Abstract
Polyalanine expansions in the nuclear RNA-binding protein PABP2 induce misfolding and aggregation of the protein into insoluble inclusions in muscle tissues and cell nuclei, leading to the disease oculopharyngeal muscular dystrophy (OPMD). We have explored the effect of solvent conditions and alanine repeat number on the propensity of fibril formation in this protein deposition disease. Three peptides mimicking the N-terminal polyalanine segment of PABP2, having the generic sequence Ac-Lys-Met-(Ala)n-Gly-Tyr with n = 7, 11, and 17 (referred to as 7-ala, 11-ala, and 17-ala, respectively), were synthesized and their conformational properties studied as a function of pH. In strongly alkaline medium (pH >10), the two longer peptides (11-ala and 17-ala, but not 7-ala) showed remarkable enhancement of β-sheet content and formed fibrils after incubation for 1–2 weeks at room temperature. Fluorescence studies suggested that tyrosyl radicals produced at high pH cross-linked to form dityrosine, which provided added stabilization for fibril growth. The kinetic progress curves for fibril formation, obtained by ThT fluorescence assay, showed exponential increase with time after an initial quiescent period (lag time) and an eventual saturation phase, all of which are indicative of a nucleation-controlled polymerization mechanism for fibrillation. Hierarchical self-assembly of the peptides led to the formation of striking fractal-shaped growth patterns on substrates, raising the possibility of designing novel materials using these peptides.
INTRODUCTION
Expansions of polyalanine stretches in proteins have been linked to several human diseases caused by the dysfunctional mutant proteins (1). Of these, oculopharyngeal muscular dystrophy is the only one that can be classified as a protein deposition disease, characterized by nuclear protein aggregation and filamentous nuclear inclusions in muscle fibers of patients (2,3). A main constituent of the deposits is the nuclear poly (A) binding protein 2 (PABP2), which stimulates polyadenylation and controls the length of poly(A) tails (4). The amino acid sequence of PABP2 has at the N-terminal a natural 12-Ala stretch that is interrupted after the 10th Ala by a single Gly residue. Trinucleotide repeat expansions in the coding sequence of the PABP2 gene result in extensions of the wild-type 10-Ala sequence preceding the Gly residue to a maximum of 17 Ala residues (5). Individuals carrying the extensions develop the disease, characterized by swallowing difficulties, eyelid drooping, and limb weakness.
Although the molecular mechanism by which Ala extensions in PABP2 lead to OPMD remains to be fully understood, biopsy material from OPMD patients exhibits the pathological hallmark of mutant PABP2 deposits in fibril form that is common to inherited diseases ascribed to misfolding and aggregation of proteins (6). Fibril formation is generally thought to be the product of conformational changes leading to β-sheet structures of the constituent proteins (7,8). We had earlier shown that increasing the length of polyalanine tracts from 7 to 11 in synthetic peptides mimicking the N-terminal segment of PABP2 produces a change of conformation from α-helix to β-sheet while increasing their apoptotic potential significantly (9). Although electron microscopic study revealed that the longer peptide formed amorphous aggregates (30–50 μm) on ageing for several days, no fibrils were detected after an extended period of incubation.
In this work we describe our efforts to simulate conditions appropriate for fibril formation by polyalanine peptides and thereby learn about the influence of the solvent environment on their conformational transitions. Three peptides having the generic sequence Ac-Lys-Met-(Ala)n-Gly-Tyr with n = 7, 11, and 17 (henceforth referred to as 7-ala, 11-ala, and 17-ala, respectively) were synthesized and their conformational properties studied as a function of pH. A remarkable enhancement of the aggregation tendency of 11-ala and 17-ala was observed when the peptides were incubated at alkaline pH (10 and above). Circular dichroic (CD) spectra showed the peptides to have substantially increased β-sheet content over that at neutral pH. Electron micrographs of peptide solutions incubated for 2–4 weeks at room temperature showed formation of well-defined fibrils. Drying the aged solutions on solid substrates produced fractal-like spatial growth patterns of rod-like peptide fibrils. The Hausdorff dimension of the patterns increased from 1.65 to 1.8 as the length of polyalanine stretch and/or period of incubation was made longer, indicating a transition in the physical nature of the assembly process.
MATERIALS AND METHODS
Synthesis of oligopeptides
Two polyalanine peptides (containing 7 and 11 successive alanine residues) mimicking the N-terminal sequence of PABP2 were synthesized with the following modifications: a lysine was added at the N-terminus to make the peptides soluble, and a tyrosine was placed at the C-terminus to enable peptide concentrations to be determined spectrophotometrically. Synthesis was performed on a LKB 4175 peptide synthesizer using the Fmoc polyamide active ester chemistry and solid phase peptide synthesis technique. They were cleaved at room temperature with a solvent mixture containing 94% trifluoroacetic acid and 2% each of anisole, ethanedithiol, and phenol, precipitated by addition of diethyl ether, purified on a reversed phase C18 high-performance liquid chromatography column and stored at −20°C in lyophilized powdered form. The purity of the peptides was confirmed by mass spectroscopic analysis on a matrix-assisted laser desorption ionization-time of flight SPEC 2E instrument (Micromass, Manchester, UK). Three such peptides, containing 7 (7-ala), 11 (11-ala), and 17 (17-ala) successive alanines, were also purchased from GenoMechanix (Gainesville, FL). Studies with peptides from the two different sources yielded identical results.
Preparation of buffers and peptide samples
For pH-dependent studies, a 50 mM aqueous solution of glycine was adjusted with NaOH to make buffers of pH 10 and above and with HCl to make one of pH 3. MOPS (3[N-morpholino] propanesulphonic acid) buffer was used between pH 7 and pH 9. Lyophilized peptides were dissolved in buffers of different pH to make stock solutions. Concentrations of peptides in these solutions were determined from tyrosine absorbance, measured on a Spectronic UNICAM UV 500 spectrophotometer and using a molar extinction coefficient of 1280 M−1cm−1 at 280 nm (10). Samples for different studies were prepared by diluting the stock solutions with corresponding buffers.
Thioflavin T binding assay and detection of dityrosine
Fluorescence emission spectra of ThT, excited at 450 nm, were recorded between 460 and 600 nm on a Hitachi (Tokyo, Japan) F-4010 spectrofluorometer using excitation and emission bandwidths of 5 nm. The kinetics of fibril formation were followed by adding 10 μl aliquots from solutions of 11-ala and 17-ala incubated in glycine-NaOH buffer (pH 11) to 1 ml of a 10 μM ThT solution at specified time points, mixing thoroughly and recording its fluorescence intensity at 490 nm (excitation 450 nm) immediately afterward (11,12). Fluorescence spectra of peptide samples incubated at high pH were collected both in excitation (monitoring emission at 410 nm) and in emission (excitation at 320 nm) to verify the formation of dityrosine.
