Abstract
Natural products are gaining increased applications in drug discovery and development. Being chemically diverse they are able to modulate several targets simultaneously in a complex system. Analysis of gene expression becomes necessary for better understanding of molecular mechanisms. Conventional strategies for expression profiling are optimized for single gene analysis. DNA microarrays serve as suitable high throughput tool for simultaneous analysis of multiple genes. Major practical applicability of DNA microarrays remains in DNA mutation and polymorphism analysis. This review highlights applications of DNA microarrays in pharmacodynamics, pharmacogenomics, toxicogenomics and quality control of herbal drugs and extracts.
Keywords: Drug discovery, evidence-based medicine, gene expression, genotyping, pharmacodynamics, transcription profiling
Introduction
Understanding the functions of genes is a major post-genomics challenge. Strategies like proteomics, transcriptomics, metabolomics are implemented to assign the role of genes in molecular networks. The gene expression profile of a cell determines its phenotype, function and response to the environment. The complement of genes expressed by a cell is very dynamic and responds rapidly to external stimuli. Therefore, analysis of gene expression becomes necessary for providing clues about regulatory mechanisms, biochemical pathways and broader cellular function. Conventional strategies for expression profiling such as northern blot, reverse northern blot, reverse transcriptase-polymerase chain reaction (RT–PCR), nuclease protection, enzyme-linked immunosorbent assay (ELISA), western blot, in situ hybridization and immunohistochemistry are optimized for single gene analysis. Although, it is possible to modify at least some of these techniques for multiplexing, the procedure becomes increasingly technically cumbersome. For genome wide expression analysis it is necessary to develop technologies having high degree of automation, since in any living organism thousands of genes and their products function in a complicated and orchestrated way. DNA microarrays were developed in response to the need for a high-throughput, efficient and comprehensive strategy that can simultaneously measure all the genes, or a large defined subset, encoded by a genome (1,2). Several different methodologies including differential display PCR, northern blots (3), quantitative PCR, serial analysis of gene expression (SAGE) (4,5) and TIGR Orthologous Gene Alignments (TOGA) (6,7) are used alongside microarrays as research tools.
With an initial focus in the post-genomic era on tracking gene expression changes for target identification, microarray applications soon widened to span the entire drug discovery pipeline (8–10). DNA microarrays are being used to study the transcriptional profile in various physiological and pathological conditions, leading to the mining of novel genes and molecular markers for diagnosis, prediction or prognosis of those specific states (11,12).
Success in the DNA microarray field has led to protein array development (13–15). Protein microarrays are mainly applied to protein function studies, screening the production of antibodies (16) and recombinant proteins (17), discovery of proteins implicated in disease or those that are potential drug targets, rapid detection or diagnosis of disease (18,19) screening for protein–protein, DNA–protein and enzyme–substrate interactions (20,21). High-density human protein arrays are undermined by the incomplete knowledge of full-length human gene clones and the vast idiosyncrasies of proteins with respect to stability and structure (22). Although protein microarrays may not have reached the stage of maturity of DNA microarrays, recent developments have shown that many of the barriers holding back the technology can be overcome (23,24).
Recently cell-based arrays employing matrices of living cells engineered to express select proteins have rapidly emerged as versatile tools for the high-throughput analysis of gene function. Such arrays can facilitate genome-scale studies on different aspects of protein function, including biochemical activities, gene disruption phenotypes and protein–protein interactions (25,26). Another version of living arrays called transfection microarray has been demonstrated as an alternative to protein microarrays for the identification of drug targets, and as an expression cloning system for the discovery of gene products that alter cellular physiology (27). A siRNA transfected cell microarray has been developed to facilitate large-scale, high-throughput functional genomics studies using RNAi (28).
Chemical microarrays, which are arrays of small organic compounds, represent a novel approach towards analysis of chemical libraries. They are widely used to analyze the interaction of proteins with organic compounds in a miniaturized and high-throughput fashion (29,30). Several modified technologies spanning wide range of macromolecular microarrays to cell arrays have opened new horizons in molecular and physiological systems.
The DNA Microarray
DNA microarray is an orderly arrangement of thousands of oligonucleotides or identified sequenced genes printed on an impermeable solid support, usually glass, silicon chips or nylon membrane. A number of terms such as DNA arrays, gene chips and biochips are often used to describe these devices. The DNA microarray field is a combination of several technologies, including automated DNA sequencing, DNA amplification by PCR, oligonucleotide synthesis, nucleic acid labeling chemistries and bioinformatics. Two most common types of DNA microarrays include one in which the DNA (in the form of a single stranded short oligonucleotide) is synthesized in situ using photolithographic or other techniques and another where the DNA (usually in the form of a cDNA or full-length ORF) is post-synthetically attached to a solid support (31,32).
Depending on the kind of probe (immobilized DNA) used to generate the array and, ultimately, the information that is derived from it, DNA microarrays are used for different applications (33,34). As a definition in the context of microarrays, a ‘probe’ is the (partial) genomic sequence of a gene deposited and fixed on the Microarray, whereas the ‘target’ is the biological sample material.
Although there are many protocols and types of microarray experiments, the basic steps involved in any microarray experiment are isolating RNA or mRNA from appropriate biological samples; applying a fluorescent tag to the RNA or cDNA copy of it; hybridizing the labeled RNA or cDNA (target) to a microarray (probe) for a period of time after which the excess is washed off; scanning the microarray under laser light and data analysis using appropriate software.
A brief description of three basic types of DNA microarray experiments and their applications (35) is provided in Table 1.
Table 1.
Basic types of DNA microarray experiments
| Microarray type | Brief description | Applications | References |
|---|---|---|---|
| Comparative genomic hybridization (CGH) | Detect chromosomal aberrations | Tumor classification Disease diagnosis Risk assessment | (36–42) |
| Expression analysis | Analysis of gene expression levels | Drug development Drug response Tracking disease progression Therapy development Drug mechanism of action | (43–49) |
| Mutation/polymorphism analysis | Detect mutations/polymorphisms in gene sequence | Drug development Tracking disease progression Disease risk assessment Genotyping Species identification Population genetics | (50–55) |
Applications of DNA Microarrays in Herbal Drug Research and Development
Natural product research is often based on ethnobotanical information and many of the drugs used today were employed in indigenous societies (56). One of the aims of ethnopharmaceutical research is better understanding of the pharmacological effects of different medicinal plants traditionally used in healthcare (57). Plants are regarded as a promising source of novel therapeutic agents due to their higher structural diversity as compared to standard synthetic chemistry. Plants have applications in the development of therapeutic agents: as a source of bioactive compounds for possible use as drugs. There are three approaches to natural product-based drug discovery: screening of crude extracts; screening of pre-fractionated extracts; screening of pure compounds (58).
In the past decade there has been a paradigm shift from single-target drugs to multi-target drugs (59). Multi-target approaches are directed toward activation of defence, protective and repair mechanisms of the body rather than destruction of the damage-causing agent. This may be achieved by the use of a combination of drugs as in AIDS, cancer, atherosclerosis or use of single compounds that affect many targets simultaneously, e.g. Gleevec™, salicylate, metformin, Pemetrexed (Alimta) and a multi-targeted antifolate (60,61). The concept of multi-targeted therapy exists in traditional medical treatments that employ multi-component extracts of natural products. With the exception of pure substances, botanical derivatives obtained from medicinal plants usually contain several classes of compounds, which simultaneously act on multiple targets. However, very few herbal medicinal products have documentation comparable to synthetic drugs. Preparation of highly standardized herbal products with respect to chemical composition and biological activity is complicated due to the myriad range of chemicals. This also makes it difficult to precisely predict the molecular targets, mechanism of action and side effects of such products. Use of validated analytical methods to quantify specific chemical markers remains the key point to obtain chemically standardized extracts (62). However many minor components with potential biological effects remain neglected from quantitative analysis of chemical markers. In the past years the development of assays on the level of molecules (receptor binding and enzyme inhibition) opened complete new perspectives for plant-based products as source of new drugs (63). Advances in high-throughput experimentation have resulted in massive databases of genomic, proteomic and chemical data which in combination with efficient separation methods and powerful spectrometric methods for identification and structure elucidation can be used for identification of active compounds. In addition, the past two decades has witnessed a number of citations that describe a diverse range of molecular mechanisms that govern cellular behavior. A systems biology approach that integrates such large and diverse sources of information together will serve to make useful biological predictions about the pharmacological effects of natural products (64). DNA microarrays may provide a suitable high-throughput platform for research and development of drugs from natural products. In natural products a broad repertoire of chemical entities act together on multiple targets that makes it necessary to study the changes in expression of multiple genes simultaneously. Novel technologies such as SAGE and DNA microarrays allow rapid and detailed analysis of thousands of transcripts, providing a revolutionary approach to the investigation of gene expression.
