Abstract
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 and enteropathogenic E. coli (EPEC) adherence to epithelial cells results in the formation of actin pedestals. Pedestal formation requires the bacterial protein Tir, which is inserted into the epithelial cell plasma membrane by the type III secretion system. EPEC and EHEC use different Tir-based mechanisms for pedestal formation, and the EPEC Tir residues required have been well described. In contrast, little is known about the regions of EHEC O157:H7 Tir that are essential for pedestal formation. Additionally, EHEC O157:H7 Tir is serine/threonine phosphorylated, although the residues involved and their role in pedestal formation are not known. In this study, we describe two regions within the carboxy terminus of EHEC O157:H7 Tir that are required for phosphorylation and pedestal formation. Serines 436 and 437 are substrates for protein kinase A phosphorylation, although this is not required to form pedestals. Using a series of internal deletion mutants, we found that amino acids 454 to 463 are required for efficient pedestal formation. Deleting this region resulted in a significant decrease in the recruitment of both filamentous actin and the actin binding protein α-actinin. As α-actinin binds directly to the EHEC O157:H7 amino terminus, these data suggest that its recruitment is dependent on pedestal formation.
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 is an important cause of food- and waterborne illness in North America, as evidenced by the recent outbreak in Walkerton, Ontario, which affected close to 2,000 individuals and resulted in seven deaths (17, 26). EHEC O157:H7 infection initially results in watery diarrhea, which can progress to severe bloody diarrhea (hemorrhagic colitis) in patients of all ages (30). In susceptible individuals, infection can result in a potentially fatal systemic complication, the hemolytic uremic syndrome, characterized by hemolytic anemia, thrombocytopenia, and renal failure (16, 30). EHEC is a member of a family of pathogens that cause attaching and effacing lesions. Attaching and effacing lesions are characterized by the degeneration of the epithelial cell microvilli and the formation of actin-rich pedestals within the host enterocytes beneath the adherent bacteria (25, 29).
Pedestal formation requires two EHEC O157:H7 proteins, Tir and EspFu/TccP, injected into the host cell by the bacterial type III secretion system (8, 14). EHEC O157:H7 Tir is a 558-amino-acid protein inserted into the host cell plasma membrane in a hairpin loop conformation (12). The extracellular domain binds to the EHEC O157:H7 outer membrane protein intimin, which is essential for clustering Tir and initiating pedestal formation (9). Considerably less is known about the contribution of the intracellular domains, although the amino-terminal domain has been shown to bind directly to the actin binding and cross-linking protein α-actinin (28). In the related pathogen enteropathogenic E. coli (EPEC), pedestal formation occurs by a different Tir-based mechanism. EPEC Tir is phosphorylated on Y474, contained within a 12-amino-acid carboxy-terminal domain shown to be required for efficient pedestal formation (5, 11, 23). Y474 phosphorylation by the Src family kinase member Fyn promotes the direct binding of the cellular adaptor protein Nck. Nck activates the N-WASP-Arp 2/3 pathway to modulate actin polymerization (7, 18, 33).
In contrast, EHEC O157:H7 Tir is not tyrosine phosphorylated and lacks the 12-amino-acid region within the carboxy terminus containing Y474 (11, 22). Additionally, pedestal formation by EHEC O157:H7 is Nck independent (7, 18). Instead, EHEC O157:H7 injects EspFu/TccP, which can bind to and activate N-WASP and is required for efficient pedestal formation (8, 14). Although EspFu/TccP can be coimmunoprecipitated with Tir, it has not been demonstrated to bind directly, suggesting the contribution of other proteins (8, 14). Little is known about the carboxy-terminal residues of EHEC O157:Tir that contribute to pedestal formation. A recent study using Tir transfected into mammalian cells suggested that 12 carboxy-terminal residues were involved in pedestal formation (4). We previously reported that EHEC can use EPEC Tir Y474F to form pedestals, and Campellone and colleagues demonstrated that this requires EspFu (8, 11). Collectively, these results suggest that similar regions in the two Tir proteins may be involved. Therefore, in this study, we generated a series of EHEC O157:H7 Tir internal deletion mutants to define the residues required for Tir phosphorylation and pedestal formation. Our results indicate that EHEC O157:H7 Tir is phosphorylated by protein kinase A (PKA). Additionally, we found that a 10-amino-acid region was required for both pedestal formation and α-actinin recruitment, a result which complements those obtained using transfected Tir- and EspFu/TccP-deficient strains (4, 14).
MATERIALS AND METHODS
Bacterial strains and HeLa cell cultures.
The strains used in this study are listed in Table 1. EHEC strains used to infect HeLa cells were grown in Luria-Bertani broth supplemented with appropriate antibiotics as standing overnight cultures at 37°C. HeLa cells (CCL2; American Type Culture Collection) were cultured in Dulbecco's minimal essential medium supplemented with 10% fetal calf serum and grown at 37°C in 5% CO2.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference |
|---|---|---|
| Strains | ||
| EHEC O157:H7 86/24 | E. coli O157:H7 wild type | 16 |
| CVD451 | escN deletion in EHEC O157:H7 86/24 | 21 |
| EHEC O157:H7Δtir | tir deletion in EHEC O157:H7 86/24 | 12 |
| EHECΔtir/Tir | EHEC O157:H7 expressing WT EHEC Tir or Tir internal deletion mutants | This study |
| EPEC E2348/69Δtir | tir deletion in E. coli O127:H6 wild type | 24 |
| Plasmids | ||
| pEH86 | EHEC O157:H7 tir in pACYC184, parental plasmid for EHEC Tir internal deletion mutants | 11 |
| pEP23 | EPEC tir in pACYC184, parental plasmid for EPEC Tir internal deletion mutants | 11 |
| pAA511.1 | His6-EHEC Tir-CesT, Tir-CesT in bacterial expression vector, parental plasmid for production of recombinant WT or mutant EHEC Tir | 1 |
Construction of in-frame EHEC O157:H7 tir internal deletion mutants.
Inverse PCR was used to construct the in-frame deletions used for this study. Briefly, PCR was performed using pEH86 or pEP23 as template (Table 1), and specific oligonucleotide primers were designed to introduce, by silent mutation, a unique restriction site flanking the region to be deleted or mutagenized (Table 2). Amplicons were digested with the chosen restriction endonuclease and then self-ligated and used to transform E. coli DH10B. Candidate clones were confirmed by sequence analysis, and then these constructs were used to transform either EHEC O157:H7Δtir strain.
TABLE 2.