Circular dichroism spectroscopy
CD spectra between 190–250 nm were recorded on a JASCO J-720 spectropolarimeter using cylindrical quartz cuvettes (Hellma, Jena, Germany) of path length 1 or 2 mm. Each spectrum represents the average of five successive scans performed at a scan speed of 20 nm/min and a bandwidth of 1 nm. Appropriate baseline subtraction and noise reduction analysis were performed. The mean residue molar ellipticity, [θ], was calculated by using the relation
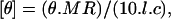 |
where θ is the measured ellipticity in millidegrees, MR is the mean residue mass (molecular weight of the peptide divided by the number of amino acid residues), l is the optical path length in cm, and c is the protein concentration in mg/ml.
Transmission electron microscopy
Solutions of 11-ala (at typical concentrations of ∼100 μM or ∼0.2 mg/ml) in glycine-NaOH buffer, pH 11, were incubated at room temperature for 20 days.
Drops of this solution were spread on carbon-coated copper grids, stained with 2% (w/v) uranyl acetate in water, air-dried, and visualized in a Hitachi H-600 transmission electron microsocpy operating at 75 kV.
Generation of fractal patterns and measurement of fractal dimension
Peptide solutions were incubated at pH 11 at room temperature, and 20-μl aliquots of these solutions were spread on a glass slide and allowed to air dry. The dried samples formed fractal-like growth patterns that could be observed under an optical microscope (mag. ∼200×). These patterns were also observed in a fluorescence microscope by adding the aggregation-specific dye Congo red. Kinetic studies of nucleation for fractal-like pattern formation were carried out by generating patterns from peptide solutions incubated for various lengths of time.
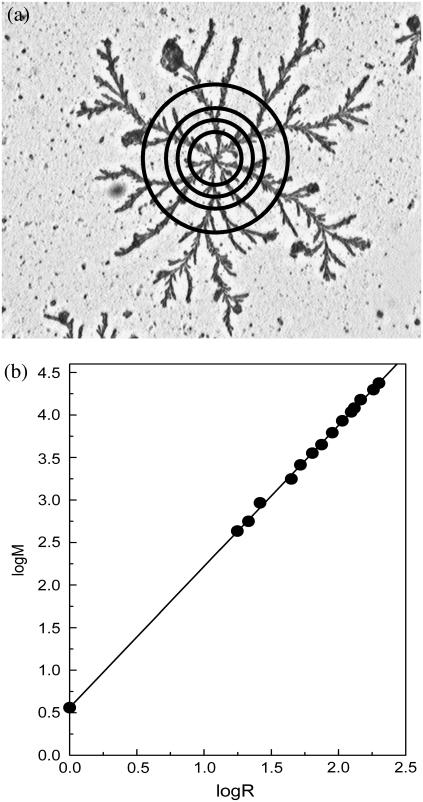
To measure the fractal dimension of a pattern, a number of concentric circles of increasing radius (R) were drawn on its scanned image, with the center of the circles coinciding with the center of the pattern. The mass (M) of the pattern enclosed by a circle of radius R was taken to be proportional to its total area (sum total of the area of the dark regions where the peptide deposits are located, see Fig. 8 a) lying within the circle. Using the formula M ∼ Rdf, the Hausdorff dimension (df) was calculated as the slope of best-fit straight line to the log(M) versus log(R) plot, as illustrated in Fig. 8 b (13).
FIGURE 8.
Dimensional analysis of fractal pattern obtained with an 11-ala sample. (a) The mass (M) of polymers deposited within a circle of given radius (R) was taken to be proportional to the area occupied by the darkened regions within that circle. This area was measured using the graphics software Image-Pro Plus supplied with an Olympus BX60 microscope (Tokyo, Japan). (b) A log-log graph of M versus R. The slope of the best-fit straight line on this graph yielded the fractal dimension df, which in this case is 1.65 ± 0.02.
Scanning electron microscopy
Scanning electron microscopy was performed in a Quanta 200 FEG machine manufactured by FEI (Eindhoven, The Netherlands) operating at 50–100 kV under 0.43–0.48 torr pressure, producing a magnification of 50–5000×. Patterns formed by dried peptide solutions on glass coverslips were visualized by placing the latter directly in the sample chamber.
RESULTS
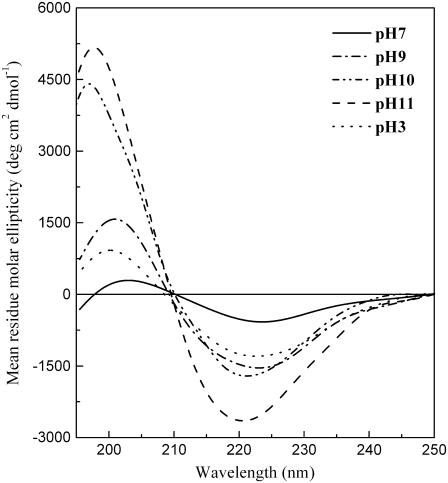
Fig. 1 shows the CD spectra of 11-ala in buffers of varying pH (from 3 to 11). Even at neutral pH, the peptide showed a weak propensity for formation of β-structures, mostly β-turns. As the solvent became increasingly alkaline, i.e., with increasing pH above 7, the ellipticity at the positive peak below 200 nm grew drastically, whereas the peak position shifted continuously to lower wavelengths (by 4.5 nm from pH 7 to pH 11). The evolution of these two features indicated substantial enhancement of β-sheet content over that at neutral pH. Similar changes were also observed in the CD spectra at acidic pH, viz. at pH 3, but the effect was nowhere near as pronounced. These results underscore the importance of solution pH in inducing β-sheet structure in polyalanine peptides, as mentioned briefly in an earlier report (14). pH-dependent structural (α-helix → β-sheet or random coil → β-sheet), as well as morphological, changes have also been observed in amyloid-forming proteins and peptides such as the Aβ, Vkappa-III Bence Jones protein, transthyretin and α-synuclein, although they all occurred at acidic pH (15–18). Such helix → sheet or coil → sheet transitions are thought to be the early events in the production of toxic protein aggregates.
FIGURE 1.
pH-dependent CD spectra of 11-ala.