In this review, we discuss the applications of DNA microarray technology in herbal drug research and development with suitable examples.
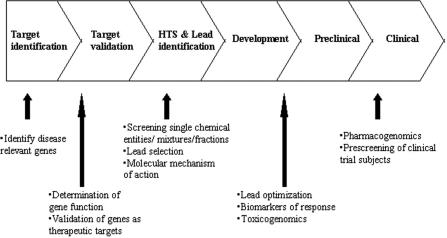
There are three main applications of DNA microarrays: first, in pharmacodynamics for discovery of new diagnostic and prognostic indicators and biomarkers of therapeutic response; elucidation of molecular mechanism of action of a herb, its formulations or its phytochemical components and identification and validation of new molecular targets for herbal drug development. Second, in pharmacogenomics for prediction of potential side-effects of the herbal drug during preclinical activity and safety studies; identification of genes involved in conferring drug sensitivity or resistance and prediction of patients most likely to benefit from the drug and use in general pharmacogenomic studies. Third, in pharmacognosy for correct botanical identification and authentication of crude plant materials as part of standardization and quality control. These applications are described here with some examples (65–68). Figure 1 shows applications of DNA microarrays at various stages of a typical drug discovery pipeline.
Figure 1.
DNA microarray: applications in natural product drug discovery and development.
DNA microarrays in pharmacodynamics
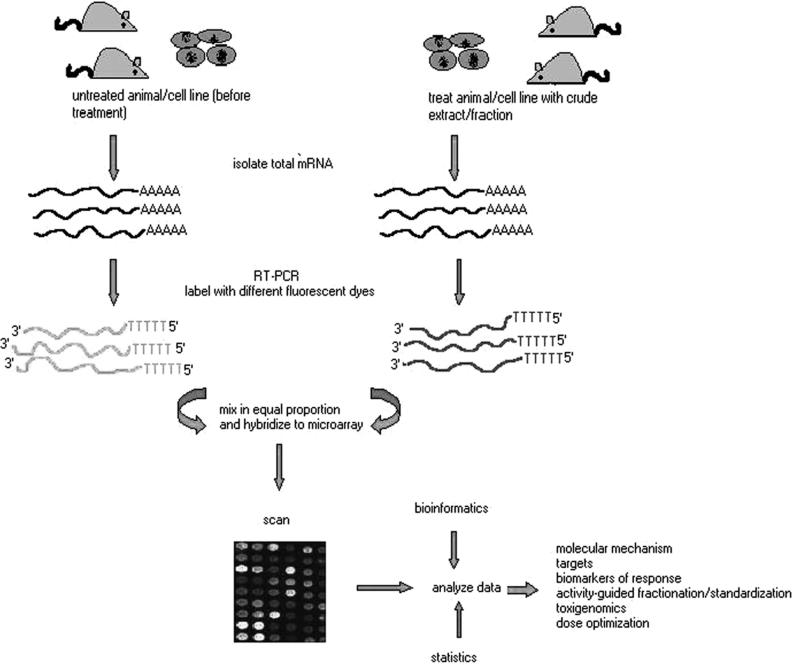
Herbal products are usually whole herbs, their formulations or extracts consisting of several bioactive compounds. With the increased demand for scientifically validated and standardized herbal product there is a need for better understanding of the molecular mechanisms underlying their biological activity. Although, the physiological actions of many herbal drugs are being studied at the molecular level, it remains unclear what are the targets of individual phytochemical components of herbs and how these molecules contribute to biological activities. Examples of the applications of microarray-based gene expression studies in elucidating molecular mechanism of action of pure compounds, different phytochemical groups and herbal extracts are discussed here. Figure 2 shows microarray based gene expression analysis of multi-component mixtures and the information that can be obtained from such studies.
Figure 2.
Microarray-based gene expression analysis of multi-component mixtures.
Purified compounds/specific phytochemical groups
The molecular pathways underlying diverse biological activity of the triterpenoid compounds isolated from the tropical medicinal plant Centella asiatica when studied with gene microarrays showed that Centella triterpenes evoke a gene-expression response consistent with their prevailing medical uses in the treatment of connective tissue disorders such as wound healing and microangiopathy. The identification of genes modulated by these compounds provides the basis for a molecular understanding of Centella's bioactivity, and opportunities for the quantitative correlation of this activity with clinical effectiveness at a molecular level (69). Similarly, the antiproliferative activity of Coptidis rhizoma, a medicinal herb, and its major component berberine was investigated in human pancreatic cancer cell lines. Gene expression patterns associated with sensitivities to each agent were analyzed with oligonucleotide arrays that comprised approximately 11,000 genes. It was possible to identify common and distinct genes related to anti-proliferative activities of purified berberine and C. rhizoma (70).
Involvement of genes in apoptosis inducing activity of alkaloids from Chinese medicinal herb Tripterygium hypoglaucum (levl.) Hutch (Celastraceae) root was examined using cDNA microarrays containing 3000 human genes derived from a leukocyte cDNA library. Induction of apoptosis by Tripterygium hypoglaucum alkaloids was found to be through c-myc and NF-kappa B signaling pathways (71).
Gene expression pattern of inferior colliculus from DBA/2J mice with audiogenic seizure and those treated with Qingyangshenylycosides, a traditional Chinese medicine, were examined. Gene expression analysis using Agilent oligo microarray showed that total of 134 genes were either up- or down-regulated during audiogenic seizure. Qingyangshenylycosides prevented many of the audiogenic seizure induced gene expression changes. Nevertheless, some of the audiogenic seizure induced genes were further enhanced or reversed by Qingyangshenylycosides treatment. The data provided important information regarding the molecular mechanisms of audiogenic seizure and the mechanism of action of Qingyangshenylycosides (72).
Oligonucleotide microarrays consisting of 87 probes representing 87 human cardiovascular disease-related genes were used to investigate the molecular mechanisms of saponins from the rhizome of Anemarrhena asphodeloides Bunge. Gene expression patterns in saponin treated and untreated human umbilical vein endothelial cells were analyzed. The results suggest that saponins may have beneficial effect on cardiovascular diseases by modulating the function of vein endothelial cells (73).
A gene chip, (Rat Genome U34A) was used to elucidate the gene regulatory pattern of Epimedium flavonoids in immune homeostasis remodeling in the aged rats. Expression pattern characterized by up-regulation of apoptosis promoting gene expression and down-regulation of apoptosis inhibiting genes expression, is the important gene background of immuno-homeostasis imbalance in aged. Epimedium flavonoids reverse these abnormal changes to reconstruct a beneficial equilibrium of gene expression and thus to further remodel the immuno-homeostasis in the aged (74).
Mechanism of herbal glycoside recipes retrieving deficient ability of spatial learning memory in mice suffering from cerebral ischemia/repurfusion was studied using DNA micoarray system. Gene expression pattern was analyzed in the groups that showed increased ability of spatial learning. A 1.8-fold increase in expression was observed for many genes (38–46) including genes in cell cycle regulation, signal transduction, nerve system transcription factors, DNA-binding protein, etc. Nine genes related to retrieving deficient ability of spatial learning memory when treated with glycoside recipes were found in this study (75).
Chronic cocaine use is known to elicit changes in the pattern of gene expression within the brain. The hippocampus plays a critical role in learning and memory and may also play a role in mediating behaviors associated with cocaine abuse. To profile the gene expression response of the hippocampus to chronic cocaine treatment, cDNA hybridization arrays were used to illuminate cocaine-regulated genes in rats treated non-contingently with a binge model of cocaine (76). Similarly, using DNA microarray analysis gene expression changes with chronic morphine and antagonist-precipitated withdrawal in two brain regions involved in behavioral effects of morphine in both mice and rats, were characterized (77).
Si-Jun-Zi decoction (SJZD), a traditional Chinese herbal prescription, has been used clinically for treating patients with disorders of the digestive system. Previous studies indicated that the polysaccharides of SJZD are the active components contributing towards its pharmacological effects in improving gastrointestinal function and immunity. SJZD polysaccharide was found to have protective effect and enhanced re-epithelialization on wounded IEC-6 cells. To elucidate the modulatory effect of polysaccharides of SJZD on wounded IEC-6 cells at the molecular level, an oligonucleotide microarray was employed to study differential gene expression of treated IEC-6 cells and the candidate genes were validated by RT–PCR. There was increased expression of genes coding for ion channels and transporters, which are critical to cell migration and restoration of wounded intestinal cells, suggesting a possible mechanism for re-epithelialization (78).
