be relatively avirulent compared with their parental strains (15, 22, 33).
In the present study, inactivation of the galE gene of P. gingivalis resulted in a shortened O antigen and a significant increase in biofilm formation, which demonstrates the relationship between the galE gene and biofilm formation in P. gingivalis.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli DH5α was grown in LB broth or on LB agar plates under aerobic conditions. P. gingivalis was grown in brain heart infusion (BHI) broth supplemented with hemin and menadione (HM) or on BHI-HM blood agar plates in an anaerobic chamber (miniMACS anaerobic workstation; Don Whitley Scientific Ltd., Shipley, United Kingdom) in 80% N2, 10% H2, and 10% CO2. For E. coli, ampicillin and erythromycin were supplemented at 100 μg/ml and 300 μg/ml, respectively, when required. For P. gingivalis, erythromycin and tetracycline were supplemented at 5 μg/ml and 0.7 μg/ml, respectively, when required. Recombinant plasmids derived from pUC19 or pBR322 were transformed into E. coli DH5α.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain/plasmid | Relevant phenotype, description, and/or selective markera | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli DH5α | F′ φ80dlacZ ΔM15 Δ(lacZYA · argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ−thi-1 gyrA96 relA1 | Takara Bio, Otsu, Japan |
| Porphyromonas gingivalis | ||
| ATCC 33277 | Wild type | American Type Culture Collection |
| ATCC 33277 htpG strain | Ermr | This study |
| ATCC 33277 wecA strain | Ermr | This study |
| ATCC 33277 wbaP strain | Ermr | This study |
| ATCC 33277 galE strain | Ermr | This study |
| ATCC 33277 galE-c strain | Tetr Erms | This study |
| ATCC 33277 wzt strain | Ermr | This study |
| Plasmids | ||
| pUC19 | Cloning vector; Ampr | Takara Bio, Otsu, Japan |
| pBR322 | Cloning vector; Ampr | Takara Bio, Otsu, Japan |
| pHS19 | Contains the ermF-ermAM cassette between EcoRI and BamHI sites of pBR322 without bla; Ermr | 10 |
| pBR322-erm | Contains the ermF-ermAM cassette between EcoRI and BamHI sites of pBR322; Ampr Ermr | This study |
| pRN2 | Contains PG0045 between KpnI and AvaI sites of pUC19; Ampr | This study |
| pRN2-erm | Contains the ermF-ermAM cassette in PmaCI site within PG0045 of pRN2; Ampr Ermr | This study |
| pRN3 | Contains PG0106 between XbaI and BamHI sites of pUC19; Ampr | This study |
| pRN3-erm | Contains the ermF-ermAM cassette in BglII site within PG0106 of pRN3; Ampr Ermr | This study |
| pRN4 | Contains PG1964 between EcoRI and XbaI sites of pUC19; Ampr | This study |
| pRN4-erm | Contains the ermF-ermAM cassette in Bsp1407I site within PG1964 of pRN4; Ampr Ermr | This study |
| pRN6 | Contains PG0347 between EcoRI and BamHI sites of pUC19; Ampr | This study |
| pRN6-erm | Contains the ermF-ermAM cassette in BseRI site within PG0347 of pRN6; Ampr Ermr | This study |
| pRN7 | Contains PG1225 between XbaI and BamHI sites of pUC19; Ampr | This study |
| pRN7-erm | Contains the ermF-ermAM cassette in ApaI site within PG1225 of pRN7; Ampr Ermr | This study |
| pKD375 | Contains tetQ cassette in pUC19; Ampr Tetr | 25 |
| pUC19Q | Contains tetQ cassette between SmaI and BamHI sites in pUC19; Ampr Tetr | This study |
| pQG | Contains an entire region of galE and the 0.3-kb downstream region between BamHI and XbaI sites of pUC19Q; Ampr Tetr | This study |
| pGQG | Contains a region of 0.8 kb upstream of galE between KpnI and SmaI sites of pQG; Ampr Tetr | This study |
Ampr, ampicillin resistant; Ermr, erythromycin resistant; Erms, erythromycin sensitive; Tetr, tetracycline resistant.
Sequence analysis.
The publicly available sequences from the P. gingivalis genome project (http://www.tigr.org) were examined for the presence of the target genes; wecA (glycosyl transferase group 4 family protein), wbaP (bacterial sugar transferase), wzt (an ATP-binding protein subunit), and galE (UDP-galactose 4-epimerase). The amino acid sequences encoded by these genes of Salmonella enterica serovar Typhi or Bacteroides thetaiotaomicron, which were obtained from the GenBank database, were used as query sequences. Homology analysis was performed with BLAST.
Construction of mutants.