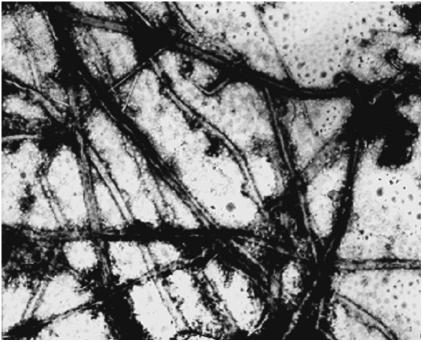
Further investigation of structural properties of these polyalanine peptides using electron microscopy revealed remarkable changes occurring in them over long periods of time. Solutions of 11-ala and 17-ala incubated at pH 11 for a few weeks showed formation of fibrillar structures in them. For example, uniform fibrils of width ∼200 nm are clearly visible in a transmission electron micrograph (Fig. 2) of an 11-ala solution incubated for 20 days at room temperature. Longer incubation led to formation of insoluble precipitates, presumably through large-scale aggregation of fibrils. However, no fibril formation was detected even after prolonged incubation at acidic pH (pH 3).
FIGURE 2.
Electron micrograph (magnification = 10,000) of the peptide 11-ala after incubation for 20 days at pH 11 at room temperature. Average width of fibrils ∼200 nm.
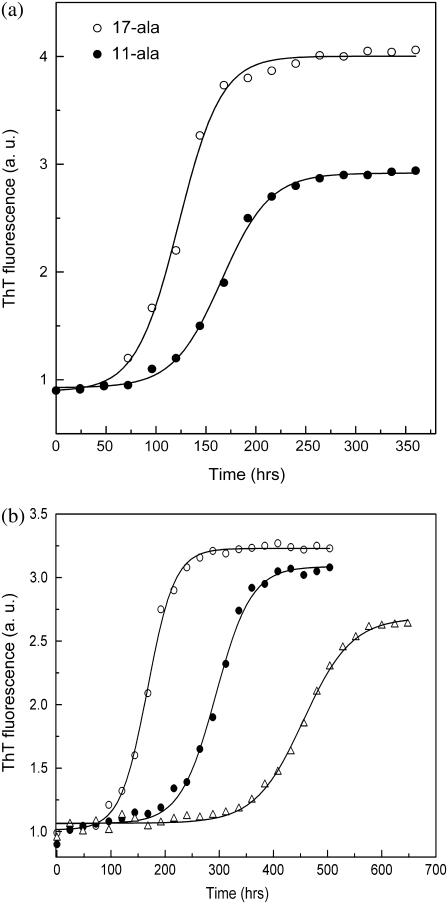
Thioflavin T (ThT) is a histological dye, which interacts with fibrils relatively specifically and is widely used for their detection (11,12). The binding of ThT to the fibrils results in fluorescence emission with a peak at ∼495 nm upon excitation at 450 nm, whereas unbound ThT does not fluoresce at this wavelength. The kinetics of fibril formation by the polyalanine peptides was studied by measuring the increase of ThT fluorescence at 490 nm upon addition of aliquots periodically withdrawn from peptide solutions incubated for varying lengths of time. Fig. 3 a shows the progress curves for fibril formation thus obtained for 11-ala and 17-ala (each at an initial concentration of 100 μM). Both samples show an initial quiescent period of several (2–4) days when no fibrils are detected, indicating existence of a lag time for fibril formation. Thereafter, the concentration of fibrils in solution increases sharply and eventually attains a saturation value. The final concentration of aggregated peptides, as given by the intensity of ThT fluorescence in the saturation phase, is higher for 17-ala than for 11-ala. The sigmoidal shapes of these curves point to a nucleation-dependent elongation process being responsible for fibril formation (19–21). The lag times are 1 day (25 h) and 3 days (75 h), respectively, for 17-ala and 11-ala, whereas the midpoints of their ThT fluorescence transition curves occur at 5 days (120 h) and 7 days (170 h). No increase of ThT fluorescence was observed for 7-ala even after incubation for 6 months.
FIGURE 3.
Polymerization progress curves, as measured by thioflavine T fluorescence intensity, at 25°C for (a) 11-ala (solid circles) and 17-ala (open circles) at 100 μM initial concentration and (b) 11-ala at three different initial concentrations: 24 μM (open triangles), 120 μM (solid circles), and 240 μM (open circles).
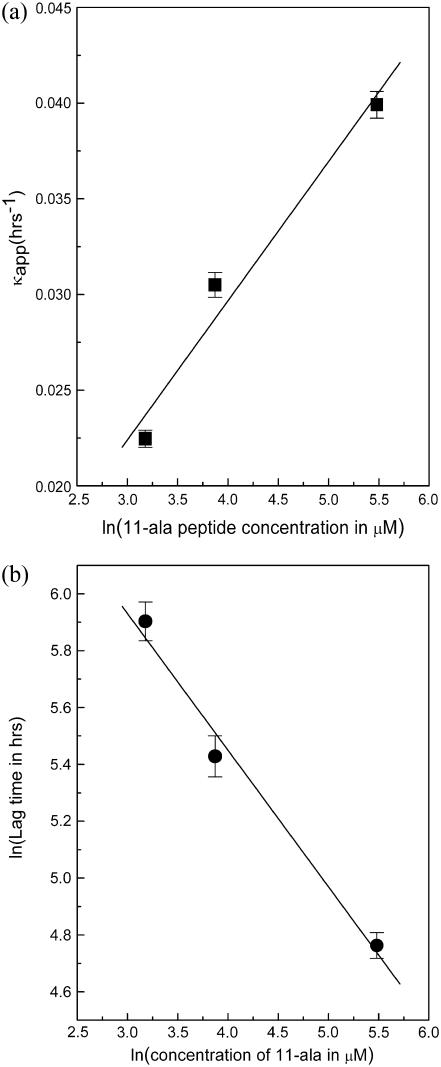
Fig. 3 b shows the dependence of the kinetics of fibril formation on the initial concentration of 11-ala in solution. As the concentration increases, the lag time becomes shorter and the rate of fibril formation increases, as shown by a steeper rise of the polymerization progress curve. These results are also consistent with the nucleation-dependent polymerization mechanism of fibril formation (13). The progress curves in Fig. 3 b were fit to an empirical relation describing nucleation-dependent kinetics (22):
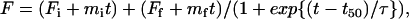 |
(1) |
where F is the ThT fluorescence intensity and t50 is the time to 50% of maximal fluorescence. The apparent rate constant, kapp, for fibril growth is given by 1/τ and the lag time is taken as t50 − 2τ. Fig. 4, a and b, shows that whereas the lag time decreases linearly with the logarithm of the initial concentration of peptide monomers in solution, the rate constant for fibril growth increases linearly with the concentration. These results suggest that formation of a critical nucleus—a small oligomer of the peptide—acts as the rate-limiting step during the polymerization process, which itself follows first-order kinetics as shown by the dependence of the lag time and kapp on concentration. Simply put, a higher concentration of peptide leads to increased probability of nucleus formation (shorter lag time) and faster growth of fibrils by monomer addition (higher kapp). Fibril formation has been shown to follow first-order kinetics for other proteins such as insulin, amyloid β-protein, and α-synuclein (23).