Clinical trials on antimicrotubule agents, docetaxel and estramustine combination treatment have suggested improved antitumor activity in hormone-refractory prostate cancer. However, the molecular mechanisms involved in the combination treatment with docetaxel and estramustine have not been fully elucidated. In order to establish such molecular mechanisms in both hormone insensitive and sensitive prostate cancer cells, gene expression profiles of docetaxel- and estramustine-treated prostate cancer cells were obtained by using Affymetrix Human Genome U133A Array. Clustering analysis based on biological function showed that docetaxel and estramustine combination treatment down-regulated some genes that are known to regulate cell proliferation, transcription, translation, and oncogenesis. In contrast, docetaxel and estramustine combination treatment up-regulated some genes related to induction of apoptosis, cell cycle arrest, and tumor suppression. Docetaxel and estramustine also showed differential effects on gene expression between mono- and combination treatment. Combination treatment with docetaxel and estramustine caused alternations of a large number of genes, many of which may contribute to the molecular mechanisms by which docetaxel and estramustine inhibit the growth of prostate cancer cells. These results provide novel molecular targets of docetaxel and estramustine combination treatment in prostate cancer cells (79).
Extracts
High-density oligonucleotide microarrays have been used for pioneer studies on the multiple gene expression effects exhibited by Ginkgo biloba leaf extract EGb 761, changing traditional pharmacology and medicine concepts (80). EGb 761 has been known to have neuroprotective effects ranging from molecular and cellular, to animal and human studies, however, the mechanisms remain unclear. DNA microarray based analyses has largely helped in identifying its targets and mechanism of action. High-density oligonucleotide microarrays were used to define the transcriptional effects in the cortex and hippocampus of mice whose diets were supplemented with the herbal extract. This study reveals that diets supplemented with Ginkgo biloba extract have notable neuromodulatory effects in vivo and illustrates the utility of genome-wide expression monitoring to investigate the biological actions of complex extracts (81). Further, DNA microarray analyses revealed that transcription of multiple apoptosis-related genes is either up- or down-regulated in cells treated with EGb 761. These results suggest that inhibition of apoptotic machinery may, at least in part, mediate multiple neuroprotective effects of EGb 761 (82). Mechanism of cytostatic action of EGb 761 was elucidated with the help of DNA microarray and the genes important for tumor growth were identified (83).
Ginkgo extract and one of its terpenoid constituents, ginkgolide B, inhibited the proliferation of a highly aggressive human breast cancer cell line and xenografts of this cell line in nude mice. cDNA microarray analyses have shown that exposure of human breast cancer cells to a Ginkgo extract altered the expression of genes that are involved in the regulation of cell proliferation, cell differentiation or apoptosis, and that exposure of human bladder cancer cells to a Ginkgo extract produced an adaptive transcriptional response that augments antioxidant status and inhibits DNA damage (84).
A human genome screen for TNFα-inducible genes was used to identify the genetic basis of the anti-inflammatory effects of standardized Boswellia serrata extract, 5-Loxin in microvascular endothelial cells. Acutely, TNFα-induced 522 genes; of which 113 genes were clearly sensitive to 5-Loxin treatment. Such genes directly related to inflammation, cell adhesion, and proteolysis. The robust 5-Loxin -sensitive candidate genes were then subjected to further processing which led to the recognition of the primary 5-Loxin -sensitive TNFα-inducible pathways. One mechanism by which TNFα, causes inflammation is by potently inducing the expression of adhesion molecule VCAM-1. 5-Loxin was found to completely inhibit VCAM-1 expression (85).
Water-extract of Paeoniae Radix has inhibitory effect on the growth of both HepG2 and Hep3B cell lines. Using cDNA microarray technology and RT–PCR analysis the drug targets of Paeoniae Radix in inhibition of tumor cells growth were elucidated (86).
Several studies have indicated that extracts of S. barbata have growth inhibitory effects on a number of human cancers. However, the mechanism underlying the antitumor activity was unclear. cDNA microarray analysis showed that 16 genes, involved in DNA damage, cell cycle control, nucleic acid binding and protein phosphorylation, underwent more than 5-fold change. These data indicated that these processes are involved in S. barbata-mediated killing of cancer cells (87).
The effect of Syzygium aromaticum (L.) Merrill and Perry (clove) extract on tissues that regulate glucose metabolism was studied by analysis of gene expression using DNA microarray. The data showed that the extract acts like insulin in hepatocytes and hepatoma cells by reducing phosphoenolpyruvate carboxykinase and glucose 6-phosphatase gene expression. Much like insulin, clove-mediated repression was reversed by PI3K inhibitors and N-acetylcysteine. Moreover, clove and insulin were found to regulate the expression of many of the same genes in a similar manner. These results indicate a potential role for compounds derived from clove as insulin-mimetic agents (88).
A cDNA microarray study demonstrated significant inhibition of inducible endothelial CD36 expression, a novel cardioregulatory gene, by IH636 grape seed proanthocyanidin extract (89).
Both the prototypic tricyclic antidepressant imipramine (IMI) and the extract of St John's wort (SJW) can be effective in the treatment of major depressive disorder. Affymetrix chips were used to study hypothalamic gene expression in rats treated with SJW or IMI to test the hypothesis that chronic antidepressant treatment by various classes of drugs results in shared patterns of gene expression that may underlie their therapeutic effects. SJW treatment differentially regulated 66 genes and expression sequence tags (ESTs) and IMI treatment differentially regulated 74 genes and ESTs. Six common transcripts in response to both treatments were found. Both treatments also affected different genes that are part of the same cell function processes, such as glycolytic pathways and synaptic function. The data support the hypothesis that chronic antidepressant treatment by drugs of various classes may result in a common, final pathway of changes in gene expression in a discrete brain region (90).
Recently, a rapid throughput screening system involving gene expression analysis was developed in order to investigate the potential of bioactive chemicals contained in natural health products as effective drug therapy, in particular the ability of these chemicals to alleviate the inflammatory response in human airway epithelial cells (91).
The pharmacogenomic activities of Anoectochilus formosanus extract as a crude phytocompound mixture were compared to those conferred by the single-compound drug, plumbagin in MCF-7 cancer cells. This study offers evidence to support the search for fractionated medicinal herb extracts or phytocompound mixtures, in addition to single-compound drugs, as defined therapeutic agents (92). DNA microarray can be used for activity-guided fractionation of herbal extracts in order to determine active principles.
DNA microarrays in Pharmacogenomics
Pharmacogenomics is the study of genes and the gene products (proteins) essential for pharmacological or toxicological responses to pharmaceutical agents. Oligonucleotide-based DNA chip technology or cDNA microarray can be used to analyze gene expression profiles that are induced or repressed by xenobiotics (93).
An attempt has been made to develop microarray genotyping system for multiplex analysis of a panel of single nucleotide polymorphisms (SNPs) in genes encoding proteins involved in blood pressure regulation, and to apply this system in a pilot study demonstrating its feasibility in the pharmacogenetics of anti-hypertensive drug response (94). DNA microarray techniques might prove to be a reliable basis for predicting response (or lack of response) of individuals to herbal drugs.
Technologies designed to characterize genes and their products on a discovery scale are now having an impact on many areas of biology, including toxicology. Toxicogenomics is the sub-discipline that merges genomics with toxicology. In toxicology research, gene expression profiling facilitates mechanism-based research on toxicant action by comparing results for an experimental compound with a database. Recently several studies have demonstrated the utility of microarray analysis for studying genome-wide effects of xenobiotics and the rapid identification of toxic hazards for novel drug candidates (95,96).
An example of such a platform is ToxBlot II, a custom microarray containing cDNAs representing 12564 human genes chosen on the basis of their potential relevance to a broad range of toxicities. ToxBlot II allows the simultaneous expression profiling of genes representing entire cellular pathways, facilitating a very detailed investigation of potential mechanisms of toxicity (97). cDNA microarray analysis was used to study the expression level of genes in oral fibroblast cell lines in response to exposure to ripe areca nut extract. The results showed up-regulation of IL-6 expression and down-regulation of PDGFR, APP-1 and KGF-1 expressions in multiple cell lines assayed. The down-regulation of KGF-1 expression in oral fibroblast cell lines potentially impairs the proliferation of overlying keratinocytes, which could partially explain the frequent epithelial atrophy observed in chronic areca chewers in vivo. This study established a novel toxicogenomic database for areca nut extract (98). cDNA microarray analysis was also used to analyze the mRNA expression patterns of 1177 genes in ten oral cancer patients with betel quid chewing history. This study provides pilot data for understanding the pathogenesis of oral cancer in countries like Taiwan where betel quid chewing is prevalent (99).