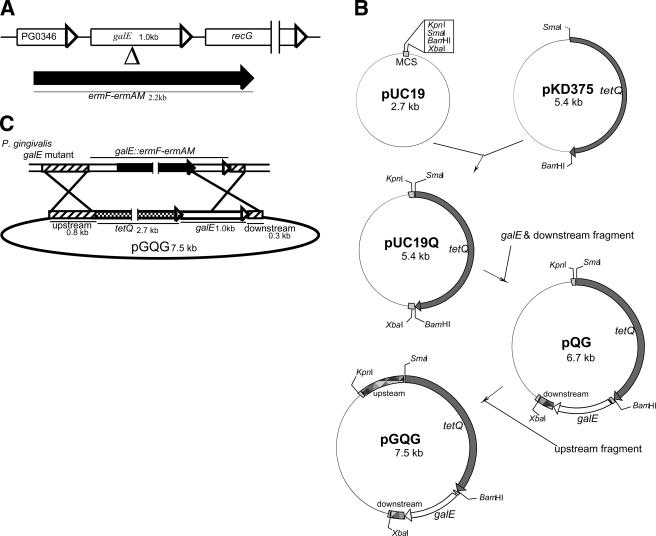
All DNA modifications and manipulations were carried out using standard methods (25, 34). An overview of the construction of the galE mutant is shown in Fig. 1A. The oligonucleotides used in this study are listed in Table 2. A 2.2-kb ermF-ermAM cassette obtained from pHS19 (10) was cloned between the EcoRI and BamHI sites of pBR322, resulting in pBR322-erm. The ermF-ermAM cassette of pBR322-erm was used as a selection marker. A 1.0-kb galE region was PCR amplified from the chromosomal DNA extracted from P. gingivalis ATCC 33277 by using the high-fidelity DNA polymerase Pyrobest (Takara Bio Inc., Shiga, Japan). The amplified region was cloned between the BamHI and XbaI sites of pUC19, resulting in pRN6. The ermF-ermAM cassette of pBR322-erm was subcloned into the BseRI site of galE of pRN6 to yield pRN6-erm. The ermF-ermAM cassette and the flanking galE regions were retrieved by BamHI and XbaI digestions and then electrotransformed into P. gingivalis ATCC 33277 to generate an insertion mutation in the galE gene. Construction of the other mutant strains was carried out in a similar manner: the BglII, NaeI, and ApaI sites in the wecA, wbaP, and wzt regions, respectively, were utilized for insertion of the ermF-ermAM cassette. Construction of the htpG mutant as a control strain was also performed, since HtpG, which is a homolog of the human heat shock protein Hsp90, was reported to have no effect on the growth of P. gingivalis or on its adherence to other bacterial and human cells (21, 36). The PmaCI site in the htpG region was utilized for insertion of the ermF-ermAM cassette.
FIG. 1.
Construction of the galE mutant and complementation of the galE mutation. (A) Strategy for construction of the galE mutant by allelic exchange. The galE gene interrupted by a 2.2-kb ermF-ermAM cassette was introduced into P. gingivalis by electroporation. ermF-ermAM conferred Ermr in P. gingivalis and E. coli. (B) Construction of the pGQG vector used for complementation of the galE strain. A 2.7-kb tetQ fragment obtained from pKD375 was ligated into the pUC19 vector, resulting in pUC19Q. A 1.3-kb fragment containing the complete sequence of the galE gene and its downstream region was coamplified from the chromosomal DNA of P. gingivalis and cloned between the BamHI and XbaI sites of pUC19Q, resulting in pQG. Finally, a 0.8-kb upstream fragment of the galE gene was amplified from the chromosomal DNA of P. gingivalis and cloned between the KpnI and SmaI sites of pQG, resulting in pGQG. (C) The 4.8-kb fragment (upstream-tetQ-galE-downstream) was retrieved from pGQG and electrotransformed into the P. gingivalis galE strain. A reciprocal recombination event occurred between the areas of homology, which are represented by two slashed bars (0.8-kb upstream and 0.3-kb downstream regions of the galE gene) and by the latter half of the white bar (galE gene) on the chromosome of the galE strain. The tetQ gene conferred Tetr in P. gingivalis.
TABLE 2.
Primer pairs used for gene cloning
| Target region | Name | Sequence (5′-3′)a |
|---|---|---|
| htpG | htpGp2-f | GGGGTACCTGAGGAGAATAGCGAGTT |
| htpGp2-r2 | CCCCCGGGTTGGGTTTTCAGCACAAC | |
| wecA | PG0106-D | GCTCTAGATGCGACCTCAATCCTTC |
| PG0106-E | CGGGATCCCAACCAGTGTTTGTCTCT | |
| wbaP | PG1964-A | GGAATTCGGGATACTACAACAAACC |
| PG1964-C | GCTCTAGAAGGCTCATATTCTCGTAG | |
| galE | PG0347-A | GGAATTCGGATCCCATACGACTGTG |
| PG0347-C | GCTCTAGAACGACTCCAAAGCTTTCC | |
| wzt | PG1225-A | GCTCTAGAGATCGGACTGCTTCTGGT |
| PG1225-B | CGGGATCCTTTCGACGGTCTGCCTTTC | |
| tetQ | SmaI-tetQ-F | TCCCCCGGGCGTTCCATTGGCCCTCAAAC |
| BamHI-tetQ-R | CGGGATCCCTCCTGCCATTCATAGAGGC | |
| galE and its downstream | BamHI-down-F | CGGGATCCGAAACCGAAATGAAACAAAAG |
| XbaI-down-R | GCTCTAGAAGGACACCGCGCAGCTGGAT | |
| Upstream of galE | KpnI-up-F | GGGGTACCCTCTCCAATGCCAGACTTTG |
| SmaI-up-R | TCCCCCGGGCGGTTTCTTATTTCGTAGC |
Restriction enzyme sites incorporated into oligonucleotides for subcloning are italicized.
Complementation of galE mutation.
The construction of the vectors used for complementation of the galE mutation is shown in Fig. 1B, while an overview of the complementation of galE mutation is shown in Fig. 1C. A 2.7-kb tetQ cassette was used as a selection marker for the complementation of the galE mutant. First, the tetQ cassette, which was tagged with additional sequences for restriction enzyme SmaI and BamHI sites, was amplified from pKD375, kindly gifted by K. Nakayama of Nagasaki University (25). The amplified DNA fragment was digested by the enzymes and cloned between the SmaI and BamHI sites of pUC19, producing pUC19Q. Next, a 1.0-kb cassette of galE gene with the 0.3-kb downstream region was coamplified from the chromosomal DNA of P. gingivalis 33277 and cloned between the BamHI and XbaI sites of pUC19Q, producing pQG. Finally, a 0.8-kb cassette of the upstream region of the galE gene was amplified from the chromosomal DNA of P. gingivalis and cloned between the KpnI and SmaI sites of pQG, producing pGQG. The 4.8-kb cassette (upstream-tetQ-galE-downstream) of pGQG was retrieved by KpnI and XbaI digestions and then electrotransformed into the P. gingivalis galE strain. Transformants that grew on tetracycline plates and not on erythromycin plates were considered to be potential target colonies. One of the many transformants that had lost erythromycin resistance and retained tetracycline resistance was named the galE-c strain. To confirm the integration of the target region into the chromosome of the galE strain, generated following the predicted recombination event, PCR analysis and partial sequencing of DNA from P. gingivalis ATCC 33277 and the galE and galE-c strains were performed (ABI Prism 310 genetic analyzer; Applied Biosystems, California).