FIGURE 4.
Plots against log(concentration of 11-ala) of the (a) apparent rate constant of polymerization and (b) lag time, obtained by fitting data of Fig. 3 b to Eq. 1 of text.
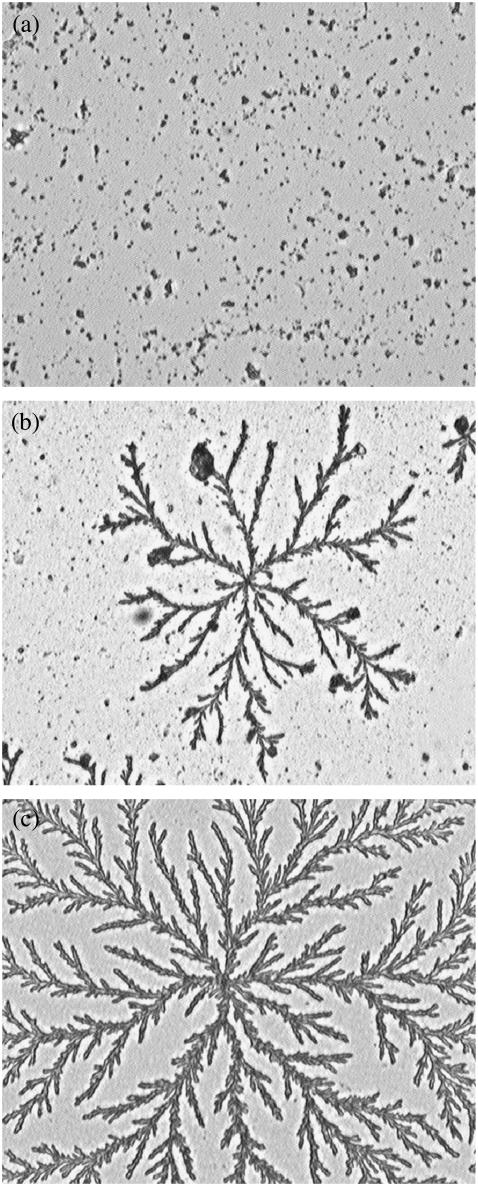
The fibrils formed in solution by 11-ala and 17-ala were found to be capable of forming higher order organized structures when exposed to suitable conditions. Peptide solutions, after incubation at high pH (10 and above) for appropriately long periods, when deposited on a glass slide and allowed to dry in air-formed highly ordered aggregates of fibrils that were visible even under an optical microscope (at magnification ∼200×). Fig. 5 shows optical micrographs of such patterns formed by drying solutions of (a) 7-ala (the control), (b) 11-ala, and (c) 17-ala. Both 11-ala and 17-ala, which formed fibrils in solution, formed patterns indicating further ordering of the fibrils on the glass surface, whereas 7-ala did not show any evidence of formation of fibrils or patterns.
FIGURE 5.
Patterns observed in an optical microscope (mag. ∼200×) for air-dried samples of (a) 7-ala, (b) 11-ala, and (c) 17-ala. Peptide solutions were incubated at pH 11 for 20 days at room temperature, spread on a coverslip, dried, and observed.
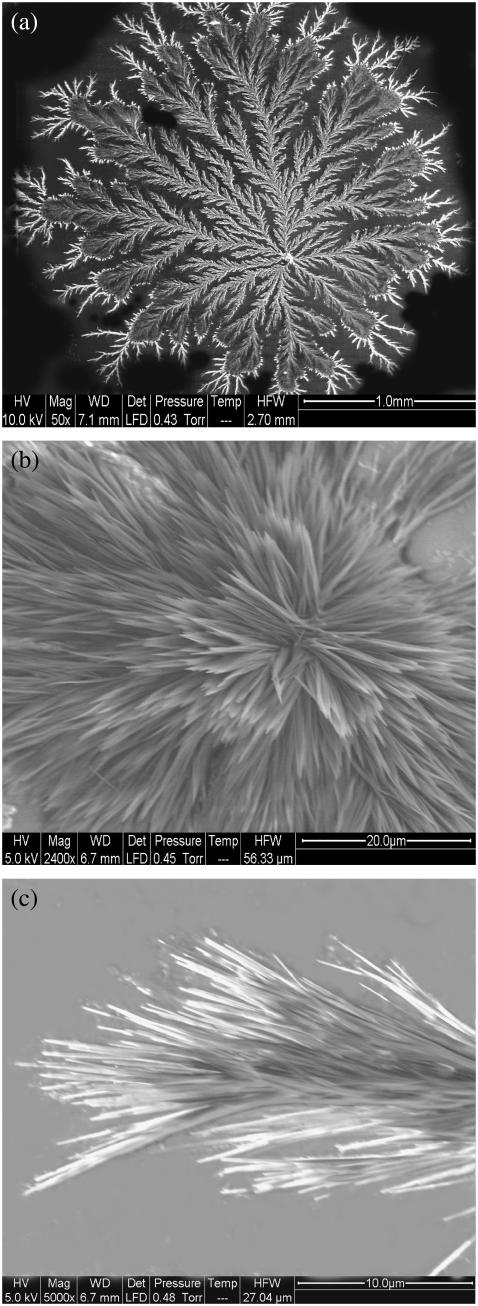
The structural details of the patterns were examined by viewing them under higher magnification under a scanning electron microscope. Fig. 6 a shows the scanning electron micrograph of a pattern formed by drying a pH 11 solution of 17-ala after prior incubation for 2 weeks, and Fig. 6, b and c, shows magnified images of its central part and the edge of one of its numerous branches. It is clearly seen that the center and the edge of the pattern are made up of ordered assemblies of individual rod-shaped objects, which can be taken to be the peptide fibrils shown in the transmission electron micrograph (Fig. 2). The average width of the rods in Fig. 6 c was estimated to be ∼200 nm, the same as that of the fibrils in Fig. 2. To test for the presence of aggregated peptides in the dried patterns, they were treated with the aggregation-specific dye Congo red and observed under a fluorescence microscope. Regions occupied by the pattern on a slide were found to emit a bright red fluorescence against a dark background (Fig. 7), confirming the presence of aggregated peptides.