Recent studies have highlighted that concurrent use of herbs may mimic, magnify, or oppose the effect of drugs (100,101). DNA microarray can be used for studying herb–drug interactions, and the mechanisms underlying these interactions.
Several botanical constituents in PC-SPES, a botanical preparation, inhibit tumor growth through cell cycle arrest and apoptosis. LNCaP prostate carcinoma cells were treated with PC-SPES, and changes in gene expression were determined by complementary DNA (cDNA) microarray hybridization and northern blot analyses. mRNA levels of α-tubulin decreased sevenfold. The results show that PC-SPES may interfere with microtubule polymerization. This activity has implications for the clinical management of patients with advanced prostate cancer who may be taking PC-SPES concurrently with microtubule-modulating chemotherapeutic agents, such as paclitaxel (102). cDNA microarray analysis was used to identify gene expression changes in LNCaP prostate carcinoma cells exposed to PC-SPES and estrogenic agents including diethylstilbestrol. Transcripts encoding cell cycle-regulatory proteins, α- and β-tubulins, and the androgen receptor were down-regulated by PC-SPES. A comparison of gene expression profiles resulting from these treatments indicates that PC-SPES exhibits activities distinct from those attributable to diethylstilbestrol and suggests that alterations in specific genes involved in modulating the cell cycle, cell structure, and androgen response may be responsible for PC-SPES-mediated cytotoxicity (103).
DNA microarray in pharmacognosy
Use of authentic herbal materials is the first step to ensure quality, safety and efficacy of herbal medicines. DNA polymorphism-based assays have been developed for the identification of herbal medicines (104,105). In this approach, small amounts of DNA are amplified by the polymerase chain reaction and the reaction products are analyzed by gel electrophoresis, sequencing, or hybridization with species-specific probes. Recently, microarrays have been applied for the DNA sequence-based identification of medicinal plants (106,107).
To utilize DNA microarrays for identification and authentication of herbal material, it is necessary to identify a distinct DNA sequence that is unique to each species of medicinal plant. The DNA sequence information is then used to synthesize a corresponding probe on a silicon-based gene chip. These probes are capable of detecting complementary target DNA sequences if present in the test sample being analyzed.
Oligonucleotide probes specific for polymorphisms in the D2 and D3 regions of 26S rDNA gene of several Fritillaria species were designed and printed on the poly-lysine coated slides to prepare a DNA chip. Differentiation of the various Fritillaria species was accomplished based on hybridization of fluorescent labeled PCR products with the DNA chip. The results demonstrated the reliability of using DNA chips to identify different species of Fritillaria, and that the DNA chip technology can provide a rapid, high throughput tool for genotyping and plant species authentication (108).
Similarly, using fluorescence-labeled ITS2 sequences as probes, distinctive signals were obtained for the five medicinal Dendrobium species listed in the Chinese Pharmacopoeia. The established microarray was able to detect the presence of D. nobile in a Chinese medicinal formulation containing nine herbal components (109).
A silicon-based DNA microarray using species-specific oligonucleotide probes is designed and fabricated to identify multiple toxic traditional Chinese medicinal plant species by parallel genotyping (110). Chip-based authentication of medicinal plants may be useful as economical, precise tool for quality control and safety monitoring of herbal pharmaceuticals and neutraceuticals.
Identification of herbal materials, which commonly consist of dried or processed parts, is difficult. This is particularly true for similar looking herbal materials that can often vary greatly in their medicinal properties and market value. DNA microarray based technology can provide an efficient, accurate and cheaper means of testing the authenticity of hundreds of samples simultaneously while conventional chemical methodologies usually take several days for verification. Chip-based authentication of medicinal plants can be useful as a tool for quality control and safety monitoring of herbal pharmaceuticals and neutraceuticals and will significantly add to the medical potential and commercial profitability of herbal products.
This application of DNA microarrays will not only benefit the herbal drug industry but can also facilitate the identification of herbal products by regulatory authorities. An international initiative, the Consortium for the Barcode of Life (CBOL) is devoted to developing DNA barcoding as a tool for correct taxonomic identification of species, including medicinal plants. DNA microarrays can provide a suitable technology platform for such an initiative.
Perspective
Traditionally, the pharmaceutical industry has focused on modifying one gene or protein at a time as the basis for identifying new drugs. The quest for a molecule to modify a single key factor in a disease process may not provide a solution for many complex diseases where multiple cell types and/or multiple pathways contribute to disease. Herbal extracts or mixtures represent combinatorial chemistry of nature with vast repertoire of chemical entities that have a complex effect on numerous cellular components and functions. They have great potential in the multi-target approach to diseases. However, potential of herbal drugs as defined therapeutic agents is undermined by the difficulty in standardization, pharmacodynamics and pharmacokinetics of these multi-component mixtures. Microarray analysis of gene expression can be useful for elucidating the molecular mechanisms and networks underlying the complex pharmacological function of herbal extracts and mixtures. Studying the patterns of gene expression at many stages during the treatment process can reveal mechanism from a modern genetic perspective and will help to identify biomarkers of adverse or favorable response. A positive correlation between the transcriptional response induced by an herbal drug and a database profile of an existing therapeutic agent can provide insight into target specificity, mechanism of action, as well as facilitate analysis of pathways downstream of the target. It will also help in identification of novel therapeutic applications of a herbal drug. DNA microarrays can also be used for activity-guided fractionation of herbal extracts thereby helping in narrowing down upon the active principle producing the desired effect. Microarrays have applications in the entire drug discovery pipeline to improve selection of biological targets and lead compounds. Correlation of gene expression data of herbal drug candidates with clinical outcome or biomarkers of response in biological system will facilitate selection of best candidate for drug development.
In the area of pharmacogenomics, DNA microarrays will facilitate development of individually optimized drugs based on differential gene expression patterns. Genetic polymorphism studies can be done to classify individuals according to their drug metabolizing capacities or response to disease (111). SNP screening of patients undergoing treatment with a particular herbal drug and correlating these data with recovery profile would help to understand which individuals or groups are likely to be benefited by the drug. Microarray analysis of gene expression has become a suitable approach for the biological effects of drugs and other chemicals. Gene expression profile screens focused on genes that are relevant to toxicity can be useful for prediction of potential herbal drug side effects during pre-clinical development and toxicology studies. They can also be applied for studying herb–drug interactions and their underlying mechanisms.
Medicinal plants are an integral part of TM/CAM such as Ayurvedic, Chinese, Native American and many other traditional systems of medicine. Clinical experiences, observations or documented data becomes a starting point for traditional knowledge based drug development. Although the clinical existence of traditional medicine spans several generations, it may be hazardous to extrapolate from such uses to current practice, since herbs and or their formulations used may be very different from the commercial herbal products available today. Many questions remain concerning quality, efficacy, safety, bioavailability and mechanisms of action of even the most widely used herbs. These questions can be answered by rigorous application of scientific principles in high quality basic and clinical research. There is a need for scientific validation of safety, efficacy, therapeutic claims, bioavailability and mechanism of action of these medicines that are being used in the population since centuries. Microarray based approach can facilitate high throughput, fast track evidence-based herbal drug development following Reverse Pharmacology path. Phytomics, a technology platform for characterization of herbal compositions wherein, Herbal Bio Response Arrays (HBR Arrays) are used to determine bioactive constituents and biological activities of an herbal composition has recently been developed and patented (112).
Further, CAM disciplines owing to their holistic nature may be impacting gene expression and signaling pathways (113). Application of such novel expression analysis tools may help to understand and characterize the genes and signaling pathways involved in CAM modalities.
DNA microarrays have the potential for application in different phases of herbal drug discovery and development. This includes quality control and standardization of the herbal drugs, identification and validation of new targets, the profiling of on-target and off-target effects during the optimization of new therapeutic agents, understanding molecular mechanisms of action, structure–activity relationships (114) and the prediction of side-effects, and the discovery of diagnostic, prognostic, and pharmacodynamic biomarkers.
