Abstract
Neutrophil-activating protein (NapA) has been well documented to play roles in human neutrophil recruitment and in stimulating host cell production of reactive oxygen intermediates (ROI). A separate role for NapA in combating oxidative stress within H. pylori was implied by studies of various H. pylori mutant strains. Here, physiological analysis of a napA strain was the approach used to assess the iron-sequestering and stress resistance roles of NapA, its role in preventing oxidative DNA damage, and its importance to mouse colonization. The napA strain was more sensitive to oxidative stress reagents and to oxygen, and it contained fourfold more intracellular free iron and more damaged DNA than the parent strain. Pure, iron-loaded NapA bound to DNA, but native NapA did not, presumably linking iron levels sensed by NapA to DNA damage protection. Despite its in vitro phenotype of sensitivity to oxidative stress, the napA strain showed normal (like that of the wild type) mouse colonization efficiency in the conventional in vivo assay. By use of a modified mouse inoculation protocol whereby nonviable H. pylori is first inoculated into mice, followed by (live) bacterial strain administration, an in vivo role for NapA in colonization efficiency could be demonstrated. NapA is the critical component responsible for inducing host-mediated ROI production, thus inhibiting colonization by the napA strain. An animal colonization experiment with a mixed-strain infection protocol further demonstrated that the napA strain has significantly decreased ability to survive when competing with the wild type. H. pylori NapA has unique and separate roles in gastric pathogenesis.
Helicobacter pylori, a gram-negative microaerophilic bacterium, infects the stomachs of more than 50% of the human population. Infection by H. pylori causes active chronic gastritis in many subjects and in some individuals leads to the development of ulcer diseases or even gastric adenocarcinoma (14). H. pylori infection of the gastric mucosa induces the appearance of inflammatory infiltrates, consisting mainly of neutrophils and monocytes, which contribute to H. pylori-induced gastritis (4, 16). A correlation exists between severity of the disease and the neutrophil-activating capacity of H. pylori isolates (10, 16, 32).
One of the bacterial factors shown to mediate neutrophil activation is the H. pylori neutrophil-activating protein encoded by the gene napA (HP0243 in the H. pylori genome). H. pylori expresses a large amount of NapA, especially in vivo (6), so that NapA is a major antigen in the human immune response to H. pylori (2, 33). NapA is a cytosolic protein expressed by virtually all H. pylori isolates, and it is oftentimes detected in the culture medium after bacterial growth (7, 36). NapA has the capacity to bind to carbohydrates, which may mediate H. pylori binding to both host cells and the stomach mucus (25, 35). NapA is chemotactic for human neutrophils and monocytes, and it plays a major role in recruiting these cells to the site of infection (7, 15). NapA is a powerful stimulant of reactive oxygen intermediate (ROI) production by human neutrophils and monocytes (15, 33). NapA-induced ROI production involves a cascade of intracellular activation events, including an increase in the cytosolic calcium ion concentration and phosphorylation of proteins, leading to assembly of the superoxide-forming NADPH oxidase on the neutrophil plasma membrane (24, 27, 33).
The primary sequence and structure of NapA are similar to those of the proteins in the Dps family which have functions in iron binding and DNA protection (17). The 17-kDa NapA protein forms a dodecameric complex with a central cavity that sequesters large amounts of iron (nearly 500 atoms of iron per dodecamer) (37, 48). Thus, NapA could be involved in iron sequestering and storage by H. pylori. However, the expression of NapA in H. pylori was shown not to be regulated in response to iron content in the medium nor to play a part in metal resistance of H. pylori; these findings led to the proposal that the primary role of NapA in vivo may not be as an iron binding and storage protein (12, 13).
Proteins within the Dps family are expressed in most bacterial species and accumulate to high levels under conditions of oxidative or nutritional stress. A role of some Dps family proteins is in protecting bacteria from oxidative stress damage. Such a role for H. pylori NapA was first implied by the observation that loss of AhpC (alkyl hydroperoxide reductase, an important antioxidant enzyme) leads to a concomitant increase in NapA expression (28). More recent studies demonstrated that increased NapA production is the most frequent up-expression change associated with the apparent compensatory response to loss of major oxidative stress resistance factors in H. pylori (30). Like other Dps proteins, H. pylori NapA production is maximal in stationary-phase cells, and an H. pylori napA mutant survived less well than the wild-type strain upon exposure to oxidative stress conditions, suggesting that NapA plays a role in protecting H. pylori from oxidative stress damage (9).
To maintain long-term persistent infection in the host, H. pylori combats oxidative stress via a battery of diverse detoxification enzymes, including superoxide dismutase (SOD), catalase (KatA), alkyl hydroperoxide reductase (AhpC), and NADPH quinone reductase (MdaB). Disruption of any of these genes in H. pylori resulted in a clear mouse stomach colonization deficiency of the strain (20, 29, 34, 43). Although NapA was assigned putative roles as an important virulence factor and as antioxidant protein, the ability of an H. pylori napA mutant to colonize the host has not been investigated. In addition, the importance of NapA to H. pylori was ambiguous, as napA mutants appeared to have much weaker oxidative stress-related phenotypes in vitro compared to other oxidative stress resistance mutants such as sodB or ahpC mutants (9, 28). Thus, the roles and mechanisms of NapA in protecting H. pylori from oxidative stress damage are not clear.
In this study, we reassessed the DNA- and iron-binding abilities of the H. pylori NapA protein. A napA mutant was constructed with mouse-adapted strain X47, and the in vitro phenotypes of the wild type and the mutant were compared, including sensitivity to oxidative stress, the level of intracellular free iron, and the extent of DNA damage. Most importantly, the effect of NapA on host colonization efficiency was examined in a mouse infection model by using a napA gene disruption mutant. Assessing an in vivo role for NapA required inoculating mice first with nonviable H. pylori as a source of NapA in order to allow for host-mediated ROI production. Only then could a role for NapA in combating oxidative stress via a mutant analysis approach be ascertained. The results demonstrate that H. pylori NapA has unique properties, with dual roles related to oxidative stress; these are to induce oxidative stress mounted by the host cells and then to protect the bacterial cells from oxidative DNA damage.