Oligonucleotides used in this studya
| Oligonucleotide | Sequence |
|---|---|
| Tir 403+ | GG ACT AGT GCA CGT ACG GTA |
| Tir 394− | GG ACT AGT TTG TTC TAC CGG |
| Tir 427+ | GG ACT AGT GAA GAT ACC ATG |
| Tir 405− | GG ACT AGT AGT TGT AGT TGT |
| Tir 442+ | GG ACT AGT TCG ACT TTC TTT GAC ACT T |
| Tir 425− | GG ACT AGT ATC TAC ATT GCC CTG TGC A |
| Tir 463+ | TTT ACT AGTCTG CAT GAT TCG CAG GTG CCG |
| Tir 442− | TTT ACT AGT GCT AGC CAT CGA GCT ACG TCT |
| Tir 472+ | TTT ACT AGT AAT TCT AAT ACG TCT GTT CAG AA |
| Tir 464− | TTT ACT AGT TTT AAC ATC AGC ATA CGG ATT C |
| Tir 490+ | GG 0ACT AGT ACC ATT CAA CAT |
| Tir 473− | GG ACT AGT CGG CAC CTG CGA |
| Tir 511+ | GG ACT AGT AGT GCG GGG ATT |
| Tir 486− | GG ACT AGT TGT ATT CCC CAT |
| Tir 449− | TTT ACT AGT CAG GTG CCG ACT TCT AAT TC |
| Tir 454− | TTT ACT AGT CCC TAT GCT GGA AGT GTC AA |
| Tir 467+ | TTT ACT AGT CAG GTG CCG ACT TCT AAT TC |
| Tir 463+ | TTT ACT AGT CTG CAT GAT TCG CAG GTG C |
| Tir 458+ | AAA GGT ACC GCT GAT GTT AAA ACA TCC C |
| Tir 454− | AAA GGT ACC TAT GCT GGA AGT GTC AAA G |
| Tir 450+ | AAA ACT AGT ATA GGG ACC GTG CAG AAT CCG |
| Tir 443− | AAA ACT AGT GCT AGC CAT CGA GCT ACG TCT GC |
| Tir 462+ | GG ACT AGT CTG CAT GAT TCG CAG GTG |
| Tir 458− | GG ACT AGT CGG ATT CTG CAC GGT CCC TAT GCT GG |
| Tir 453+ | AAA ACT AGT GTG CAG AAT CCG TAT GCT GAT G |
| Tir 450− | GG ACT AGT GTC AAA GAA AGT CGA CGA GG |
| TirΔPKA+ | TTT TCT AGA CGA GCC GCG ATG GCT AGC AC |
| TirΔPKA− | TTT TCT AGA CTC CAT GGT ATC TTC TGA CCC |
| TirY458F+ | ATG CAG ACT AGT TTT AAC ATC AGC AAA CGG ATT CTG |
| TirY458F− | GTT AAA ACT AGT CTG CAT GAT TCG CAG GTG |
| EPEC 451+ | TTT GCG GCG TGA CCA GTG TGT CGA TGC AAC |
| EPEC 459− | TTT GCG CGC CAC TTC GCT AGA GGA ACT TGA CC |
The sequences of introduced restriction sites are underlined.
We used pAA511.1 (Table 1) as the template to construct the His6-Tir proteins used in the in vitro phosphorylation assay. We used inverse PCR to construct His6-TirΔ425-442 (oligonucleotides Tir 442+ and Tir 425−) (Table 2) and His6-Tir S436A/S437A (TirΔPKA+ and TirΔPKA). The resulting constructs were transformed into E. coli BL21DE3, and expression was induced using IPTG (isopropyl-β-d-thiogalactopyranoside) (1).
In vitro phosphorylation assay.
His6-Tir constructs were purified using nickel-agarose chromatography following the manufacturer's instructions (Novagen). For each reaction, 0.25 μg His6-Tir (12.5 μg/ml) was mixed with 3 μl of 10× reaction buffer (500 mM Tris [pH 7.5], 100 mM MgCl2, 2 mM ATP) and 0 to 5 units of recombinant PKA catalytic subunit (10 units/μl; Sigma) and brought up to a volume of 30 μl with H2O. The reaction was then incubated for 30 min at 30°C and stopped by adding 5 μl sodium dodecyl sulfate (SDS) sample buffer. In studies using the PKA inhibitor H89 (Sigma), 20 or 40 μM H89 was added at the start of the incubation period. Samples were resolved by 8% SDS-polyacrylamide gel electrophoresis prior to transfer to nitrocellulose and immunoblotting with rat anti-EHEC Tir antisera as described previously (1).
Cellular fractionation and immunoblotting.
HeLa cells were plated at a starting density of 2 × 106 cells/10-cm culture dish. The next day, the cultures were infected with EHECΔtir strains for 6 h, and monolayers were washed in phosphate-buffered saline (PBS) prior to solubilization with lysis buffer containing 50 mM Tris (pH 7.4)-1% Triton X-100 (TX-100) supplemented with protease inhibitors (Complete EDTA-free; Calbiochem) and phosphatase inhibitors (1 mM sodium vanadate, 100 nM Microcystin LR). TX-100-soluble fractions (membranes, cytosol, and solubilized bacterial proteins) were separated from the insoluble fraction containing most of the bacteria and host cytoskeleton by centrifugation. Samples prepared for alkaline phosphatase treatment were solubilized in 50 mM Tris (pH 8.0)-1% TX-100 without protease and phosphatase inhibitors and incubated with 30 units of calf alkaline phosphatase (10,000 U/ml; NEB) for 60 min at 37°C. Samples were resolved by 8% SDS-polyacrylamide gel electrophoresis and analyzed by immunoblotting with rat anti-EHEC Tir (1).
Immunofluorescence microscopy.
One milliliter of 2 × 104 HeLa cells was added to each well of a 24-well plate containing a 12-mm glass coverslip and grown overnight. The next day, monolayers were infected with 5 μl of broth-grown EHECΔtir strains and incubated for either 6 h or 4 h, followed by the addition of gentamicin (50 μg/ml) and incubation for an additional 3 h to maximize pedestal elongation (11). Cells were fixed in 2.5% paraformaldehyde, permeabilized with 0.5% TX-100-PBS, and blocked in 10% normal goat serum-0.1% TX-100-PBS prior to incubation with primary and secondary antisera as previously described (11). Antisera were used at the following dilutions: rat anti-EHEC Tir, 1:5,000; mouse anti-EPEC Tir monoclonal antibody clone 2A8, 1:50; mouse anti-α-actinin, 1:200; rabbit anti-Nck, 1:200; rabbit anti-O157, 1:200; goat anti-mouse Alexa 488 (Molecular Probes), 1:400; donkey anti-rat Cy3 (Jackson Labs), 1:800; and Alexa 488 or 568 phalloidin (Molecular Probes), 1:100. Samples were examined using a Leica DMIRE2 inverted microscope, and images were acquired with a Hammamatsu ORCA-ER digital camera and analyzed using OpenLab imaging software (Improvision).
Bacterial adherence, α-actinin recruitment, and the efficiency of pedestal formation were assessed using immunofluorescence microscopy. Bacterial adherence was measured by determining the percentage of cells showing at least five adhering bacteria that were associated with focused Tir. We counted three replicate coverslips with at least 100 cells/coverslip. The level of actin and α-actinin recruitment was measured by determining the percentage of adhering EHEC O157:H7 with focused Tir that colocalized with actin or α-actinin labeling. Samples were photographed for anti-O157, anti-Tir, actin, and/or α-actinin, and images were pseudocolored and then overlaid using OpenLab. Adhering bacteria colabeling for Tir and actin were considered to recruit actin, and Tir and α-actinin were considered to recruit α-actinin. Between 100 and 250 adhering bacteria were assessed in each experiment, and three independent experiments were performed for each strain. In all cases, samples were counted in a blinded manner and by multiple investigators. Data are expressed as the means ± standard errors of the means for three independent experiments. This method is adapted from the protocol published by Campellone et al. to assess pedestal formation by Tir mutant and EspFu-deficient strains and has been used extensively by our laboratory (1, 2, 4-7). Statistical differences between groups were determined by analysis of variance followed by Tukey's multiple range test.
RESULTS
EHEC O157:H7 Tir is a substrate for PKA.