FIGURE 6.
Scanning electron micrographs of a fractal pattern formed by the peptide 17-ala. Panels a, b, and c represent the whole pattern and its center and edge, respectively. Average width of individual fibers ∼200 nm.
FIGURE 7.
Pattern observed in a fluorescence microscope after addition of the aggregation-specific dye Congo red to a sample of 11-ala air-dried on glass coverslip. The dye specifically binds to the aggregated peptides, emitting a bright red fluorescence against a dark background devoid of any dye molecules.
Visual inspection of microscopic images of the aggregates showed them to have self-similar, fractal-like structures, similar to those formed by kinetic ordering of microscopic species ranging from gold nanoparticles to DNA fragments. The formation of such fractal patterns was not an isolated observation in a particular experiment; they were formed consistently and distributed copiously over the entire slide. The Hausdorff dimension (df) of each pattern was evaluated from its scanned image by the standard procedure (13), whereby the mass M of the aggregating species contained within a circle of radius R drawn around the center of the pattern is given by the relation M ∼ Rdf. An example of calculation of df is provided in Fig. 8, a and b. The slope of the line in Fig. 8 b yielded a value of 1.65 ± 0.02 for df, which agrees well with that found for similar patterns generated by drying ultraviolet-irradiated DNA solution kept in a glass petri dish (24). This value of df points to a nonuniversal diffusion-limited aggregation (DLA) characterized by a distribution of particle sizes (a range of fibril lengths), as predicted by Ossadnik et al. (25). However, a few patterns like the one generated by 17-ala after prolonged incubation of ∼12 weeks (Fig. 6 a) showed a higher value of 1.84 for df, indicating that the nature of the aggregation process in these cases may be somewhat different from that producing the patterns shown in Figs. 5, 9, and 10.
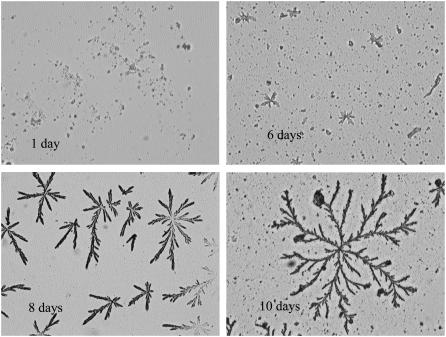
FIGURE 9.
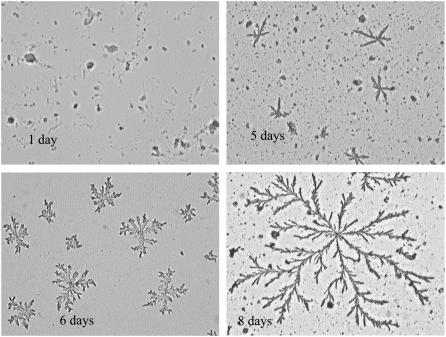
Patterns generated by 11-ala incubated for various lengths of time (magnification ∼200×).
FIGURE 10.
Patterns generated by 17-ala incubated for various lengths of time (magnification ∼200×).
Pattern formation by the peptides was subject to the same kinetic process as that controlling fibril formation in solution. That is, the size and space-filling density of the fractal patterns (i.e., df) are determined by the extent of fibril formation that, in turn, is governed by kinetic progress curves similar to those shown in Figs. 3 and 4. Figs. 9 and 10 show the patterns observed on drying solutions of 11-ala and 17-ala aged for different lengths of time. No discernible pattern was observed after very short periods of incubation (Figs. 9 a and 10 a). Patterns formed from 11-ala (17-ala) solutions incubated for 6 days (5 days) consisted of rudimentary domains with a single center and only a few linear branches (Figs. 9 b and 10 b). The size of such domains was obviously restricted by the limited supply of fibrils in solution. Larger space-filling domains of more complex structure were generated from solutions incubated for longer periods and containing a larger amount of fibrils, as shown in both Fig. 9 and Fig. 10, c and d.
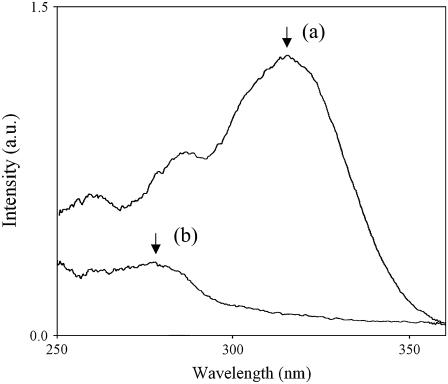
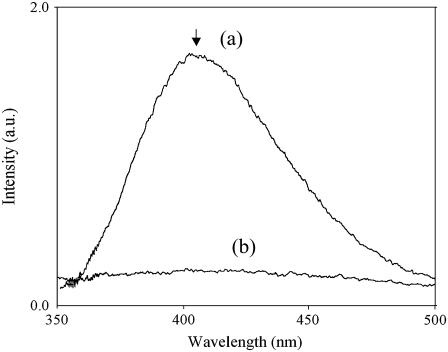
To ascertain the reason behind the ease of fibril formation at high pH, a fluorescence study of the peptides was done at different pH. Fig. 11 shows the excitation spectra of 11-ala at pH 7 and pH 11, with the emission being monitored at 410 nm. The excitation peak showed a red shift from 278 nm at pH 7 to 315 nm at pH 11 and increased in strength. Fig. 12 shows the emission spectra of 11-ala obtained with excitation at 320 nm. Whereas there was hardly any fluorescence at pH 7, an intense fluorescence band at ∼410 nm was observed at pH 11. The wavelengths of the excitation and emission peaks of the high pH solution matched those of the cross-linked amino acid dityrosine, presumably formed by cross-linking of the tyrosyl groups at the C-termini of pairs of 11-ala peptide chains (26). The pH dependence of the fluorescence spectra of the other two peptides (7-ala and 17-ala) were similar to those shown in Figs. 11 and 12, confirming that dityrosine formation also occurs for them at alkaline pH.
FIGURE 11.
Excitation spectra of 11-ala at (a) pH 11 and at (b) pH 7. The peaks marked with the arrows occur at (a) 315 nm and (b) 278 nm.
FIGURE 12.
Emission spectra of 11-ala obtained using excitation at 320 nm at (a) pH 11 and (b) pH 7. The peak of curve (a) occurs at 407 nm.