MATERIALS AND METHODS
H. pylori culture conditions.
H. pylori was cultured on brucella agar (Difco) plates supplemented with 10% defibrinated sheep blood or 5% fetal bovine serum (called BA plates). Chloramphenicol (50 μg/ml) or kanamycin (40 μg/ml) was added to the medium for culturing mutants. Cultures of H. pylori were grown microaerobically at 37°C in an incubator under continuously controlled levels of oxygen (1 to 12% partial pressure as indicated, 5% CO2, and balanced with N2).
napA mutant construction.
A plasmid containing the H. pylori napA gene sequence disrupted with a chloramphenicol resistance cassette (pNapA::Cm from reference 28) was used to transform H. pylori wild-type strains, including SS1, 26695, 43504, and X47, by natural transformation. The mutant was selected on blood agar plates supplemented with chloramphenicol under 2% O2 conditions. Genomic DNA was prepared from the mutant clones, and the disruption of the napA gene on the genome was confirmed by an 800-bp increment of the PCR amplicon because of the insertion of the antibiotic resistance cassette into the gene.
Protein gel electrophoresis and protein identification by N-terminal sequencing.
Bacteria were harvested from the plates and resuspended in phosphate-buffered saline (PBS) buffer containing 20 mM sodium phosphate and 150 mM NaCl, pH 8.0. After one wash with the buffer, cells were resuspended in the same buffer and broken by two passages through a French pressure cell (SLM instruments, Inc.) at 138,000 kPa. Cell lysates were obtained by centrifugation (8,000 rpm for10 min), and the supernatant was transferred to a clean tube. The protein concentration of the cell extract obtained was determined by Bradford protein assay (Bio-Rad). Seven micrograms of cell extract was mixed with sodium dodecyl sulfate (SDS) buffer and incubated at 90°C for 5 min. Proteins were then separated by 12.5% SDS-polyacrylamide gel electrophoresis (PAGE) for 1.5 h at 100 V.
The suspected NapA protein band, which was clear in extracts resolved by SDS-PAGE from the wild-type strain but was missing from the mutant, was subjected to N-terminal sequencing; this was done upon transferring and excising the band from a polyvinylidene difluoride membrane (38) (Protein Sequencing Lab, Georgia State University).
Disk assay for oxidative stress sensitivity.
The napA mutant was tested for sensitivity to different oxidative stress reagents in comparison to the wild type. The BA plates were streaked uniformly with 0.1 ml of liquid culture at an optical density (OD) of 0.8. Sterile 7.5-mm filter paper disks individually saturated with 10 μl of a stress reagent (1 M cumene hydroperoxide, 1 M t-butyl hydroperoxide, or 0.1 M paraquat separately) were then placed onto the plates. The cells were cultured under microaerophilic conditions (5% O2) for 2 days before the zones of inhibition were measured (see Table 1).
TABLE 1.
Disk sensitivity assay results
| Strain | Zone of growth inhibition diam (mm)a
|
||
|---|---|---|---|
| 1 M t-Butyl hydroperoxide | 1 M Cumene hydroperoxide | 0.1 M Paraquat | |
| Wild-type X47 | 14.3 ± 1.0 | 9.2 ± 1.5 | 18.9 ± 2.8 |
| napA mutant X47 | 14.1 ± 1.4 | 12.3 ± 2.2 | 25.8 ± 3.6 |
Zones of growth inhibition were measured around filter paper disks saturated with 10 μl of the indicated compounds. Water (as a control) did not yield any zones of growth inhibition. Results are averages of five independent experiments with standard deviations.
Growth sensitivity to oxygen.
The wild-type or mutant strain was streaked onto BA plates. These plates were placed into the CO2 incubator under continuously controlled O2 levels (from 2 to 12% partial pressure) and incubated for 2 days (44). The growth of the bacteria on the plates was then scored (see Table 2).
TABLE 2.
Growth sensitivity to oxygen
| Strain | Growtha at O2 concn (%) of:
|
||||
|---|---|---|---|---|---|
| 2 | 5 | 7.5 | 10 | 12 | |
| Wild-type X47 | ++ | ++ | ++ | ++ | ++ |
| napA mutant X47 | ++ | + | ± | − | − |
Cells were inoculated onto plates and incubated under different concentrations of oxygen as indicated, and growth was scored after 2 days as follows: ++, healthy growth; +, slow growth; ±, slight growth at the site of heaviest inoculum; −, no growth at all. The experiments were repeated three times with similar results.
Determination of intracellular free-iron level by electron paramagnetic resonance (EPR) spectroscopy.
The protocol used for determination of intracellular free-iron levels by EPR was described previously (41). Briefly, a 5-ml cell suspension in PBS (OD at 600 nm [OD600] = 8.0) was incubated with 20 mM desferrioxamine at 37°C for 15 min. The cells were then centrifuged, washed with cold 20 mM Tris-HCl (pH 7.4), resuspended in a final volume of 0.4 ml of the same buffer, and frozen in 3-mm quartz EPR tubes by immersion in liquid nitrogen. Samples were stored at −78°C for EPR analysis. X-band (∼9.6 GHz) EPR spectra were recorded on a Bruker ESP-300E EPR spectrometer equipped with an ER-4116 dual-mode cavity and an Oxford Instruments ESR-9 flow cryostat. The relative amounts of intracellular free iron were assessed on the basis of the doubly integrated intensity of the g = 4.3 EPR signal in desferrioxamine-treated samples after normalization to the cell concentration.
DNA damage and fragmentation analysis by electrophoresis.