Preparation of cell extracts and UDP-galactose 4-epimerase assay.
One-milliliter portions of fresh P. gingivalis wild-type, galE, and galE-c strains at an optical density at 660 nm (OD660) of 1.0 were harvested by centrifugation (10,000 × g for 2 min at 4°C) and then washed twice and suspended with 20 mM of phosphate buffer (pH 6.5) containing 50 mM NaCl, 10 mM MgCl2, and 1 mM dithiothreitol (1). The cells were disrupted ultrasonically in an ice bath for 60 cycles of 1 second each at 100 W (IKASONIC U50 control; Janke & Kunkel GmbH & Co. KG, Staufen, Germany). To obtain the cell extract, debris was removed by centrifugation (10,000 × g for 2 min at 4°C). The protein concentration of the cell extract was determined using a Bio-Rad protein assay based on the method of Bradford (3). UDP-galactose 4-epimerase activity was assayed as described by Maxwell et al. (23). The procedure was executed as follows. The reaction mixture contained 50 μl of glycine-NaOH buffer (1 M, pH 8.7), 20 μl of β-NAD+ (25 μmol/ml; ORIENTAL YEAST Co., Osaka, Japan), 25 μl of UDP-glucose dehydrogenase (2 U/ml; EMD Biosciences, Inc., San Diego, CA), 475 μl of distilled water, and 20 μl of cell extract. The reaction was started by the addition of 30 μl of UDP-galactose (2.2 μmol/ml; EMD Biosciences, Inc.). After 30 min, the formation of NADH was determined by measuring the increase in absorbance at 340 nm. Enzyme activity was expressed as nmol per minute per 1 mg of total cell extract protein. The blank consisted of the reaction mixture without the cell extract.
Quantification of intracellular carbohydrates.
P. gingivalis wild type or the galE strain was cultured in 0.5-liter portions of BHI-HM broth supplemented with and without galactose (0%, 0.01%, 0.05%, and 0.1%). The cells were collected and washed with phosphate-buffered saline (PBS) (pH 7.4) three times (3,000 × g for 20 min at 4°C) and then lyophilized. Forty milligrams of lyophilized cells was suspended in 5 ml of phosphate buffer (pH 7.2) with 1% Triton X-100 and then disrupted ultrasonically in an ice bath for five cycles of 30 seconds each at 100 W (IKASONIC U50 control). Cell debris was removed by centrifugation (10,000 × g for 2 min at 4°C). The supernatant was centrifuged at 100,000 × g for 2 h at 10°C. The soluble fraction was collected as intracellular components. The quantity of carbohydrates was determined using a phenol-sulfuric acid colorimetric method (12) with glucose as the control sugar.
LPS analysis.
P. gingivalis wild-type and mutant strains were cultured in 0.5 liter BHI-HM broth. The cells were collected and washed with PBS (pH 7.4) three times (3,000 × g for 20 min at 4°C) and then lyophilized. LPS was extracted from the same weight (40 mg) of the lyophilized cells by using a hot phenol extraction method (39). The aqueous phase was separated by centrifugation (9,000 × g for 30 min at 4°C) and then collected and dialyzed at 4°C for 12 h against distilled water to remove phenol contamination. To aid in the elimination of nucleic acids, the dialyzed solution was brought to 0.15 M NaCl-NaOH (pH 6.5) and treated with RNase A (0.02 mg/ml; QIAGEN, GmbH, Hilden, Germany) for 2 h at room temperature, followed by the addition of MgCl2 to 4 mM Mg2+, and then treated with DNase I (5 μg/ml; Sigma, St. Louis, MO) for 6 h at room temperature. Next, the solution was dialyzed against distilled water overnight at 4°C. To aid in the elimination of protein, the solution was brought to 30 mM Tris-Cl (pH 8.0) and treated with proteinase K (0.4 mg/ml; QIAGEN) for 1 h at 60°C and dialyzed with distilled water. Finally, the dialyzed solution was centrifuged twice at 100,000 × g for 12 h at 10°C, and the pellet was lyophilized as purified LPS. An equivalent weight of the lyophilized LPS (5 μg) was electrophoresed on a 15% sodium dodecyl sulfate-polyacrylamide gel (17) and subjected to silver staining (38). The band densities of the O-antigen ladder in the wild-type, galE, and galE-c strains were evaluated using densitometric scanning (CS analyzer 1.0; ATTO Co., Tokyo, Japan).
Biofilm formation assay.
Biofilm formation by the P. gingivalis wild-type and mutant strains was assayed using a method described previously (20) with some modifications. To produce biofilms, 2 × 107 CFU of P. gingivalis in 200 μl of BHI-HM broth (1 × 108 CFU/ml) was added to the wells of 96-well flat-bottom polystyrene microtiter plates (Corning, New York, NY). After the plates were anaerobically incubated at 37°C for 34 and 48 h, planktonic cells in liquid medium were discarded and the plates were washed twice with distilled water. The plates were then air dried, and attached biofilms were stained with 0.25% safranin for 30 min. Then, the plates were rinsed twice with distilled water to remove excess dye and air dried. All dye associated with the attached biofilms was dissolved with 200 μl of 100% ethanol, and then OD492/620 absorbance was measured by use of a microplate reader (Multiskan Ascent; Thermo Electron Oy, Vantaa, Finland) to determine the amount of biofilm formation.
Scanning electron microscopy.