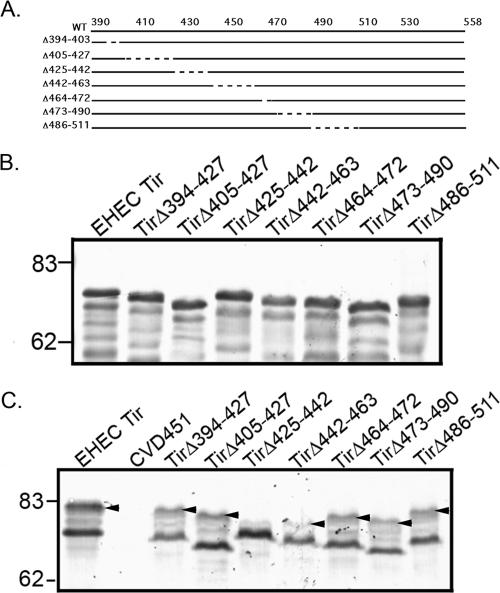
In order to determine the residues within the EHEC O157:H7 Tir carboxy terminus that are involved in Tir phosphorylation and pedestal formation, we constructed a series of in-frame deletions ranging from 9 to 25 amino acids long, spanning residues 394 to 511 (Fig. 1A). EHEC TirΔ394-403 removes 9 of 11 consecutive threonine residues, whereas EHEC TirΔ473-490 deletes an 18-amino-acid region that is absolutely not conserved between EHEC and EPEC Tir proteins (11). In EPEC, this region contains the residues essential for Tir tyrosine phosphorylation (Y474) and Nck binding (7, 11). The other deletions were designed to span the EHEC Tir carboxy terminus. The resulting constructs were transformed into EHEC O157:H7Δtir. All mutant Tir proteins were produced, as demonstrated by anti-Tir immunoblots of bacterial lysates (Fig. 1B).
FIG. 1.
The EHEC O157:H7 carboxy terminus is important for Tir phosphorylation. (A) Deletion mutations described in the text. (B) Anti-Tir immunoblot of bacterial pellets derived from EHECΔtir expressing WT Tir or the deletion mutants shown in panel A. (C) Anti-Tir immunoblot of TX-100-soluble fractions from HeLa cells incubated with EHECΔtir expressing WT Tir or the deletion mutants shown in panel A. CVD451 is a type III secretion mutant and does not translocate Tir. Arrowheads indicate phosphorylated Tir. In panels B and C, apparent molecular masses are in kilodaltons.
EHEC O157:H7 Tir is serine/threonine phosphorylated upon delivery into the host cell, but the role of these events in pedestal formation is not known (12). Phosphorylation results in an increase in the apparent molecular mass of Tir found in the membrane, from ∼72 kDa to ∼83 kDa, and is easily assessed by anti-Tir immunoblotting (12). We infected HeLa cells with EHEC O157:H7Δtir expressing the mutant Tir proteins and prepared TX-100-soluble lysates containing HeLa cell membrane and cytosolic proteins as described in Materials and Methods. Anti-Tir immunoblotting revealed that all the mutant Tir constructs could be detected in the TX-100-soluble fraction (Fig. 1C). As expected, the type III secretion mutant CVD451 did not deliver Tir into HeLa cells. Only one mutant protein, TirΔ425-442, did not show evidence of phosphorylation. These data suggest that the site(s) for Tir phosphorylation resides within amino acids 425 to 442.
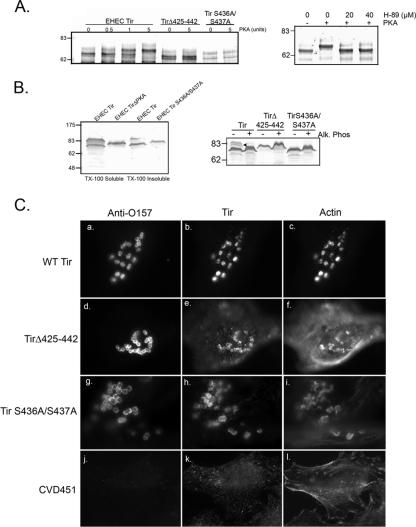
EHEC O157:H7 TirΔ425-442 contains consensus sites for phosphorylation by both PKA (amino acids 434 to 437, RRSS) and PKC (amino acids 433 to 435, SRR) (PROSITE at ExPASy). Tir from the related pathogen EPEC is phosphorylated by PKA, which affects EPEC Tir multimerization and enhances the rate of pedestal formation (19, 37). We examined whether EHEC O157:H7 Tir is phosphorylated by PKA by performing in vitro phosphorylation assays. His6-Tir translational fusions of wild-type (WT) Tir, TirΔ425-442, or Tir S436A/S437A, which has alanine residues in place of the predicted PKA substrates Ser436 and Ser437, were incubated with PKA as described in Materials and Methods. EHEC O157:H7 Tir is phosphorylated by PKA, as shown by the increase in apparent molecular mass occurring after incubation with either 1 or 5 units of purified enzyme (Fig. 2A, left panel). In contrast, EHEC O157:H7 Tir proteins lacking amino acids 425 to 442 or the PKA substrate serines (Tir S436A/S437A) did not show a shift in apparent molecular mass, suggesting that they are not phosphorylated by PKA. In addition, Tir phosphorylation was blocked by the selective PKA inhibitor H89 at both 20 and 40 μM (Fig. 2A, right panel).
FIG. 2.
EHEC O157:H7 Tir is a substrate for PKA. (A) In vitro phosphorylation assay. Left panel, anti-Tir immunoblot of WT or mutant Tir incubated with increasing concentrations of PKA. Right panel, WT Tir phosphorylation is blocked by the PKA inhibitor H-89. (B) Tir S4366A/S437A is translocated into the host cell but not phosphorylated. Left panel, anti-Tir immunoblot of TX-100-soluble and insoluble fractions derived from EHEC-infected HeLa cells as described in the text. Right panel, anti-Tir immunoblot of TX-100-soluble fractions from EHEC-infected HeLa cells treated with calf alkaline phosphatase (Alk. Phos). (C) PKA phosphorylation is not required for pedestal formation. EHEC-infected HeLa cells were triple labeled for anti-O157, anti-Tir, and phalloidin (actin).
In order to determine whether Tir S436A/S437A is phosphorylated within the host cell, TX-100-soluble (membrane and cytosol) and insoluble (cytoskeleton and bacteria) fractions from HeLa cells infected with EHECΔtir/Tir S436A/S437A were examined by anti-Tir immunoblotting. As shown in Fig. 2B (left panel), Tir S436A/S437A resolved as a single band in both fractions, suggesting that Ser436 and Ser437 are required for Tir phosphorylation in vitro. We were unable to determine whether this phosphorylation was sensitive to H89, due to its cytotoxicity over the 6-h infection period. To confirm that Tir S436A/S437A was not phosphorylated within the host cell, TX-100-soluble samples were treated with a nonspecific phosphatase, calf intestinal phosphatase, as described in Materials and Methods. Neither TirΔ425-442 nor Tir S436A/S437A demonstrated a decrease in apparent molecular mass, indicating they were not phosphorylated (Fig. 2B, right panel). In contrast, the apparent molecular mass of WT EHEC Tir was reduced to ∼70 kDa from ∼83 kDa, as reported previously (12).
We next examined whether the removal of the residues necessary for PKA phosphorylation affected the efficiency of pedestal formation. HeLa cells were infected for 6 h with either EHECΔtir/Tir, EHECΔtir/Tir S436A/S437A, EHECΔtir/TirΔ425-442, or the type III secretion mutant CVD451, fixed, and labeled with anti-EHEC O157, anti-EHEC Tir, and phalloidin to visualize filamentous actin. We detected no differences in the abilities of the mutant strains to form pedestals (Fig. 2C), and pedestal formation was equally efficient for the mutant strains and for EHECΔtir/Tir. Pedestals formed beneath 76.4 ± 2.6% of adherent EHECΔtir/Tir (panels a to c), 81.1 ± 8.4% of EHECΔtir/TirΔ425-442 (panels d to f), and 80.6 ± 6.6% of EHECΔtir/Tir S436A/S437A (panels g to i) (n = 3, P = 0.9 by analysis of variance) strains. As expected, CVD451 did not deliver Tir or form pedestals (Fig. 2C, j to l). Collectively, our data suggest that EHEC Tir can be phosphorylated by PKA in vitro and in vivo and that phosphorylation is not required for pedestal formation by EHEC O157:H7.