Wild-type and napA mutant cells were harvested and suspended in PBS buffer to an OD600 of 0.5. Cell suspensions were exposed to air for 2 h. Analysis of DNA fragmentation was performed as described by Zirkle and Krieg (50), with the following minor modifications. A 500-μl volume of the sample was centrifuged for 1 min at 15,000 × g, the pellet was washed in Tris-EDTA (TE) buffer (50 mM Tris HCl, 5 mM EDTA, pH 8) at 4°C, and the final pellet was suspended in 10 μl of TE buffer. This suspension was then added to 50 μl of 1% low-melting-point agarose (Fisher) at 37°C. Agarose and cells were mixed thoroughly, and 60-μl blocks were made by pipetting the mixture onto Parafilm. After solidification, the blocks were placed in a lysing solution (0.25 mM EDTA, 0.5% [wt/vol] Sarkosyl, 0.5 mg/ml proteinase K) and incubated at 55°C for 1 h, followed by overnight incubation at room temperature. The next day, the blocks were washed three times for 10 min each in cold TE buffer. Agarose plugs were then submerged in a 0.8% agarose gel. Samples were subjected to gel electrophoresis for 7 h at 30 V. Gels were then stained for 30 min with ethidium bromide (0.5 μg/ml), destained in H2O, and then visualized under UV light.
Construction of plasmid for overproduction of H. pylori NapA.
A 458-bp DNA fragment containing the napA gene was PCR amplified with primers NapA-F (5′CGCGCGGCATATGAAAACATTTGAAATT3′) and NapA-R (5′CGATATACTCGAGTTAAGCCAAATGGGC3′) and H. pylori strain X47 genomic DNA as the template. The PCR product was digested with NdeI and XhoI and cloned into vector pET-21a (Novagen) treated with the same restriction enzymes. The recombinant plasmid (pET-NapA) was then transformed into Escherichia coli BL21 Origami competent cells (Novagen).
Overexpression and purification of NapA.
E. coli BL21 Origami cells containing pET-NapA were grown at 37°C in 500 ml Luria-Bertani medium supplemented with 100 μg/ml ampicillin to an OD600 of 0.5. Induction of NapA expression was achieved by adding 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to the medium, followed by extended incubation for 2 h. Cells were then harvested by centrifugation at 5,000 rpm for 15 min, washed one time in cold buffer A (20 mM NaCl, 10 mM Tris-Cl, pH 8.0), and suspended in 5 ml of the same buffer. The cells were then broken by two passages through a French pressure cell, and cell debris was removed by centrifugation at 10,000 rpm. To further remove some residual insoluble proteins, the supernatant was subjected to a high-speed spin at 45,000 rpm for another 2.5 h. The supernatant (∼5 ml) obtained was then applied to a HiTrap Q HP anion-exchange column (Amersham Biosciences) that had been equilibrated with buffer A. Proteins were eluted with an NaCl gradient generated by mixing buffer A (see above) with buffer B (1 M NaCl, 10 mM Tris Cl, pH 8.0) by use of the fast protein liquid chromatography system (Amersham Biosciences). The eluted fractions were resolved by SDS-PAGE, and two partially purified fractions were pooled and further purified by gel filtration chromatography with a Sephacryl S-200 column (Amersham Biosciences). Proteins were eluted with buffer A, and the purified NapA was mixed with 20% glycerol and stored at −80°C. The concentration of the protein was determined by the Bradford assay method (Pierce).
Preparation of Fe-loaded NapA protein.
A protocol described previously (21) was used to prepare Fe-loaded NapA protein. Briefly, ferrous ammonium sulfate (Sigma) was added to a 100-μg/ml NapA preparation in 0.1 mM morpholinepropanesulfonic acid (MOPS)-NaOH buffer (pH 7.0) at a final concentration of 1 mM. The mixture was kept under a nitrogen atmosphere and incubated at room temperature for 1 h. The Fe-loaded NapA was then purified on a PD-10 column (Amersham Pharmacia Biotech) equilibrated with 20 mM MOPS-NaOH buffer (pH 7.0). The eluted protein was concentrated by an Centricon YM-50 (Millipore Corp.).
DNA-binding assay.
The DNA-binding assay was done as described by Almirón et al. (1). According to the method, 30 μg of native NapA, Fe-loaded NapA, or bovine serum albumin (BSA; negative control) was added to 200 ng of plasmid pGEMT DNA (Promega) in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) and incubated at 37°C for 30 min. The reaction mixture was then run on 1.0% agarose gel. DNA was detected by staining with ethidium bromide and observed under UV light.
Mouse colonization.
Mouse colonization assays were performed essentially as described previously (43, 44). Briefly, wild-type or napA mutant X47 cells were harvested after 48 h of growth on BA plates (37°C, 5% oxygen) and suspended in PBS to an OD600 of 1.7. The headspace in the tube was sparged with argon gas to minimize oxygen exposure, and the tube was tightly sealed. The bacterial suspensions were administered to C57BL/6J mice (3 × 108 H. pylori cells/mouse) twice, with the oral deliveries made 2 days apart. Three weeks after the first inoculation, the mice were sacrificed and the stomachs were removed, weighed, and homogenized in argon-sparged PBS (34) to avoid O2 exposure. Stomach homogenate dilutions were plated on BA plates supplemented with bacitracin (100 μg/ml), vancomycin (10 μg/ml), and amphotericin B (10 μg/ml), and the plates were rapidly transported into an incubator containing sustained 5% partial O2 pressure. After incubation for 5 to 7 days, the fresh H. pylori colonies were enumerated and the data were expressed as CFU per gram of stomach.
This conventional colonization assay was necessarily modified here in order to assess the effect of NapA disruption on mouse colonization. The modification was at the initial inoculation step; the inoculation on each day (day 1 and day 3) included first injecting 2 × 108 dead wild-type cells as a source of NapA and then (4 h later) injecting 2 × 108 test cells (i.e., either live wild-type cells or live napA mutant cells). Dead cells were prepared by three cycles of freezing (−80°C) and thawing (room temperature) of the wild-type cell suspension. The cells were proven to be dead by their culture on BA plates; there were no colonies. The remaining steps were like the conventional assay procedure. The rationale for the above modification is that dead wild-type cells contain the functional (secreted) NapA protein able to induce oxidative stress from the host; this permitted assessment of the role of NapA via a mutant-versus-wild type colonization comparison. As a control for this modified protocol, mice were first inoculated with dead napA mutant cells, followed (4 h later) by inoculation with either wild-type or napA mutant cells.
