Abstract
We followed 68 cellular metabolites after carbon or nitrogen starvation of Escherichia coli and Saccharomyces cerevisiae, using a filter-culture methodology that allows exponential growth, nondisruptive nutrient removal, and fast quenching of metabolism. Dynamic concentration changes were measured by liquid chromatography–tandem mass spectrometry and viewed in clustered heat-map format. The major metabolic responses anticipated from metabolite-specific experiments in the literature were observed as well as a number of novel responses. When the data were analyzed by singular value decomposition, two dominant characteristic vectors were found, one corresponding to a generic starvation response and another to a nutrient-specific starvation response that is similar in both organisms. Together these captured a remarkable 72% of the metabolite concentration changes in the full data set. The responses described by the generic starvation response vector (42%) included, for example, depletion of most biosynthetic intermediates. The nutrient-specific vector (30%) included key responses such as increased phosphoenolpyruvate signaling glucose deprivation and increased α-ketoglutarate signaling ammonia deprivation. Metabolic similarity across organisms extends from the covalent reaction network of metabolism to include many elements of metabolome response to nutrient deprivation as well.
Keywords: Escherichia coli, metabolomics, nitrogen/carbon metabolism, Saccharomyces cerevisiae, starvation response
The pathways of cellular metabolism are close to identical across widely divergent organisms (1). The prokaryote Escherichia coli and the eukaryote Saccharomyces cerevisiae share essentially the same metabolic network (2) despite radically different compartmentation. The concentrations and fluxes of metabolites depend on the interactions among this conserved network structure, the cellular environment, and species-specific factors, such as the location, activities, and regulation of metabolic enzymes. Except for a few classic examples, e.g., central carbon metabolism in E. coli and yeast (3–5) and nitrogen assimilation in bacteria (6–8) the effect of environmental nutrient perturbations on the cellular metabolome have not been directly measured.
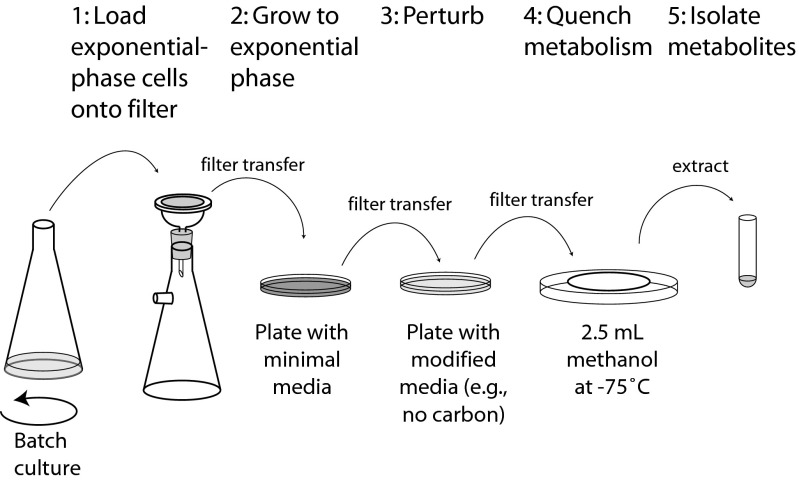
We explored exponentially growing E. coli and S. cerevisiae cultures suddenly deprived of their carbon or nitrogen sources. Metabolic composition can change in as little as a few seconds (9, 10). To get accurate snapshots of the metabolome, we devised a method of growing cells directly on filters to avoid time-consuming procedures (e.g., centrifugation or filtration) before quenching of biochemical activity (11). Transfer of a filter from a plate containing growth medium into cold organic solvent quickly quenches metabolism; transfer to a plate with a different medium composition allows a quick change of the nutrient environment (Fig. 1).
Fig. 1.
Schematic of the filter-culture approach.
For measurement of a number of metabolites simultaneously, we used a liquid chromatography–electrospray ionization–triple quadrupole mass spectrometry (LC–MS/MS) method tailored to known, water-soluble cellular metabolites (12). The method's strengths include the use of hydrophilic interaction chromatography (HILIC) to enable effective separation of polar analytes (13, 14), and the detection of metabolites in selected reaction-monitoring mode, which provides superior sensitivity and specificity as compared with simple MS analysis (15).
In addition to finding basic features of the response to carbon and nitrogen starvation that recapitulate literature based on traditional compound-by-compound methods, we observed global trends that are remarkably conserved between E. coli and S. cerevisiae.
Results and Discussion
Effect of Nutrient Removal on Cell Growth Rate and Atomic Composition.
