Abstract
This study provides evidence of a novel function for mitochondrial creatine kinase (MtCK) and nucleoside diphosphate kinase (NDPK-D). Both are basic peripheral membrane proteins with symmetrical homo-oligomeric structure, which in the case of MtCK was already shown to allow crossbridging of lipid bilayers. Here, different lipid dilution assays clearly demonstrate that both kinases also facilitate lipid transfer from one bilayer to another. Lipid transfer occurs between liposomes mimicking the lipid composition of mitochondrial contact sites, containing 30 mol % cardiolipin, but transfer does not occur when cardiolipin is replaced by phosphatidylglycerol. Ubiquitous MtCK, but not NDPK-D, shows some specificity in the nature of the lipids transferred and it is not active with phosphatidylcholine alone. MtCK can undergo reversible oligomerization between dimeric and octameric forms, but only the octamer can bridge membranes and promote lipid transfer. Cytochrome c, another basic mitochondrial protein known to bind to anionic membranes but not crosslinking them, is also incapable of promoting lipid transfer. The lipid transfer process does not involve vesicle fusion or loss of the internal contents of the liposomes.
INTRODUCTION
Morphological analysis of mitochondria with different methods revealed connections between the outer and peripheral inner membranes of mitochondria, the so-called contact sites, e.g., in chemically fixed mitochondria where the intermembrane space is enlarged (1), as jumps in the fracture planes of freeze-fractured mitochondria (2), or in three-dimensional electron tomography (3). These contacts fulfill various functions, like the import of mitochondrial precursor proteins (4), the channeling of high-energy phosphates from mitochondria to the cytosol (5), and the formation of the mitochondrial permeability transition pore that is involved in apoptotic signaling (6). Contact sites are regulated, dynamic structures, since their number depends on the metabolic activity of the cell (7). Because of the close propinquity of the two membranes, these contact sites would be expected to favor interbilayer transfer of lipids. Two oligomeric kinases located at a specific type of contact sites, and probably contributing to their formation are the mitochondrial isoenzymes of creatine kinase (MtCK) and mitochondrial isoform of nucleotide diphosphate kinase (NDPK-D) (8–10). In this work, we have studied whether these kinases are able to facilitate lipid transfer between membranes, as this could be a novel functional property of the two proteins. Transfer of lipids between bilayers is only known to be facilitated by specific lipid transfer proteins (11–13). There is a report indicating that t-Bid has lipid transfer activity (14), but this is suggested to be a result of its structural similarity to other lipid transfer proteins (15). Such lipid transfer is known to occur in intact mitochondria, e.g., during apoptosis, in the form of cardiolipin transfer from the inner to the outer mitochondrial membrane (16–19).
Creatine kinase (CK) plays an important role in forming a circuit or shuttle for phosphocreatine between microcompartments in the cell (20). Most vertebrate tissues express two isoforms of CK, a dimeric cytosolic and a mostly octameric mitochondrial isoform (MtCK). The latter occurs either as the sarcomeric form in striated muscle, or as the ubiquitous form (uMtCK) in most other tissues including brain and kidney (21). MtCK undergoes concentration-dependent reversible oligomerization between octameric and dimeric forms with octameric uMtCK dissociating 23–24 times slower, compared with the other mitochondrial isoform (22). Mainly the octameric form shows fast binding to anionic phospholipids, while much slower binding of dimers probably occurs through octamerization at the membrane surface (23). Similarly, mainly octameric MtCK mediates intermembrane contact between liposomes of isolated inner and outer mitochondrial membranes. This membrane binding is largely stabilized by electrostatic interactions with anionic lipids (24). The crystal structure of both octameric MtCKs were determined (25,26) and it was subsequently demonstrated that several C-terminal lysine residues of the protein facilitated this interaction (27). Using surface plasmon resonance (28,29), MtCK membrane binding was shown to occur already at 16% cardiolipin and to involve two independent binding sites with similar affinities (80–100 nM). The basic mitochondrial protein, cytochrome c, which we also use in this work, also bound to the immobilized lipid under these conditions. Binding of the MtCK to lipid is accompanied by a small conformational change in the protein as well as by a change in the ordering of the lipids (30).
MtCK is localized between the inner and outer mitochondrial membranes, the so-called peripheral intermembrane space, as well as along the cristae membranes (31), mostly bound to cardiolipin at the inner membrane and enriched in contact sites (32). Recent evidence indicates that uMtCK induces and stabilizes contact sites between inner and outer mitochondrial membranes (33). Liver mitochondria, mainly devoid of MtCK, were compared to those from transgenic mice expressing uMtCK in their livers. Only transgenic mitochondria showed an increased number of contact sites as revealed by electron microscopy, and an improved resistance against lysis by detergent or oxidative agent.
NDPK-D is part of a large family of hexameric isoenzymes that are encoded by different nm23 genes and exert multiple functions in cellular energetics, signaling, proliferation and differentiation. Although NDPK activity was reported in mitochondria with different localization depending on the species or the organ (34) and was located in various submitochondrial fractions, only NDPK-D has a mitochondrial targeting sequence and has been unambiguously localized as bound to the inner mitochondrial membrane, in particular to contact sites (10). NDPK-D shows similar properties to uMtCK, insofar as it also binds to anionic phospholipids like cardiolipin and is capable of crosslinking two lipid bilayers, probably due to the symmetry of its hexameric structure (M. Tokarska-Schlattner, U. Schlattner, and M.-L. Lacombe, unpublished data). However, unlike uMtCK, NDPK-D remains hexameric at any protein concentration (35). There is particular interest in the products from the nm23 gene family because they are involved in tumor progression and metastasis (36). NDPK-D was found to be overexpressed in a majority of gastric and colon cancers (37), possibly linking NDPK-D to the development of tumors.
Interbilayer transfer of lipid between the inner and outer membranes of mitochondria may play an important role in the regulation and initiation of apoptosis. It is known that the proapoptotic Bcl-2 proteins t-Bid and Bax will bind more readily to membranes with exposed cardiolipin (38–41). However, the major fraction of cardiolipin in cells is present on the inner leaflet of the inner mitochondrial membrane (42). Cardiolipin must first become exposed to the outer surface of the outer mitochondrial membrane to interact with Bcl-2 proteins to maximally induce apoptosis. In fact, at least with some forms of apoptosis, there is increased exposure of cardiolipin at the mitochondrial surface (16–18). Such a transfer has to include an exchange from the inner mitochondrial membrane to the outer membrane, as well as transbilayer diffusion in each of the two mitochondrial membranes. It has been shown that Bcl-2 proteins themselves can accelerate transbilayer diffusion of phospholipids (43,44). In addition, a lipid transfer protein was shown to promote cytochrome c release in isolated mitochondria, demonstrating a direct link between lipid transfer and apoptosis (19). Finally, intermembrane cardiolipin transfer would also be essential for a recently suggested regulation of the VDAC channel (45) in the mitochondrial outer membrane and part of the contact site complexes that often involve uMtCK and NDPK-D.
In this work we wish to determine if phospholipid transfer is facilitated by proteins present in mitochondrial sites in which the inner and outer mitochondrial membranes become juxtaposed, i.e., by uMtCK and/or NDPK-D. With a lipid composition resembling that of mitochondrial contact sites (46), in which each of these two proteins can bridge vesicles and allow their close approach, we find a marked increase in the rate of phospholipid transfer facilitated by uMtCK and NDPK-D, without concomitant fusion of the vesicles. The transfer process is specific for the oligomeric state of the protein that can bridge membranes and does not occur with the dimeric form of uMtCK.
EXPERIMENTAL PROCEDURES
Materials
Unlabeled phospholipids, as well as n-(lissamine Rhodamine B sulfonyl) phosphatidylethanolamine (Rh-PE) and n-(7-nitro-2,1,3-benzoxadiazol-4-yl) phosphatidylethanolamine (NBD-PE), were purchased from Avanti Polar Lipids (Alabaster, AL). The CL is the tetraoleoyl form. The tetraoleoyl form is abundant in human lymphoblasts and in yeast (47). All other fluorescently-labeled phospholipids were from Molecular Probes (Eugene, OR). The structures of the fluorescently-labeled phospholipids used in this work are shown in Fig. 1. Poly-L-lysine (21 kDa) and cytochrome c were obtained from Sigma (St. Louis, MO). Recombinant mature uMtCK was expressed and purified as described earlier (26). For some experiments, the final preparation of uMtCK was concentrated fourfold using a 10 kDa cutoff Microcon centrifugal filter YM-10 (Millipore, Billerica, MA). It was found that lipid transfer assays in which uMtCK was added from a more concentrated protein solution exhibited higher activity. Recombinant NDPK-D, missing the mitochondrial targeting sequence (first 33 amino acids) and fused to a hexahistidine tag, was expressed and purified as described in Milon et al. (10).