EHEC O157:H7 Tir residues 454 to 463 are required for pedestal formation.
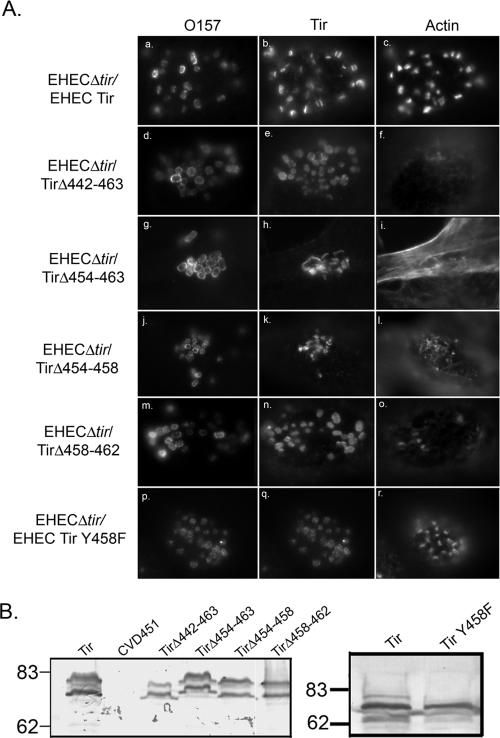
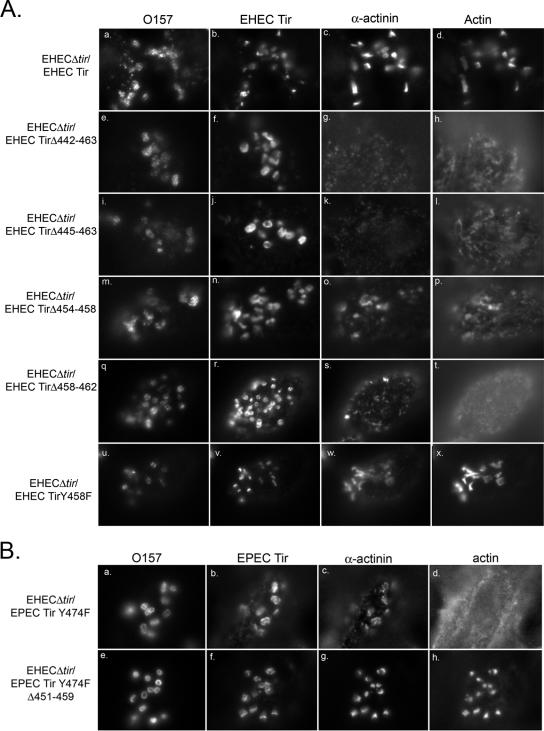
The ability of the mutant Tir proteins shown in Fig. 1A to support pedestal formation was examined by immunofluorescence microscopy. In order to promote pedestal elongation, HeLa cells were infected for 4 h, followed by incubation in Dulbecco's minimal essential medium/gentamicin for an additional 3 h (11, 35). HeLa cells were infected with EHEC O157:H7Δtir expressing the mutant Tir proteins and labeled with anti-EHEC Tir; phalloidin was used to label filamentous actin. WT EHEC Tir is found in the HeLa cell membrane at the tip of the actin pedestals that form beneath adhering bacteria (Fig. 3A, panels a to c). Only one EHEC Tir mutant protein, TirΔ442-463, was unable to form pedestals within the host cell (Fig. 3A, panels d to f). Although TirΔ442-463 is focused beneath the adhering bacteria, only 1.1 ± 0.6% of the adhering EHEC O157:H7Δtir/TirΔ442-463 colocalized with filamentous actin (Table 3). Actin labeling was less intense, and elongated pedestals did not form. These results are similar to those we reported previously for EPEC Tir mutants deficient for pedestal formation (11). All the other mutant Tir proteins, including the mutant lacking the nonconserved region (TirΔ473-490), supported pedestal formation at an efficiency equal to that observed with WT Tir (data not shown, P > 0.05).
FIG. 3.
EHEC O157:H7 Tir amino acids 452 to 463 are required for pedestal formation. (A) Immunofluorescence micrograph of HeLa cells infected with EHEC Tir mutants deficient for pedestal formation. Cells are triple labeled with anti-O157, anti-EHEC Tir, and phalloidin. (B) Anti-Tir immunoblot of TX-100-soluble fractions from HeLa cells infected with EHECΔtir expressing WT Tir or the deletion mutants. Apparent molecular mass markers are in kilodaltons.
TABLE 3.
EHEC O157:H7 adherence and actin and α-actinin recruitmenta
| Tir construct | Adherence | Recruitment of:
|
|
|---|---|---|---|
| Actin | α-Actinin | ||
| WT EHEC Tir | 27.9 ± 3.4 | 76.4 ± 2.6 | 77.7 ± 1.8 |
| EHEC TirΔ442-463 | 18.4 ± 2.6 | 1.1 ± 0.6b | 8.9 ± 3.6b |
| EHEC TirΔ454-463 | 18.1 ± 2.0 | 11.3 ± 1.6b,c | 17.9 ± 2.3b |
| EHEC TirΔ454-458 | 19.7 ± 2.1 | 7.3 ± 1.5b,c | 18.4 ± 2.6b,c |
| EHEC TirΔ458-462 | 23.9 ± 1.8 | 13.1 ± 1.4b,c | 21.5 ± 4.3b,c |
| EHEC Tir Y458F | 29.6 ± 2.3 | 86.8 ± 0.5 | 87.1 ± 1.6 |
| EPEC Tir Y474F | 21.8 ± 2.3 | 91.6 ± 3.4b | 91.4 ± 2.9b |
| EPEC Tir Y474F Δ451-459 | 20.6 ± 2.1 | 15.6 ± 6.1 | 73.8 ± 3d |
Adherence represents the percentage of cells with ≥5 adhering EHEC O157:H7 bacteria. Actin and α-actinin recruitment were measured as described in Materials and Methods. Data are the means ± standard errors of the means from at least three independent experiments.
Significantly different from WT EHEC Tir; P < 0.05.
Significantly different from EHEC TirΔ442-463; P < 0.05.
Significantly different from EPEC Tir Y474F; P < 0.05.
These data suggested that the residues required for pedestal formation reside within the 22 amino acids deleted from TirΔ442-463. In order to further define this region, we constructed a second series of internal deletion mutations and transformed the resulting constructs into EHEC O157:H7Δtir. The sequences of these mutants are listed in Table 4. The ability of the mutant Tir proteins to support pedestal formation was examined by immunofluorescence microscopy. Eight mutant Tir proteins were unable to support pedestal formation, although Tir was focused beneath the adhering bacteria (Table 4). These proteins were all deficient for amino acids 454 to 463, suggesting that this region plays an important role in pedestal formation. We selected three mutant Tir proteins with deletions within this region for further study and compared them with WT EHEC O157:H7 Tir and TirΔ442-463.
TABLE 4.