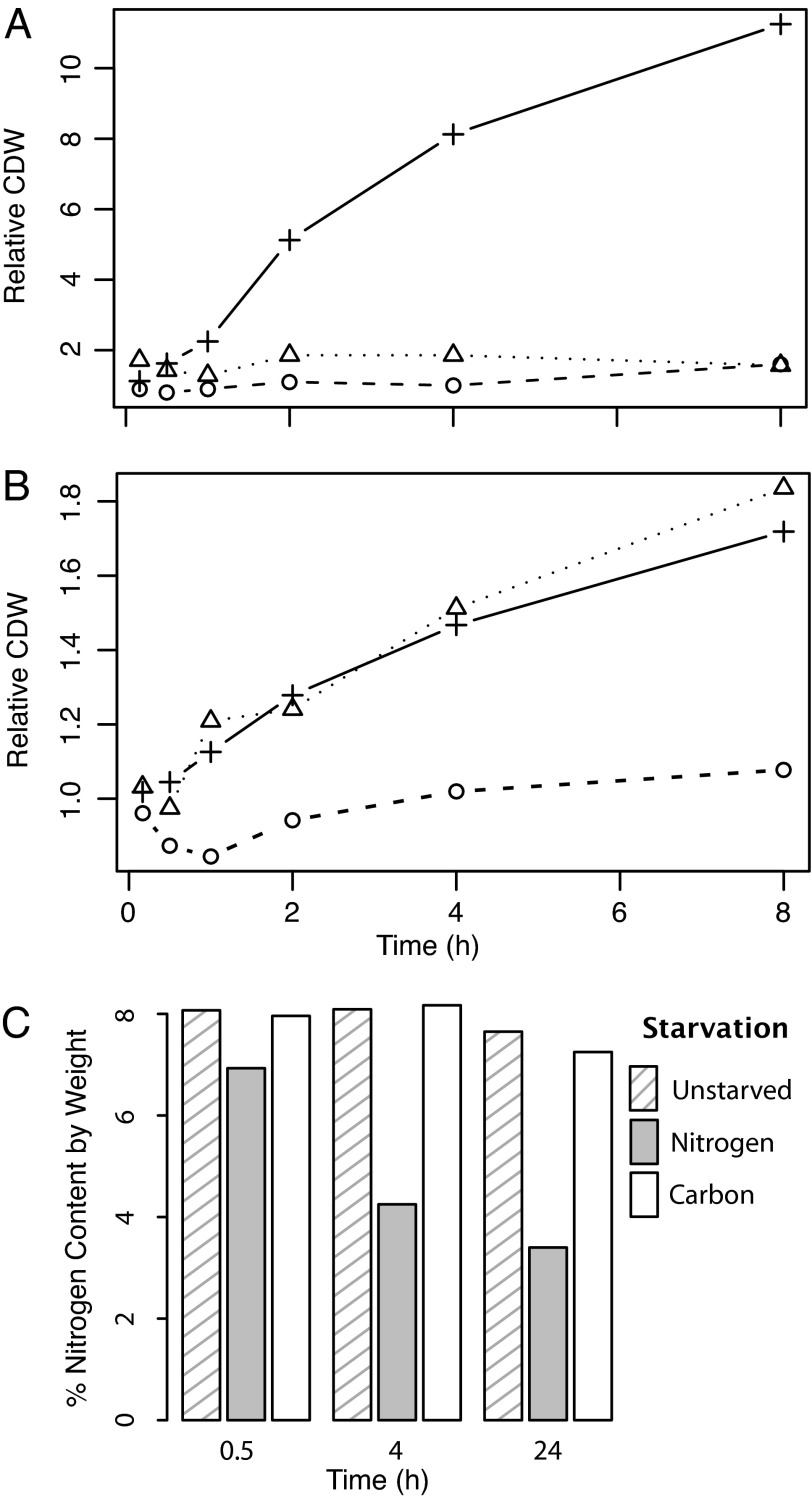
The filter-culture methodology effectively enables exponential cell growth; doubling times on the filters were 1.2 h (vs. 1.4 h in comparable liquid media) for E. coli (16) and 3.0 h (vs. 3.1 h) for S. cerevisiae. The transfer of E. coli to glucose- or ammonia-free media stopped their growth almost immediately (Fig. 2A). Transfer of S. cerevisiae to carbon-deficient media similarly blocked growth (Fig. 2B). In contrast, transfer of S. cerevisiae to nitrogen-deficient media did not immediately alter the growth rate (Fig. 2B), although growth of nitrogen-starved cells did stop before cells in complete minimal media (data not shown). The continued yeast growth in the absence of nitrogen also occurred in liquid culture, confirming that it is not an artifact of the filter-culture methodology (17).
Fig. 2.
Cell growth and elemental composition response to nutrient deprivation. (A and B) Cell dry weight (CDW) is plotted relative to the starting weight, for cultures that are grown on complete minimal media (+) vs. starved for carbon (o) or for nitrogen (Δ) in E. coli (A) and S. cerevisiae (B). (C) Nitrogen content of starved yeast.
An implication of the increase in yeast biomass after removal of the external nitrogen source is that the fraction of the biomass consisting of nitrogen decreases. Elemental analysis of nitrogen-starved yeast revealed this to be the case (Fig. 2C). None of the other starvation conditions resulted in substantial changes in cellular elemental composition [see supporting information (SI) Table 1]. Thus, a major metabolic difference between E. coli and S. cerevisiae is that yeast can replicate in the absence of environmental nitrogen by using internal nitrogen stores. Because yeast are not known to contain any specialized storage form of nitrogen, the nitrogen likely comes from biopolymers, presumably primarily protein (18).
Metabolome Changes Induced by Nutrient Removal.