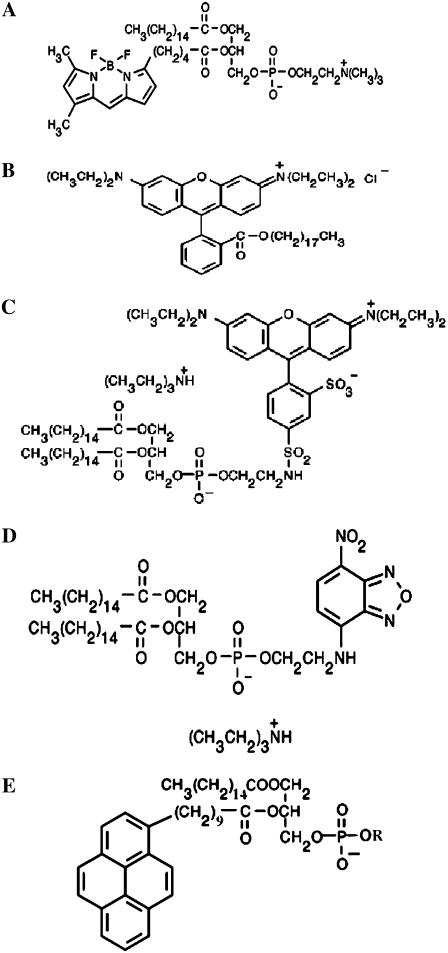
FIGURE 1.
Structures of fluorescently labeled lipids used in this work. (A) BODIPY-FL-C5-HPC; (B) R18; (C) Rh-PE; (D) NBD-PE; and (E) generic pyrene phospholipid, where R completes the headgroup of PC, PE, or PG.
Preparation of large unilamellar vesicles (LUVs)
Lipids were codissolved in chloroform/methanol (2:1, v/v). The solvent was then evaporated under a stream of nitrogen with constant rotation of a test tube so as to deposit a uniform film of lipid over the bottom third of the tube. Last traces of solvent were removed by placing the tube under high vacuum for 3 h. The lipid film was then hydrated with buffer, as indicated for specific experiments. Large unilamellar vesicles (LUVs) were made from this suspension by five freeze-thaw cycles and extruded 10 times through two polycarbonate filters with either 50- or 200-nm pore size, in a Lipex barrel extruder under nitrogen pressure (Lipex Biomembranes, Vancouver, BC, Canada). Vesicles were kept on ice under Argon gas and used within a few hours of preparation. Lipid concentration was determined with the phosphate assay of Ames (48).
BODIPY-FL-C5-HPC transfer assay
The transfer of BODIPY-labeled lipid between donor and acceptor vesicles was monitored by a lipid dilution assay based on the relief of self-quenching similar to that recently described by Esposti and co-workers (14). Donor vesicles were prepared from lipid films containing 2-(4,4-difluoro-5,7-dimethyl-4-bora-3a, 4a-diaza-s-indacene-3-pentanoyl)-1-hexadecanoyl-sn-glycero-3-phosphocholine (BODIPY-FL-C5-HPC)/1-palmitoyl-2-oleoyl phosphatidylethanolamine (POPE)/cardiolipin (CL) in equimolar amounts and were made into 50-nm diameter LUVs by extrusion. Acceptor LUVs were made of an equimolar mixture of 1-palmitoyl-2-oleoyl phosphatidylcholine (POPC)/POPE/CL and made into 200-nm diameter LUVs. The LUV were prepared in 10 mM HEPES, 1 mM β-mercaptoethanol, pH 7.0 and kept in ice under nitrogen, in the dark, before use. The assay was carried out in 1 cm × 1 cm path-length disposable acrylic fluorimeter cuvettes at 25°C, with magnetic stirring using an SLM-Aminco Bowman AB-2 spectrofluorimeter (Urbana, IL). Donor vesicles were added to 2 mL of 10 mM HEPES and 1 mM β-mercaptoethanol, pH 7.0 to a lipid concentration of 300 nM. After equilibration for 30 s, acceptor LUVs were added to a concentration of 4 μM. Fluorescence emission was measured at 515 nm, using an excitation wavelength of 485 nm and a 4 nm bandpass in both excitation and emission. After a few hundred seconds, octameric uMtCK was added as a solution that had been dialyzed against 10 mM HEPES and 1 mM β-mercaptoethanol, pH 7 and fluorescence recording was continued. To obtain the fluorescence intensity corresponding to 100% transfer, 20 μL 20% Lubrol PX was added as a detergent to the mixture in the cuvette.
General procedure for other lipid transfer assays
We describe here the elements that are common to the various lipid mixing assays used in the course of this study. Details specific for each of the methods are given subsequently. The lipid composition of both donor and acceptor vesicles was POPC/POPE/CL (1:1:1). Donor vesicles with fluorescent label were extruded to make 50-nm diameter LUV (or sonicated to make SUV in some pyrene label assays) and the acceptor vesicles extruded to 200 nm diameter LUV. The donor LUVs were placed in 2 mL of 10 mM HEPES, 0.1 mM EDTA, and 0.14 M NaCl, pH 6.9 (HEPES buffer) at 37°C with magnetic stirring. In experiments where Ca2+ was required, the buffer composition was calculated so that in the presence of 0.1 mM EDTA and 1 mM Mg2+ there would be 200 nM free Ca2+ (HEPES-Ca2+ buffer) using the program WINMAXC v2.50 (Chris Patton, Stanford University). This HEPES-Ca2+ buffer still contains 10 mM HEPES, 0.1 mM EDTA, and 0.14 M NaCl, pH 6.9. LUVs were kept on ice, under Argon, in the dark. Labeled donor vesicles were added to 2 mL HEPES buffer in the 1 cm × 1 cm path-length fluorimeter cuvette, with magnetic stirring, to give a final lipid concentration of 300 nM. After a few seconds equilibration, acceptor LUVs were added (3 μM) and the fluorescence intensity was recorded for a length of time before and after addition of protein to the cuvette. Emission scans were also performed before and after addition of protein. Experiments were repeated at least twice. Molarities were always calculated using a molecular mass corresponding to the octamer for uMtCK and to the hexamer for NDPK-D.
Octadecyl rhodamine B (R18) transfer assay
The octadecyl rhodamine B (R18) assay has been extensively used to monitor the fusion of enveloped viruses to target membranes (49). A solution of R18 in ethanol was added to the donor vesicles to give 10% R18 in the vesicles (the final concentration of ethanol in the 2 mL total volume in the cuvette was 0.1%). The mixture was subjected to an incubation period of several hours at room temperature, under argon, with occasional gentle shaking to allow incorporation of the label into the LUVs. The labeled liposomes were then passed through a 1.5 × 25 cm column of Sephadex G-75 (Amersham Biosciences, Piscataway, NJ), collecting the vesicles in the void volume. It was found that almost all of the R18 had incorporated into the liposomes; therefore, this last step was skipped in subsequent experiments. The excitation wavelength was 565 nm and emission was 595 nm. Cutoff filters used were 550 nm in excitation and 570 nm in emission. A 4-nm bandpass in excitation and in emission was used. A fluorescence emission spectrum was also run for each sample, both before and after addition of protein. Controls in the absence of acceptor LUVs were also run. The value for 100% transfer was obtained with the addition of 20 μL 20% Lubrol PX followed by brief sonication or by adding an amount of R18 to LUVs corresponding to the complete dilution into donor plus acceptor vesicles (∼1%).
Transfer of pyrene-labeled lipids between liposomes
A lipid dilution assay, based on the loss of intermolecular excimer fluorescence, was employed to measure lipid transfer (50,51). Donor vesicles were made with 10 mol % of either 1-hexadecanoyl-2-(1-pyrenedecanoyl) (Pyrene) phosphatidylcholine (PC), Pyrene phosphatidylethanolamine (PE), or Pyrene phosphatidylglycerol (PG) incorporated into films of POPC/POPE/CL (1:1:1) or POPC/POPE/POPG (1:1:1) and made into SUVs or into 50 nm LUVs after hydration with HEPES-Ca2+ buffer, pH 6.9. Acceptor vesicles were unlabeled 200-nm LUVs of the same composition, but without the presence of pyrene-labeled lipid. Fluorescence was recorded for a few seconds, and then protein was added. The decrease in excimer fluorescence, that occurred only after protein was added, was followed over time. The excitation wavelength was 344 nm and emission was monitored as a ratio (Ie/Im) of the intensities of excimer emission at 476 nm (Ie) and monomer emission at 397 nm (Im). A 4-nm bandpass was used in both excitation and emission. In addition, a wavelength emission scan was performed before and after protein addition. Runs were repeated in the absence of acceptor to assess the effect of the protein on the fluorescence properties of the pyrene lipid in the absence of net lipid transfer. A constant stream of nitrogen was passed continually through the cell compartment to prevent oxygen quenching of pyrene.
A calibration curve was established for the dependence of Ie/Im on the mol fraction of pyrene-labeled lipid in the membrane. This standard curve was then used to correlate the observed Ie/Im with the extent of transfer of labeled lipid, assuming that the labeled lipid in the inner monolayer does not get transferred within the time of the experiment.