For scanning electron microscopy (SEM) examinations, P. gingivalis wild type or the galE strain was developed on nontreated plastic sheets (Wako Chemical Ltd., Osaka, Japan) placed in six-well polystyrene cell culture plates (Corning). P. gingivalis (2 × 108 CFU) in 2 ml of BHI-HM broth per well was added and incubated at 37°C under an anaerobic condition. After 12 h, the sheets were rinsed twice with PBS to remove any planktonic cells. Attached cells on the sheets were fixed with 2.5% glutaraldehyde and 2% paraformaldehyde in PBS for 30 min at room temperature and subsequently washed three times in PBS. Then, secondary fixation with 1% osmium tetraoxide in PBS was performed. The samples were washed in PBS, dehydrated in 50% ethanol to absolute ethanol, immersed in isoamyl acetate, dried by critical point drying, coated with osmium vapor by an osmium plasma coater, and observed by SEM (S5200; HITACHI Corporation, Hitachi, Japan).
Determination of MIC.
Etest benzylpenicillin, oxacillin, ampicillin, cefotaxime, ceftriaxone, imipenem, tetracycline, doxycycline, erythromycin, clindamycin, tobramycin, kanamycin, and vancomycin strips (AB Biodisk, Solna, Sweden) were used to determine MICs. An inoculum of each P. gingivalis strain adjusted to a standard turbidity of McFarland 0.5 was applied to BHI-HM blood agar plates. The Etest strips with continuous gradients of the antibiotics were placed on the surface of the agar. After a 5-day incubation anaerobically, the MICs were determined by the regions where the zones of inhibition intersected the MIC scale on the strips.
Statistical analysis.
Statistical analysis was performed using the Mann-Whitney U test. P values of 0.05 or less were considered to indicate statistical significance.
Nucleotide sequence accession numbers.
The nucleotide sequences of the PG0106 (wecA), PG1964 (wbaP), PG0347 (galE), PG1225 (wzt), and PG0045 (htpG) regions of P. gingivalis W83 strain are listed under accession nos. AAQ65351, AAQ66941, AAQ65558, AAQ66314, and AAQ65296, respectively.
RESULTS
Construction of P. gingivalis mutants.
To determine whether P. gingivalis possesses homologs of wecA, wbaP, galE, and wzt genes, we performed BLAST search analysis using the genome sequence entries of S. enterica serovar Typhi or B. thetaiotaomicron as query sequences. All of those homologs were found in the genome of P. gingivalis, and the amino acid sequences corresponding to the wecA, wbaP, galE, and wzt genes in P. gingivalis showed 25%, 44%, 40%, and 38% identity, respectively, to those of the corresponding genes of S. enterica serovar Typhi, and 35%, 43%, 66%, and 52% identity, respectively, to those of B. thetaiotaomicron. In order to analyze the biological roles of wecA, wbaP, galE, and wzt genes, we constructed each target gene mutant by use of P. gingivalis ATCC 33277. Each gene disrupted by insertion of the ermF-ermAM cassette was introduced into P. gingivalis by electroporation (Fig. 1A). We also generated a galE-complemented mutant, the galE-c strain, by cis replacement of the ermF-ermAM cassette-inserted galE mutation to the intact galE gene with the tetQ gene (Fig. 1B and C) in order to confirm that inactivation of the galE gene results in the phenotypes of the galE strain.
Loss of UDP-galactose 4-epimerase activity and accumulation of intracellular carbohydrates by galE mutation.
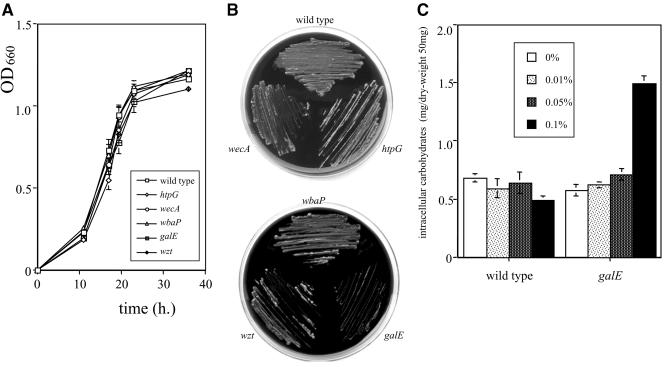
First, we examined the growth of the wild-type and mutant strains. All mutant strains grew to same extent as the parental strain in BHI-HM broth (Fig. 2A). Since it has been reported that galE mutants of a number of species accumulated intracellular Gal-1-P and UDP-Gal in the presence of galactose, leading to bacteriostasis and bacteriolysis (8, 26, 30), we examined their growth in BHI-HM broth supplemented with galactose. Among the mutant strains, only the growth of the galE strain was delayed in the presence of more than 0.1% galactose (data not shown). The galE strain did not grow in the presence of more than 1% galactose (Fig. 2B). In contrast, the complemented strain, the galE-c strain, was able to grow in the presence of 1% galactose (data not shown). Moreover, the galE strain accumulated intracellular carbohydrates in large quantities in the presence of 0.1% galactose (Fig. 2C). In order to confirm the defect of GalE activity in the galE strain, a UDP-galactose 4-epimerase assay was carried out. As expected, the galE strain lost the enzyme activity, while that of the parental strain was active (0 versus 111.8 ± 2.3 nmol/mg protein · min). The galE-c strain demonstrated that activity at an intermediate level compared to the wild-type and galE strains (6.7 ± 0.6 nmol/mg protein · min).
FIG.2.