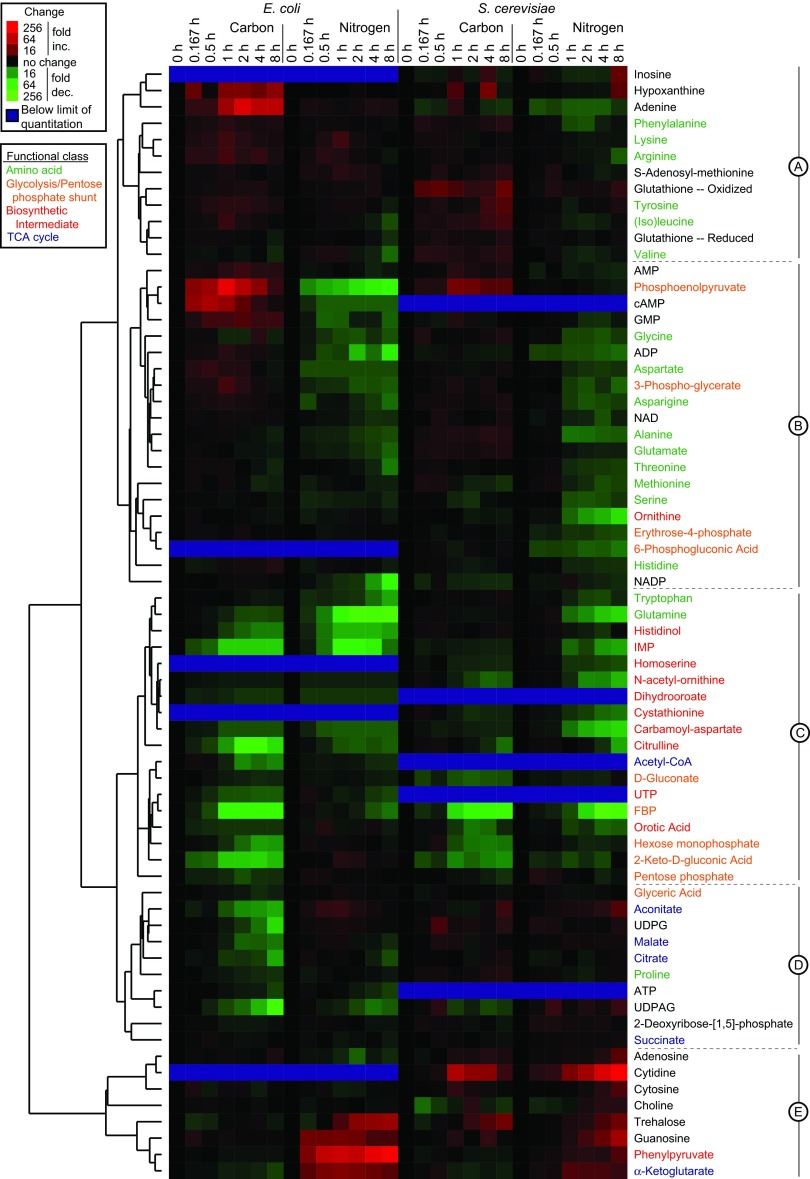
For metabolome sampling, filter-grown cells were quenched by dropping them into −75°C methanol. Extraction of metabolites was conducted by using a series of methanol/water steps (19, 20). LC–MS/MS analysis included selected reaction-monitoring scans for ≈170 metabolites; of these, 68 gave quantitative data in at least one organism (the others were below the limit of quantitation or too unstable to yield reliable information). The dynamics of these metabolites after glucose or ammonia removal are provided in full at http://genomics-pubs.princeton.edu/StarvationMetabolomics/Download.shtml and are shown in false-color format after hierarchical clustering of the metabolites in Fig. 3. A unique aspect of this hierarchical clustering was simultaneous use of data from E. coli and S. cerevisiae; the ability to cocluster these data to yield a well organized map provides a first line of evidence that metabolome response to nutrient deprivation is similar across both organisms.
Fig. 3.
Clustered heat map of metabolome dynamics during nutrient starvation. Columns represent different starvation time points. Fold-change is relative to exponentially growing cells (t = 0). Cluster A contains 12 metabolites that tend to rise in all forms of starvation, R2 = 0.485; cluster B contains 20 metabolites that rise in carbon but fall in nitrogen starvation, R2 = 0.575; cluster C contains 18 metabolites that fall in all forms of starvation, R2 = 0.677; cluster D contains 10 metabolites that rise in yeast but all in E. coli starvation, R2 = 0.507; and cluster E contains 8 metabolites that rise in nitrogen but fall in carbon starvation, R2 = 0.248.
A striking feature of the data in Fig. 3 is the dynamic range of the metabolite concentration changes (≈100-fold increased to ≈100-fold decreased). Concentration changes exceeded 4-fold under at least one starvation condition for 90% of the metabolites and exceeded 16-fold for 37% of metabolites. The magnitude and speed of these changes is generally greater in E. coli than in yeast. The metabolite concentration changes substantially exceeded the experiment-to-experiment variability: the median relative standard deviation of duplicate independent experiments was 26%. The observed effects were not due to ion suppression or other MS artifacts, because similar data were obtained at the 2-h time point by isotope-ratio-based measurements, which correlated substantially with the direct measurements reported here; R2 = 0. 704; SI Fig. 7.
Characteristic Metabolic Responses to Carbon Starvation.
Our methods are aimed at gaining a global view of metabolism. An important test of the validity of our approach is its ability to recapitulate literature findings on specific compounds. Among the best established of these is increased cAMP in E. coli upon glucose removal (21, 22). Our data capture this rise in cAMP, which peaked during the first hour after glucose removal and fell off thereafter. In yeast, where cAMP is not a starvation signal, levels were below the limit of quantitation at all time points.
Clustering directly adjacent to cAMP in Fig. 3, AMP and phosphoenolpyruvate (PEP) also increased during carbon starvation. The rise in AMP in both organisms presumably reflects energy limitation: AMP is in equilibrium with the ADP-to-ATP ratio because of the action of adenylate kinase, which buffers falling ATP by increasing AMP. Consistent with the observed rise in AMP is the falling level of ATP in E. coli. As expected because of the action of adenylate kinase and consistent with ref. 23, the fall in ATP occurs relatively late in carbon starvation. ATP levels are not provided for S. cerevisiae, because our methanol/water procedure did not effectively extract triphosphates from filter-grown yeast.