Fluorescence resonance energy transfer (FRET)-based lipid transfer assay
The resonance energy transfer assay of Struck et al. (52) was used. Two populations of LUVs were prepared, one unlabeled and one labeled. The 50-nm diameter donor vesicles were labeled with 1 mol % each of NBD-PE and Rh-PE. The excitation wavelength was 450 nm and emission 530 nm, with a 4-nm bandpass in excitation and emission, and a 500-nm cutoff filter in the emission path. Fluorescence was recorded before and after addition of protein, as a function of time. An emission scan was performed for each run, before and after addition of protein.
Leakage studies with ANTS/DPX
Leakage of aqueous contents from liposomes was determined using the ANTS-DPX assay (53). Lipid films of POPC/POPE/CL (1:1:1) were hydrated with 12.5 mM ANTS, 45 mM DPX, and 10 mM HEPES, at pH 6.9. The osmolarity of this solution was adjusted to 300 mOsm with NaCl using a cryoosmometer (Advanced Model 3MOplus Micro-Osmometer, Advanced Instruments, Norwood, MA). LUVs of 50-nm diameter were prepared by extrusion as described above. The LUVs were then passed through a Sephadex G-75 column (Amersham Biosciences) previously equilibrated with the same buffer, to remove unentrapped dyes. Phosphate analysis was then carried out with the method of Ames (48). A second population of LUVs of 200-nm diameter were prepared with the same lipid composition suspended in 10 mM HEPES, 0.1 mM EDTA, and 0.14 M NaCl, pH 6.9 adjusted to 300 mOsm, but without fluorescent probes. LUVs were kept on ice under nitrogen, in the dark, before measurement. The fluorescence measurements were performed in 2 mL of the HEPES buffer, pH 6.9, in a 1 cm × 1 cm path-length quartz cuvette equilibrated at 37°C with magnetic stirring. When Ca2+ was required, the HEPES-Ca2+ buffer, pH 6.9, was used. Aliquots of LUVs were added to the cuvette to a final lipid concentration of 2.5 μM LUVs with entrapped dye and 25 μM unlabeled LUVs. The fluorescence was recorded as a function of time with an SLM Aminco Bowman Series II spectrofluorimeter, using an excitation wavelength of 344 nm and an emission wavelength of 544 nm with an 8-nm bandwidth slit in excitation and 4 nm in emission. A 500-nm cutoff filter was placed in the emission path. The value for 100% leakage was obtained by adding 20 μL of a 20% Lubrol PX solution to the cuvette, followed by brief sonication. Measurements were performed in duplicate.
Quasi-elastic light scattering (QUELS)
The size distributions of the LUVs were determined with quasi-elastic light scattering using a Brookhaven Model B1 9000AT digital correlator equipped with a BI-200sm goniometer, version 2.0 and a BI-900AT Digital Correlator System (Brookhaven, Holtsville, NY). The temperature of the sample compartment was maintained at 25°C with fluid circulating from a thermostated bath. LUVs of 50- and 200-nm diameters and of the same lipid composition and concentrations as used in transfer assays were mixed in the cuvette. The scattering from the sample was measured at 90° over a period of a few minutes. Size distribution was calculated with a nonnegatively constrained least-squares algorithm, with software provided by the instrument manufacturer.
RESULTS
BODIPY-FL-C5-HPC lipid mixing assay with uMtCK
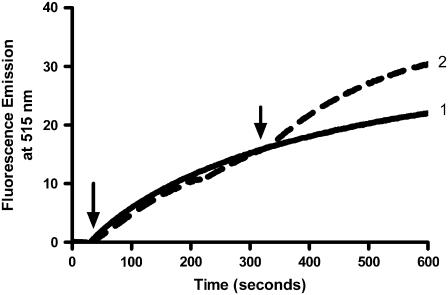
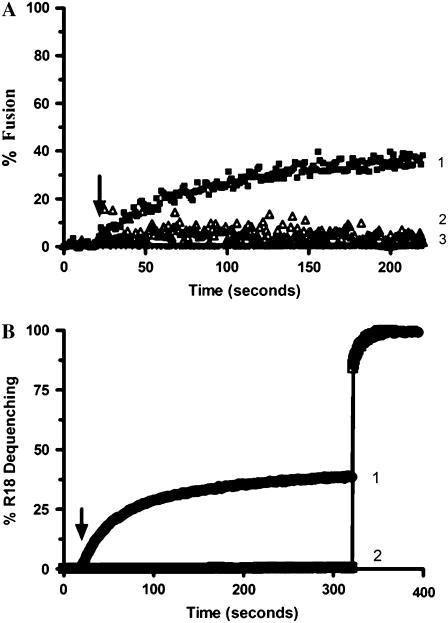
The acyl chain-labeled zwitterionic probe BODIPY-FL-C5-HPC (Fig. 1 A) was incorporated into the donor vesicles together with POPE and CL, all three lipids being at equimolar amounts. A similar assay based on the relief of self-quenching was used previously by Esposti (14) to study interbilayer lipid transfer promoted by the pro-apoptotic protein, tBid. Application of this assay to our particular set of conditions resulted in a high spontaneous rate of lipid transfer in the presence of acceptor vesicles (Fig. 2). Nevertheless, a significant additional increase in the rate of lipid dilution was clearly observed after the addition of octameric uMtCK, a basic protein of the mitochondrial intermembrane space known to crosslink membranes. A control in the absence of acceptor vesicles showed that there was no direct effect of uMtCK on the fluorescence of the probe (not shown).
FIGURE 2.
Phospholipid transfer assay using BODIPY-FL-C5-HPC and uMtCK. The assay mixture contained 300 nM donor vesicles composed of BODIPY-FL-C5-HPC/POPE/CL in 10 mM HEPES buffer, 1 mM mercaptoethanol, pH 7.0. (1) Addition of 4 μM acceptor vesicles of POPC/POPE/CL (1:1:1) at 30 s (arrow) shows spontaneous lipid transfer; (2) Supplementation with additional 300 nM octameric uMtCK at 300 s (arrow) further increases the rate of lipid transfer.
R18 lipid mixing assay with uMtCK
Dilution of R18, a cationic amphiphile (Fig. 1 B), to relieve self-quenching can be a result of transfer between membranes (54), although for many assay conditions the exchange process is small (55,56). In the absence of membrane fusion R18 has already been used in some systems to assay lipid exchange processes (57,58). We have found that in our assay system R18 did not give a spontaneous rate of lipid transfer in the absence of protein. Addition of octameric uMtCK caused a marked increase in the rate of R18 lipid transfer. Without protein, no lipid transfer was seen. In this assay, the uMtCK is added into the assay cuvette from a protein stock solution of a concentration above 4 mg/mL. The protein is therefore in its octameric form, but once diluted into the cuvette it would dissociate into dimers over time. However, this is a slow process exceeding the time required for lipid transfer (23). As a control, we first allowed uMtCK to dissociate into dimers by dilution and incubation overnight in the refrigerator before being mixed with the liposomes. When uMtCK is added in the predissociated dimeric form it is almost completely inactive in promoting transfer of R18 lipid (not shown), in contrast to the case in which the protein is added in the octameric form from concentrated solution. It is also known that uMtCK binds anionic lipids more strongly at low ionic strengths (25). However, we observed that at very low ionic strengths the rate of transfer is very small. This low transfer of R18 lipid at low ionic strength may be the result of electrostatic repulsion keeping the two liposomes apart as well as to changes in surface pH or changes in the nature of the binding of uMtCK. This behavior is similar to that reported previously with myelin basic protein (59). Consequently, most of the assays were carried out at 140 mM NaCl. Addition of 1 mM Mg2+ subsequent to the addition of uMtCK caused a more rapid rate of R18 transfer.
Role of divalent cations in the exchange process
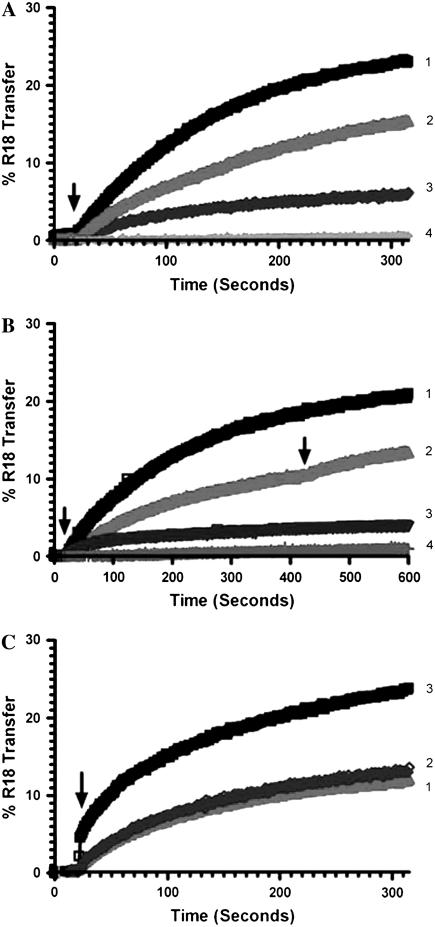
It has been suggested that Ca2+ plays an important role in localizing uMtCK to mitochondrial contact sites (60). We observed that millimolar concentrations of Ca2+ markedly stimulated the transfer of R18 lipid in the presence of uMtCK (data not presented). Mg2+ can also bind to anionic lipids and reduce the concentration of Ca2+ required for certain processes. Unlike Ca2+, physiological concentrations of Mg2+ are in the millimolar range and not markedly regulated. We therefore investigated if much lower concentrations of Ca2+ in the presence of Mg2+, which is closer to physiological situations, could still have some stimulatory effect in liposomal assays. A dose response curve showed that the extent of R18 lipid exchange is strongly dependent on the amount of octameric uMtCK added (Fig. 3 A). An analysis of these curves gives a reasonable fit as a first-order rate process with half-times of 90, 120, and 80 s for 200, 100, and 50 nM uMtCK, respectively. The calculated extent of maximal transfer is 32, 23, and 8% for the three concentrations of uMtCK used. When utilizing a HEPES buffer with EDTA, adjusted to contain 200 nM free Ca2+, synergistic effects in the promotion of lipid transfer were observed upon the addition of 1 mM Mg2+ (Fig. 3 B, curves 1 and 2), and thus this buffer composition was used throughout this study.