Growth assays and quantitation of intracellular polysaccharide in the absence or presence of galactose. (A) P. gingivalis wild-type and mutant strains were grown in BHI-HM broth, and OD660 absorbance was measured at different time points. Data shown are representative of three independent experiments. The results are expressed as the mean ± standard deviation (SD) from a triplicate assay. Similar results were obtained in three independent experiments. (B) The strains were also grown on BHI-HM agar plates supplemented with 1% galactose. (C) P. gingivalis wild-type and galE strains were grown in BHI-HM broth supplemented with or without galactose (0%, 0.01%, 0.05%, and 0.1%). Quantities of intracellular polysaccharides, which were extracted from every 50 mg of lyophilized cells, were determined by a phenol-sulfuric acid colorimetric method with glucose as the control. Total intracellular carbohydrates are expressed as mg per 50 mg of dry weight. The results are expressed as the mean ± SD of a triplicate assay. Similar results were obtained in two independent experiments.
Modification of LPS O antigen in wecA and galE strains.
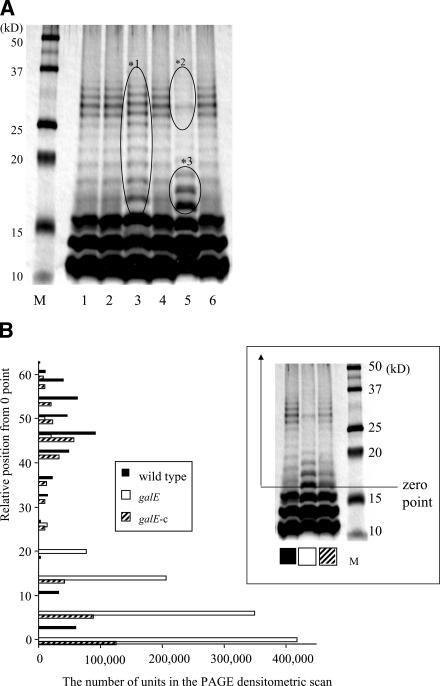
We hypothesized that the defect of the proposed genes would have an effect on the synthesis of LPS in this organism. To confirm this hypothesis, we analyzed the LPS profiles of each strain by using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) and silver staining. Although the distribution of the bands of LPS in the htpG, wbaP, and wzt strains scarcely changed relative to those of the parental strain, LPS in both the wecA and galE strains showed an altered profile (Fig. 3A). A minor shift of the O-antigen ladder was observed for the wecA strain; this shift was denoted by *1. For the galE strain, though the lowest three bands of O antigen scarcely change, the densities of the O-antigen bands at high molecular mass, from 25 to 35 kDa, nearly disappeared (*2), while those at low molecular mass, from 16 to 19 kDa, became more intense (*3). The galE-c strain recovered production of LPS at an intermediate level compared to the wild-type and galE strains, as shown in the inset in Fig. 3B. When the densities of the O-antigen ladder bands were quantitated using gel scanning, those of the galE strain were higher at a lower molecular mass (Fig. 3B), indicating that the O-antigen length of the galE strain was shorter than that of the wild type.
FIG. 3.
LPS profiles of P. gingivalis wild-type and mutant strains. LPS samples extracted from the P. gingivalis wild-type and mutant strains were analyzed by sodium dodecyl sulfate-PAGE and silver staining. Each lane contains 5 μg of LPS. (A) Lanes: 1, wild type; 2, the htpG strain; 3, the wecA strain; 4, the wbaP strain; 5, the galE strain; and 6, the wzt strain. M: molecular mass marker. The areas shown with ovals represent a minor shift of the O-antigen ladder (*1) in the wecA strain and a decrease of the high molecular band of O antigen (*2) and an increase of the low molecular band of O antigen (*3) in the galE strain. (B) The band densities of the O-antigen ladders after silver staining in the wild-type, galE, and galE-c strains were evaluated using densitometric scanning. The y axis of the histogram indicates the relative position from 0 point, whose position, denoted in the inset, was determined to be approximately 16 kDa.
Biofilm formation and SEM analysis.
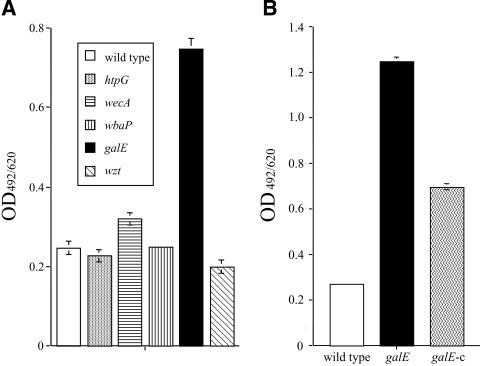
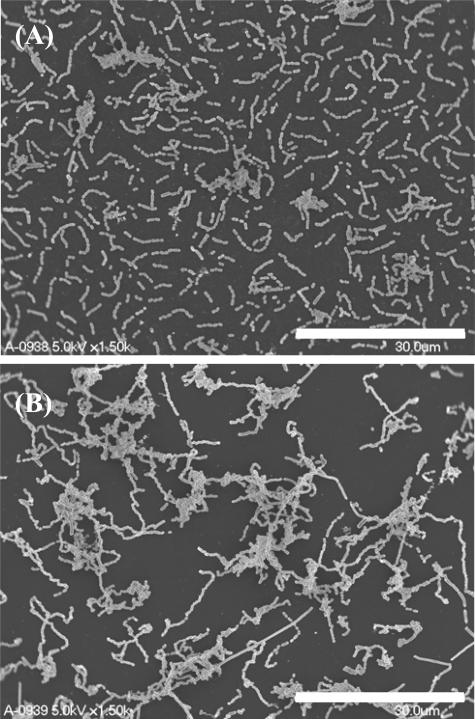
To confirm the involvement of the proposed genes in biofilms organized from P. gingivalis, biofilm formation assays were carried out using 96-well microplates. No major change in biofilm mass was observed in any of the mutants, except for the galE strain. After a 34-hour incubation, biofilms organized from the galE strain showed an intensity more than threefold greater than those from the wild-type strain (Fig. 4A). After a 48-hour incubation, biofilms related to the galE strain showed greater than 4.5 times the mass of the wild-type strain (Fig. 4B). Further, the galE-c strain formed biofilms of an intermediate level compared to those of the wild-type and galE strains (Fig. 4B). We also examined the biofilm structure of the galE strain by SEM. After a 12-hour incubation on plastic surfaces, there was no difference between the wild-type and galE strains in terms of the number of attached cells. However, the galE strain was found to have cell-aggregative and long-chain phenotypes, in contrast to the wild type (Fig. 5A and B). After 24 h, the number of attached cells for the galE strain was greatly increased compared with that for the parental strain (data not shown).