At present the major practical applicability of DNA microarrays remains in SNP mapping, genotyping and pharmacogenetics. In recent years, array-based sequencing (minisequencing/resequencing arrays) that combines target hybridization with enzymatic primer extension reactions has emerged as a powerful means to scan for all possible DNA sequence variations (115–117). Highly multiplexed microarray systems offering potential for mutation analysis (118,119) and genome-wide SNP mapping (120) have been developed. DNA microarrays are a effective new strategy for global gene expression profiling, but reliable detection of expression changes and understanding the functional relevance of these expression profiles is a challenge. Other problems in microarray experiments include non-reproducibility, difficulty in statistical analysis and comparability of data from different experiments (121). Inconsistencies in data may arise due to low specificity of cDNA probes, discrepancy in fold-change calculations, variations in laboratories, technicians, probe labeling, biochemical reactions, scanners, lasers etc (122). Also the sensitivity and specificity of DNA microarrays depends on the nucleotide composition of the target and probe. Repetitive sequence elements and highly structured nucleic acids can greatly decrease the sensitivity of hybridization based analysis. In its current stage, a microarray experiment needs to be validated by independent confirmation of results by laboratory-based analysis and in silico comparison with information available in literature and expression databases (123). Although DNA microarrays have huge potential for pharmacodynamics and toxigenomics applications, these are still in exploratory stage and need validation by other biological experiments. Bioinformatics and statistical tools have a major role to play in the microarray field, whereby data from multiple experiments can be integrated to answer complex biological questions. Another factor limiting microarray application is the cost of the technology. The challenge today is to provide standardized, sensitive, reproducible microarray platforms, databases and visualization methods for expression profiles that are also affordable to scientists. With the development of new, uniform and more sophisticated experimental designs, data management systems (124,125) statistical tools and algorithms for data analysis (126,127) DNA microarrays can be optimally used in herbal drug research. In spite of the huge potential offered by microarray technology, the importance of in vitro biological assays, cell-line studies and in vivo animal studies cannot be ignored. A comprehensive strategy integrating information from diverse scientific experiments and technologies will lead to molecular evidence-based herbal medicine.
Acknowledgments
The authors thank CSIR, India for financial assistance (PC) during writing of this review.
References
- 1.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–70. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 2.Schena M, Shalon D, Heller R, Chai A, Brown PO, Davis RW. Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc Natl Acad Sci USA. 1996;93:10614–19. doi: 10.1073/pnas.93.20.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taniguchi M, Miura K, Iwao H, Yamanaka S. Quantitative assessments of DNA microarrays-comparison with northern blot analyses. Genomics. 2001;71:34–9. doi: 10.1006/geno.2000.6427. [DOI] [PubMed] [Google Scholar]
- 4.Yamamoto M, Wakatsuki T, Hada A, Ryo A. Use of serial analysis of gene expression (SAGE) technology. J Immunol Methods. 2001;250:45–66. doi: 10.1016/s0022-1759(01)00305-2. [DOI] [PubMed] [Google Scholar]
- 5.Bertelsen AH, Velculescu VE. High-throughput gene expression analysis using SAGE. Drug Discov Today. 1998;3:152–9. [Google Scholar]
- 6.Lee Y, Sultana R, Pertea G, Cho J, Karamycheva S, Tsai J, et al. Cross-referencing eukaryotic genomes: TIGR Orthologous Gene Alignments (TOGA) Genome Res. 2002;12:493–502. doi: 10.1101/gr.212002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sogayar MC, Camargo AA, Bettoni F, Carraro DM, Pires LC, Parmigiani RB. Ludwig-FAPESP transcript finishing initiative: a transcript finishing initiative for closing gaps in the human transcriptome. Genome Res. 2004;14:1413–23. doi: 10.1101/gr.2111304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Debouck C, Goodfellow PN. DNA microarrays in drug discovery and development. Nat Genet. 1999;21:48–50. doi: 10.1038/4475. [DOI] [PubMed] [Google Scholar]
- 9.Gerhold DL, Jensen RV, Gullans SR. Better therapeutics through microarrays. Nat Genet. 2002;32:547–51. doi: 10.1038/ng1042. [DOI] [PubMed] [Google Scholar]
- 10.Reynolds MA. Microarray technology GEM microarrays and drug discovery. J Ind Microbiol Biotechnol. 2002;28:180–5. doi: 10.1038/sj/jim/7000136. [DOI] [PubMed] [Google Scholar]
- 11.Koppal T. Microarrays: migrating from discovery to diagnostics. Drug Discov Dev. 2004;7:30–4. [Google Scholar]
- 12.Gebauer M. Microarray applications: emerging technologies and perspectives. Drug Discov Today. 2004;9:915–17. doi: 10.1016/S1359-6446(04)03242-8. [DOI] [PubMed] [Google Scholar]
- 13.Huang JX, Mehrens D, Wiese R, Lee S, Tam SW, Daniel S, et al. High-throughput genomic and proteomic analysis using microarray technology. Clin Chem. 2001;47:1912–16. [PubMed] [Google Scholar]
- 14.Merchant M, Weinberger SR. Recent advancements in surface enhanced laser desorption/ionization time-of-flight mass spectrometry. Electrophoresis. 2000;21:1164–77. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1164::AID-ELPS1164>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 15.Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–5. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 16.Stoll D, Templin MF, Bachmann J, Joos TO. Protein microarrays: applications and future challenges. Curr Opin Drug Discov Dev. 2005;8:239–52. [PubMed] [Google Scholar]
- 17.Kersten B, Feilner T, Kramer A, Wehrmeyer S, Possling A, Witt I, et al. Generation of Arabidopsis protein chips for antibody and serum screening. Plant Mol Biol. 2003;52:999–1010. doi: 10.1023/a:1025424814739. [DOI] [PubMed] [Google Scholar]
- 18.Haab BB. Advances in protein microarray technology for protein expression and interaction profiling. Curr Opin Drug Discov Dev. 2001;4:116–23. [PubMed] [Google Scholar]
- 19.Kumble KD. Protein microarrays: new tools for pharmaceutical development. Anal Bioanal Chem. 2003;377:812–19. doi: 10.1007/s00216-003-2088-6. [DOI] [PubMed] [Google Scholar]
- 20.MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–3. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 21.Templin MF, Stoll D, Schrenk M, Traub PC, Vohringer CF, Joos TO. Protein microarray technology. Drug Discov Today. 2002;7:815–22. doi: 10.1016/s1359-6446(00)01910-2. [DOI] [PubMed] [Google Scholar]
- 22.Kodadek T. Protein microarrays: prospects and problems. Chem Biol. 2001;8:105–15. doi: 10.1016/s1074-5521(00)90067-x. [DOI] [PubMed] [Google Scholar]
- 23.Schweitzer B, Predki P, Snyder M. Microarrays to characterize protein interactions on a whole-proteome scale. Proteomics. 2003;3:2190–9. doi: 10.1002/pmic.200300610. [DOI] [PubMed] [Google Scholar]
- 24.Lueking A, Cahill DJ, Mullner S. Protein biochips: a new and versatile platform technology for molecular medicine. Drug Discov Today. 2005;10:789–94. doi: 10.1016/S1359-6446(05)03449-5. [DOI] [PubMed] [Google Scholar]
- 25.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight, et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–7. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 26.Qureshi AE, Cagney G. Large-scale functional analysis using peptide or protein arrays. Nat Biotechnol. 2000;18:393–7. doi: 10.1038/74442. [DOI] [PubMed] [Google Scholar]
- 27.Junaid Z, David MS. Microarrays of cells expressing defined cDNAs. Nature. 2001;411:107–10. doi: 10.1038/35075114. [DOI] [PubMed] [Google Scholar]
- 28.Mousses S, Caplen NJ, Cornelison R, Weaver D, Basik M, Hautaniemi S, et al. RNAi microarray analysis in cultured mammalian cells. Genome Res. 2003;13:2341–7. doi: 10.1101/gr.1478703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kit SL, Manat R. From combinatorial chemistry to chemical microarray. Curr Opin Chem Biol. 2002;6:353–8. doi: 10.1016/s1367-5931(02)00326-5. [DOI] [PubMed] [Google Scholar]
- 30.Dickopf S, Frank HD, Junker S, Maier G, Metz H, Ottleben H, et al. Custom chemical microarray production and affinity fingerprinting for the S1 pocket of factor VIIa. Anal Biochem. 2004;335:50–7. doi: 10.1016/j.ab.2004.08.033. [DOI] [PubMed] [Google Scholar]
- 31.Barret JC, Kawasaki ES. Microarrays: the use of oligonucleotides and cDNA for the analysis of gene expression. Drug Discov Today. 2003;8:134–41. doi: 10.1016/s1359-6446(02)02578-3. [DOI] [PubMed] [Google Scholar]
- 32.Sinibaldi R. Gene expression analysis and drug R&D. Drug Discov World. 2003;5:37–43. [Google Scholar]
- 33.Sheils O, Finn S, O'Leary J. Nucleic acid microarrays: an overview. Curr Diagn Pathol. 2003;9:155–8. [Google Scholar]
- 34.Sauter G, Simon R. Predictive molecular pathology. N Engl J Med. 2002;347:1995–6. doi: 10.1056/NEJMp020155. [DOI] [PubMed] [Google Scholar]
- 35. http://www.ncbi.nlm.nih.gov/About/primer/microarrays.html.