Summary of Tir internal deletion mutant sequences and phenotypesa
| Mutant | Sequence | Focused Tir | Pedestal formation |
|---|---|---|---|
| TirΔ442-463 | MASTSSTFFDTSSIGTVQNPYADVKTSLHDSQVPTS | Yes | No |
| TirΔ449-472 | MASTSSTFFDTSSIGTVQNPYADVKTSLHDSQVPTS | Yes | No |
| TirΔ449-467 | MASTSSTFFDTSSIGTVQNPYADVKTSLHDSQVPTS | Yes | No |
| TirΔ454-472 | MASTSSTFFDTSSIGTVQNPYADVKTSLHDSQVPTS | Yes | No |
| TirΔ449-463 | MASTSSTFFDTSSIGTVQNPYADVKTSLDHSQVPTS | Yes | No |
| TirΔ454-467 | MASTSSTFFDTSSIGTVQNPYADVKTSLHDSQVPTS | Yes | No |
| TirΔ464-467 | MASTSSTFFDTSSIGTVQNPYADVKTSLHDSQVPTS | Yes | Yes |
| TirΔ442-450 | MASTSSTFFDTSSIGTVQNPYADVKTSLDHSQVPTS | Yes | Yes |
| TirΔ454-463 | MASTSSTFFDTSSIGTVQNPYADVKTSLDHSQVPTS | Yes | No |
| TirΔ458-462 | MASTSSTFFDTSSIGTVQNPYADVKTSLHDSQVPTS | Yes | No |
| TirΔ454-458 | MASTSSTFFDTSSIGTVQNPYADVKTSLDHSQVPTS | Yes | No |
| TirΔ450-453 | MASTSSTFFDTSSIGTVQNPYADVKTSLHDSQVPTS | Yes | Yes |
| EPECΔ451-459 | QGSVASTHWSDSSSEVVNPYAEVGGARNSLSAHQ | Yes | Yes |
| EPEC Y474F Δ451-459 | QGSVASTHWSDSSSEVVNPYAEVGGARNSLSAHQ | Yes | No |
All constructs were expressed from a plasmid harbored by EHECΔtir. Deleted residues are underlined, and the residues that distinguish functional and nonfunctional Tir mutants are shown in boldface type.
All of the mutant Tir proteins were translocated into the host cell and phosphorylated, as demonstrated by the appearance of both the higher (83-kDa) and lower (72-kDa) molecular mass bands (Fig. 3B, left panel). We observed no differences in the levels of adherence to HeLa cells between EHEC O157:H7Δtir expressing the mutant Tir proteins and EHEC O157:H7Δtir/Tir (Table 3, P > 0.05). Both TirΔ442-463 and TirΔ454-463 were focused beneath adhering EHEC O157:H7, but only weakly recruited actin (Fig. 3A, panels d to h; Table 4). Only 11.3 ± 1.6% of the adhering EHEC expressing TirΔ454-463 was associated with foci of filamentous actin (Table 4, Fig. 3B). We never observed the formation of elongated actin pedestals with these strains. These results are similar to those we reported previously for EPEC Tir mutants deficient for pedestal formation (11). We were unable to further subdivide this region, as mutant Tir proteins lacking either amino acids 454 to 458 or 458 to 462 also inefficiently recruited, suggesting the entire region is involved (Fig. 3B, panels j to o; Table 4). Although Tir proteins lacking amino acids 454 to 463, 454 to 458, and 458 to 462 were impaired in their ability to recruit actin, they did this significantly more efficiently than the original mutant TirΔ442-463 (Table 3). These results complement data reported by Campellone et al. (4), who used transfected Tir. Collectively, these data suggest that EHEC O157:H7 amino acids 454 to 463 are required for pedestal formation.
We noticed that all the mutants that were deficient in pedestal formation contained one common residue, Y458 (Table 4). This residue is homologous to EPEC Tir Y454, which is phosphorylated and involved in Nck-independent pedestal formation (6). In order to determine whether Y458 is the only residue involved in pedestal formation, we constructed EHEC O157:H7 Tir Y458F. EHECΔtir/EHEC Tir Y458F adhered and formed pedestals within the host cell with an efficiency equal to that of WT EHEC Tir (Fig. 3A, panels p to r; Table 3). Additionally, Tir Y458F was phosphorylated upon translocation into the host cell (Fig. 3B, right panel). These data suggest that although Y458 is common to all the constructs deficient in pedestal formation, it is not the only residue required.
EPEC Tir residues 451 to 459 are required for Nck-independent pedestal formation.
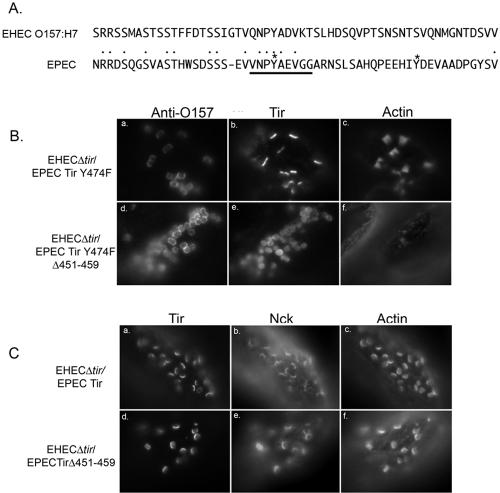
High-efficiency pedestal formation by the related pathogen EPEC requires Tir tyrosine 474 phosphorylation (11, 22). This event is required for the direct binding of the adapter protein Nck (5, 18). Replacement of Y474 with phenylalanine significantly decreases the level of pedestal formation observed in response to EPEC infection (6, 11). In contrast, pedestal formation by EHEC O157:H7 occurs independently of Nck recruitment and Tir tyrosine phosphorylation, and EPEC Tir Y474F is fully functional when expressed by EHEC O157:H7. Therefore, we wanted to determine the region of EPEC Tir required for Nck-independent pedestal formation when expressed by EHEC O157:H7. EPEC Tir contains a region with high homology to EHEC O157:H7 Tir residues 454 to 463 (EHEC, VQNPYADVKT; EPEC 451 to 459, VNPYAEVGG) (Fig. 4A, Table 4). We deleted this region from wild-type EPEC Tir and EPEC Tir Y474F and expressed the resulting constructs in EHEC O157:H7Δtir.
FIG. 4.
Amino acids 451 to 459 are required for Nck-independent pedestal formation by EPEC Tir. (A) Alignment of EHEC O157:H7 and EPEC C-terminal Tir sequences within the regions of the deletions described within the text. Phosphorylated tyrosines within EPEC Tir are indicated by asterisks, while the conserved region deleted within EPEC Tir is underlined. (B) Immunofluorescence micrograph of HeLa cells infected with either EHECΔtir/EPEC Tir Y474F or EHECΔtir/EPEC Tir Y474F Δ451-459 and labeled for EPEC Tir, actin, and anti-O157. (C) HeLa cells infected with EHECΔtirEPEC Tir or EHECΔtirEPEC TirΔ451-459 and labeled for EPEC Tir, actin, and Nck.
Deleting residues 451 to 459 from EPEC Tir Y474F significantly reduced Nck-independent pedestal formation without affecting adherence or Tir delivery. Actin recruitment decreased from 91.6 ± 3.4% for EHECΔtir/EPEC Tir Y474F (Fig. 4B, panels a to c; Table 4) to 15.6 ± 6.1% for EHECΔtir/EPEC Tir Y474F Δ451-459 (Fig. 4A, panels d to f; Table 4). Interestingly, pedestal formation by EHECΔtir/EPEC Tir Y474F was significantly more efficient than that by EHECΔtir/EHEC Tir (Table 3). Although we infrequently observed actin condensation beneath EHECΔtir/EPEC Tir Y474F Δ451-459, we never saw elongated actin pedestal formation. In contrast, deleting residues 451 to 459 from WT EPEC Tir had no effect on Nck-dependent pedestal formation. Nck was recruited to the pedestal tip and colocalized with Tir in HeLa cells infected with EHECΔtir/EPEC Tir (Fig. 4C, panels a to c) or EHECΔtir/EPECTirΔ451-459 (Fig. 4C, panels d to f). Taken together, these data suggest that EPEC Tir residues 451 to 459 are involved in Nck-independent pedestal formation and play a role similar to that of EHEC Tir amino acids 454 to 463.
EHEC O157:H7 Tir residues 454 to 463 are involved in α-actinin recruitment.