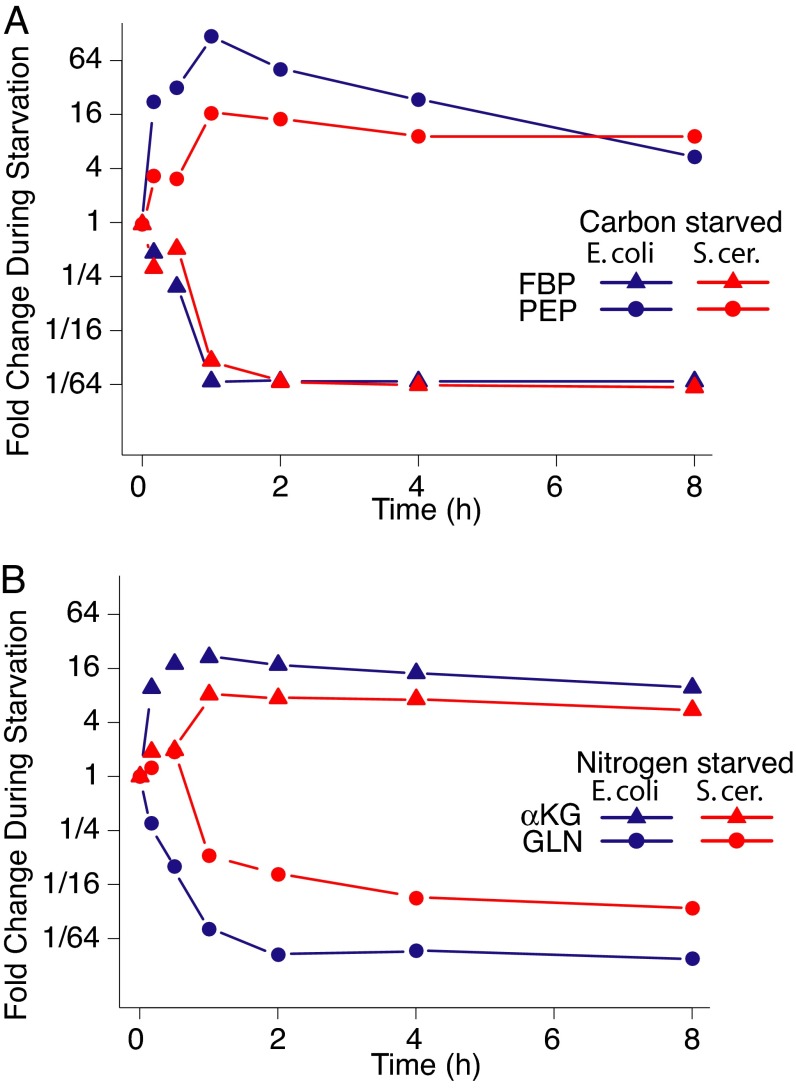
Given the almost identical role of AMP in bacteria and yeast, the similarity of the AMP response across these organisms is not surprising. Less intuitive is the similar rise in PEP during carbon starvation of both species (Figs. 3 and 4A). In E. coli, this rise may be primarily explained by the role of PEP as the phosphate donor for the phosphotransferase system (PTS) responsible for glucose import. When glucose is not present, this route of PEP consumption is cut off. The PTS system is not found in eukaryotes, however. Instead, the dramatic rise in PEP in yeast carbon starvation may arise from the effects of fructose-1,6-bisphosphate (FBP) as a positive regulator of pyruvate kinase, which is the major enzyme consuming PEP. Glucose removal resulted in marked decreases in FBP (Figs. 3 and 4A). These decreases presumably down-regulated the activity of pyruvate kinase, leading to PEP accumulation. Increased PEP in turn is anticipated to inhibit phosphofructokinase, reinforcing the blockade of glycolysis. As expected given the regulatory function of phosphofructokinase in both organisms, FBP levels changed much more dramatically during carbon starvation than did the levels of hexose phosphates (comprising the analytically identical glucose–phosphates and fructose–phosphates).
Fig. 4.
Key metabolic signals of carbon vs. nitrogen starvation. (A) FBP and PEP levels during carbon starvation. (B) α-Ketoglutarate and glutamine during nitrogen starvation.
Primary Metabolic Responses to Nitrogen Starvation.
Ammonia is assimilated in both enteric bacteria and yeast into glutamine and glutamate, which subsequently distribute nitrogen to other biomolecules (24). Previous studies in enteric bacteria (7, 25) provided evidence for markedly decreased levels of cellular glutamine during nitrogen starvation, and for comparatively less profound changes in cellular glutamate. Our results (Figs. 3 and 4B) confirm these findings in E. coli and extend them to S. cerevisiae, showing that glutamine is decreased by >40-fold in E. coli and by 13-fold in S. cerevisiae at 2 h into nitrogen starvation. In contrast, glutamate is decreased by only 2-fold in E. coli and hardly at all in yeast. The similarity of these responses involving glutamate and glutamine between E. coli and yeast is particularly interesting given that glutamine synthetase is regulated by covalent modification in enteric bacteria but not yeast, and that nitrogen flow into glutamate is largely via glutamine in E. coli but direct in S. cerevisiae.
The carboxylic acid skeleton of glutamine and glutamate, α-ketoglutarate, is also integral to nitrogen assimilation. Although previous technologies had been adequate to monitor glutamine and glutamate, monitoring α-ketoglutarate had been more challenging, largely because of the difficulty of differentiating excreted from intracellular α-ketoglutarate without separating cells from media and risking the alteration of the cellular α-ketoglutarate pools. Our filter-culture approach resolved this problem, enabling direct measurement of profoundly increased α-ketoglutarate during nitrogen starvation. Although α-ketoglutarate was detected in the substrate media of nitrogen-starved cells, indicating that the metabolite is indeed excreted (26), the measured α-ketoglutarate levels from the cellular samples were not strongly influenced by the extent of α-ketoglutarate accumulation in the substrate media, confirming that the measured α-ketoglutarate is indeed largely intracellular.
Responses of Specific Biosynthetic Pathways.
The data shown in Fig. 3 are rich in specific observations. We highlight two cases here, the first involving nucleotide synthesis and the second involving aromatic amino acid synthesis.
Nucleotide biosynthesis has been studied extensively, both biochemically and genetically; however, assays sensitive enough to quantitate biosynthetic intermediates from wild-type cells have only recently been developed. We found that the concentrations of all measured de novo nucleotide biosynthetic intermediates (carbamoyl aspartate, dihydroorotate, and orotate on the pyrimidine pathway and IMP on the purine pathway) dropped substantially and quickly upon starvation in both E. coli and S. cerevisiae (Fig. 3). These observations are consistent with recent evidence for decreased flux down these pathways in carbon-starved E. coli (16). The molecular basis for this decreased flux is not yet clear, but appears to exceed simple feedback inhibition by end product, because nucleotide levels decreased during nitrogen starvation of both organisms.