FIGURE 3.
Transfer of R18 lipid promoted by uMtCK or NDPK-D at 37°C. (A) Dose response curve for uMtCK added at 40 s (arrow) together with 1 mM Mg2+ to a HEPES-Ca2+ buffer pH 6.9. Labeled donor vesicles with a 50-nm diameter, of composition POPC/POPE/CL (1:1:1) at 300 nM concentration, were mixed in cuvette with 3 μM of 200 nm diameter acceptor LUVs, of the same lipid composition. (1) 200 nM uMtCK; (2) 100 nM uMtCK; (3) 50 nM uMtCK; (4) No protein added. (B) Conditions for transfer promoted by uMtCK in HEPES-Ca2+ buffer, pH 6.9. (1) 1 mM Mg2+ added at 40 s together with 100 nM uMtCK; (2) Protein added at 40 s and 1 mM Mg2+ added at 420 s (see arrow); (3) Removal of CL in donor vesicles. Donor vesicle composition here is POPC/POPE (1:1). Mg2+ added at 40 s together with uMtCK; (4) Control without protein, but with donor and acceptor vesicles. Mg2+ added at 40 s. (C). Transfer of R18 lipid promoted by 100 nM NDPK-D, at 37°C. (1) Protein added at 40 s (arrow), no Mg2+ added to a HEPES-Ca2+ buffer (containing 0.14 M NaCl), pH 6.9. (2) NDPK-D added at 40 s with 1 mM Mg2+, in 10 mM HEPES, 0.14 M NaCl, pH 6.9. (3) NDPK-D added at 40 s together with 1 mM Mg2+, in a HEPES-Ca2+ buffer, pH 6.9.
R18 lipid mixing assay with NDPK-D
Another oligomeric basic kinase of the mitochondrial intermembranous space is hexameric NDPK-D, which can also bridge the two mitochondrial membranes. Like uMtCK, NDPK-D also promotes the exchange of R18 lipid (Fig. 3 C), which is enhanced by a HEPES-Ca2+ buffer in the presence of 1 mM Mg2+ (Fig. 3 C, curve 3). The hexameric form has a high stability, and low concentrations of the protein are sufficient for promoting the transfer.
Role of CL in lipid transfer process
Cardiolipin is a marker lipid for mitochondria. This lipid, present predominantly in the inner membrane, is also enriched in contact sites and its biophysical properties in model membranes are modulated by divalent cations. We therefore determined if CL affected the transfer of lipid promoted by uMtCK or by NDPK-D. When we changed the lipid composition of the liposomes in the R18 lipid transfer assay from POPC/POPE/CL (1:1:1) to POPC/POPE/CL (41.5:41.5:17.5), lipid transfer was essentially completely blocked, even in the presence of 200 nM Ca2+ and 1 mM Mg2+ (not shown). The lower CL concentration of 17.5% is just above the 16% required for the protein to bind to membranes (29), while the higher concentration corresponds to the estimated amount of CL in contact sites (46). We also removed CL from the donor vesicles, making them with the lipid composition POPC/POPE (1:1), while retaining the composition of the acceptor vesicles as POPC/POPE/CL (1:1:1) (Fig. 3 B, curve 3). Again, lipid transfer was completely blocked. Finally, we replaced CL with POPG to make both donor and acceptor vesicles of POPC/POPE/POPG (1:1:1); again lipid transfer was blocked (not shown).
Membrane fusion and aqueous content leakage with uMtCK and NDPK-D
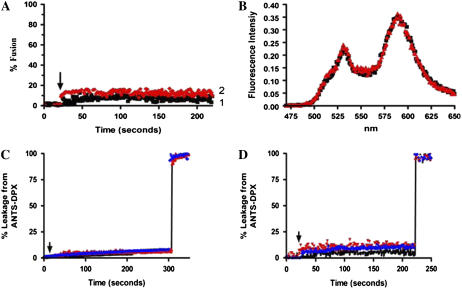
As a control for membrane fusion, we used a lipid dilution assay based on FRET between Rh-PE and NBD-PE. There was no lipid mixing measured by this assay with either uMtCK or with NDPK-D in the presence or absence of 1 mM Mg2+ and 200 nM Ca2+ (Fig. 4, A and B). This assay can also detect lipid transfer, but it does not occur with these headgroup-labeled lipids.
FIGURE 4.
(A) No fusion observed with Rh-PE/NBD-PE FRET assay. (1) 100 nM uMtCK. (2) 100 nM NDPK-D added with 1 mM Mg2+ at 25 s (arrow) to donor and acceptor vesicles of composition DOPC/DOPE/CL (1:1:1). LUVs as described in Experimental Procedures. (B) Fluorescence emission scans for the assay shown in panel A, before (black curve) and after (red curve) addition of 100 nM NDPK-D. (C). Leakage of ANTS/DPX assay carried out in 10 mM HEPES, 0.14 M NaCl, pH 6.9, as described in Experimental Procedures. (Black curve) LUVs alone. (Red curve) 120 nM uMtCK added at 20 s (arrow). (Blue curve) 240 nM uMtCK added at 20 s. The value for 100% release was obtained by adding 20 μL of a 20% Lubrol PX solution to the cuvette at 300 s. (D). Leakage assay carried out in HEPES-Ca2+ buffer. 1 mM Mg2+ added at 40 s (arrow). (Black curve) LUVs alone. (Red curve) 100 nM uMtCK added at 25 s. (Blue curve) 100 nM NDPK-D added at 25 s. For 100% release, 20 μL of 20% Lubrol PX was added at 220 s.
We also determined the ability of uMtCK or NDPK-D to induce leakage of aqueous contents with LUV of POPC/POPE/CL (1:1:1) in the presence of Ca2+ and Mg2+. No leakage of aqueous contents was detected using the ANTS/DPX dequenching assay with either uMtCK (Fig. 4 C) or with NDPK-D (Fig. 4 D). This finding provides further evidence against R18 lipid dilution being the result of membrane fusion, since protein or peptide-promoted membrane fusion in model liposome systems is always accompanied by some leakage of aqueous contents. In addition, this finding also eliminates the possibility that probe transfer was a result of bilayer destabilization and vesicle collapse.
Comparison with other basic proteins
Comparison of uMtCK was made with another mitochondrial protein that associates with anionic lipids, that is cytochrome c, as well as the basic polypeptide, poly-L-lysine. In contrast to both uMtCK and NDPK-D (Fig. 4, A and B), poly-L-lysine causes the promotion of membrane fusion as assessed by the fluorescence resonance energy transfer (FRET) assay (Fig. 5 A, curve 1), thus eliminating the possibility that association of the positive charge with a negative probe prevented lipid exchange. Cytochrome c was unable to produce fusion of vesicles (Fig. 5 A, curve 2). It had also been shown that a synthetic peptide corresponding to the presequence of cytochrome c oxidase subunit IV from yeast inserted itself into lipid monolayers and strongly promoted the formation of close contacts with large unilamellar lipid vesicles present in the subphase, a property which was also specific for cardiolipin, resulting in the flow of lipids from the vesicles to the monolayer (61). Although cytochrome c binds to anionic lipids, it does not cross-link vesicles and it does not promote the exchange of R18 lipid, either in the presence (Fig. 5 B, curve 2) or absence (not shown) of divalent cations. In comparison, the highly basic polypeptide, poly-L-lysine, that is known to aggregate and destabilize liposomes (62), does promote transfer of R18 lipid (Fig. 5 B, curve 1).
FIGURE 5.
(A) Rh-PE/NBD-PE FRET assay. (1) 100 nM poly-L-lysine; (2) 100 nM cytochrome c; (3) control without protein and with 1 mM Mg2+ added at 25 s (arrow). Vesicles as described in Experimental Procedures. (B) R18 lipid transfer assay. (1) 100 nM poly-L-lysine; (2) 100 nM cytochrome c. LUVs as described in Fig. 3 A. The assay was carried out in a HEPES-Ca2+ buffer at pH 6.9 and 37°C; proteins were added together with 1 mM Mg2+ at 25 s (arrow). No effect of Mg2+ was detected when the assay was repeated in the absence of Mg2+. At 320 s, 20 μL of 20% Lubrol PX was added.