α-Actinin is an actin binding and cross-linking protein that binds directly to the amino terminus of EHEC O157:H7 and EPEC Tirs (28). In EPEC, α-actinin recruitment to Tir occurs independently of Tir phosphorylation and pedestal formation and is observed after infection with EPECΔtir/EPEC Tir Y474F (15). Considerably less is known about the requirements for α-actinin recruitment to EHEC Tir; therefore, we examined it with HeLa cells infected with EHEC O157:H7Δtir expressing either WT Tir or the Tir internal deletion mutants that did not support pedestal formation. Strains expressing Tir proteins lacking residues 442 to 463 (Fig. 5A, panels e to h), 454 to 463 (panels i to l), 454 to 458 (panels m to p), or 458 to 462 (panels q to t) inefficiently recruited α-actinin, although Tir was focused beneath the adherent bacteria. The efficiency of α-actinin recruitment between the different Tir mutants varied, ranging from ∼9 to 22% of adhering EHEC O157:H7 (Table 3). WT EHEC Tir (5A, panels a to d) and EHEC Tir Y458F (panels u to x) recruited α-actinin to the pedestal with equal efficiency (Table 3). These data complement what is observed with strains lacking EspFu/TccP, which also fail to recruit α-actinin (14).
FIG. 5.
The requirements for α-actinin recruitment differ between EHEC and EPEC Tirs. (A) Immunofluorescence micrograph of HeLa cells infected with EHECΔtir expressing either WT EHEC Tir or mutant Tir proteins unable to support pedestal formation. Cells were quadruple labeled with anti-O157, anti-EHEC Tir, anti-α-actinin, and phalloidin. (B) Immunofluorescence micrograph of HeLa cells infected with either EHECΔtir/EPEC Tir Y474F or the non-pedestal-forming mutant EHECΔtir/EPEC Tir Y474FΔ451-459. Cells were quadruple labeled for anti-O157, anti-EPEC Tir, anti-α-actinin, and phalloidin.
One possible explanation for these data is that the decrease in α-actinin recruitment is due solely to a nonspecific effect of the deletion of between 5 and 22 amino acids from the EHEC O157:H7 Tir carboxy terminus. We examined this by determining the efficiency of α-actinin recruitment to EPEC Tir Y474F Δ451-459, which does not support pedestal formation (Fig. 4A). As α-actinin recruitment to EPEC Tir occurs independently of pedestal formation, we anticipated that α-actinin would be recruited to EPEC Tir Y474FΔ451-459. Indeed, this was the case, although there was a slight but significant decrease in α-actinin recruitment between cells infected with EHECΔtir/EPEC Tir Y474FΔ451-459 and EHECΔtir/EPEC Tir Y474F (Fig. 5B, Table 4). α-Actinin was recruited to 73.8 ± 3% of cells with adhering EHECΔtir/EPEC Tir Y474F Δ451-459 (5B, panels a to d). This level of recruitment was significantly higher than what we observed with the corresponding EHEC Tir deletion mutant, EHEC TirΔ454-463, which recruited α-actinin to only 17.9 ± 2.3% of the adhering bacteria. Taken together, these data indicate that residues in the EHEC Tir C terminus are important for both actin and α-actinin recruitment, illustrating another difference between EHEC O157:H7 and EPEC Tirs.
DISCUSSION
Little is known about the functional domains of EHEC O157:H7 Tir. We previously reported that amino acids 519 to 524 are required for efficient Tir type III secretion but are uninvolved in pedestal formation (1). In this study, we have identified two additional domains required for Tir phosphorylation and efficient pedestal formation. EHEC O157:H7 Tir is phosphorylated by PKA, which involves serines 436 and 437, whereas Nck-independent pedestal formation and α-actinin recruitment involves a 10-amino-acid domain containing residues 454 to 463. Additionally, we have identified a similar domain in EPEC Tir, which supports Nck-independent pedestal formation when expressed in EHEC O157:H7. Our data, obtained using mutant Tir proteins delivered by EHEC O157:H7, are highly complementary to those reported by Campellone et al. using Tir mutants ectopically expressed in mammalian cells (4).
Our data demonstrate that EHEC O157:H7 Tir is a substrate for PKA, although we were unable to detect an effect of Tir phosphorylation on pedestal formation. This is in contrast to what was observed with Tir from the related pathogen EPEC. EPEC Tir is phosphorylated by PKA at two sites (Ser434 and Ser463). Ser434 is in a sequence context similar to the EHEC O157:H7 Tir Ser437 contained within Tir S436A/S437A, whereas Ser463 is unique to EPEC Tir. EPEC Tir mutants unable to be phosphorylated on Ser434 form shorter pedestals, suggesting that this site is involved in efficient pedestal elongation (37). Further experiments are required to determine which serine residue within EHEC O157:H7 Tir is phosphorylated, although it is tempting to speculate that it is Ser437, as it is in a sequence context similar to that of EPEC Tir Ser434. PKA activation requires the generation of cyclic AMP, and cyclic AMP-dependent chloride secretion was observed in response to the attaching/effacing pathogen Citrobacter rodentium (36). This chloride secretion may be due to the activity of the CFTR, which opens in response to PKA phosphorylation (3). Intriguingly, both CFTR expression and chloride secretion are enhanced after infection with C. rodentium (36). Whether the same occurs in response to EHEC O157:H7 is not known. Tir phosphorylation may be a response to a general activation of PKA by EHEC O157:H7 or may suggest a role for Tir in pedestal-independent signaling.
In this study, we identified a 10-amino-acid domain within the EHEC O157:H7 carboxy terminus (454 to 463) which, when deleted, causes a significant decrease in the efficiency of pedestal formation. Interestingly, this region cannot be subdivided, suggesting the entire area is involved. Surprisingly, Tir proteins deficient in residues 454 to 463, 454 to 458, and 458 to 462 recruited actin and α-actinin with a significantly higher efficiency than did the parental mutant TirΔ442-463 (Table 3). This may represent a partial phenotype, with cytoskeletal components targeted to Tir, in the absence of pedestal elongation. It is intriguing to speculate that cytoskeletal proteins shown to be required for pedestal formation, such as NWASP, EspFu/TccP, and the Arp 2/3 complex, either are not recruited or remain inactive in cells infected with EHEC O157:H7 strains carrying these mutant Tir proteins (4, 14). We identified a homologous region in EPEC Tir (amino acids 451 to 459). EPEC Tir proteins deficient in this region are unable to support pedestal formation when expressed by EHEC O157:H7. In EPEC Tir, this region contains Y454, which Campellone and colleagues demonstrated was involved in Nck-independent pedestal formation by EPEC (6). Similarly to Y474, Y454 is tyrosine phosphorylated, which is required for actin assembly. EHEC O157:H7 Tir contains a similar tyrosine (Y458), which is absent from all of our constructs that did not support pedestal formation. Substituting Y458 with phenylalanine had no effect on pedestal formation, suggesting this residue plays a different role in EHEC and EPEC Tirs. It is tempting to speculate that there may be more than one way to form Nck-independent pedestals. Pedestal formation was more efficient with EHEC O157:H7Δtir/EPEC Tir Y474F than with EHEC O157:H7 expressing WT EHEC Tir. Although EHEC O157:H7 and EPEC Tirs share homology within the 10-amino-acid region identified to be involved in pedestal formation, they are quite divergent outside that region and show only 41% identity throughout their carboxy termini. This suggests that there may be other regions within EPEC Tir that contribute to Nck-independent pedestal formation. Additionally, the contribution of EPEC Tir Y454 may promote the more efficient pedestal formation observed with EHEC O157:H7Δtir/EPEC Tir Y474F. Our data are complementary to those seen by Campellone et al., who used Tir transfected into HeLa cells (4). Those studies identified a 12-amino-acid region which was sufficient for EspFu/TccP recruitment and the initiation of Nck-independent actin assembly and overlapped with the region identified in this study.