Synthesis of the aromatic amino acids phenylalanine, tyrosine, and tryptophan follow a common pathway up to the key intermediate chorismate, which contains no nitrogen. Chorismate then can acquire nitrogen from glutamine to follow the tryptophan pathway or, alternatively, can form the nitrogen-free precursor to phenylalanine and tyrosine, prephenate. Prephenate then forms phenylpyrvuate and hydroxyphenylpyruvate, which finally receive nitrogen from glutamate to form phenylalanine and tyrosine. The compounds on these pathways yielding quantitative data with the current analytical method were the amino acid end products and phenylpyruvate. During carbon starvation, none of them changed markedly in concentration in either organism. During nitrogen starvation, however, tryptophan dropped greatly in both organisms, but phenylalanine and tyrosine did not. This finding is consistent with tryptophan's receiving nitrogen from glutamine, which decreases profoundly during nitrogen starvation, whereas phenylalanine and tyrosine receive nitrogen from glutamate, which is much less sensitive to nitrogen starvation.
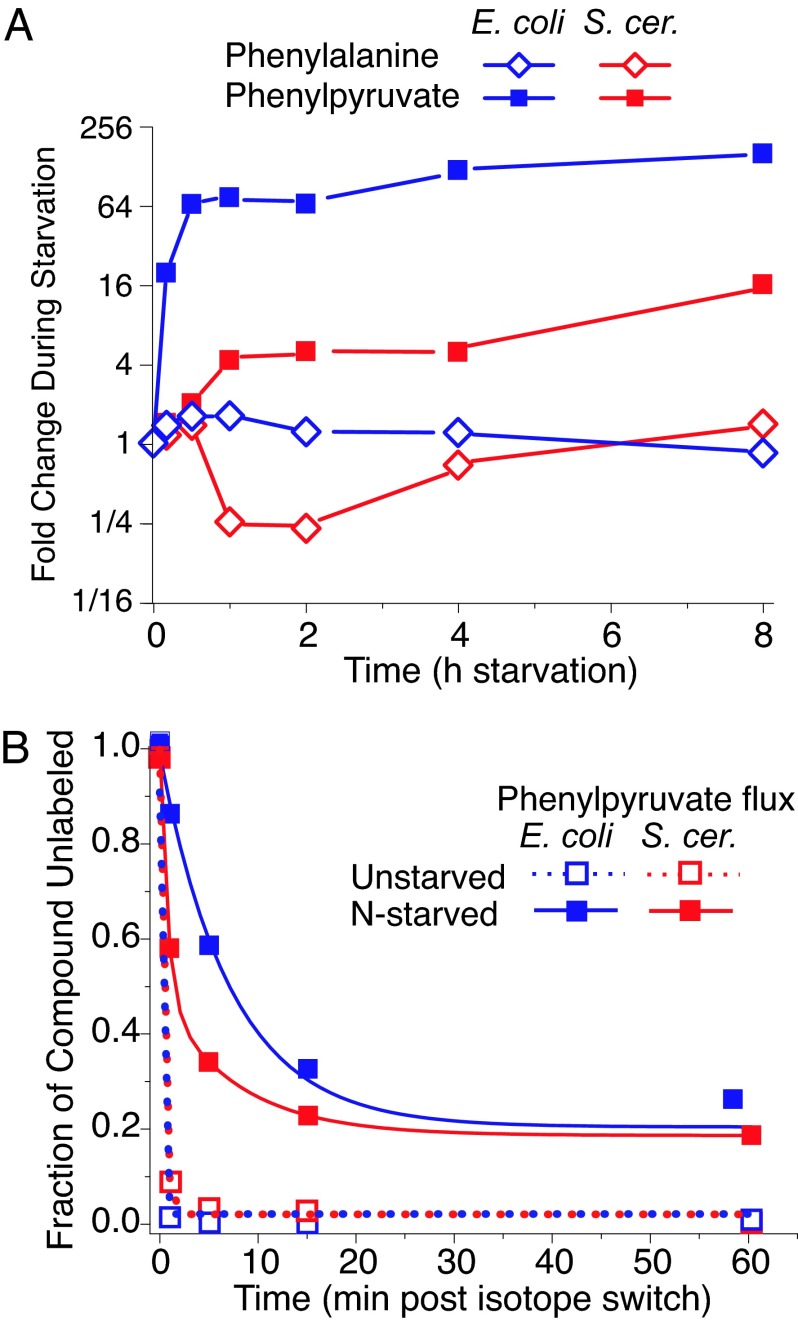
Although phenylalanine levels changed relatively little during nitrogen starvation, phenylpyruvate levels increased greatly in both organisms (Figs. 3 and 5A). To determine whether the origin of the phenylpyruvate was catabolic or anabolic, we measured the rate of incorporation of isotopic carbon into phenylpyruvate in unstarved and nitrogen-starved cultures (Fig. 5B). If aromatic amino acid biosynthesis were shut off during nitrogen starvation and the phenylpyruvate accumulation was due solely to increased protein catabolism and phenylalanine degradation, the incorporation of labeled carbon into phenylpyruvate should have ceased upon nitrogen starvation. Although the phenylpyruvate pool of nitrogen-starved cells became labeled somewhat less rapidly and completely than that of unstarved cells,¶ most of the phenylpyruvate was nevertheless labeled within 30 min, demonstrating that most of the accumulated phenylpyruvate arose from thwarted de novo biosynthesis.
Fig. 5.
During nitrogen starvation, phenylpyruvate accumulates because of thwarted de novo phenylalanine biosynthesis. (A) Phenylpyruvate and phenylalanine levels. (B) Kinetics of isotope labeling of phenylpyruvate. Plotted is the disappearance of unlabeled phenylpyruvate upon switching from unlabeled to uniformly [13C]glucose in unstarved cells vs. cells starved for nitrogen for 2 h before the isotope switch. The x axis represents minutes after isotope switching; the y axis represents the fraction of the observed phenylpyruvate signal attributable to uniformly 12C form.