Specificity of lipids that can be exchanged
Lipid dilution assays were also carried out with several lipids that were labeled with pyrene on the acyl chain. This assay monitors the transfer of diacyl phospholipids labeled on the acyl chain and it can be readily applied to several commercially available pyrene-labeled phospholipids. It is based on the loss of intermolecular excimer emission because of lipid dilution. Excimers form because of collision between two pyrene moieties, one in the excited state and the other in the ground state. Fluorescence emission from an excimer occurs at a different wavelength than from the monomer. In our assay, the pyrene-labeled lipid was incorporated into the lipid film and hence was on both monolayers of the extruded vesicle. This is different from the R18 assay, in which the more soluble probe was introduced to preformed liposomes and hence was only in the outer monolayer. A standard curve comparing the Ie/Im ratio observed for a series of liposomes containing different mol fractions of pyrene-labeled lipid was established, taking into account the fact that the label on the inner monolayer could not undergo exchange without crossing the bilayer. In addition, protein effects on the Ie/Im ratio were controlled by measuring changes to donor vesicles in the absence of acceptor vesicles.
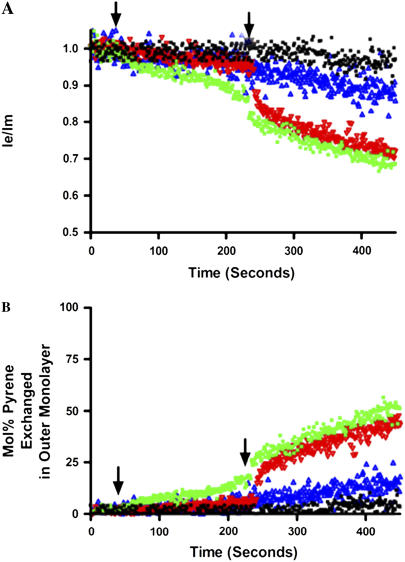
The same conditions as for the R18 assay (i.e., POPC/POPE/CL vesicles), except for the change in probe, were utilized. Lipid exchange promoted by uMtCK was very dependent on the presence of divalent cations and showed specificity for PE and PG over PC (Fig. 6). There was little effect of the uMtCK on the probe fluorescence in the absence of acceptor vesicles. With NDPK-D, there was less transfer of pyrene lipid and no specificity in the lipid transferred (not shown). The effect of NDPK-D on probe fluorescence was not due to direct effects of the protein on the probe distribution in the membrane or its mobility, since controls lacking acceptor vesicles did not show any decrease in Ie/Im over time. In addition, fluorescence emission scans before and after addition of NDPK-D revealed the characteristic decrease in excimer and increase in monomer emission, which is indicative of dilution of the pyrene-labeled lipid, rather than a consequence of protein binding. No transfer of pyrene lipids was observed when CL was removed from the donor and acceptor vesicles by using POPC/POPE/POPG (1:1:1) with either uMtCK or NDPK-D (data not shown).
FIGURE 6.
(A) Dilution of pyrene-labeled lipids with uMtCK. LUVs as described in Fig. 3 A but containing pyrene labels. The assay was carried out in HEPES-Ca2+ buffer at pH 6.9 and 37°C. 100 nM uMtCK was added at 30 s (arrow). Mg2+ was added at 240 s (arrow). (Black curve) Control with donor vesicles containing Pyrene PG but without acceptor vesicles (controls for other pyrene lipids without acceptor were similar and are not shown). (Blue curve) Ie/Im in the presence of 10 mol % Pyrene PC. (Red curve) Ie/Im in the presence of 10 mol % Pyrene PE. (Green curve) Ie/Im in the presence of 10 mol % Pyrene PG. (B) Percentage transfer in the outer monolayer calculated from a standard curve constructed with different mol % of pyrene lipids.
Change in vesicle size as measured by QUELS
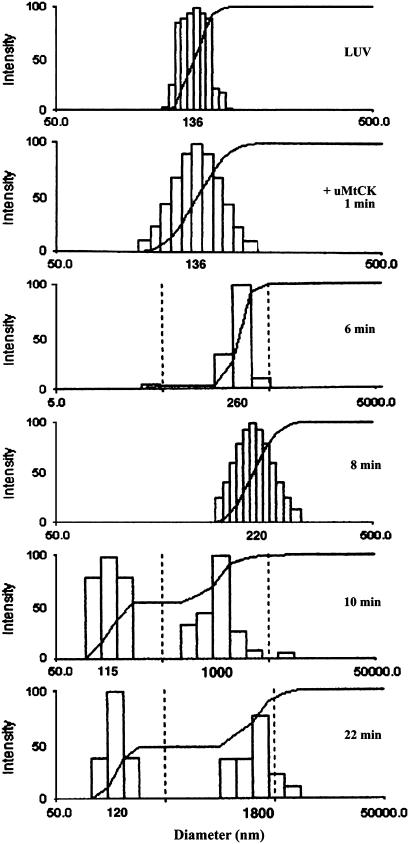
QUELS is a phenomenon that is dependent on the rate of diffusion of the particles that scatter light. It is dependent on the size and state of aggregation of vesicles. We observe initially a slower, gradual increase in the size of the liposome aggregates over the first 6 min. Thereafter, a split into two vesicle populations becomes apparent, with the reappearance of small vesicles, similar in size to the original liposomes, and a fraction of large vesicles even further increasing in size (Fig. 7). This time-course is the result of initial protein-induced bridging of vesicles, followed by a partial dissociation of octameric uMtCK into the dimeric form that no longer can bridge the liposomes (28).
FIGURE 7.
QUELS analysis of liposome size distribution after addition of 120 nM uMtCK to donor and acceptor LUVs of POPC/POPE/CL (1:1:1) in HEPES-Ca2+ buffer pH 6.9, together with 1 mM Mg2+. (1) LUVs. (2) One min after protein addition. (3) Six min after protein addition. (4) Eight min after protein addition. (5) 10 min after protein addition. (6) 22 min after protein addition. Note that the scales on the abscissa are not all the same, nor are they aligned.
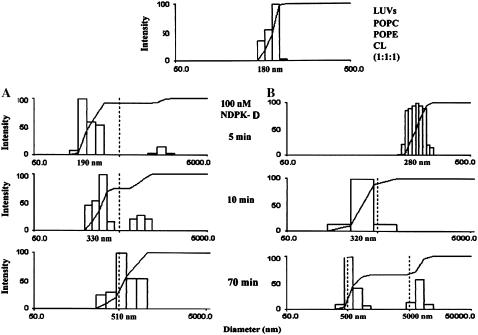
With NDPK-D (Fig. 8), there is no reversal in the size of the liposomes. Aggregation to particles of ∼300-nm diameter (vesicle dimers) occurs rapidly and does not increase greatly with time, although at longer times of 70 min the major population increases in size to 500–600 nm and there is a small fraction of very large aggregates. There is little difference in the state of aggregation in the presence (Fig. 8 A) or absence (Fig. 8 B) of divalent cations.
FIGURE 8.
QUELS analysis of liposome size distribution: after addition of 100 nM NDPK-D to donor and acceptor LUVs of POPC/POPE/CL (1:1:1). (Top curve) The LUVs were made in HEPES-Ca2+ pH 6.9 and showed no change in size in the presence or absence of added Mg2+. No protein added. (A) In HEPES-Ca2+ pH 6.9 buffer. (B) In 10 mM HEPES, 0.1 mM EDTA, 0.14 M NaCl, pH 6.9. (1) Five min after protein addition. (2) 10 min after protein addition. (3) 70 min after protein addition. Note that the scales on the abscissa are not all the same, nor are they aligned.
DISCUSSION
Both protein kinases, uMtCK and NDPK-D, exhibited the ability to accelerate the dilution of labeled lipids from donor to acceptor liposomes. We have demonstrated this phenomenon by several alternative methods including the transfer of BODIPY-FL-C5-HPC, of R18 lipid and of pyrene-labeled lipids. All of these assays showed similar effects, indicating that the phenomenon is not sensitive to the nature of the fluorescent amphiphile used. These assays by themselves do not reveal the mechanism by which transfer of lipid occurs. It could be a consequence of fusion, hemifusion, collapse of vesicle morphology, or lipid transfer. However, additional assays (membrane fusion, leakage, and QUELS) indicate that the lipid probes are diluted into acceptor vesicles by lipid transfer. The only proteins known to enhance this process are specific lipid transfer proteins, except for t-Bid, which is suggested to resemble a lipid transfer protein (15). In addition, the process is specific for the oligomeric forms of uMtCK and NDPK-D that can bridge membranes and does not occur with dimeric uMtCK or with the basic mitochondrial protein, cytochrome c.