- 36.Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–21. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 37.Yu W, Ballif BC, Kashank CD, Heilstedt HA, Howard LA, Cai WW, et al. Development of a comparative genomic hybridization microarray and demonstration of its utility with 25 well-charcterized 1p36 deletions. Hum Mol Genet. 2003;12:2145–52. doi: 10.1093/hmg/ddg230. [DOI] [PubMed] [Google Scholar]
- 38.Pettus JA, Cowley BC, Maxwell T, Milash B, Stephenson RA, Rohr LR, et al. Multiple abnormalities detected by dye reversal genomic microarrays in prostate cancer: a much greater sensitivity than conventional cytogenetics. Cancer Genet Cytogenet. 2004;154:110–18. doi: 10.1016/j.cancergencyto.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 39.Shaw CJ, Shaw CA, Yu W, Stankiewicz P, White LD, Beaudet AL, et al. Comparative genomic hybridisation using a proximal 17pBAC/PAC array detects rearrangements responsible for four genomic disorders. J Med Genet. 2004;41:113–19. doi: 10.1136/jmg.2003.012831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strohmeyer DM, Berger AP, Moore DH, II, Bartsch G, Klocker H, Carroll PR, et al. Genetic aberrations in prostate carcinoma detected by comparative genomic hybridization and microsatellite analysis: association with progression and angiogenesis. Prostate. 2004;59:43–58. doi: 10.1002/pros.20028. [DOI] [PubMed] [Google Scholar]
- 41.Roerig P, Nessling M, Radlwimmer B, Joos S, Wrobel G, Schwaenen C, et al. Molecular classification of human gliomas using matrix-based comparative genomic hybridization. Int J Cancer. 2005;117:95–103. doi: 10.1002/ijc.21121. [DOI] [PubMed] [Google Scholar]
- 42.Oostlander A, Meijer G, Ylstra B. Microarray-based comparative genomic hybridization and its applications in human genetics. Clin Genet. 2004;66:488–95. doi: 10.1111/j.1399-0004.2004.00322.x. [DOI] [PubMed] [Google Scholar]
- 43.Diosdado B, Wapenaar MC, Franke L, Duran KJ, Goerres MJ, Hadithi M, et al. A microarray screen for novel candidate genes in coeliac disease pathogenesis. Gut. 2004;53:944–51. doi: 10.1136/gut.2003.018374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kittleson MM, Hare JM. Molecular signature analysis: using the myocardial transcriptome as a biomarker in cardiovascular disease. Trends Cardiovasc Med. 2005;15:130–8. doi: 10.1016/j.tcm.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 45.Gunther EC, Stone DJ, Gerwien RW, Bento P, Melvyn PH. Prediction of clinical drug efficacy by classification of drug-induced genomic expression profiles in vitro. Proc Natl Acad Sci USA. 2003;100:9608–13. doi: 10.1073/pnas.1632587100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ganter B, Tugendreich S, Pearson CI, Ayangolu E, Baumhueter S, Bostian KA, et al. Development of a large-scale chemogenomics database to improve drug candidate selection and to understand mechanisms of chemical toxicity and action. J Biotechnol. 2005;119:219–44. doi: 10.1016/j.jbiotec.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 47.Villuendas R, Steegmann JL, Pollan M, Tracey L, Granda A, Fernandez-Ruiz E, et al. Identification of genes involved in imatinib resistance in CML: a gene-expression profiling approach. Leukemia. 2006;20:1047–54. doi: 10.1038/sj.leu.2404197. [DOI] [PubMed] [Google Scholar]
- 48.Boyer J, Allen WL, McLean EG, Wilson PM, McCulla A, Moore S, et al. Pharmacogenomic identification of novel determinants of response to chemotherapy in colon cancer. Cancer Res. 2006;66:2765–77. doi: 10.1158/0008-5472.CAN-05-2693. [DOI] [PubMed] [Google Scholar]
- 49.Tsai MF, Wang CC, Chang GC, Chen CY, Chen HY, Cheng CL, et al. A new tumor suppressor DnaJ-like heat shock protein, HLJ1, and survival of patients with non-small-cell lung carcinoma. J Natl Cancer Inst. 2006;98:825–38. doi: 10.1093/jnci/djj229. [DOI] [PubMed] [Google Scholar]
- 50.Hacia JG. Resequencing and mutational analysis using oligonucleotide microarrays. Nat Genet. 1999;21:42–7. doi: 10.1038/4469. [DOI] [PubMed] [Google Scholar]
- 51.Lindroos K, Sigurdsson S, Johansson K, Ronnblom L, Syvanen AC. Multiplex SNP genotyping in pooled DNA samples by a four-colour microarray system. Nucleic Acids Res. 2002;30:e70. doi: 10.1093/nar/gnf069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi MM. Technologies for individual genotyping: detection of genetic polymorphisms in drug targets and disease genes. Am J Pharmacogenomics. 2002;2:197–205. [PubMed] [Google Scholar]
- 53.Irving JA, Bloodworth L, Bown NP, Case MC, Hogarth LA, Hall AG. Loss of heterozygosity in childhood acute lymphoblastic leukemia detected by genome-wide microarray single nucleotide polymorphism analysis. Cancer Res. 2005;65:3053–8. doi: 10.1158/0008-5472.CAN-04-2604. [DOI] [PubMed] [Google Scholar]
- 54.Klevering BJ, Yzer S, Rohrschneider K, Zonneveld M, Allikmets R, van den Born LI, et al. Microarray-based mutation analysis of the ABCA4 (ABCR) gene in autosomal recessive cone-rod dystrophy and retinitis pigmentosa. Eur J Hum Genet. 2004;12:1024–32. doi: 10.1038/sj.ejhg.5201258. [DOI] [PubMed] [Google Scholar]
- 55.Janecke AR, Thompson DA, Utermann G, Becker C, Hubner CA, Schmid E, et al. Mutations in RDH12 encoding a photoreceptor cell retinol dehydrogenase cause childhood-onset severe retinal dystrophy. Nat Genet. 2004;36:850–4. doi: 10.1038/ng1394. [DOI] [PubMed] [Google Scholar]
- 56.Patwardhan B. Ethnopharmacology and drug discovery. J Ethnopharmacol. 2005;100:50–2. doi: 10.1016/j.jep.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 57.Heinrich M. Ethnobotany and natural products: the search for new molecules, new treatments of old diseases or a better understanding of indigenous cultures? Curr Top Med Chem. 2003;3:141–54. doi: 10.2174/1568026033392570. [DOI] [PubMed] [Google Scholar]
- 58.Vuorela P, Leinonen M, Saikku P, Tammela P, Rauha JP, Wennberg T, et al. Natural products in the process of finding new drug candidates. Curr Med Chem. 2004;11:1375–89. doi: 10.2174/0929867043365116. [DOI] [PubMed] [Google Scholar]
- 59.Wermuth CG. Multitargeted drugs: the end of the ‘one-target-one-disease’ philosophy? Drug Discov Today. 2004;9:826–7. doi: 10.1016/S1359-6446(04)03213-1. [DOI] [PubMed] [Google Scholar]
- 60.Csermely P, Agoston V, Pongor S. The efficiency of multi-target drugs: the network approach might help drug design. Trends Pharmacol Sci. 2005;26:178–82. doi: 10.1016/j.tips.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 61.Cappuzzo F, Finocchiaro G, Trisoloini R, Toschi L, Bartolini S, Metro G, et al. Perspectives on salvage therapy for non-small-cell lung cancer. Oncology. 2005;19:989–95. [PubMed] [Google Scholar]
- 62.Amagaya S, Iizuka A, Makino B, Kubo M, Komatsu Y, Cheng FC, et al. General pharmacological properties of Sho-seiryu-to (TJ-19) extracts. Phytomedicine. 2001;8:338–47. doi: 10.1078/0944-7113-00061. [DOI] [PubMed] [Google Scholar]
- 63.Ye F, Wang H, Jiang S, Wu J, Shao J, Cheng X, et al. Quality evaluation of commercial extracts of Scutellaria baicalensis. Nutr Cancer. 2004;49:217–22. doi: 10.1207/s15327914nc4902_14. [DOI] [PubMed] [Google Scholar]
- 64.Wagner H. New approaches in phytopharmacological research. Pure Appl Chem. 1999;71:1649–54. [Google Scholar]