Persistent phenylpyruvate biosynthesis during nitrogen starvation, although seemingly wasteful, has no obvious cost to microbes whose growth is limited by nitrogen availability. To the contrary, it may have an important function in yeast, where a byproduct of phenylpyruvate (phenylethanol) is a key intercellular quorum-sensing signal for nitrogen starvation (27).
Global Trends in the Metabolome Map During Nutrient Starvation.
One advantage of high throughput analytical technologies is the ability to quickly generate novel compound-specific discoveries, as illustrated by the example of phenylpyruvate. The large number of simultaneously measured compounds is also amenable to global analyses such as are commonly done on microarray and other genome-wide measurements (28). For example, hierarchical clustering identifies major groups of metabolites with common responses to starvation (Fig. 3).
Several functional categories of compounds show highly correlated behavior. Amino acids (P < 0.0001), biosynthetic intermediates (P < 0.0001), and tricarboxylic acid (TCA) cycle intermediates (P < 0.001) cluster much more closely than would be expected by chance. In general, the pattern of the concentration changes in these metabolites can be rationalized based on their biochemical roles:
Amino acids (clusters A and B in Fig. 3) tend to accumulate during carbon starvation, presumably as the result of starvation-induced protein catabolism (29). A subset of these compounds (cluster B) decrease during nitrogen starvation, consistent with their being deaminated to yield usable nitrogen.
Most biosynthetic intermediates (cluster C) decrease in concentration over all starvation conditions, consistent with cells turning off de novo biosynthesis as an early, strong, and consistent response to nutrient deprivation.
TCA cycle compounds (cluster D) are among the few metabolites to show opposing effects in E. coli vs. yeast. Both carbon and nitrogen limitation have been shown to enhance respiration relative to fermentation in yeast, consistent with the increased levels of TCA cycle components found here during yeast starvation (30).
Conserved Nature of the Metabolome Response.
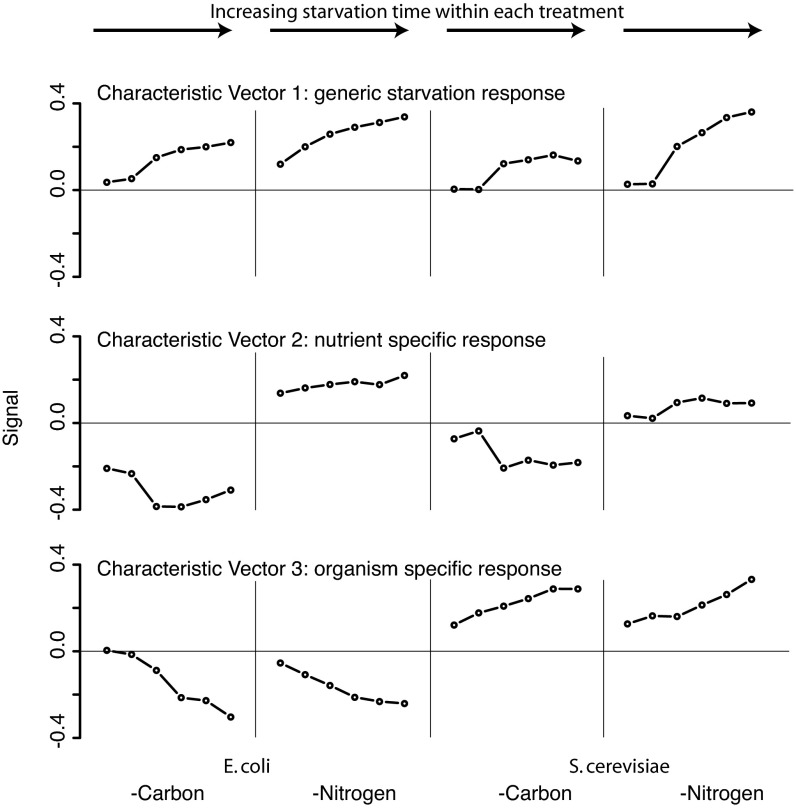
Singular value decomposition (SVD) is a process for extracting major signals from complex data sets (31). SVD factorization of the normalized metabolite concentration data resulted in three characteristic vectors, each able to account for >10% of the information in the overall data set (Fig. 6; also SI Fig. 8). The first (42% of the information) represents a generic starvation response that increases in a nearly monotonic fashion as the starvation time course proceeds, regardless of the nature of the starvation or the species in which it occurs. The second (30%) represents a nutrient-specific response and shows the opposing pattern for carbon vs. nitrogen starvation but a similar pattern across both organisms. The third (11%) represents an organism-specific response and shows the same pattern for carbon and nitrogen starvation but an opposite sign in E. coli vs. yeast. The largest contributions to these three most significant characteristic vectors are made by metabolites whose identities accord well with the responses to which they contribute (SI Table 2). For example, key contributors to the generic starvation response include intermediates in glucose and ammonia assimilation (FBP, glutamine) and in purine and pyrimidine biosynthesis (IMP, carbamoyl-aspartate), consistent with general down-regulation of nutrient use during starvation. For the nutrient-specific response, the strongest contributor is PEP, consistent with its rise during glucose but not ammonia deprivation; another substantial contributor is α-ketoglutarate, consistent with its rise during nitrogen but not carbon starvation. Together the three key characteristic vectors account for 83% of the metabolite concentration changes in the data shown in Fig. 3; the first two, which both represent evolutionarily conserved metabolome responses, account for 72%.
Fig. 6.
Singular value decomposition of the metabolome starvation matrix shown in Fig. 3. Each characteristic vector includes entries for each starvation time point (t = 10 and 30 min and 1, 2, 4, and 8 h) in each of the four tested starvation conditions (two nutrients times two organisms). The fraction of total information in the metabolome starvation matrix captured by the characteristic vectors is 42%, 30%, and 11% for vectors 1, 2, and 3 respectively.