Notably, lipid dilution via lipid transfer is supported by the absence of membrane fusion in the NBD-PE/Rh-PE FRET assay. If lipid dilution would occur via membrane fusion or hemifusion, it should just depend on protein-induced formation of new morphologies, and not on properties of the applied probe as in our assays. A large degree of membrane fusion is also excluded by the QUELS assays. Small vesicles comparable in size to the original donor vesicles reappear from larger aggregates after dissociation of octameric uMtCK into dimers in dilute solution (Fig. 7). This would not occur if fusion had taken place, although it does not exclude the possibility that there is some percentage of fusion.
Lipid transfer between the two mitochondrial membranes is known in cells. Cardiolipin is synthesized on the mitochondrial inner membrane (63) but moves to the surface of mitochondria early in apoptosis (17). This finally leads to transfer of cardiolipin to other membranes in the cell (64). Such transfer likely happens at contact sites where cardiolipin is enriched (46) and where part of uMtCK and NDPK-D are located (8). The bridging of the two membranes by these proteins can be explained by their common feature of exposing identical top and bottom faces, which specifically bind to lipid membranes, preferentially to those containing cardiolipin; this brings these membranes into close proximity where they can participate in lipid transfer. The biological importance of this lipid transfer is shown by the fact that a promyelocytic leukemia cell line that exhibits somewhat less exposure of cardiolipin is resistant to apoptosis (16). Hence, there is a clear association between apoptosis and the exposure of cardiolipin on the surface of mitochondria. In addition, tumor cells that are poorly differentiated have fewer contact sites (65). Such cells are also more resistant to apoptosis (66), again suggesting a relationship between apoptosis and the presence of contact sites where the proteins we are studying are known to be located.
The conditions used in these liposomal assays mimic many of the properties one would find in mitochondria. The pH and ionic strength of these assays is similar to that found in mitochondria. It is known that the pH in the intermembrane space in mitochondria is 0.2–0.6 units below that in the cytosol and thus similar to the pH 6.9 that we used, as is the ionic strength, which is 100–150 mM both in our assays and in mitochondria (67). The lipid composition of our liposomes are similar to that found with isolated mitochondrial contact sites (46). Liposomes can be bridged by addition of MtCK or NDPK-D similar to the process leading to contact sites in mitochondria. The mechanism of lipid transfer may be analogous to the one demonstrated with myelin basic protein (59). There is some specificity in the lipid transfer carried out by uMtCK in that pyrene-PC is not transferred. This is in accord with the fact that this enzyme does not bind to PC alone (29). As compared to the oligomeric kinases, monomeric cytochrome c also binds to CL-rich membranes and is enriched in contact sites, but it does not promote lipid transfer because it cannot bridge two membranes.
Although an anionic membrane is required to bind to the cationic residues near the C-terminus of uMtCK, the requirement for CL is not solely on the basis of its charge. CL carries approximately one negative charge at neutral pH (68,69), yet another anionic lipid, phosphatidylglycerol, cannot replace CL. Furthermore, CL binding of uMtCK is necessary but not sufficient for lipid transfer. Although 16% CL in the membrane is sufficient for uMtCK to bind (29), the fraction of CL in the membrane has to be >16% to promote lipid transfer in liposomes.
A marked difference between CL and PG is the greater tendency of CL to form nonlamellar phases. It is known that when CL loses its negative charge it does not form stable bilayers. However, in this study, no hexagonal phases are formed, as indicated by the lack of liposome contents leakage as well as from microscopy studies using GUVs that show that the vesicle morphology is retained in the presence of protein. In addition, it has been shown that bilayers of PG are less readily dehydrated by proteins than those of CL (70). The binding of MtCK to the membrane changes the interfacial properties, as measured by the fluorescent probe Laurdan (30), in a manner consistent with the protein causing dehydration of the membrane surface. Membrane dehydration is often accompanied by rigidification of the membrane (71). It is possible that binding of these mitochondrial proteins to CL-rich membranes induces loss of interfacial water and allows more facile lipid transfer if there are two juxtaposed bilayers. Such a mechanism would be very different from that of lipid transfer proteins that bind to lipids.
The structures of uMtCK and NDPK-D do not contain features such as hydrophobic segments or sequence homology with lipid transfer proteins that would necessarily indicate that they function as lipid transfer proteins. However, there is a stretch of some hydrophobic residues in the C-terminal phospholipid-binding domain of MtCK, which could perturb lipid organization or partially penetrate the bilayer (5,20). Furthermore, except for their oligomeric structures and their capability to bind to and to crosslink membranes containing anionic phospholipids, these two proteins are unrelated, yet both can function to promote lipid transfer. uMtCK and NDPK-D have in common that they associate into highly symmetrical oligomeric entities with identical top and bottom faces that are able to interact with and cross-link membranes. It is this structural arrangement, as well as the effects of these proteins on membrane properties that result in them being able to promote the transfer of lipids. Nevertheless, these protein oligomers do not allow the bilayers to come into direct contact and it is not clear what path the lipid takes to traverse the length of the protein oligomer. This is an issue under current investigation.
Finally, there is potential for a regulation of the lipid transfer rate between mitochondrial membranes by these kinases. The dimeric form of uMtCK is not active in lipid transfer and cannot bridge two membranes. Thus, any change in the octamer/dimer ratio would affect lipid transfer. With NDPK-D, the level of expression varies considerably among different cell types and may provide another mechanism of regulation. The interbilayer movement of lipid in mitochondria likely has several functional consequences, among which is apoptotic signaling. For example, exposure of CL at the mitochondrial surface can promote apoptosis (18). Furthermore, uMtCK is downregulated in oral squamous cell carcinoma (72) possibly making these cells more resistant to apoptosis.
uMtCK and NDPK-D are also present at contact sites in cells not undergoing apoptosis, yet little cardiolipin is translocated to the mitochondrial surface. A small amount of cardiolipin is possibly associated with proteins at contact sites of the outer membrane, but difficult to detect. However, for such cardiolipin translocation in vivo, one has to consider the net flux of cardiolipin, which is a result of the balance of several processes. There is a mechanism for the rapid inward movement of lipids in mitochondria (73), and a mitochondrial lipid scramblase is also known to promote the redistribution of lipids between mitochondrial membranes (74). Thus, there are multiple pathways for the movement of lipid between mitochondrial membranes and the relative importance of each of these will depend on the expression level of the relevant proteins, as well as on the regulation of their lipid transfer activity, e.g., by factors like mitochondrial calcium. It is interesting in this context that the substrate of MtCK, creatine, significantly protects the mitochondrial permeability transition pore against opening, a trigger of apoptosis induction by mitochondria (see (75)). As already stated above, in apoptosis there must be a change in the balance of rates among all lipid transfer processes to favor net outward movement of cardiolipin. The novel, rather striking function of MtCK (ubiquitous and sarcomeric) and NDPK-D, in being capable of mediating lipid transfer between two membranes has been demonstrated in vitro. Elucidation of its contribution in vivo and its detailed mechanism will be a challenge for future experiment.
Acknowledgments
This work was supported by the Canadian Institutes for Health Research, grant No. MT-7654 and the Swiss National Science Foundation SNF grant No. 3100AO-102075, as well as the Swiss Society for Research on Muscle Diseases, the Germaine de Stael program for Franco-Swiss collaboration, the Institut National de la Santé et de la Recherche Médicale (INSERM), and the France-Canada Research Foundation.