In a competition assay, each mouse was inoculated with a mixture of 2 × 108 wild-type cells and 2 × 108 napA mutant cells twice (2 days apart). The results of H. pylori colonization were examined after 3 weeks as described above. Stomach homogenate dilutions were plated on plates either without or with chloramphenicol (50 μg/ml). The number of H. pylori cells on plates without chloramphenicol represents the total number of cells colonizing the mouse stomach, and the number of cells on plates with chloramphenicol represents the number of napA mutant cells (X47 napA:CAT).
RESULTS
Construction of a napA mutant with mouse-adapted strain X47.
The primary goal of this study was to investigate the effect of NapA disruption on H. pylori's mouse colonization ability. However, attempts to construct an H. pylori napA mutant strain in the mouse-adapted strain SS1 background were unsuccessful, probably because of the lower transformation frequency in this particular strain. The napA mutants were obtained in other strains such as 43504 and 26695, but these parent strains were not useful for the mouse colonization assay (data not shown). Finally, the napA mutant was constructed with another mouse-adapted strain, X47. PCR analysis (not shown) indicated the correct insertion of a cat cassette into the napA gene on the genome.
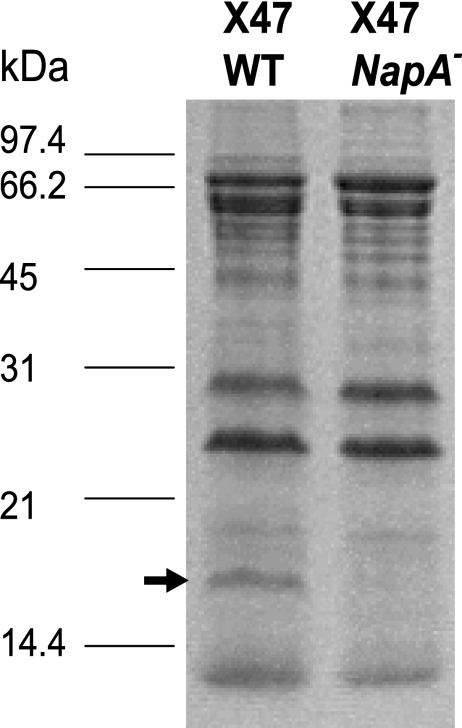
Crude extracts from wild-type and napA mutant cells were resolved by SDS-PAGE to examine the protein profiles. A band of 17 kDa that is present in wild-type X47 and is lacking in the mutant was confirmed to be the NapA protein by N-terminal sequencing (Fig. 1).
FIG. 1.
Protein profiles of wild-type (WT) and napA mutant X47 cells. The 12.5% polyacrylamide gel shown was stained with Coomassie brilliant blue. The sizes of protein standards are labeled on the left. The arrow indicates the 17-kDa NapA band which is present in wild-type cells but lacking in mutant cells.
napA mutant is more sensitive to oxidative stress.
The phenotypes of napA mutants of certain H. pylori strains in relation to oxidative stress resistance were reported previously (9, 28). We characterized the in vitro oxidative-stress-related phenotypes of the X47 napA mutant before performing the mouse colonization assay. We conducted paper disk assays with 1 M cumene hydroperoxide, 1 M t-butyl hydroperoxide, and 0.1 M paraquat (Table 1). Inhibition zones around disks previously saturated with the reagents were measured. According to the Student t distribution test, the napA mutant had significantly (P < 0.01) greater inhibition zones around cumene hydroperoxide and paraquat than did the parent strain.
An oxygen sensitivity assay was conducted next by culturing both wild-type and mutant cells under different continuous oxygen levels (2 to 12%) for 2 days (Table 2). Wild-type cells grew well under all of these O2 conditions. The napA mutant grew like the parent strain only at a 2% oxygen partial pressure and slower than the parent at a 5% oxygen partial pressure. The napA mutant did not grow at all in an environment where the oxygen concentration was above 7.5%. These results indicate that the napA mutant is more O2 sensitive than its parent, as expected for a strain containing an oxidative-stress-related mutation.
napA mutant cells contain higher levels of intracellular free iron and damaged DNA.
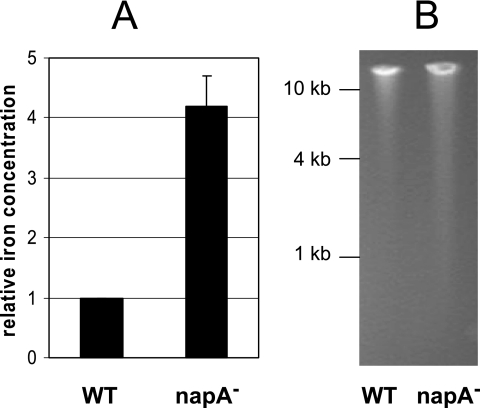
An excess amount of intracellular free iron promotes the generation of reactive oxygen species that attack important cellular components such as DNA. As a Dps homolog, a proposed function of NapA is to sequester intracellular free iron and to protect against oxidative DNA damage. Previously, we determined the intracellular free-iron level in H. pylori (wild type and katA, sodB, and ahpC mutants) by EPR spectroscopy and determined the level of DNA damage in these strains with a DNA fragmentation assay (41). In the present study, we applied these techniques to H. pylori X47 and its targeted napA mutant cells. The cells were grown under 2% controlled oxygen conditions to late log phase and then suspended in PBS. The cell suspension was exposed to air for 2 h, followed by EPR analysis to determine intracellular free-iron levels and also by agarose gel electrophoresis to determine the level of DNA fragmentation (Fig. 2). As shown in Fig. 2A, X47 napA mutant cells contain approximately fourfold more free iron than wild-type cells do. This result is clearly in agreement with a role for NapA in sequestering intracellular free iron. Figure 2B shows the extent of DNA fragmentation within wild-type H. pylori X47 and the napA mutant strain. DNA fragmentation could potentially arise because of a series of physical or chemical processes such as radiation, oxidation, or endonuclease activities. A small amount of fragmented DNA was observed for the wild-type sample, with the smallest fragment being about 4 kb. In contrast, a significantly greater amount of fragmented DNA was observed for the mutant sample, with the smallest fragment being about 1 kb. Therefore, compared to wild-type cells, napA mutant cells contain higher levels of both intracellular free iron and damaged DNA, suggesting a physiological role for NapA in protection of H. pylori DNA from iron-mediated damage.