The ability to account for such a large fraction of the metabolome response to nutrient starvation with two characteristic vectors, which show such similar patterns across the two studied organisms, is quite surprising, especially given the divergent gross behavior of E. coli vs. yeast during nitrogen starvation (quiescence vs. growth), the existence of some nontrivial differences in the covalent metabolic map of these organisms (e.g., use of PEP vs. ATP to drive glucose import), and the fact that E. coli and S. cerevisiae lie in distinct kingdoms of life associated with markedly different subcellular organization. One interpretation of the conserved nature of the metabolome response across these organisms is that there are very few schemes for regulating metabolism that meet all of the requirements of microbes, given the variable environmental conditions they face. Another is that the evolution of metabolome response to the environment has become stuck in one of many possible local optima but that escaping to a different local optimum is difficult given the upstream position of metabolism in the complex sequence of events (involving metabolism, transcription, cell growth decisions, etc.) required to react appropriately to changing environmental conditions. Dynamic metabolomic data from other divergent microbes, e.g., photosynthetic alga (32) or Gram-positive bacteria (6), or from E. coli or S. cerevisiae mutants, may shed light on the generality and validity of these hypotheses.
Materials and Methods
Strains and Media.
E. coli K-12 strain NCM 3722 and S. cerevisiae strain DBY11069, a Mata derivative of S288c, were cultured in minimal complete media: for E. coli, minimal salts media (33) with 10 mM NH4Cl and 0.4% glucose at 37°C; for S. cerevisiae, yeast nitrogen base without amino acids (Difco, Detroit, MI) supplemented with 2% glucose at 30°C. Plates contained, in addition to the appropriate media, 1.5% ultrapure agarose washed three times with cartridge-filtered water to remove trace organic contaminants.
Culture Conditions and Metabolite Extraction.
Filter cultures were prepared by passing cells growing exponentially in liquid minimal media culture through nitrocellulose membranes. The filters were then placed on top of agarose loaded with identical minimal media. To induce nutrient starvation, filters were transferred to agarose loaded with media lacking glucose or ammonia. At various time points thereafter, metabolism was quenched by dropping the filters directly into −75°C methanol, metabolites were extracted by using methanol/water (20) as described in ref. 16, the 10 isotope-labeled internal standards listed in ref. 12 were added, and the resulting extract was stored at 4°C until analyzed by LC-MS/MS. To prevent substantial sample contamination with excreted material, filters collected at starvation times >30 min were transferred to a fresh plate 30 min before sample harvesting. One filter was used for each time point, and replicates were prepared to allow duplicate metabolite extractions and measurements of other culture parameters. Further details on cell manipulations and metabolite extraction, as well as measurement of cell dry weight and elemental composition, are provided as SI Supporting Methods. For media composition details, see SI Table 3.
Metabolome Quantitation.
LC-MS/MS analyses were performed on a Quantum Ultra triple quadrupole MS (Thermo Scientific, Waltham, MA). LC separation used a 250 × 2-mm aminopropyl column (Luna; Phenomenex, Torrance, CA) at pH 9.4 in hydrophilic interaction chromatography mode (12). All samples were analyzed within 16 h of their preparation. Because internal standard signals did not vary substantially or systematically between samples within a given starvation time course, the reported metabolite concentration changes are between-sample ratios of the peak heights of the selected reaction-monitoring chromatograms without correction for the internal standard response.
Phenylpyruvate Flux Measurement.
Cells were transferred to plates with unlabeled [12C]glucose replaced by uniformly [13C]glucose (>99%; Cambridge Isotope, Andover, MA). At various time points thereafter, cells were harvested and extracted. The eight forms of labeled phenylpyruvate that can be produced from mixtures of [12C]- and [13C]glucose without extensive carbon shuffling were analyzed (see SI Supporting Methods and SI Table 4 for details). Reported values are (signal[unlabeled phenylpyruvate]) / (sum of signals[all forms of phenylpyruvate]) and are fit to y = a + (1 − a) e−kt (16).
Hierarchical Clustering and Singular Value Decomposition Analysis.
Metabolite-concentration changes induced by starvation (expressed as log2 ratios) were clustered by metabolite (28) by using the Princeton Microarray Database (http://puma.princeton.edu). The statistical significance of clustering of selected metabolite functional categories (amino acids, biosynthetic intermediates, TCA cycle compounds) was evaluated in two ways: χ2 test of functional categories times metabolite clusters and calculation of the average pair-wise Pearson correlation within each functional class (which was compared with a bootstrap-derived null distribution). Both gave similar results.
The matrix of log-transformed relative metabolite concentration changes was mathematically decomposed by SVD (31) by using the “Eigensystem” function of Mathematica (Wolfram Research, Champaign, IL). Compounds that were missing data for one organism were omitted from the matrix before SVD.
Supplementary Material
Acknowledgments
We thank Ned Wingreen, Bernhard Palsson, and Sydney Kustu for useful feedback on this work. This work was supported by the National Institutes of Health (NIH) Center for Quantitative Biology at Princeton University (P50 GM071508), NIH Grant R01 GM046406, the Beckman Foundation, and the National Science Foundation Dynamic, Data-Driven Application System–Small Multidisciplinary Research Projects (DDDAS-SMRP) program.
Abbreviations
- FBP
fructose-1,6-bisphosphate
- PEP
phosphoenolpyruvate
- SVD
singular value decomposition
- TCA
tricarboxylic acid.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609508103/DC1.
For fixed pathway flux, a larger pool results in slower labeling (16); accordingly, the decreased rate of labeling in the nitrogen-starved cells can be attributed largely to the larger pool size.