References
- 1.Hackenbrock, C. R. 1968. Chemical and physical fixation of isolated mitochondria in low-energy and high-energy states. Proc. Natl. Acad. Sci. USA. 61:598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Venetie, R., and A. J. Verkleij. 1982. Possible role of non-bilayer lipids in the structure of mitochondria. A freeze-fracture electron microscopy study. Biochim. Biophys. Acta. 692:397–405. [DOI] [PubMed] [Google Scholar]
- 3.Frey, T. G., and C. A. Mannella. 2000. The internal structure of mitochondria. Trends Biochem. Sci. 25:319–324. [DOI] [PubMed] [Google Scholar]
- 4.Reichert, A. S., and W. Neupert. 2002. Contact sites between the outer and inner membrane of mitochondria-role in protein transport. Biochim. Biophys. Acta. 1592:41–49. [DOI] [PubMed] [Google Scholar]
- 5.Schlattner, U., M. Forstner, M. Eder, O. Stachowiak, K. Fritz-Wolf, and T. Wallimann. 1998. Functional aspects of the x-ray structure of mitochondrial creatine kinase: a molecular physiology approach. Mol. Cell. Biochem. 184:125–140. [PubMed] [Google Scholar]
- 6.Crompton, M. 2000. Mitochondrial intermembrane junctional complexes and their role in cell death. J. Physiol. 529:11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knoll, G., and D. Brdiczka. 1983. Changes in freeze-fractured mitochondrial membranes correlated to their energetic state. Dynamic interactions of the boundary membranes. Biochim. Biophys. Acta. 733:102–110. [DOI] [PubMed] [Google Scholar]
- 8.Adams, V., W. Bosch, J. Schlegel, T. Wallimann, and D. Brdiczka. 1989. Further characterization of contact sites from mitochondria of different tissues: topology of peripheral kinases. Biochim. Biophys. Acta. 981:213–225. [DOI] [PubMed] [Google Scholar]
- 9.Schlattner, U., M. Tokarska-Schlattner, and T. Wallimann. 2006. Mitochondrial creatine kinase in human health and disease. Biochim. Biophys. Acta. 1762:164–180. [DOI] [PubMed] [Google Scholar]
- 10.Milon, L., P. Meyer, M. Chiadmi, A. Munier, M. Johansson, A. Karlsson, I. Lascu, J. Capeau, J. Janin, and M. L. Lacombe. 2000. The human nm23–H4 gene product is a mitochondrial nucleoside diphosphate kinase. J. Biol. Chem. 275:14264–14272. [DOI] [PubMed] [Google Scholar]
- 11.Alpy, F., and C. Tomasetto. 2005. Give lipids a START: the StAR-related lipid transfer (START) domain in mammals. J. Cell Sci. 118:2791–2801. [DOI] [PubMed] [Google Scholar]
- 12.Arondel, V., and J. C. Kader. 1990. Lipid transfer in plants. Experientia. 46:579–585. [DOI] [PubMed] [Google Scholar]
- 13.Rueckert, D. G., and K. Schmidt. 1990. Lipid transfer proteins. Chem. Phys. Lipids. 56:1–20. [DOI] [PubMed] [Google Scholar]
- 14.Esposti, M. D., J. T. Erler, J. A. Hickman, and C. Dive. 2001. Bid, a widely expressed proapoptotic protein of the Bcl-2 family, displays lipid transfer activity. Mol. Cell. Biol. 21:7268–7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esposti, M. D. 2002. Sequence and functional similarities between pro-apoptotic Bid and plant lipid transfer proteins. Biochim. Biophys. Acta. 1553:331–340. [DOI] [PubMed] [Google Scholar]
- 16.Garcia Fernandez, M., L. Troiano, L. Moretti, J. Pedrazzi, S. Salvioli, I. Castilla-Cortazar, and A. Cossarizza. 2000. Changes in intramitochondrial cardiolipin distribution in apoptosis-resistant HCW-2 cells, derived from the human promyelocytic leukemia HL-60. FEBS Lett. 478:290–294. [DOI] [PubMed] [Google Scholar]
- 17.Garcia Fernandez, M., L. Troiano, L. Moretti, M. Nasi, M. Pinti, S. Salvioli, J. Dobrucki, and A. Cossarizza. 2002. Early changes in intramitochondrial cardiolipin distribution during apoptosis. Cell Growth Differ. 13:449–455. [PubMed] [Google Scholar]
- 18.Qi, L., N. D. Danielson, Q. Dai, and R. M. Lee. 2003. Capillary electrophoresis of cardiolipin with on-line dye interaction and spectrophotometric detection. Electrophoresis. 24:1680–1686. [DOI] [PubMed] [Google Scholar]
- 19.Crimi, M., A. Astegno, G. Zoccatelli, and M. D. Esposti. 2006. Pro-apoptotic effect of maize lipid transfer protein on mammalian mitochondria. Arch. Biochem. Biophys. 445:65–71. [DOI] [PubMed] [Google Scholar]
- 20.Schlattner, U., and T. Wallimann. 2004. Metabolite channeling: creatine kinase microcompartments. In Encyclopedia of Biological Chemistry, Vol. 2. Elsevier, NY. 646–651.
- 21.Wallimann, T., M. Wyss, D. Brdiczka, K. Nicolay, and H. M. Eppenberger. 1992. Intracellular compartmentation, structure and function of creatine kinase isoenzymes in tissues with high and fluctuating energy demands: the “phosphocreatine circuit” for cellular energy homeostasis. Biochem. J. 281:21–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schlattner, U., and T. Wallimann. 2000. Octamers of mitochondrial creatine kinase isoenzymes differ in stability and membrane binding. J. Biol. Chem. 275:17314–17320. [DOI] [PubMed] [Google Scholar]
- 23.Rojo, M., R. Hovius, R. Demel, T. Wallimann, H. M. Eppenberger, and K. Nicolay. 1991. Interaction of mitochondrial creatine kinase with model membranes. A monolayer study. FEBS Lett. 281:123–129. [DOI] [PubMed] [Google Scholar]
- 24.Vacheron, M. J., E. Clottes, C. Chautard, and C. Vial. 1997. Mitochondrial creatine kinase interaction with phospholipid vesicles. Arch. Biochem. Biophys. 344:316–324. [DOI] [PubMed] [Google Scholar]
- 25.Fritz-Wolf, K., T. Schynder, T. Wallimann, and W. Kabsch. 1996. Structure of mitochondrial creatine kinase. Nature. 381:341–345. [DOI] [PubMed] [Google Scholar]
- 26.Eder, M., K. Fritz-Wolf, W. Kabsch, T. Wallimann, and U. Schlattner. 2000. Crystal structure of human ubiquitous mitochondrial creatine kinase. Proteins. 39:216–225. [DOI] [PubMed] [Google Scholar]
- 27.Schlattner, U., F. Gehring, N. Vernoux, M. Tokarska-Schlattner, D. Neumann, O. Marcillat, C. Vial, and T. Wallimann. 2004. C-terminal lysines determine phospholipid interaction of sarcomeric mitochondrial creatine kinase. J. Biol. Chem. 279:24334–24342. [DOI] [PubMed] [Google Scholar]
- 28.Stachowiak, O., M. Dolder, and T. Wallimann. 1996. Membrane-binding and lipid vesicle cross-linking kinetics of the mitochondrial creatine kinase octamer. Biochemistry. 35:15522–15528. [DOI] [PubMed] [Google Scholar]
- 29.Schlattner, U., and T. Wallimann. 2000. A quantitative approach to membrane binding of human ubiquitous mitochondrial creatine kinase using surface plasmon resonance. J. Bioenerg. Biomembr. 32:123–131. [DOI] [PubMed] [Google Scholar]
- 30.Granjon, T., M. J. Vacheron, C. Vial, and R. Buchet. 2001. Mitochondrial creatine kinase binding to phospholipids decreases fluidity of membranes and promotes new lipid-induced beta structures as monitored by red edge excitation shift, Laurdan fluorescence, and FTIR. Biochemistry. 40:6016–6026. [DOI] [PubMed] [Google Scholar]
- 31.Wegmann, G., R. Huber, E. Zanolla, H. M. Eppenberger, and T. Wallimann. 1991. Differential expression and localization of brain-type and mitochondrial creatine kinase isoenzymes during development of the chicken retina: Mi-CK as a marker for differentiation of photoreceptor cells. Differentiation. 46:77–87. [DOI] [PubMed] [Google Scholar]
- 32.Muller, M., R. Moser, D. Cheneval, and E. Carafoli. 1985. Cardiolipin is the membrane receptor for mitochondrial creatine phosphokinase. J. Biol. Chem. 260:3839–3843. [PubMed] [Google Scholar]
- 33.Speer, O., N. Back, T. Buerklen, D. Brdiczka, A. Koretsky, T. Wallimann, and O. Eriksson. 2005. Octameric mitochondrial creatine kinase induces and stabilizes contact sites between the inner and outer membrane. Biochem. J. 385:445–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muhonen, W. W., and D. O. Lambeth. 1995. The compartmentation of nucleoside diphosphate kinase in mitochondria. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 110:211–223. [DOI] [PubMed] [Google Scholar]
- 35.Lascu, L., A. Giartosio, S. Ransac, and M. Erent. 2000. Quaternary structure of nucleoside diphosphate kinases. J. Bioenerg. Biomembr. 32:227–236. [DOI] [PubMed] [Google Scholar]
- 36.Lacombe, M. L., L. Milon, A. Munier, J. G. Mehus, and D. O. Lambeth. 2000. The human Nm23/nucleoside diphosphate kinases. J. Bioenerg. Biomembr. 32:247–258. [DOI] [PubMed] [Google Scholar]
- 37.Seifert, M., C. Welter, Y. Mehraein, and G. Seitz. 2005. Expression of the nm23 homologues nm23–H4, nm23–H6, and nm23–H7 in human gastric and colon cancer. J. Pathol. 205:623–632. [DOI] [PubMed] [Google Scholar]
- 38.Epand, R. F., J. C. Martinou, S. Montessuit, and R. M. Epand. 2002. Membrane perturbations induced by the apoptotic Bax protein. Biochem. J. 367:849–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gonzalvez, F., F. Pariselli, P. Dupaigne, I. Budihardjo, M. Lutter, B. Antonsson, P. Diolez, S. Manon, J. C. Martinou, M. Goubern, X. Wang, S. Bernard, and P. X. Petit. 2005. tBid interaction with cardiolipin primarily orchestrates mitochondrial dysfunctions and subsequently activates Bax and Bak. Cell Death Differ. 12:614–626. [DOI] [PubMed] [Google Scholar]
- 40.Newmeyer, D. D., and S. Ferguson-Miller. 2003. Mitochondria: releasing power for life and unleashing the machineries of death. Cell. 112:481–490. [DOI] [PubMed] [Google Scholar]
- 41.Zamzami, N., and G. Kroemer. 2003. Apoptosis: mitochondrial membrane permeabilization—the (w)hole story? Curr. Biol. 13:R71–R73. [DOI] [PubMed] [Google Scholar]
- 42.Daum, G. 1985. Lipids of mitochondria. Biochim. Biophys. Acta. 822:1–42. [DOI] [PubMed] [Google Scholar]
- 43.Epand, R. F., J. C. Martinou, S. Montessuit, and R. M. Epand. 2003. Transbilayer lipid diffusion promoted by Bax: implications for apoptosis. Biochemistry. 42:14576–14582. [DOI] [PubMed] [Google Scholar]
- 44.Terrones, O., B. Antonsson, H. Yamaguchi, H. G. Wang, J. Liu, R. M. Lee, A. Herrmann, and G. Basanez. 2004. Lipidic pore formation by the concerted action of proapoptotic BAX and tBID. J. Biol. Chem. 279:30081–30091. [DOI] [PubMed] [Google Scholar]
- 45.Rostovtseva, T. K., N. Kazemi, and S. M. Bezrukov. 2006. The role of cardiolipin in VDAC channel regulation. Biophys J. 90:333a.1609. (Abstr.)