FIG. 2.
Levels of intracellular free iron and DNA fragmentation in H. pylori cells. Wild-type (WT) or napA mutant H. pylori X47 cells were grown under 2% oxygen (partial pressure) conditions to late log phase and suspended in PBS. The cell suspension was exposed to air for 2 h, followed by EPR analysis to determine the intracellular free-iron level and by agarose gel electrophoresis to determine the level of DNA fragmentation. (A) The free-iron concentration in the cells was determined by measuring the magnitude of the g = 4.3 EPR signal, and the relative values were determined by comparing the napA mutant cells to the wild-type cells. The data are the mean from three experiments with the standard deviation indicated. (B) Agarose gel electrophoresis showing genomic DNA fragmentation of wild-type or napA mutant H. pylori X47 cells. The sizes of DNA standards are labeled on the left. The experiments were repeated three times with similar results.
Iron-loaded NapA protein binds DNA.
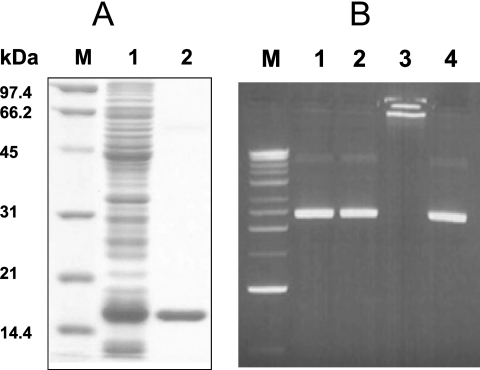
It was reported that H. pylori NapA does not bind DNA on the basis of an in vitro DNA-binding assay (37). However, another study demonstrated that NapA colocalizes with nuclear material, suggesting that it could interact with DNA (9). Our recent study of H. hepaticus Dps demonstrated that the iron-loaded form of Dps protein has much greater DNA-binding ability than native or iron-free Dps (21). In the present study, we reassessed the DNA-binding ability of H. pylori NapA. The protein was overexpressed in E. coli and purified by using anion- exchange and gel filtration approaches (Fig. 3A). Fe-loaded NapA was prepared by incubating the purified protein with ferrous ammonium sulfate as described in Materials and Methods. A DNA-binding assay was performed by incubating purified NapA (native form) or iron-loaded NapA with plasmid DNA and resolving the reaction mixture on agarose gel (Fig. 3B). The native form of NapA did not bind DNA (lane 2). In contrast, Fe-loaded NapA showed a strong ability to bind DNA, as DNA was totally retained in the well of the agarose gel (lane 3). As a negative control, the BSA protein did not change the electrophoresis pattern of plasmid DNA on the gel, indicating that BSA did not associate with DNA. The DNA used in the binding assay was either plasmid pGEMT (3 kb, from E. coli) or the same plasmid carrying an additional 1-kb H. pylori DNA sequence (pGEMT-mdaB) (43). The same results were obtained with both experiments, indicating that NapA binds to DNA nonspecifically.
FIG. 3.
Purification of H. pylori NapA protein and analysis of its DNA-binding ability. (A) Twelve percent polyacrylamide gel containing 1% SDS stained with Coomassie brilliant blue showing the expression and purification of the H. pylori NapA protein. Lane 1, cell extract of IPTG-induced E. coli BL21 Origami containing pET-NapA; lane 2, purified NapA protein; lane M, protein standards (Bio-Rad). (B) Purified NapA or BSA control protein was incubated with plasmid pGEMT DNA at 37°C for 30 min before being subjected to agarose gel electrophoresis. DNA on the gel was stained with ethidium bromide. Lanes: 1, DNA (200 ng) alone; 2, native NapA (30 μg) and DNA (200 ng); 3, Fe-loaded NapA (30 μg) and DNA (200 ng); 4, BSA (30 μg) and DNA (200 ng).
H. pylori NapA has dual effects within the host.
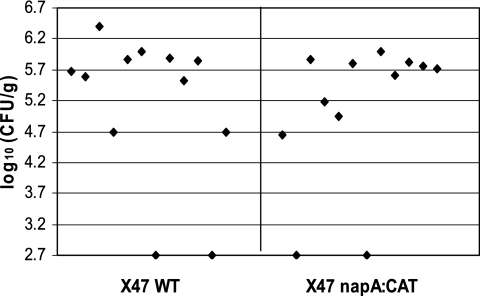
On the basis of the in vitro phenotypes of the napA mutant characterized above, we would expect that the napA mutant may have a defective or attenuated ability to colonize the host. With a well-established mouse colonization assay (29, 40, 43), the relative abilities of the wild-type and napA mutant X47 strains to colonize the mouse stomach were evaluated. Twelve C57BL/6J mice were inoculated with the wild-type strain, and another 12 mice were inoculated with the napA mutant. H. pylori cells were recovered from the mouse stomachs 3 weeks after oral administration. Surprisingly, in contrast to other oxidative stress gene disruption strains (29, 34, 43), the napA mutant showed a colonization efficiency similar to that of the wild type (Fig. 4).
FIG. 4.