FIG. 4.
Biofilm formation by P. gingivalis wild-type and mutant strains. Biofilm formation was examined after 34 (A) and 48 (B) hours of culturing in BHI-HM broth. OD492/620 absorbance was measured to determine the biofilm mass. Data shown are representative of three independent assays. The results are expressed as the mean ± SD of a triplicate assay. Similar results were obtained in three independent experiments.
FIG. 5.
SEM images of P. gingivalis wild-type and galE strains. P. gingivalis wild-type (A) and galE (B) strains were grown on plastic sheets for 12 h, after which attached cells were investigated using SEM. Scale bars are shown at the lower right of each electron micrograph.
High sensitivity to antibiotics by galE mutation.
Etest strips were used to examine whether the galE strain had an altered sensitivity to antibiotics. The galE strain was found to be significantly more susceptible to benzylpenicillin, oxacillin, cefotaxime, imipenem, and vancomycin than the wild type (Table 3).
TABLE 3.
MICs for P. gingivalis wild-type, galE, and galE-c strains
| Antibiotic | MIC (μg/ml)a for:
|
||
|---|---|---|---|
| 33277 (wild type) | galE strain | galE-c strain | |
| Benzylpenicillin | 0.011 ± 0.0023 | 0.0047 ± 0.0012b | 0.0073 ± 0.0012 |
| Oxacillin | 0.10 ± 0.020 | 0.053 ± 0.010b | 0.084 ± 0.017 |
| Ampicillin | <0.016 | <0.016 | <0.016 |
| Cefotaxime | 0.0067 ± 0.0012 | 0.002 ± 0.00b | 0.006 ± 0.002c |
| Ceftriaxone | <0.002 | <0.002 | <0.002 |
| Imipenem | 0.084 ± 0.035 | 0.024 ± 0.0080b | 0.043 ± 0.018 |
| Tetracycline | 0.042 ± 0.0087 | 0.037 ± 0.0087 | 1.7 ± 0.29d |
| Erythromycin | <0.016 | >256e | <0.016 |
| Clindamycin | <0.016 | >256e | <0.016 |
| Kanamycin | >256 | >256 | >256 |
| Tobramycin | >256 | >256 | >256 |
| Vancomycin | 12 ± 4 | 1.8 ± 0.29b | 4 ± 0c |
Values are shown as means ± standard deviations.
The galE strain had significant difference in sensitivity to the indicated antibiotic compared to the wild-type 33277.
The galE-c strain had significant difference in sensitivity to the indicated antibiotic compared to the ΔgalE strain.
Resistance was dependent on insertion of tetQ.
Resistance was dependent on insertion of ermF.
DISCUSSION
In the present study, the essential role of the galE gene of P. gingivalis in LPS biosynthesis was established following PAGE analysis of its LPS (Fig. 3). The O-antigen ladder bands of P. gingivalis galE strain were found to be distributed at molecular masses lower than those seen for the parental strain, indicating that the O polysaccharide length of the galE strain was decreased. Further, the galE-c strain partially restored the synthesis of the O antigen (Fig. 3B). The incomplete recovery observed in our complementary trial might have been dependent on the low level of activity of the tetQ promoter in comparison with that of the native promoter, because the GalE activity of the galE-c strain was also incomplete in comparison with that of the wild type.
GalE protein catalyzes the interconversion of UDP-glucose to UDP-galactose. In previous studies, the galE mutant of Vibrio cholerae, whose O antigen does not include galactose, did not have an altered LPS profile (26), while galE mutants of other bacteria, such as Neisseria meningitidis and Helicobacter pylori, were found to produce truncated LPS molecules that lacked galactose (16, 19). Bramanti et al. reported that galactose accounted for 25.3% of the total monosaccharides of LPS in P. gingivalis strain 33277 (4). Further, Paramonov et al. also showed that the O antigen of P. gingivalis strain W50 consists of the tetrasaccharide repeating unit (6)-α-d-Glcp-(1-4)-α-l-Rhap-(1-3)-β-d-GalNAc-(1-3)-α-d-Galp-(1) (31). Therefore, we speculated that the truncated O antigen in the P. gingivalis galE strain is a result of the inability of this mutant to utilize UDP-galactose, which is one of the essential building blocks of O antigen in P. gingivalis. Further, the galE strain was found to exhibit the long-chain phenotype, in contrast to the wild-type strain. This phenotype may be due to a failure of cellular segregation resulting from alteration of the cell surface, which contains a variety of glycoconjugates, such as LPS and peptidoglycan. However, a more precise analysis is required to fully understand the relationship between the galE mutation and these effects.
Inactivation of the galE gene in P. gingivalis also had an effect on biofilm formation (Fig. 4), which may have been due to the fact that galactose is one of the components of the carbohydrate polymer on the surface of P. gingivalis (35). Therefore, we considered that the diminished levels of galactose in biofilms may have an effect on the total amount of biofilms and/or chemical behavior, such as hydrophobicity, resulting in aggregation of the cells (Fig. 5D) and overproduction of biofilms (Fig. 4). Alternatively, the shortened O antigen of the galE strain might promote the property of initial attachment to the materials, followed by an increase in biofilm formation. The O antigen on the bacterial surface is located at the interface between the bacterium and its environment. When the length of the O polysaccharide chain as a physical obstacle becomes short on its surface, an essential component of the outer membrane involved in the affinity of the organism to solid surfaces or other organisms may become easily exposed to the environment. This may be true in the case of the galE strain of P. gingivalis, which was shown to promote biofilm formation. It has also been reported that O-antigen chain length has an effect on the sensitivity of bacteria to bactericidal molecules. For example, a truncated O-antigen mutant of V. cholerae raised its sensitivity to complement and cationic peptides (26). Likewise, the increased sensitivity to some antibiotics shown for the galE strain (Table 3) might be dependent on the shortened O antigen on its surface.