In addition to pedestal formation, EHEC O157:H7 Tirs lacking residues 454 to 463 are unable to recruit α-actinin. α-Actinin is an actin binding and cross-linking protein which binds directly to the EHEC O157:H7 Tir amino terminus at a site distinct from the chaperone CesT (15, 27, 28). The contribution of α-actinin to pedestal formation has been best studied for EPEC. α-Actinin has been shown to bind to both the amino and carboxy termini of EPEC Tir but does not appear to play a direct role in pedestal formation (13). Campellone and colleagues examined the effect of ectopic expression of the EPEC Tir intimin binding domain and carboxy terminus on α-actinin recruitment and pedestal formation (7). This construct, referred to as TirMC, contains Y474, which is required for Nck-dependent pedestal formation but lacks the amino terminal residues involved in α-actinin binding. TirMC supported both pedestal formation and α-actinin recruitment, which were both blocked when Y474 was mutated. Studies using full-length EPEC Tir indicate that α-actinin is efficiently recruited to EPEC Tir proteins unable to form pedestals using either Nck-dependent or Nck-independent mechanisms (Fig. 5B) (18). Collectively, these data suggest that α-actinin recruitment to the EPEC Tir amino terminus is independent of pedestal formation, whereas recruitment to the carboxy terminus is a result of Tir-dependent actin assembly. The mechanism of α-actinin recruitment by EHEC O157:H7 Tir is different. α-Actinin recruitment is dependent on actin assembly and does not occur, even in the presence of an intact amino terminus, in Tir proteins unable to support pedestal formation. Additionally, α-actinin recruitment to Tir is not observed after infection with strains lacking EspFu/TccP (14). Whether α-actinin recruitment is required for pedestal formation by EHEC O157:H7 remains an open question.
Classically, EPEC pedestal formation has been used as a model system to draw conclusions about other attaching and effacing organisms. However, it is increasingly recognized that the mechanisms for pedestal formation in EHEC O157:H7 differ from those of EPEC, some non-O157 EHEC strains, and C. rodentium, as summarized in Table 5. Previous studies have determined that EHEC pedestal formation occurs independently of Tir tyrosine phosphorylation, Nck, and lipid rafts, whereas EPEC pedestals depend on all three (2, 5, 11, 18, 20, 22, 34). C. rodentium and some non-O157:H7 Tirs show a pattern of tyrosine phosphorylation similar to that of EPEC Tir and are thought to use the same mechanism for pedestal formation as EPEC (10, 11). Our data illustrate yet another difference in the mechanism of pedestal formation between EHEC O157:H7 and EPEC. Compelling questions are why EHEC O157:H7 evolved a distinct mechanism for pedestal formation and whether that mechanism provides an advantage during infection. These differences may be due to differences in tissue tropism, as EHEC O157:H7 adheres preferentially to Peyer's patches, whereas EPEC adheres within the small intestine (31, 32). Additionally, unlike EPEC, EHEC O157:H7 can colonize both ruminant and human hosts, with differing effects. EHEC O157:H7 can cause life-threatening illness in humans, whereas infection of adult ruminants is mainly asymptomatic (30). It is tempting to speculate that the differences in the mechanisms of pedestal formation evolved to maximize colonization of two very different hosts.
TABLE 5.
Summary of differences in the mechanisms of pedestal formation by EPEC and EHEC O157:H7
| Property | Characteristic of indicated Tir (reference[s])
|
|
|---|---|---|
| EPEC | EHEC O157:H7 | |
| Tir tyrosine phosphorylation | Y474 and Y454; required for Nck-dependent and independent pedestal formation, respectively (6, 23) | None detected |
| Host cell adaptors | Nck (5, 18) | None |
| Additional bacterial factors | None | EspFu/TccP (8, 14) |
| α-Actinin recruitment | Independent of pedestal formation (15) | Dependent on pedestal formation (14, this study) |
| Membrane cholesterol | Required for pedestal formation (2, 20, 34) | May depend on strain or cell type used (2, 34) |
| Serine phosphorylation | Involved in pedestal elongation (19, 34) | No role in pedestal formation (this study) |
Acknowledgments
We thank Carolyn Southward and Bryan Jones for critical readings of the manuscript.
Research in R.D.'s laboratory is supported by the Canadian Institutes for Health Research (CIHR MOP44089). E.A.-V. is a Canadian Association for Gastroenterology-CIHR fellow, and R.D. is an Alberta Heritage Foundation for Medical Research Senior Scholar.
Editor: F. C. Fang
Footnotes
Published ahead of print on 5 September 2006.
REFERENCES
- 1.Allen-Vercoe, E., M. C. Toh, B. Waddell, H. Ho, and R. DeVinney. 2005. A carboxy-terminal domain of Tir from enterohemorrhagic Escherichia coli O157:H7 (EHEC O157:H7) required for efficient type III secretion. FEMS Microbiol. Lett. 243:355-364. [DOI] [PubMed] [Google Scholar]
- 2.Allen-Vercoe, E., B. Waddell, S. Livingstone, J. Deans, and R. Devinney. 2006. Enteropathogenic Escherichia coli Tir translocation and pedestal formation requires membrane cholesterol in the absence of bundle-forming pili. Cell. Microbiol. 8:613-624. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, M. P., R. J. Gregory, S. Thompson, D. W. Souza, S. Paul, R. C. Mulligan, A. E. Smith, and M. J. Welsh. 1991. Demonstration that CFTR is a chloride channel by alteration of its anion selectivity. Science 253:202-205. [DOI] [PubMed] [Google Scholar]
- 4.Campellone, K., M. Brady, J. Alamares, D. Rowe, B. Skehan, D. Tipper, and J. Leong. 2006. Enterohemorrhagic Escherichia coli Tir requires a C-terminal 12-residue peptide to initiate EspFU-mediated actin assembly and harbours N-terminal sequences that influence pedestal length. Cell. Microbiol. 8:1488-1503. [DOI] [PubMed] [Google Scholar]
- 5.Campellone, K., A. Giese, D. Tipper, and J. Leong. 2002. A tyrosine-phosphorylated 12-amino-acid sequence of enteropathogenic Escherichia coli Tir binds the host adaptor protein Nck and is required for Nck localization to actin pedestals. Mol. Microbiol. 43:1227-1241. [DOI] [PubMed] [Google Scholar]
- 6.Campellone, K. G., and J. M. Leong. 2005. Nck-independent actin assembly is mediated by two phosphorylated tyrosines within enteropathogenic Escherichia coli Tir. Mol. Microbiol. 56:416-432. [DOI] [PubMed] [Google Scholar]
- 7.Campellone, K. G., S. Rankin, T. Pawson, M. W. Kirschner, D. J. Tipper, and J. M. Leong. 2004. Clustering of Nck by a 12-residue Tir phosphopeptide is sufficient to trigger localized actin assembly. J. Cell Biol. 164:407-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campellone, K. G., D. Robbins, and J. M. Leong. 2004. EspFU is a translocated EHEC effector that interacts with Tir and N-WASP and promotes Nck-independent actin assembly. Dev. Cell 7:217-228. [DOI] [PubMed] [Google Scholar]