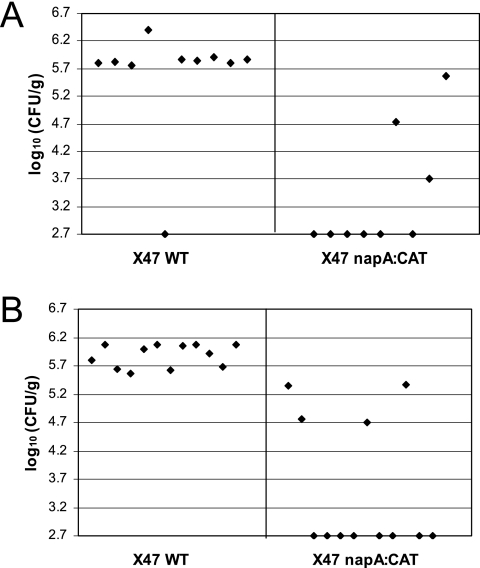
Conventional assay of mouse colonization by wild-type (WT) and isogenic napA mutant H. pylori X47. Mice were inoculated with H. pylori two times (2 days apart) with a dose of 3 × 108 cells. Colonization of mouse stomachs by H. pylori was examined 3 weeks after the first inoculation. Data are presented as a scatter plot of numbers of CFU per gram of stomach as determined by plate counts. Each spot represents the CFU count from one mouse. The baseline (log10 2.7 CFU/g) is the detection limit of the assay, which represents the count below 500 CFU/g of stomach. According to Wilcoxon rank test analysis, there is no significant difference between the wild type and the napA mutant in mouse colonization ability.
It is well known that H. pylori NapA induces host cells to produce ROIs; the purified NapA protein or the NapA in the cell extract also retains this ability (15, 33). The dual roles of NapA as an oxidative stress inducer and an oxidative damage defender could explain the above mouse colonization results. Presumably, wild-type H. pylori induces ROI production of the host via NapA (as well as other factors), and the bacterium itself is protected by functional NapA, as well as other antioxidant proteins. Compared to wild-type cells, the napA mutant likely has a decreased ability to induce ROI production; thus, upon infection the mutant strain would encounter a much less hostile oxidative environment than the parent strain. This environment would presumably be equivalent to the low-oxygen condition in vitro, where the napA mutant survives and grows like the wild type. To distinguish a role for NapA by use of a mutant-versus-wild type comparison, we modified the mouse colonization procedure to include a NapA-mediated host stimulation response. First, dead wild-type H. pylori cells were inoculated into the mouse stomach, which is expected to induce a regular level of ROI production. After 4 h, live wild-type or napA mutant cells were inoculated. As shown in Fig. 5A, H. pylori was recovered from the stomachs of 9 of 10 mice inoculated with the wild-type strain, with numbers of around 106 CFU/g of stomach. However, only three of nine mice that were inoculated with the napA mutant strain were found to harbor H. pylori. In addition, the titers of colonization (i.e., viable numbers) for these H. pylori-positive stomachs were much lower than those of mice inoculated with the wild-type strain. The experiment was repeated with 12 mice for the wild type and 12 mice for the mutant, and similar results were obtained (Fig. 5B).
FIG. 5.
Modified inoculation procedure for mouse colonization by wild-type (WT) and isogenic napA mutant H. pylori X47. Mice were inoculated with H. pylori two times, with the second inoculation 2 days after the first. Each time, the inoculum contained 2 × 108 dead wild-type cells and 4 h later 2 × 108 live test cells (i.e., either live wild-type cells or live napA mutant cells). Colonization was examined 3 weeks after the first inoculation. Data collection and analysis were done as described in the legend to Fig. 4. Panels A and B show results from two independent experiments. In both experiments, the napA mutant strain data are significantly lower (less colonization) than those of the wild type at a 99% confidence level, according to Wilcoxon rank test analysis.
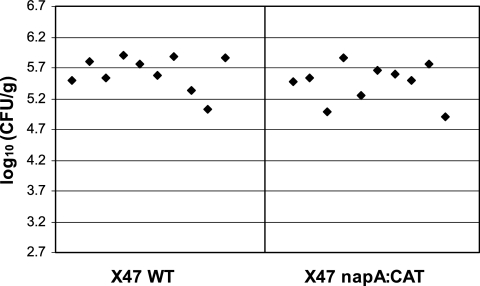
As a control for this modified procedure, we conducted a reverse experiment in which mice were first inoculated with dead napA mutant cells, followed (4 h later) by inoculation with either wild-type or napA mutant cells. As shown in Fig. 6, with this inoculation regimen, both strains were equally efficient in mouse colonization. By comparing these results to those in Fig. 5, we conclude that NapA, which is present in the preparation of nonviable H. pylori wild-type cells, is the critical active component inducing ROI production; it must be the trigger ultimately responsible for killing the napA mutant cells.
FIG. 6.
Results of the reverse experiment to Fig. 5 but still with the modified inoculation procedure for mouse colonization. Mice were inoculated with H. pylori two times, with the second inoculation 2 days after the first. Each time, the inoculum contained 2 × 108 dead napA mutant cells and 4 h later 2 × 108 live test cells (i.e., either live wild-type [WT] cells or live napA mutant cells). Colonization was examined 3 weeks after the first inoculation. Data collection and analysis were done as described in the legend to Fig. 4. According to Wilcoxon rank test analysis, there is no significant difference between the wild type and the napA mutant in mouse colonization ability under these conditions.
To further confirm the role of NapA in host colonization, a competition assay was conducted in which a mixture of cells (wild-type and napA mutant cells mixed in a 1:1 ratio) was inoculated into mice. Three weeks after inoculation, each mouse stomach homogenate was examined for the total number of H. pylori cells and separately for the number of napA mutant cells; the latter carried a chloramphenicol resistance marker. As shown in Table 3, at stomach harvest, all 10 of the mice contained 7.5 × 104 to 8.0 × 105 H. pylori cells per g of stomach. However, napA mutant cells were the clear minority in all of the stomachs in which they were present at all and this strain was not detectable in 6 of 10 stomachs.
TABLE 3.