By measuring the optical density (OD) of the peptide solutions at pH 11 and using the extinction coefficients (ɛ) of tyrosine and dityrosine at 284 nm (10,26), the final concentrations of tyrosine (CTyr) and dityrosine (CDiTyr) could be determined from the following relations:
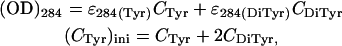 |
where (CTyr)ini is the initial concentration of tyrosine and was obtained by measuring the OD at pH 7, where very little of the tyrosine is expected to be dimerized. It was thus found that for both the peptides 11-ala and 17-ala, ∼45% of the initial population of tyrosines at pH 7 formed dityrosine at pH 11, whereas the rest (55%) existed in the monomeric state. Spectroscopic studies also ruled out formation of dityrosine at pH 3.
DISCUSSION
Aggregation of proteins leads to a variety of diseases, many of which are neurodegenerative and are characterized by the occurrence of β-sheet motifs in the pathological protein deposits. Among the first to study polyalanine-based peptides as disease models were Blondelle and co-workers, who showed that polyalanine tracts containing >10 alanines formed β-pleated-sheet complexes in solution (27,28). High levels of intracellular aggregates and nuclear fragmentation were also observed in cells expressing green fluorescent protein (GFP) fused to 19–37 alanines, when compared to cells expressing GFP alone or GFP fused to seven alanines, suggesting an association between aggregate formation and cell death (29). In our study of the effect of solution conditions on the aggregation properties of peptides mimicking the N-terminal polyalanine segment of PABP2, we found that those containing alanine stretches longer than the disease-causing threshold of 10 show high propensity for fibril formation in alkaline medium. The ionization states of the amino acid side chains are considered to be the crucial factors in such pH-dependent aggregation behavior (15–18). Fluorescence studies reported here indicate that at high pH, where formation of tyrosyl radicals is enhanced, cross-linking of tyrosyl side chains at the C-termini of peptides occur through the production of dityrosine. Although formation of such interpeptide links can provide anchoring between two neighboring peptide chains, the fact that the lysine side chains at the other end lose their positive charge at alkaline pH (pKa = 10.8) eliminates the electrostatic repulsion between their N-terminal ends, enabling amide hydrogens and carbonyl oxygens of their alanine residues to come together in the proper orientation for hydrogen-bond formation and development of intermolecular β-sheet structure. The change in CD spectra of 11-ala signaling a transition from β-turn to β-sheet, observed on increasing the pH (Fig. 1), is consistent with this scenario.
Although fluorescence spectra indicate dityrosine formation in all three peptides, even a month-long incubation of 7-ala did not lead to fibril formation. Thus other structural mechanisms are at work causing the transition from β-sheet to the mature fibrils formed after incubation of 11-ala and 17-ala, typically for ∼2 weeks (Fig. 2). The failure of 7-ala to aggregate may lie in the observed preference of this peptide for the α-helical conformation, which is an unlikely motif for oligomerization. The longer peptides, on the other hand, form β-sheets that undergo further aggregation into nuclei for fibril formation through the nucleation-controlled elongation route, as indicated by ThT fluorescence assay experiments (Figs. 3 and 4).
A recent study found that a recombinant protein with sequence identical to residues 1–125 of wild-type PABPN1 (containing 10 alanines) and its variant containing 7 additional alanine residues (i.e., 17 altogether) in the N-terminal polyalanine stretch form fibrils similar to those reported here with lag times of 27 days and 7 days, respectively, when 2 mM of each protein was incubated at 37°C (30). Decreasing the incubation temperature of the longer variant from 37°C to 20°C increased the lag time from 7 days to 84 days. In this study, the bare polyalanine peptides showed faster polymerization kinetics in high pH solution condition: the lag times were 4 days and 2.5 days, respectively, for 100 μM of 11-ala and 17-ala incubated at 25°C (Fig. 3 a). Two factors might have contributed to this accelerated kinetics: the larger size of the full protein molecule PABPN1 (compared to 11-ala and 17-ala) could hinder nucleation due to the excluded volume effect (20) and slow down fibril formation, whereas the added stabilization provided by dityrosine formation between polyalanine peptides would assist polymerization.
The kinetics of fibril formation by polyalanine peptides (Ac-KA14K-NH2) were investigated by Hall and co-workers using an off-lattice intermediate resolution simulation method (31,32). They found evidence for conformational conversion of the peptides from small amorphous aggregates to β-sheets and then into ordered nuclei, followed by rapid growth of stable fibrils or protofilaments. Fibril formation was nucleation dependent, occurring after a lag time that decreased more or less exponentially with increasing peptide concentration. The main results of these simulations are thus in good agreement with our measurements on the kinetics of fibril formation, as represented in Figs. 3 and 4.
The majority of fractal patterns created by the peptide fibrils are the result of a process akin to DLA in two dimensions, caused by drying the solvent in which the peptides were dissolved. In agreement with the physical nature of such processes, the fractal dimension (df) of these aggregates (Figs. 8 and 9) was found to be ∼1.65 (13). Generation of very similar fractal patterns by the pH-dependent hierarchical self-assembly of a cross-linkable coiled-coil peptide has recently been reported (33). However, molecular dynamics simulations indicated that those patterns were formed by aggregation of globules and clusters made up of hexameric coiled-coils, unlike in the case in this study where scanning electron micrographs (Fig. 6) show the aggregating objects to be rod-shaped polymers of the peptides. In the case of the denser patterns (such as the one in Fig. 6) the measured value of df (∼1.84) is much closer to the limiting value of 2, indicating transition to the ballistic limit where strain fields created by solvent evaporation dictate the pattern formation process (13).
A recent study of the self-assembly of synthetic polyalanine peptides by Shinchuk et al., although confirming the β-sheet-mediated aggregation propensity of peptides containing 10–20 alanine residues, did not observe any ThT fluorescence or birefringence with Congo red in the presence of the peptide fibrils (34). On the whole their results agree with ours, except in the reported lack of ThT binding and the fact that the observed fibril widths (25–75 Å) were nearly 30–100 times narrower than the fibril widths (200 nm) observed by us. However, it should be noted that fibrils obtained from polymerization of the N-terminal domain (residues 1–125) of the protein PABPN1 containing 10–17 alanines (30) bind both ThT and 1-anilinonapthalene-8-sulfonic acid strongly and are considerably wider (15 nm) than those reported by Shinchuk et al. A possible explanation for these differences in physical characteristics might be that fibrils of the PABPN1 fragment, and especially those reported here, are larger assemblies of more elementary polymers that were reported by the authors of Shinchuk et al. (34), somewhat similar to the ‘macrofibers’ formed from 14-stranded helical polymers of sickle hemoglobin (35). During the assembly of these higher-order organized structures from the ‘elementary’ polymer units, hydrophobic pockets can be created due to packing considerations. The existence of such regions facilitates binding of ThT or Congo red, leading to enhancement of ThT fluorescence and the appearance of a birefringence signal on being placed between crossed polarizers.