We also examined the LPS profiles of two glycosyltransferase mutants, the wecA and wbaP strains, and of an ABC transporter mutant, the wzt strain (Fig. 3A). For the wecA strain, a minor shift in the O-antigen ladder was observed; this might have been because WecA functions in the first step of O polymerization involved in the formation of lipid-linked GlcNAc by transfer of GlcNAc-P residue to the lipid anchor (24). In contrast, the distribution of the O-antigen ladder in the wbaP strain did not change in relation to that of the parental strain. It is possible that there is an alternative gene product that compensates for the loss of function by WbaP. For example, PG1135 is paralogous to the wbaP gene (PG1964) (42% amino acid identity) and may fulfill the role of WbaP in the wbaP strain. Further, the defect of the wzt gene, which encodes the ABC transporter subunit, also did not have an effect on the LPS profile. In the same way, P. gingivalis possesses two paralogs of the wzt gene (PG1225), PG2199 and PG2206 (31% and 30% amino acid identities, respectively). Since both PG2199 and PG2206 encode the ABC transporter, they may fulfill the role of Wzt in the wzt strain. Otherwise, another pathway for O-antigen transport in the wzt strain might compensate for the ability. The three currently known pathways for O-antigen transport are distinguished by their respective export mechanisms. The pathways are called ABC transporter dependent, Wzy dependent, and synthase dependent (32). Thus, it is possible that the Wzy-dependent and/or synthase-dependent pathway might be active in P. gingivalis.
In conclusion, our results suggest that P. gingivalis galE plays an important role in both the synthesis of O antigen and the formation of biofilms, which were found to be closely related with its virulence. Although the effects following the galE mutation are insufficient to explain the detailed mechanism, we suggest that the phenotypes related to the galE mutation are associated with polysaccharide changes that occur at the bacterial surface. Further studies to elucidate the precise mechanism of these phenomena are in progress. Finally, our conclusive evidence for the relationship between the P. gingivalis galE mutation and these virulence factors may provide important clues for understanding the mechanism involved in the progress of periodontal diseases.
Acknowledgments
We thank Howard K. Kuramitsu for his critical reading of the manuscript and Noriko Saito for technical support.
This work was supported in part by grants-in-aid from Development Scientific Research of the Ministry of Education, Culture, Sports, Science, and Technology of Japan (15390571 and 17791348) and the Ministry of Health, Labor and Welfare (H16-Medical Services-014).
Editor: V. J. DiRita
Footnotes
Published ahead of print on 5 September 2006.
REFERENCES
- 1.Barreto, M., E. Jedlicki, and D. S. Holmes. 2005. Identification of a gene cluster for the formation of extracellular polysaccharide precursors in the chemolithoautotroph Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 71:2902-2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boman, H. G. 1995. Peptide antibiotics and their role in innate immunity. Annu. Rev. Immunol. 13:61-92. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-256. [DOI] [PubMed] [Google Scholar]
- 4.Bramanti, T. E., G. G. Wong, S. T. Weintraub, and S. C. Holt. 1989. Chemical characterization and biologic properties of lipopolysaccharide from Bacteroides gingivalis strains W50, W83, and ATCC 33277. Oral Microbiol. Immunol. 4:183-192. [DOI] [PubMed] [Google Scholar]
- 5.Camprubi, S., S. Merino, J. F. Guillot, and J. M. Tomas. 1993. The role of the O-antigen lipopolysaccharide on the colonization in vivo of the germfree chicken gut by Klebsiella pneumoniae. Microb. Pathog. 14:433-440. [DOI] [PubMed] [Google Scholar]
- 6.Cutler, C. W., J. R. Calmer, J. R., and C. A. Genco. 1995. Pathogenic strategies of the oral anaerobe, Porphyromonas gingivalis. Trends Microbiol. 3:45-51. [DOI] [PubMed] [Google Scholar]
- 7.Farquharson, S. I., G. R. Germaine, and G. R. Gray. 2000. Isolation and characterization of the cell-surface polysaccharides of Porphyromonas gingivalis ATCC 53978. Oral Microbiol. Immunol. 15:151-157. [DOI] [PubMed] [Google Scholar]
- 8.Fukusawa, T., and H. Nikaido. 1961. Galactose sensitive mutants of Salmonella. II. Bacteriolysis induced by galactose. Biochim. Biophys. Acta 48:470-479. [DOI] [PubMed] [Google Scholar]
- 9.Grenier, D., and D. Mayrand. 1987. Functional characterization of extracellular vesicles produced by Bacteroides gingivalis. Infect. Immun. 55:111-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haake, S. K., S. C. Yoder, G. Attarian, and K. Podkaminer. 2000. Native plasmids of Fusobacterium nucleatum: characterization and use in development of genetic systems. J. Bacteriol. 182:1176-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harvill, E. T., A. Preston, P. A. Cotter, A. G. Allen, D. J. Maskell, and J. F. Miller. 2000. Multiple roles for Bordetella lipopolysaccharide molecules during respiratory tract infection. Infect. Immun. 68:6720-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodge, J. E., and B. T. Hofreiter. 1962. Phenol-sulfuric acid colorimetric method. Methods Carbohydr. Chem. 1:388-389. [Google Scholar]