Mouse colonization after 1:1 mixed inoculation of wild-type and napA mutant H. pylori cells
| Mouse no. | No. of CFU/g of stomach
|
napA:CAT/ total (%) | |
|---|---|---|---|
| Totala | napA:CAT | ||
| 1 | 2.7 × 105 | 2.25 × 104 | 8.3 |
| 2 | 7.5 × 104 | 0 | 0 |
| 3 | 8.0 × 105 | 3.5 × 104 | 4.4 |
| 4 | 6.25 × 105 | 0 | 0 |
| 5 | 3.25 × 105 | 0 | 0 |
| 6 | 2.13 × 105 | 5.0 × 103 | 2.3 |
| 7 | 4.5 × 105 | 0 | 0 |
| 8 | 2.55 × 105 | 7.5 × 103 | 2.9 |
| 9 | 6.0 × 105 | 0 | 0 |
| 10 | 3.0 × 105 | 0 | 0 |
Wild type plus napA:CAT.
DISCUSSION
The Dps family members constitute a group of ferritin-like iron-binding proteins that are widespread in eubacteria and archaea and are implicated in oxidative stress resistance and virulence (8, 22, 45, 47, 49). As a member of the Dps family, H. pylori NapA was previously shown to be involved in oxidative stress resistance (9, 28, 30). Distinct from other proteins of the Dps family, however, H. pylori NapA has a documented ability to induce a series of signal transduction events in eukaryotic host cells, resulting in the production of reactive oxygen species (15, 27, 33). Originally NapA was described as an important virulence factor because of its function in neutrophil activation, and now it is also recognized as an antioxidant protein. In this study, we sought to assess the colonization ability of a napA mutant and investigate the roles and mechanisms of NapA in protecting H. pylori from oxidative stress damage. For these purposes, a napA gene disruption mutant of mouse-adapted H. pylori strain X47 was generated and characterized.
So far, all of the Dps family proteins have been reported to bind iron. The sequestration of intracellular free iron was proposed as a mechanism affording oxidative stress by Dps family proteins. Yamamoto et al. (46) demonstrated that Streptococcus mutans dpr mutant cells contain higher levels of intracellular free iron than do wild-type cells when subjected to oxidative stress conditions. With a similar method of whole-cell EPR, we have shown that H. pylori napA mutant cells contain about fourfold greater amounts of intracellular free iron than do wild-type cells. At the same time, H. pylori napA mutant cells contain more DNA damage. Thus, H. pylori NapA has a major role in protecting H. pylori DNA from oxidative damage via sequestering free iron. H. pylori has a “canonical” ferritin, Pfr, which is the major iron storage protein playing a significant role in iron homeostasis (5, 39). H. pylori Pfr confers resistance to killing by iron toxicity but appears not to confer significant protection against oxidative stress (5). With the same EPR method, we found that H. pylori pfr mutant cells contain the same level of intracellular free iron as do wild-type cells (data not shown), suggesting that the sequestration of free iron under oxidative stress is not the role of Pfr. Thus, the role of ferritin as iron storage may not require rapid iron sensing, while NapA is probably able to quickly sequester fluxes of free iron. A mechanism of rapid iron binding would be advantageous to prevent generation of oxygen-related radicals during fluxes of oxidative stress.
Some specific Dps family proteins, for example, those from E. coli and Mycobacterium smegmatis, have DNA-binding capacity. These proteins were shown to interact with DNA and condense it, so that the DNA is “sheltered” from the attack of free oxidative radicals (18, 49). Other Dps family proteins were reported not to have DNA-binding ability, such as S. mutans Dpr (47), Agrobacterium tumefaciens Dps (8), and H. pylori NapA (37). However, another study demonstrated that H. pylori NapA, when expressed in E. coli cells, colocalizes with the nuclear material, suggesting that it can interact with DNA in vivo (9). The contradictory results may be due to a nonoptimized in vitro binding condition used to study H. pylori NapA; it may need a more favorable (i.e., in vivo) environment to associate with DNA. In this study, we showed that the newly purified H. pylori NapA does not bind DNA, confirming the result of Tonello et al. (37). However, when fully loaded with iron, H. pylori NapA has the ability to bind DNA. Recently we showed that, under oxidative stress, H. pylori cells accumulate free iron because of destruction of iron-sulfur cluster proteins (41, 42). Since NapA quickly sequesters free iron, which in turn promotes DNA binding, we propose that protection of DNA from iron-mediated damage under oxidative stress is a major role of NapA. This is supported by the direct evidence from the DNA fragmentation assays.
Oxidative damage to pathogenic bacteria constitutes a key part of the immune response of the host. Many studies show that H. pylori infection in human gastric mucosa elicits a strong oxidative stress response by the host (3, 11, 26, 31). To survive the effects of the production of reactive oxygen by the host, H. pylori depends on a significant repertoire of detoxification enzymes such as SOD, catalase, AhpC, and MdaB, as well as some repair enzymes. Disruption of any of the genes for these enzymes in H. pylori resulted in deficiency or attenuation of the ability to colonize the host stomach (20, 29, 34, 43). Among them, loss of AhpC or SOD exhibited the most severe oxidative stress-related phenotypes and resulted in almost complete deficiency in host colonization (29, 34). The catalase mutant is hypersensitive to hydrogen peroxide in vitro (19), but katA gene disruption has a much weaker effect on effective host colonization (20), compared to ahpC or sodB mutants. This could be explained by the fact that H. pylori catalase itself can induce oxidative stress production by host cells (31).