- 46.Ardail, D., J. P. Privat, M. Egret-Charlier, C. Levrat, F. Lerme, and P. Louisot. 1990. Mitochondrial contact sites. Lipid composition and dynamics. J. Biol. Chem. 265:18797–18802. [PubMed] [Google Scholar]
- 47.Schlame, M., M. Ren, Y. Xu, M. L. Greenberg, and I. Haller. 2005. Molecular symmetry in mitochondrial cardiolipins. Chem. Phys. Lipids. 138:38–49. [DOI] [PubMed] [Google Scholar]
- 48.Ames, B. N. 1966. Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 8:115–118. [Google Scholar]
- 49.Hoekstra, D., and K. Klappe. 1986. Use of a fluorescence assay to monitor the kinetics of fusion between erythrocyte ghosts, as induced by Sendai virus. Biosci. Rep. 6:953–960. [DOI] [PubMed] [Google Scholar]
- 50.Jones, J. D., and T. E. Thompson. 1989. Spontaneous phosphatidylcholine transfer by collision between vesicles at high lipid concentration. Biochemistry. 28:129–134. [DOI] [PubMed] [Google Scholar]
- 51.Lalanne, F., and G. Ponsin. 2000. Mechanism of the phospholipid transfer protein-mediated transfer of phospholipids from model lipid vesicles to high density lipoproteins. Biochim. Biophys. Acta. 1487:82–91. [DOI] [PubMed] [Google Scholar]
- 52.Struck, D. K., D. Hoekstra, and R. E. Pagano. 1981. Use of resonance energy transfer to monitor membrane fusion. Biochemistry. 20:4093–4099. [DOI] [PubMed] [Google Scholar]
- 53.Ellens, H., J. Bentz, and F. C. Szoka. 1985. H+- and Ca2+-induced fusion and destabilization of liposomes. Biochemistry. 24:3099–3106. [DOI] [PubMed] [Google Scholar]
- 54.Ohki, S., T. D. Flanagan, and D. Hoekstra. 1998. Probe transfer with and without membrane fusion in a fluorescence fusion assay. Biochemistry. 37:7496–7503. [DOI] [PubMed] [Google Scholar]
- 55.Arbuzova, A., T. Korte, P. Muller, and A. Herrmann. 1994. On the validity of lipid dequenching assays for estimating virus fusion kinetics. Biochim. Biophys. Acta. 1190:360–366. [DOI] [PubMed] [Google Scholar]
- 56.Nunes-Correia, I., A. Eulálio, S. Nir, N. Düzgünes, J. Ramalho-Santos, and M. C. P. De Lima. 2002. Fluorescent probes for monitoring virus fusion kinetics: comparative evaluation of reliability. Biochim. Biophys. Acta BioMembr. 1561:65–75. [DOI] [PubMed] [Google Scholar]
- 57.Arrastua, L., E. San Sebastian, A. F. Quincoces, C. Antony, and U. Ugalde. 2003. In vitro fusion between Saccharomyces cerevisiae secretory vesicles and cytoplasmic-side-out plasma membrane vesicles. Biochem. J. 370:641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuwana, T., B. M. Mullock, and J. P. Luzio. 1995. Identification of a lysosomal protein causing lipid transfer, using a fluorescence assay designed to monitor membrane fusion between rat liver endosomes and lysosomes. Biochem. J. 308:937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cajal, Y., J. M. Boggs, and M. K. Jain. 1997. Salt-triggered intermembrane exchange of phospholipids and hemifusion by myelin basic protein. Biochemistry. 36:2566–2576. [DOI] [PubMed] [Google Scholar]
- 60.Bakker, A., I. Bernaert, M. De Bie, T. Ravingerova, A. Ziegelhoffer, H. Van Belle, and W. Jacob. 1994. The effect of calcium on mitochondrial contact sites: a study on isolated rat hearts. Biochim. Biophys. Acta. 1224:583–588. [DOI] [PubMed] [Google Scholar]
- 61.Torok, Z., R. A. Demel, J. M. Leenhouts, and B. de Kruijff. 1994. Presequence-mediated intermembrane contact formation and lipid flow. A model membrane study. Biochemistry. 33:5589–5594. [DOI] [PubMed] [Google Scholar]
- 62.Epand, R. M., and W. Lim. 1995. Mechanism of liposome destabilization by polycationic amino acids. Biosci. Rep. 15:151–160. [DOI] [PubMed] [Google Scholar]
- 63.Schlame, M., D. Rua, and M. L. Greenberg. 2000. The biosynthesis and functional role of cardiolipin. Prog. Lipid Res. 39:257–288. [DOI] [PubMed] [Google Scholar]
- 64.Sorice, M., A. Circella, I. M. Cristea, T. Garofalo, L. Di Renzo, C. Alessandri, G. Valesini, and M. Degli Esposti. 2004. Cardiolipin and its metabolites move from mitochondria to other cellular membranes during death receptor-mediated apoptosis. Cell Death Differ. 11:1133–1145. [DOI] [PubMed] [Google Scholar]
- 65.Denis-Pouxviel, C., I. Riesinger, C. Buhler, D. Brdiczka, and J. C. Murat. 1987. Regulation of mitochondrial hexokinase in cultured HT 29 human cancer cells. An ultrastructural and biochemical study. Biochim. Biophys. Acta. 902:335–348. [DOI] [PubMed] [Google Scholar]
- 66.Pastorino, J. G., and J. B. Hoek. 2003. Hexokinase II: the integration of energy metabolism and control of apoptosis. Curr. Med. Chem. 10:1535–1551. [DOI] [PubMed] [Google Scholar]
- 67.Cortese, J. D., A. L. Voglino, and C. R. Hackenbrock. 1992. The ionic strength of the intermembrane space of intact mitochondria is not affected by the pH or volume of the intermembrane space. Biochim. Biophys. Acta. 1100:189–197. [DOI] [PubMed] [Google Scholar]
- 68.Haines, T. H., and N. A. Dencher. 2002. Cardiolipin: a proton trap for oxidative phosphorylation. FEBS Lett. 528:35–39. [DOI] [PubMed] [Google Scholar]
- 69.Nichols-Smith, S., and T. Kuhl. 2005. Electrostatic interactions between model mitochondrial membranes. Colloids Surf. B Biointerfaces. 41:121–127. [DOI] [PubMed] [Google Scholar]
- 70.Choi, S., and J. M. Swanson. 1995. Interaction of cytochrome c with cardiolipin: an infrared spectroscopic study. Biophys. Chem. 54:271–278. [DOI] [PubMed] [Google Scholar]
- 71.Schneider, A. S., C. R. Middaugh, and M. D. Oldewurtel. 1979. Role of bound water in biological membrane structure: fluorescence and infrared studies. J. Supramol. Struct. 10:265–275. [DOI] [PubMed] [Google Scholar]
- 72.Onda, T., K. Uzawa, Y. Endo, H. Bukawa, H. Yokoe, T. Shibahara, and H. Tanzawa. 2006. Ubiquitous mitochondrial creatine kinase downregulated in oral squamous cell carcinoma. Br. J. Cancer. 94:698–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gallet, P. F., A. Zachowski, R. Julien, P. Fellmann, P. F. Devaux, and A. Maftah. 1999. Transbilayer movement and distribution of spin-labelled phospholipids in the inner mitochondrial membrane. Biochim. Biophys. Acta. 1418:61–70. [DOI] [PubMed] [Google Scholar]
- 74.Liu, J., Q. Dai, J. Chen, D. Durrant, A. Freeman, T. Liu, D. Grossman, and R. M. Lee. 2003. Phospholipid scramblase 3 controls mitochondrial structure, function, and apoptotic response. Mol. Cancer Res. 1:892–902. [PubMed] [Google Scholar]
- 75.Dolder, M., B. Walzel, O. Speer, U. Schlattner, and T. Wallimann. 2003. Inhibition of the mitochondrial permeability transition by creatine kinase substrates. Requirement for microcompartmentation. J. Biol. Chem. 278:17760–17766. [DOI] [PubMed] [Google Scholar]