- 9.de Grado, M., A. Abe, A. Gauthier, O. Steele-Mortimer, R. DeVinney, and B. B. Finlay. 1999. Identification of the intimin binding domain of Tir of enteropathogenic E. coli (EPEC). Cell. Microbiol. 1:7-18. [DOI] [PubMed] [Google Scholar]
- 10.Deng, W., B. A. Vallance, Y. Li, J. L. Puente, and B. B. Finlay. 2003. Citrobacter rodentium translocated intimin receptor (Tir) is an essential virulence factor needed for actin condensation, intestinal colonization and colonic hyperplasia in mice. Mol. Microbiol. 48:95-115. [DOI] [PubMed] [Google Scholar]
- 11.DeVinney, R., J. Puente, A. Gauthier, D. L. Goosney, and B. B. Finlay. 2001. Enterohaemorrhagic and enteropathogenic Escherichia coli use a different Tir-based mechanism for pedestal formation. Mol. Microbiol. 41:1445-1458. [DOI] [PubMed] [Google Scholar]
- 12.DeVinney, R., M. Stein, D. Reinscheid, A. Abe, S. Ruschkowski, and B. B. Finlay. 1999. Enterohemorrhagic Escherichia coli O157:H7 produces Tir, which is translocated to the host cell membrane but is not tyrosine phosphorylated. Infect. Immun. 67:2389-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freeman, N. L., D. V. Zurawski, P. Chowrashi, J. C. Ayoob, L. Huang, B. Mittal, J. M. Sanger, and J. W. Sanger. 2000. Interaction of the enteropathogenic Escherichia coli protein, translocated intimin receptor (Tir), with focal adhesion proteins. Cell Motil. Cytoskeleton 47:307-318. [DOI] [PubMed] [Google Scholar]
- 14.Garmendia, J., A. D. Phillips, M. F. Carlier, Y. Chong, S. Schuller, O. Marches, S. Dahan, E. Oswald, R. K. Shaw, S. Knutton, and G. Frankel. 2004. TccP is an enterohaemorrhagic Escherichia coli O157:H7 type III effector protein that couples Tir to the actin-cytoskeleton. Cell. Microbiol. 6:1167-1183. [DOI] [PubMed] [Google Scholar]
- 15.Goosney, D. L., R. DeVinney, R. A. Pfuetzner, E. A. Frey, N. C. Strynadka, and B. B. Finlay. 2000. Enteropathogenic E. coli translocated intimin receptor, Tir, interacts directly with α-actinin. Curr. Biol. 10:735-738. [DOI] [PubMed] [Google Scholar]
- 16.Griffin, P., S. Ostroff, R. Tauxe, K. Greene, J. Wells, J. Lewis, and P. Blake. 1988. Illnesses associated with Escherichia coli O157:H7 infections. Ann. Intern. Med. 109:705-712. [DOI] [PubMed] [Google Scholar]
- 17.Griffin, P. M., and R. V. Tauxe. 1991. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol. Rev. 13:60-98. [DOI] [PubMed] [Google Scholar]
- 18.Gruenheid, S., R. DeVinney, F. Bladt, D. L. Goosney, S. Gelkop, G. Gish, T. Pawson, and B. B. Finlay. 2001. Enteropathogenic E. coli Tir binds Nck to initiate actin pedestal formation in host cells. Nat. Cell Biol. 3:856-859. [DOI] [PubMed] [Google Scholar]
- 19.Hawrani, A., C. E. Dempsey, M. J. Banfield, D. J. Scott, A. R. Clarke, and B. Kenny. 2003. Effect of protein kinase A-mediated phosphorylation on the structure and association properties of the enteropathogenic Escherichia coli Tir virulence protein. J. Biol. Chem. 278:25839-25846. [DOI] [PubMed] [Google Scholar]
- 20.Hayward, R. D., R. J. Cain, E. J. McGhie, N. Phillips, M. J. Garner, and V. Koronakis. 2005. Cholesterol binding by the bacterial type III translocon is essential for virulence effector delivery into mammalian cells. Mol. Microbiol. 56:590-603. [DOI] [PubMed] [Google Scholar]
- 21.Jarvis, K., and J. Kaper. 1996. Secretion of extracellular proteins by enterohemorrhagic Escherichia coli via a putative type III secretion system. Infect. Immun. 64:4826-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kenny, B. 2001. The enterohaemorrhagic Escherichia coli (serotype O157:H7) Tir molecule is not functionally interchangeable for its enteropathogenic E. coli (serotype O127:H6) homologue. Cell. Microbiol. 3:499-510. [DOI] [PubMed] [Google Scholar]
- 23.Kenny, B. 1999. Phosphorylation of tyrosine 474 of the enteropathogenic Escherichia coli (EPEC) Tir receptor molecule is essential for actin nucleating activity and is preceded by additional host modifications. Mol. Microbiol. 31:1229-1241. [DOI] [PubMed] [Google Scholar]
- 24.Kenny, B., R. DeVinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511-520. [DOI] [PubMed] [Google Scholar]
- 25.Knutton, S., T. Baldwin, P. H. Williams, and A. S. McNeish. 1989. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 57:1290-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kondro, W. 2000. E. coli outbreak deaths spark judicial inquiry in Canada. Lancet 355:2058. [DOI] [PubMed] [Google Scholar]
- 27.Lafont, F., L. Abrami, and F. G. van der Goot. 2004. Bacterial subversion of lipid rafts. Curr. Opin. Microbiol. 7:4-10. [DOI] [PubMed] [Google Scholar]
- 28.Luo, Y., M. G. Bertero, E. A. Frey, R. A. Pfuetzner, M. R. Wenk, L. Creagh, S. L. Marcus, D. Lim, F. Sicheri, C. Kay, C. Haynes, B. B. Finlay, and N. C. Strynadka. 2001. Structural and biochemical characterization of the type III secretion chaperones CesT and SigE. Nat. Struct. Biol. 8:1031-1036. [DOI] [PubMed] [Google Scholar]
- 29.Moon, H. W., S. C. Whipp, R. A. Argenzio, M. M. Levine, and R. A. Giannella. 1983. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 41:1340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nataro, J., and J. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phillips, A. D., and G. Frankel. 2000. Intimin-mediated tissue specificity in enteropathogenic Escherichia coli interaction with human intestinal organ cultures. J. Infect. Dis. 181:1496-1500. [DOI] [PubMed] [Google Scholar]
- 32.Phillips, A. D., S. Navabpour, S. Hicks, G. Dougan, T. Wallis, and G. Frankel. 2000. Enterohaemorrhagic Escherichia coli O157:H7 target Peyer's patches in humans and cause attaching/effacing lesions in both human and bovine intestine. Gut 47:377-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phillips, N., R. D. Hayward, and V. Koronakis. 2004. Phosphorylation of the enteropathogenic E. coli receptor by the Src-family kinase c-Fyn triggers actin pedestal formation. Nat. Cell Biol. 6:618-625. [DOI] [PubMed] [Google Scholar]
- 34.Riff, J. D., J. W. Callahan, and P. M. Sherman. 2005. Cholesterol-enriched membrane microdomains are required for inducing host cell cytoskeleton rearrangements in response to attaching-effacing Escherichia coli. Infect. Immun. 73:7113-7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenshine, I., S. Ruschkowski, M. Stein, D. Reinscheid, S. Mills, and B. Finlay. 1996. A pathogenic bacterium triggers epithelial signals to form a functional bacterial receptor that mediates actin pseudopod formation. EMBO J. 15:2613-2624. [PMC free article] [PubMed] [Google Scholar]
- 36.Umar, S., J. Scott, J. H. Sellin, W. P. Dubinsky, and A. P. Morris. 2000. Murine colonic mucosa hyperproliferation. I. Elevated CFTR expression and enhanced cAMP-dependent Cl− secretion. Am. J. Physiol. Gastrointest. Liver Physiol. 278:G753-764. [DOI] [PubMed] [Google Scholar]
- 37.Warawa, J., and B. Kenny. 2001. Phosphoserine modification of the enteropathogenic Escherichia coli Tir molecule is required to trigger conformational changes in Tir and efficient pedestal elongation. Mol. Microbiol. 42:1269-1280. [DOI] [PubMed] [Google Scholar]