- 65.Clarke PA, te Poele R, Wooster R, Workman P. Gene expression microarray analysis in cancer biology, pharmacology, and drug development: progress and potential. Biochem Pharmacol. 2001;62:1311–36. doi: 10.1016/s0006-2952(01)00785-7. [DOI] [PubMed] [Google Scholar]
- 66.Butte A. The use and analysis of microarray data. Nat Rev Drug Discov. 2002;1:951–60. doi: 10.1038/nrd961. [DOI] [PubMed] [Google Scholar]
- 67.Crowther DJ. Applications of microarrays in the pharmaceutical industry. Curr Opin Pharmacol. 2002;2:551–4. doi: 10.1016/s1471-4892(02)00207-2. [DOI] [PubMed] [Google Scholar]
- 68.Klapa MI, Quackenbush J. The quest for the mechanisms of life. Biotechnol Bioeng. 2003;84:739–42. doi: 10.1002/bit.10858. [DOI] [PubMed] [Google Scholar]
- 69.Coldren CD, Hashim P, Ali JM, Oh SK, Sinskey AJ, Rha C. Gene expression changes in the human fibroblast induced by Centella asiatica triterpenoids. Planta Med. 2003;69:725–32. doi: 10.1055/s-2003-42791. [DOI] [PubMed] [Google Scholar]
- 70.Iizuka N, Oka M, Yamamoto K, Tangoku A, Miyamoto K, Miyamoto T, et al. Identification of common or distinct genes related to antitumor activities of a medicinal herb and its major component by oligonucleotide microarray. Int J Cancer. 2003;107:666–72. doi: 10.1002/ijc.11452. [DOI] [PubMed] [Google Scholar]
- 71.Zhuang WJ, Fong CC, Cao J, Ao L, Leung CH, Cheung HY, et al. Involvement of NF-kappa B and c-myc signaling pathways in the apoptosis of HL-60 cells induced by alkaloids of Tripterygium hypoglaucum (levl.) Hutch Phytomedicine. 2004;11:295–302. doi: 10.1078/0944711041495128. [DOI] [PubMed] [Google Scholar]
- 72.Li X, Hu Y. Gene expression profiling reveals the mechanism of action of anticonvulsant drug QYS. Brain Res Bull. 2005;66:99–105. doi: 10.1016/j.brainresbull.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 73.Li ZS, Li DL, Huang J, Ding Y, Ma BP, Wang SQ. Investigations on the molecular mechanisms of saponins from Anemarrhena asphodeloides Bunge using oligonucleotide microarrays. Yao Xue Xue Bao. 2003;38:496–500. [Chinese] [PubMed] [Google Scholar]
- 74.Chen Y, Shen ZY, Chen WH. Molecular mechanism of epimedium flavonoids in immune homeostasis remodeling in aged rats revealed by lymphocyte gene expression profile. Zhongguo Zhong Xi Yi Jie He Za Zhi. 2004;24:59–62. [Chinese] [PubMed] [Google Scholar]
- 75.Wang Z, Du Q, Wang F, Liu Z, Li B, Wang A, et al. Microarray analysis of gene expression on herbal glycoside recipes improving deficient ability of spatial learning memory in ischemic mice. J Neurochem. 2004;88:1406–15. doi: 10.1046/j.1471-4159.2003.02258.x. [DOI] [PubMed] [Google Scholar]
- 76.Freeman WM, Brebner K, Lynch WJ, Robertson DJ, Roberts DC, Vrana KE. Cocaine-responsive gene expression changes in rat hippocampus. Neuroscience. 2001;108:371–80. doi: 10.1016/s0306-4522(01)00432-8. [DOI] [PubMed] [Google Scholar]
- 77.McClung CA, Nestler EJ, Zachariou V. Regulation of gene expression by chronic morphine and morphine withdrawal in the locus ceruleus and ventral tegmental area. J Neurosci. 2005;25:6005–15. doi: 10.1523/JNEUROSCI.0062-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu L, Han L, Wong DY, Yue PY, Ha WY, Hu YH, et al. Effects of Si-Jun-Zi decoction polysaccharides on cell migration and gene expression in wounded rat intestinal epithelial cells. Br J Nutr. 2005;93:21–9. doi: 10.1079/bjn20041295. [DOI] [PubMed] [Google Scholar]
- 79.Li Y, Hong X, Hussain M, Sarkar SH, Li R, Sarkar FH. Gene expression profiling revealed novel molecular targets of docetaxel and estramustine combination treatment in prostate cancer cells. Mol Cancer Ther. 2005;4:389–98. doi: 10.1158/1535-7163.MCT-04-0244. [DOI] [PubMed] [Google Scholar]
- 80.Christen Y, Olano-Martin E, Packer L. Egb 761 in the postgenomic era: new tools from molecular biology for the study of complex products such as Ginkgo biloba extract. Cell Mol Biol. 2002;48:593–9. [PubMed] [Google Scholar]
- 81.Watanabe CM, Wolffram S, Ader P, Rimbach G, Packer L, Maguire, et al. The in vivo neuromodulatory effects of the herbal medicine Ginkgo biloba. Proc Natl Acad Sci USA. 2001;98:6577–80. doi: 10.1073/pnas.111126298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smith JV, Burdick AJ, Golik P, Khan I, Wallace D, Luo Y. Anti-apoptotic properties of Ginkgo biloba extract EGb 761 in differentiated PC12 cells. Cell Mol Biol. 2002;48:699–707. [PubMed] [Google Scholar]
- 83.Li W, Pretner E, Shen L, Drieu K, Papadopoulos V. Common gene targets of Ginkgo biloba extract (EGb 761) in human tumor cells: relation to cell growth. Cell Mol Biol. 2002;48:655–62. [PubMed] [Google Scholar]
- 84.De Feudis FV, Papadopoulos V, Drieu K. Ginkgo biloba extracts and cancer: a research area in its infancy. Fundam Clin Pharmacol. 2003;17:405–17. doi: 10.1046/j.1472-8206.2003.00156.x. [DOI] [PubMed] [Google Scholar]
- 85.Roy S, Khanna S, Shah H, Rink C, Phillips C, Preuss H, et al. Human genome screen to identify the genetic basis of the anti-inflammatory effects of Boswellia in microvascular endothelial cells. DNA Cell Biol. 2005;24:244–55. doi: 10.1089/dna.2005.24.244. [DOI] [PubMed] [Google Scholar]
- 86.Lee SM, Li ML, Tse YC, Leung SC, Lee MM, Tsui SK, et al. Paeoniae Radix, a Chinese herbal extract, inhibit hepatoma cells growth by inducing apoptosis in a p53 independent pathway. Life Sci. 2002;71:2267–77. doi: 10.1016/s0024-3205(02)01962-8. [DOI] [PubMed] [Google Scholar]
- 87.Yin X, Zhou J, Jie C, Xing D, Zhang Y. Anticancer activity and mechanism of Scutellaria barbata extract on human lung cancer cell line A549. Life Sci. 2004;75:2233–44. doi: 10.1016/j.lfs.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 88.Prasad RC, Herzog B, Boone B, Sims L, Waltner-Law M. An extract of Syzygium aromaticum represses genes encoding hepatic gluconeogenic enzymes. J Ethnopharmacol. 2005;96:295–301. doi: 10.1016/j.jep.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 89.Bagchi D, Sen CK, Ray SD, Das DK, Bagchi M, Preuss HG, et al. Molecular mechanisms of cardioprotection by a novel grape seed proanthocyanidin extract. Mutat Res. 2003;523–524:87–97. doi: 10.1016/s0027-5107(02)00324-x. [DOI] [PubMed] [Google Scholar]
- 90.Wong ML, O'Kirwan F, Hannestad JP, Irizarry KJ, Elashoff D, Licinio J. St John's wort and imipramine-induced gene expression profiles identify cellular functions relevant to antidepressant action and novel pharmacogenetic candidates for the phenotype of antidepressant treatment response. Mol Psychiatr. 2004;9:237–51. doi: 10.1038/sj.mp.4001470. [DOI] [PubMed] [Google Scholar]
- 91.Katz S, Harris R, Tien-Yau Lau J, Chau A. The use of gene expression analysis and proteomic databases in the development of a screening system to determine the value of natural medicinal products. Evid Based Complement Alternat Med. 2006;3:1–6. doi: 10.1093/ecam/nek001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang NS, Shyur LF, Chen CH, Wang SY, Tzeng CM. Medicinal herb extract and a single-compound drug confer similar complex pharmacogenomic activities in mcf-7 cells. J Biomed Sci. 2004;11:418–22. doi: 10.1159/000077111. [DOI] [PubMed] [Google Scholar]
- 93.Rushmore TH, Kong AN. Pharmacogenomics, regulation and signaling pathways of phase I and II drug metabolizing enzymes. Curr Drug Metab. 2002;3:481–90. doi: 10.2174/1389200023337171. [DOI] [PubMed] [Google Scholar]