References
- 1.Caspi R, Foerster H, Fulcher CA, Hopkinson R, Ingraham J, Kaipa P, Krummenacker M, Paley S, Pick J, Rhee SY, et al. Nucleic Acids Res. 2006;34:D511–D516. doi: 10.1093/nar/gkj128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forster J, Famili I, Fu P, Palsson BO, Nielsen J. Genome Res. 2003;13:244–253. doi: 10.1101/gr.234503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lowry OH, Carter J, Ward JB, Glaser L. J Biol Chem. 1971;246:6511–6521. [PubMed] [Google Scholar]
- 4.Betz A, Chance B. Arch Biochem Biophys. 1965;109:585–594. doi: 10.1016/0003-9861(65)90404-2. [DOI] [PubMed] [Google Scholar]
- 5.Mashego MR, Wu L, Van Dam JC, Ras C, Vinke JL, Van Winden WA, Van Gulik WM, Heijnen JJ. Biotechnol Bioeng. 2004;85:620–628. doi: 10.1002/bit.10907. [DOI] [PubMed] [Google Scholar]
- 6.Hu P, Leighton T, Ishkhanova G, Kustu S. J Bacteriol. 1999;181:5042–5050. doi: 10.1128/jb.181.16.5042-5050.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ikeda TP, Shauger AE, Kustu S. J Mol Biol. 1996;259:589–607. doi: 10.1006/jmbi.1996.0342. [DOI] [PubMed] [Google Scholar]
- 8.Tempest DW, Meers JL, Brown CM. Biochem J. 1970;117:405–407. doi: 10.1042/bj1170405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oldiges M, Takors R. Adv Biochem Eng Biotechnol. 2005;92:173–196. doi: 10.1007/b98913. [DOI] [PubMed] [Google Scholar]
- 10.Visser D, van Zuylen GA, van Dam JC, Oudshoorn A, Eman MR, Ras C, van Gulik WM, Frank J, van Dedem GW, Heijnen JJ. Biotechnol Bioeng. 2002;79:674–681. doi: 10.1002/bit.10328. [DOI] [PubMed] [Google Scholar]
- 11.Saez MJ, Lagunas R. Mol Cell Biochem. 1976;13:73–78. doi: 10.1007/BF01837056. [DOI] [PubMed] [Google Scholar]
- 12.Bajad SU, Lu W, Kimball EH, Yuan J, Peterson C, Rabinowitz JD. J Chromatogr A. 2006;1125:76–88. doi: 10.1016/j.chroma.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 13.Olsen BA. J Chromatogr A. 2001;913:113–122. doi: 10.1016/s0021-9673(00)01063-3. [DOI] [PubMed] [Google Scholar]
- 14.Tolstikov VV, Fiehn O. Anal Biochem. 2002;301:298–307. doi: 10.1006/abio.2001.5513. [DOI] [PubMed] [Google Scholar]
- 15.Hopfgartner G, Varesio E, Tschappat V, Grivet C, Bourgogne E, Leuthold LA. J Mass Spectrom. 2004;39:845–855. doi: 10.1002/jms.659. [DOI] [PubMed] [Google Scholar]
- 16.Yuan J, Fowler WU, Kimball E, Lu W, Rabinowitz JD. Nat Chem Biol. 2006;2:529–530. doi: 10.1038/nchembio816. [DOI] [PubMed] [Google Scholar]
- 17.Lagunas R, Ruiz E. J Gen Microbiol. 1988;134:2507–2511. doi: 10.1099/00221287-134-9-2507. [DOI] [PubMed] [Google Scholar]
- 18.Jorgensen H, Olsson L, Ronnow B, Palmqvist EA. Appl Microbiol Biotechnol. 2002;59:310–317. doi: 10.1007/s00253-002-1017-5. [DOI] [PubMed] [Google Scholar]
- 19.Kimball E, Rabinowitz JD. Anal Biochem. 2006;358:273–280. doi: 10.1016/j.ab.2006.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villas-Boas SG, Hojer-Pedersen J, Akesson M, Smedsgaard J, Nielsen J. Yeast. 2005;22:1155–1169. doi: 10.1002/yea.1308. [DOI] [PubMed] [Google Scholar]
- 21.Gstrein-Reider E, Schweiger M. EMBO J. 1982;1:333–337. doi: 10.1002/j.1460-2075.1982.tb01170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Notley-McRobb L, Death A, Ferenci T. Microbiology. 1997;143(Pt 6):1909–1918. doi: 10.1099/00221287-143-6-1909. [DOI] [PubMed] [Google Scholar]
- 23.Murray HD, Schneider DA, Gourse RL. Mol Cell. 2003;12:125–134. doi: 10.1016/s1097-2765(03)00266-1. [DOI] [PubMed] [Google Scholar]
- 24.Reitzer L. Annu Rev Microbiol. 2003;57:155–176. doi: 10.1146/annurev.micro.57.030502.090820. [DOI] [PubMed] [Google Scholar]
- 25.Schmitz RA. Curr Microbiol. 2000;41:357–362. doi: 10.1007/s002840010149. [DOI] [PubMed] [Google Scholar]
- 26.Neijssel OM, Tempest DW. Arch Microbiol. 1975;106:251–258. doi: 10.1007/BF00446531. [DOI] [PubMed] [Google Scholar]
- 27.Chen H, Fink GR. Genes Dev. 2006;20:1150–1161. doi: 10.1101/gad.1411806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eisen MB, Spellman PT, Brown PO, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuroda A, Nomura K, Ohtomo R, Kato J, Ikeda T, Takiguchi N, Ohtake H, Kornberg A. Science. 2001;293:705–708. doi: 10.1126/science.1061315. [DOI] [PubMed] [Google Scholar]
- 30.Lagunas R, Dominguez C, Busturia A, Saez MJ. J Bacteriol. 1982;152:19–25. doi: 10.1128/jb.152.1.19-25.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alter O, Brown PO, Botstein D. Proc Natl Acad Sci USA. 2000;97:10101–10106. doi: 10.1073/pnas.97.18.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bolling C, Fiehn O. Plant Physiol. 2005;139:1995–2005. doi: 10.1104/pp.105.071589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gutnick D, Calvo JM, Klopotowski T, Ames BN. J Bacteriol. 1969;100:215–219. doi: 10.1128/jb.100.1.215-219.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.