Our study presents a system of model peptides that can be used to determine the nucleation probability of aggregation-prone peptides and its dependence on solution conditions (e.g., pH) and alanine repeat length. Since none of the 10 tyrosine residues in the protein PABP2 is situated in the N-terminal region where the polyalanine stretch occurs, it may be argued that the results may not have a direct bearing on formation of the fibrillar deposits found in OPMD patients. However, it serves to illustrate the importance of solvent conditions like pH and other, hitherto unknown, mechanisms that may act as triggers in the fibril formation process. Our kinetic studies showed that both fibril formation and growth of fractal patterns occur sooner and faster for 17-ala than for 11-ala, indicating a correlation between longer alanine stretches and more efficient nucleation-controlled polymerization.
This is consistent with other in vitro findings on polyalanine peptides and a PABPN1 protein fragment and can be attributed to the larger β-sheet content of longer polyalanine stretches (30,34). Similar considerations also apply in determining the pathology of a group of other protein aggregation diseases, such as Huntington's chorea, where the age of onset and the severity of the disease are crucially dependent on the number of successive glutamines in a polyglutamine sequence in the protein Huntingtin (36).
On the other hand, our finding of fractal patterns formed by drying peptide solutions on glass surfaces is both unexpected and intriguing, since no patterns were observed by Shinchuk et al. using polyalanine peptides subjected to the same procedure (34). This phenomenon is likely to be related to the formation at high pH of thick, rod-like polymers that are stiffer than rudimentary fibrils and during drying produce DLA of rigid rods on a two-dimensional (glass) surface. Although dityrosine formation (as suggested here) may play a role in the nucleation of the polymers, this study demonstrates that hydrophobic interactions between alanine residues are strong enough to hold together aggregates of much larger than microscopic dimensions (200 nm). It would be interesting to find out if such large-sized polyalanine aggregates have a role to play in the filamentous inclusions found in muscle fibers of OPMD patients (2,3). The hierarchical structures formed by self-assembling peptides on substrates can also act as templates for the design of novel materials based on directed assembly of the peptides (33).
It is interesting to speculate on the mechanism of dityrosine formation at high pH, especially since this study did not employ the usual oxidizing agents such as enzymes of the peroxidase family to obtain the cross-linked peptides. A tentative scheme for arriving at dityrosine by starting from tyrosines is suggested in the following:
At pH 11 the phenolic-OH group of Tyr (pKa = 10.8) loses its H+ ion to form the phenoxide ion. The negative charge on the oxygen atom delocalizes by resonance and moves to the C2 position, forming an intermediate. Each intermediate then loses an electron, forming a C radical on the tyrosine residue, by a mechanism such as photochemical excitation or simply by thermal excitation (37). Two such radicals form a C-C bond, releasing two protons for rearomatization to gain stability by reformation of the aromatic rings. OH− ions present in the media at high pH then take up the protons and carry forward the dimerization reaction. Since the high pH condition favors both the first (increased dissociation of the phenolic-OH of Tyr) and last (proton abstraction by OH− ions) steps of this reaction scheme, it promotes dityrosine formation. As a corollary, acidic pH should inhibit dimerization, which is indeed observed.
Acknowledgments
We thank the Electron Microscope Facility and the Surface Physics Division for allowing us generous use of the Transmission Electron Microscope and the Scanning Electron Microscope, respectively.
References
- 1.Albrecht, A., and S. Mundlos. 2005. The other trinucleotide repeat: polyalanine expansion disorders. Curr. Opin. Genet. Dev. 15:285–293. [DOI] [PubMed] [Google Scholar]
- 2.Coquet, M., C. Vital, and J. Julien. 1990. Presence of inclusion body myositis-like filaments in oculopharyngeal muscular dystrophy. Ultrastructural study of 10 cases. Neuropathol. Appl. Neurobiol. 16:393–400. [DOI] [PubMed] [Google Scholar]
- 3.Shanmugam, V., P. Dion, D. Rochefort, J. Laganiere, B. Brais, and G. A. Rouleau. 2000. PABP2 polyalanine tract expansion causes intranuclear inclusions in oculopharyngeal muscular dystrophy. Ann. Neurol. 48:798–802. [PubMed] [Google Scholar]
- 4.Calado, A., F. M. S. Tomé, B. Brais, G. A. Rouleau, U. Kühn, E. Wahle, and M. C. Fonseca. 2000. Nuclear inclusions in the oculopharyngeal muscular dystrophy consist of Poly(A) binding protein 2 aggregates which sequester poly(A) RNA. Hum. Mol. Genet. 9:2321–2328. [DOI] [PubMed] [Google Scholar]
- 5.Brais, B., J. P. Bouchard, Y. G. Xie, D. L. Rochefort, N. Chretien, F. M. Tome, R. G. Lafreniere, J. M. Rommens, E. Uyama, O. Nohira, S. Blumen, A. D. Korczyn, P. Heutink, J. Mathieu, A. Duranceau, F. Codere, M. Fardeau, G. A. Rouleau, and A. D. Korcyn. 1998. Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nat. Genet. 18:164–167. [DOI] [PubMed] [Google Scholar]
- 6.Dobson, C. M. 2003. Protein folding and misfolding. Nature. 426:884–890. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen, J., M. A. Baldwin, F. E. Cohen, and S. B. Prusiner. 1995. Prion protein peptides induce α-helix to β-sheet conformational transitions. Biochemistry. 34:4186–4192. [DOI] [PubMed] [Google Scholar]
- 8.Stefani, M., and C. M. Dobson. 2003. Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J. Mol. Med. 81:678–699. [DOI] [PubMed] [Google Scholar]
- 9.Giri, K., U. Ghosh, N. P. Bhattacharyya, and S. Basak. 2003. Caspase 8 mediated apoptotic cell death induced by β-sheet forming polyalanine peptides. FEBS Lett. 555:380–384. [DOI] [PubMed] [Google Scholar]
- 10.Gill, S. C., and P. H. Von Hipple. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319–326. [DOI] [PubMed] [Google Scholar]
- 11.Naiki, H., K. Higuchi, K. Matsushima, A. Shimada, W. H. Chen, M. Hosokawa, and T. Takeda. 1990. Fluorometric examination of tissue amyloid fibrils in murine senile amyloidosis: use of the fluorescent indicator, thioflavine T. Lab. Invest. 62:768–773. [PubMed] [Google Scholar]
- 12.Levine III, H. 1993. Thioflavine T interaction with synthetic Alzheimer's disease β-amyloid peptides: detection of amyloid aggregation in solution. Protein Sci. 2:404–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vicsek, T. 1989. Fractal Growth Phenomena. World Scientific, Singapore.