- 94.Liljedahl U, Karlsson J, Melhus H, Kurland L, Lindersson M, Kahan T, et al. A microarray minisequencing system for pharmacogenetic profiling of antihypertensive drug response. Pharmacogenetics. 2003;13:7–17. doi: 10.1097/00008571-200301000-00003. [DOI] [PubMed] [Google Scholar]
- 95.Amin RP, Hamadeh HK, Bushel PR, Bennett L, Afshari CA, Paules RS. Genomic interrogation of mechanism(s) underlying cellular responses to toxicants. Toxicology. 2002;181:555–63. doi: 10.1016/s0300-483x(02)00481-x. [DOI] [PubMed] [Google Scholar]
- 96.Waring JF, Gum R, Morfitt D, Jolly RA, Ciurlionis R, Heindel M, et al. Identifying toxic mechanisms using DNA microarrays: evidence that an experimental inhibitor of cell adhesion molecule expression signals through the aryl hydrocarbon nuclear receptor. Toxicology. 2002;181:537–50. doi: 10.1016/s0300-483x(02)00477-8. [DOI] [PubMed] [Google Scholar]
- 97.Pennie WD. Custom cDNA microarrays; technologies and applications. Toxicology. 2002;181–182:551–4. doi: 10.1016/s0300-483x(02)00479-1. [DOI] [PubMed] [Google Scholar]
- 98.Ko SY, Lin SC, Chang KW, Liu CJ, Chang SS, Lu SY, et al. Modulation of KGF-1 gene expression in oral fibroblasts by ripe areca nut extract. J Oral Pathol Med. 2003;32:399–407. doi: 10.1034/j.1600-0714.2003.00133.x. [DOI] [PubMed] [Google Scholar]
- 99.Tsai WC, Tsai ST, Ko JY, Jin YT, Li C, Huang W, et al. The mRNA profile of genes in betel quid chewing oral cancer patients. Oral Oncol. 2004;40:418–26. doi: 10.1016/j.oraloncology.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 100.Fugh-Berman A. Herb–drug interactions. Lancet. 2000;355:134–8. doi: 10.1016/S0140-6736(99)06457-0. [DOI] [PubMed] [Google Scholar]
- 101.Coxeter PD, McLachlan AJ, Duke CC, Roufogalis BD. Herb–drug interactions: an evidence based approach. Curr Med Chem. 2004;11:1513–25. doi: 10.2174/0929867043365198. [DOI] [PubMed] [Google Scholar]
- 102.Bonham MJ, Galkin A, Montgomery B, Stahl WL, Agus D, Nelson PS. Effects of the herbal extract PC-SPES on microtubule dynamics and paclitaxel-mediated prostate tumor growth inhibition. J Natl Cancer Inst. 2002;94:1641–7. doi: 10.1093/jnci/94.21.1641. [DOI] [PubMed] [Google Scholar]
- 103.Bonham M, Arnold H, Montgomery B, Nelson PS. Molecular effects of the herbal compound PC-SPES: identification of activity pathways in prostate carcinoma. Cancer Res. 2002;62:3920–4. [PubMed] [Google Scholar]
- 104.Joshi K, Chavan P, Warude D, Patwardhan B. Molecular markers in herbal drug technology. Curr Sci. 2004;87:159–65. [Google Scholar]
- 105.Warude D, Chavan P, Joshi K, Patwardhan B. Isolation of DNA from fresh and dry samples having highly acidic tissues. Plant Mol Biol Rep. 2003;21:1–6. [Google Scholar]
- 106.Carles M, Lee T, Moganti S, Lenigk R, Tsim KW, Ip NY, et al. Chips and Qi: microcomponent-based analysis in traditional Chinese medicine. Fresenius J Anal Chem. 2002;371:190–4. doi: 10.1007/s002160100964. [DOI] [PubMed] [Google Scholar]
- 107.Trau D, Lee TM, Lao AI, Lenigk R, Hsing IM, Ip NY. Genotyping on a complementary metal oxide semiconductor silicon polymerase chain reaction chip with integrated DNA microarray. Anal Chem. 2002;74:3168–73. doi: 10.1021/ac020053u. [DOI] [PubMed] [Google Scholar]
- 108.Tsoi PY, Wu HS, Wong MS, Chen SL, Fong WF, Xiao, et al. Genotyping and species identification of Fritillaria by DNA chip technology. Acta Pharm Sin. 2003;4:185–190. [PubMed] [Google Scholar]
- 109.Zhang YB, Wang J, Wang ZT, But PP, Shaw PC. DNA microarray for identification of the herb of dendrobium species from Chinese medicinal formulations. Planta Med. 2003;69:1172–4. doi: 10.1055/s-2003-818015. [DOI] [PubMed] [Google Scholar]
- 110.Carles M, Cheung MK, Moganti S, Dong TT, Tsim KW, Ip NY, et al. A DNA microarray for the authentication of toxic traditional Chinese medicinal plants. Planta Med. 2005;71:580–4. doi: 10.1055/s-2005-864166. [DOI] [PubMed] [Google Scholar]
- 111.Patwardhan B. Ayugenomics: integration for customized medicine. Indian J Nat Products. 2003;19:16–23. [Google Scholar]
- 112.Kung PC, Cheng YC. Phytomics: a genomic-based approach to herbal compositions. WIPO Patent No. WO 00/24934, 4 May 2000. [Google Scholar]
- 113.Ventura C. CAM and cell fate targeting: molecular and energetic insights into cell growth and differentiation. Evid Based Complement Alternat Med. 2005;2:277–83. doi: 10.1093/ecam/neh100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Blower PE, Yang C, Fligner MA, Verducci JS, Yu L, Richman S, et al. Pharmacogenomic analysis: correlating molecular substructure classes with microarray gene expression data. Pharmacogenomics J. 2002;2:259–71. doi: 10.1038/sj.tpj.6500116. [DOI] [PubMed] [Google Scholar]
- 115.Tonisson N, Zernant J, Kurg A, Pavel H, Slavin G, Roomere H, et al. Evaluating the arrayed primer extension resequencing assay of TP53 tumor suppressor gene. Proc Natl Acad Sci USA. 2002;99:5503–8. doi: 10.1073/pnas.082100599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hacia JG, Collins FS. Mutational analysis using oligonucleotide microarrays. J Med Genet. 1999;36:730–6. doi: 10.1136/jmg.36.10.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lin B, Wang Z, Vora GJ, Thornton JA, Schnur JM, Thach DC, et al. Broad-spectrum respiratory tract pathogen identification using resequencing DNA microarrays. Genome Res. 2006;16:527–35. doi: 10.1101/gr.4337206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gerry NP, Witowski NE, Day J, Hammer RP, Barany G, Barany F. Universal DNA microarray method for multiplex detection of low abundance point mutations. J Mol Biol. 1999;292:251–62. doi: 10.1006/jmbi.1999.3063. [DOI] [PubMed] [Google Scholar]
- 119.Favis R, Day JP, Gerry NP, Phelan C, Narod S, Barany F. Universal DNA array detection of small insertions and deletions in BRCA1 and BRCA2. Nat Biotechnol. 2000;18:561–4. doi: 10.1038/75452. [DOI] [PubMed] [Google Scholar]
- 120.Syvanen AC. Toward genome-wide SNP genotyping. Nat Genet. 2005;37:5–10. doi: 10.1038/ng1558. [DOI] [PubMed] [Google Scholar]
- 121.King HC, Sinha AA. Gene expression profile analysis by DNA microarrays: promise and pitfalls. J Am Med Assoc. 2001;286:2280–8. doi: 10.1001/jama.286.18.2280. [DOI] [PubMed] [Google Scholar]
- 122.Kothapalli R, Yoder SJ, Mane S, Loughran TP. Microarray results: how accurate are they? BMC Bioinformatics. 2002;3:22. doi: 10.1186/1471-2105-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chuaqui RF, Bonner RF, Best CJM, Gillespie JW, Flaig MJ, Hewitt SM, et al. Post analysis follow-up and validation of microarray experiments. Nat Genet. 2002;32:509–13. doi: 10.1038/ng1034. [DOI] [PubMed] [Google Scholar]
- 124.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–71. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 125.Churchill GA. Fundamentals of experimental design for cDNA microarrays. Nat Genet. 2002;32:490–5. doi: 10.1038/ng1031. [DOI] [PubMed] [Google Scholar]
- 126.Fathallah-Shaykh H, He B, Zhao L-J, Badruddin A. A mathematical algorithm for discovering states of expression from direct genetic comparison by microarrays. Nucleic Acids Res. 2004;32:3807–14. doi: 10.1093/nar/gkh718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fathallah-Shaykh H. Microarrays: applications and pitfalls. Arch Neurol. 2005;62:1669–72. doi: 10.1001/archneur.62.11.1669. [DOI] [PubMed] [Google Scholar]