- 13.Holt, S. C., L. Kesavalu, S. Walker, and C. A. Genco. 2000. Virulence factors of Porphyromonas gingivalis. Periodontol. 2000 20:168-238. [DOI] [PubMed] [Google Scholar]
- 14.Holt, S. C., and T. E. Bramanti. 1991. Factors in virulence expression and their role in periodontal disease pathogenesis. Crit. Rev. Oral Biol. Med. 2:177-281. [DOI] [PubMed] [Google Scholar]
- 15.Hone, D. M., R. Morona, S. Attridge, and J. Hackett. 1987. Construction of defined galE mutants of Salmonella for use as vaccines. J. Infect. Dis. 156:167-174. [DOI] [PubMed] [Google Scholar]
- 16.Kwon, D. H., J. S. Woo, C. L. Perng, M. F. Go, D. Y. Graham, and F. A. El-Zaatari. 1998. The effect of galE gene inactivation on lipopolysaccharide profile of Helicobacter pylori. Curr. Microbiol. 37:144-148. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 18.Lamont, R. J., and H. F. Jenkinson. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62:1244-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, F. K., D. S. Stephens, B. W. Gibson, J. J. Engstrom, D. Zhou, and M. A. Apicella. 1995. Microheterogeneity of Neisseria lipooligosaccharide: analysis of a UDP-glucose 4-epimerase mutant of Neisseria meningitidis NMB. Infect. Immun. 63:2508-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopatin, D. E., A. Combs, D. G. Sweier, J. C. Fenno, and S. Dhamija. 2000. Characterization of heat-inducible expression and cloning of HtpG (Hsp90 homologue) of Porphyromonas gingivalis. Infect. Immun. 68:1980-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maskell, D. J., M. J. Szabo, P. D. Butler, A. E. Williams, and E. R. Moxon. 1991. Molecular analysis of a complex locus from Haemophilus influenzae involved in phase-variable lipopolysaccharide biosynthesis. Mol. Microbiol. 5:1013-1022. [DOI] [PubMed] [Google Scholar]
- 23.Maxwell, E. S., K. Kurahashi, and H. M. Kalckar. 1962. Enzymes of the Leloir pathway. Methods Enzymol. 5:174-189. [Google Scholar]
- 24.Meier-Dieter, U., K. Barr, R. Starman, L. Hatch, and P. D. Rick. 1992. Nucleotide sequence of the Escherichia coli rfe gene involved in the synthesis of enterobacterial common antigen. Molecular cloning of the rfe-rff gene cluster. J. Biol. Chem. 267:746-753. [PubMed] [Google Scholar]
- 25.Nakayama, K., T. Kadowaki, K. Okamonto, and K. Yamamoto. 1995. Construction and characterization of arginine-specific cystein protease (Arg-gingipain)-deficient mutants of Porphyromonas gingivalis. Evidence for significant contribution of Arg-gingipain to virulence. J. Biol. Chem. 270:23619-23626. [DOI] [PubMed] [Google Scholar]
- 26.Nesper, J., C. M. Lauriano, K. E. Klose, D. Kapfhammer, A. Kraiss, and J. Reidl. 2001. Characterization of Vibrio cholerae O1 E1 Tor galU and galE mutants: influence on lipopolysaccharide structure, colonization, and biofilm formation. Infect. Immun. 69:435-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nevola, J. J., B. A. D. Stocker, D. C. Laux, and P. S. Cohen. 1985. Colonization of the mouse intestine by an avirulent Salmonella typhimurium strain and its lipopolysaccharide-deficient mutants. Infect. Immun. 50:152-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nevola, J. J., D. C. Laux, and P. S. Cohen. 1987. In vivo colonization of the mouse large intestine and in vitro penetration of intestinal mucus by an avirulent smooth strain of Salmonella typhimurium and its lipopolysaccharide-deficient mutant. Infect. Immun. 55:2884-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicolas, P., and A. Mor. 1995. Peptides as weapons against microorganisms in the chemical defense system of vertebrates. Annu. Rev. Microbiol. 49:277-304. [DOI] [PubMed] [Google Scholar]
- 30.Nikaido, H. 1961. Galactose sensitive mutants of Salmonella. I. Metabolism of galactose. Biochim. Biophys. Acta 48:460-469. [DOI] [PubMed] [Google Scholar]
- 31.Paramonov, N., D. Bailey, M. Rangarajan, A. Hashim, G. Kelly, M. A. Curtis, and E. F. Hounsell. 2001. Structural analysis of the polysaccharide from the lipopolysaccharide of Porphyromonas gingivalis strain W50. Eur. J. Biochem. 268:4698-4707. [DOI] [PubMed] [Google Scholar]
- 32.Raetz, C. R. H., and C. Whitfield. 2002. Lipopolysaccharide endotoxin. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robertson, B. D., M. Frosch, and J. P. van Putten. 1993. The role of galE in the biosynthesis and function of gonococcal lipopolysaccharide. Mol. Microbiol. 8:891-901. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Schifferle, R. E., M. S. Reddy, J. J. Zambon, R. J. Genco, and M. J. Levine. 1989. Characterization of a polysaccharide antigen from Bacteroides gingivalis. J. Immunol. 143:3035-3042. [PubMed] [Google Scholar]
- 36.Sweier, D. G., A. Combs, C. E. Shelburnej, J. C. Fenno, and D. E. Lopatin. 2003. Construction and characterization of a Porphyromonas gingivalis htpG disruption mutant. FEMS Microbiol. Lett. 225:101-106. [DOI] [PubMed] [Google Scholar]
- 37.Taylor, P. W. 1983. Bactericidal and bacteriolytic activity of serum against gram-negative bacteria. Microbiol. Rev. 47:46-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai, C. M., and C. E. Frasch. 1982. A sensitive stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 39.Westphal, O., and K. Jann. 1965. Bacterial lipopolysaccharides. Extraction with phenol-water and further applications of the procedure. Methods Carbohydr. Chem. 5:83-92. [Google Scholar]