- 14.Sarkar, P. K., and P. Doty. 1966. The optical rotatory properties of the β-configuration in polypeptides and proteins. Proc. Natl. Acad. Sci. USA. 55:981–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraser, P. E., J. T. Nguyen, W. K. Surewicz, and D. A. Kirschner. 1991. pH-dependent structural transitions of Alzheimer amyloid peptides. Biophys. J. 60:1190–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rostagno, A., R. Vidal, B. Kaplan, J. Chuba, A. Kumar, J. I. Elliott, B. Frangione, G. Gallo, and J. Ghiso. 1999. pH-dependent fibrillogenesis of a Vkappa III Bence Jones protein. Br. J. Haematol. 107:835–844. [DOI] [PubMed] [Google Scholar]
- 17.Lai, Z., W. Colon, and J. W. Kelly. 1996. The acid-mediated denaturation pathway of transthyretin yields a conformational intermediate that can self-assemble into amyloid. Biochemistry. 35:6470–6482. [DOI] [PubMed] [Google Scholar]
- 18.Uversky, V. N., J. Li, and A. L. Fink. 2001. Evidence for a partially folded intermediate in α-synuclein fibril formation. J. Biol. Chem. 276:10737–10744. [DOI] [PubMed] [Google Scholar]
- 19.Harper, J. D., and P. T. Lansbury Jr. 1997. Models of amyloid seeding in Alzheimer's disease and scrapie: mechanistic truths and physiological consequences of the time-dependent solubility of amyloid proteins. Annu. Rev. Biochem. 66:385–407. [DOI] [PubMed] [Google Scholar]
- 20.Ferrone, F. 1999. Analysis of protein aggregation kinetics. Methods Enzymol. 309:256–274. [DOI] [PubMed] [Google Scholar]
- 21.Perutz, M., and A. H. Windle. 2001. Cause of neural death in neurodegenerative diseases attributable to expansion of glutamine repeats. Nature. 412:143–144. [DOI] [PubMed] [Google Scholar]
- 22.Uversky, V. N., J. Li, and A. L. Fink. 2001. Metal-triggered structural transformations, aggregation, and fibrillation of human α-synuclein. A possible molecular link between Parkinson's disease and heavy metal exposure. J. Biol. Chem. 276:44284–44296. [DOI] [PubMed] [Google Scholar]
- 23.Nielsen, L., R. Khurana, A. Coats, S. Frokjaer, J. Brange, S. Vyas, V. N. Uversky, and A. L. Fink. 2001. Effect of environmental factors on the kinetics of insulin fibril formation: elucidation of the molecular mechanism. Biochemistry. 40:6036–6046. [DOI] [PubMed] [Google Scholar]
- 24.Chandra, A., M. K. Shukla, P. C. Mishra, and S. Chandra. 1995. Fractal growth in UV-irradiated DNA: evidence of nonuniversal diffusion limited aggregation. Phys. Rev. E. 51:R2767–R2768. [DOI] [PubMed] [Google Scholar]
- 25.Ossadnik, P., C. Lam, and L. M. Sander. 1994. Nonuniversal diffusion-limited aggregation and exact fractal dimensions. Phys. Rev. E. 49:R1788–R1791. [DOI] [PubMed] [Google Scholar]
- 26.Smail, E. H., P. Briza, A. Panagos, and L. Berenfeld. 1995. Candida albicans cell walls contain the fluorescent cross-linking amino acid dityrosine. Infect. Immun. 63:4078–4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forood, B., E. Pérez-Payá, R. A. Houghten, and S. E. Blondelle. 1995. Formation of an extremely stable polyalanine beta-sheet macromolecule. Biochem. Biophys. Res. Commun. 211:7–13. [DOI] [PubMed] [Google Scholar]
- 28.Blondelle, S. E., B. Forood, R. A. Houghten, and E. Pérez-Payá. 1997. Polyalanine-based peptides as models for self-associated beta-pleated-sheet complexes. Biochemistry. 36:8393–8400. [DOI] [PubMed] [Google Scholar]
- 29.Rankin, J., A. Wyttenbach, and D. C. Rubinsztein. 2000. Intracellular green fluorescent protein–polyalanine aggregates are associated with cell death. Biochem. J. 348:15–19. [PMC free article] [PubMed] [Google Scholar]
- 30.Scheuermann, T., B. Schulz, A. Blume, E. Wahle, R. Rudolph, and E. Schwarz. 2003. Trinucleotide expansions leading to an extended poly-L-alanine segment in the poly (A) binding protein PABPN1 cause fibril formation. Protein Sci. 12:2685–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguygen, H. D., and C. K. Hall. 2004. Molecular dynamics simulations of spontaneous fibril formation by random-coil peptides. Proc. Natl. Acad. Sci. USA. 101:16180–16185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguygen, H. D., and C. K. Hall. 2005. Kinetics of fibril formation by polyalanine peptides. J. Biol. Chem. 280:9074–9082. [DOI] [PubMed] [Google Scholar]
- 33.Lomander, A., W. Hwang, and S. Zhang. 2005. Hierarchical self-assembly of a coiled-coil peptide into fractal structure. Nano Lett. 5:1255–1260. [DOI] [PubMed] [Google Scholar]
- 34.Shinchuk, L. M., D. Sharma, S. E. Blondelle, N. Reixach, H. Inouye, and D. A. Kirschner. 2005. Poly-(L-alanine) expansions form core β-sheets that nucleate amyloid assembly. Proteins Struct. Funct. Bioinform. 61:579–589. [DOI] [PubMed] [Google Scholar]
- 35.Dickerson, R. E., and I. Geis. 1983. Hemoglobin: Structure, Function, Evolution and Pathology. Benjamin Cummings, Menlo Park, CA. 138–142.
- 36.Perutz, M. F. 1999. Glutamine repeats and neurodegenerative diseases: molecular aspects. Trends Biochem. Sci. 24:58–63. [DOI] [PubMed] [Google Scholar]
- 37.Fancy, D. A., and T. Kodadek. 1999. Chemistry for the analysis of protein-protein interactions: rapid and efficient cross-linking triggered by long wavelength light. Proc. Natl. Acad. Sci. USA. 96:6020–6024. [DOI] [PMC free article] [PubMed] [Google Scholar]