In this study, the dual roles of NapA as both an oxidative stress inducer and a damage defender were investigated. First, loss of NapA did not exhibit an observable phenotypic effect on host colonization when the conventional mouse colonization assay was used. In the modified procedure, nonviable wild-type H. pylori cells served as an inducer of ROI production. Four hours after nonviable-cell inoculation, either wild-type or napA mutant cells were inoculated to test for their colonizing efficiency. We arbitrarily chose a time interval of 4 h between injection of dead cells and inoculation of live cells. However, we did not conduct histological examination to assess the resulting inflammation in the mouse gastric epithelium. It was reported that such an inflammation change is most likely insignificant (23), particularly in the 4-h time frame. However, it was reported that NapA-induced ROI production by host cells is detectable within a few minutes of its addition to neutrophils or monocytes, and this response occurs in a NapA dose-dependent manner (33). Thus, the damaging effect of ROI production on bacterial cells may be detected earlier (or more easily) than a pathologic effect on host epithelial cells can be observed. The results in Fig. 5 clearly showed that the modified inoculation regimen is effective to demonstrate the dual roles of NapA in vivo. Simultaneous inoculation of wild-type and napA mutant cells in a 1:1 mixture yielded results showing that napA mutant cells have a much lower ability to compete with the wild type. From the initial data, we concluded that induction of ROI by wild-type H. pylori cell components, which may not cause detectable host cell inflammation, is responsible for eliminating proficiency of colonization by H. pylori napA mutant cells. Numerous components of H. pylori cells may have roles in stimulating ROI production or altering the gastric environment. Nevertheless, the reverse experiment with the modified inoculation protocol provided evidence that NapA is the critical active component responsible for inducing ROI production. In conclusion, NapA induces production of oxygen radicals from host cells and plays an identifiable role in protecting H. pylori from iron-mediated oxidative DNA damage. Both roles likely contribute to the bacterium's long-term infection and pathogenesis.
Acknowledgments
This work was supported by NIH grants DK60061 and DK062852 to R.J.M.
We thank Richard C. Conover and Michael K. Johnson (Department of Chemistry, UGA) for help with the EPR analysis.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 9 October 2006.
REFERENCES
- 1.Almirón, M., A. J. Link, D. Furlong, and R. Kolter. 1992. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 6:2646-2654. [DOI] [PubMed] [Google Scholar]
- 2.Amedei, A., A. Cappon, G. Codolo, A. Cabrelle, A. Polenghi, M. Benagiano, E. Tasca, A. Azzurri, M. M. D'Elios, G. Del Prete, and M. de Bernard. 2006. The neutrophil-activating protein of Helicobacter pylori promotes Th1 immune responses. J. Clin. Investig. 116:1092-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baik, S. C., H. S. Youn, M. H. Chung, W. K. Lee, M. J. Cho, G. H. Ko, C. K. Park, H. Kasai, and K. H. Rhee. 1996. Increased oxidative DNA damage in Helicobacter pylori-infected human gastric mucosa. Cancer Res. 56:1279-1282. [PubMed] [Google Scholar]
- 4.Bayerdörffer, E., N. Lehn, R. Hatz, G. A. Mannes, H. Oertel, T. Sauerbruch, and M. Stolte. 1992. Difference in expression of Helicobacter pylori gastritis in antrum and body. Gastroenterology 102:1575-1582. [DOI] [PubMed] [Google Scholar]
- 5.Bereswill, S., U. Waidner, S. Odenbreit, F. Lichte, F. Fassbinder, G. Bode, and M. Kist. 1998. Structural, functional and mutational analysis of the pfr gene encoding a ferritin from Helicobacter pylori. Microbiology 144(Pt. 9):2505-2516. [DOI] [PubMed] [Google Scholar]
- 6.Blom, K., B. S. Lundin, I. Bolin, and A. Svennerholm. 2001. Flow cytometric analysis of the localization of Helicobacter pylori antigens during different growth phases. FEMS Immunol. Med. Microbiol. 30:173-179. [DOI] [PubMed] [Google Scholar]
- 7.Brisslert, M., K. Enarsson, S. Lundin, A. Karlsson, J. G. Kusters, A. M. Svennerholm, S. Backert, and M. Quiding-Jarbrink. 2005. Helicobacter pylori induce neutrophil transendothelial migration: role of the bacterial HP-NAP. FEMS Microbiol. Lett. 249:95-103. [DOI] [PubMed] [Google Scholar]
- 8.Ceci, P., A. Ilari, E. Falvo, and E. Chiancone. 2003. The Dps protein of Agrobacterium tumefaciens does not bind to DNA but protects it toward oxidative cleavage: x-ray crystal structure, iron binding, and hydroxyl-radical scavenging properties. J. Biol. Chem. 278:20319-20326. [DOI] [PubMed] [Google Scholar]
- 9.Cooksley, C., P. J. Jenks, A. Green, A. Cockayne, R. P. Logan, and K. R. Hardie. 2003. NapA protects Helicobacter pylori from oxidative stress damage, and its production is influenced by the ferric uptake regulator. J. Med. Microbiol. 52:461-469. [DOI] [PubMed] [Google Scholar]
- 10.Davies, G. R., N. J. Simmonds, T. R. Stevens, A. Grandison, D. R. Blake, and D. S. Rampton. 1992. Mucosal reactive oxygen metabolite production in duodenal ulcer disease. Gut 33:1467-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies, G. R., N. J. Simmonds, T. R. Stevens, M. T. Sheaff, N. Banatvala, I. F. Laurenson, D. R. Blake, and D. S. Rampton. 1994. Helicobacter pylori stimulates antral mucosal reactive oxygen metabolite production in vivo. Gut 35:179-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dundon, W. G., H. Nishioka, A. Polenghi, E. Papinutto, G. Zanotti, P. Montemurro, G. G. Del, R. Rappuoli, and C. Montecucco. 2002. The neutrophil-activating protein of Helicobacter pylori. Int. J. Med. Microbiol. 291:545-550. [DOI] [PubMed] [Google Scholar]
- 13.Dundon, W. G., A. Polenghi, G. Del Guidice, R. Rappuoli, and C. Montecucco. 2001. Neutrophil-activating protein (HP-NAP) versus ferritin (Pfr): comparison of synthesis in Helicobacter pylori. FEMS Microbiol. Lett. 199:143-149. [DOI] [PubMed] [Google Scholar]
- 14.Dunn, B. E., H. Cohen, and M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans, D. J., Jr., D. G. Evans, T. Takemura, H. Nakano, H. C. Lampert, D. Y. Graham, D. N. Granger, and P. R. Kvietys. 1995. Characterization of a Helicobacter pylori neutrophil-activating protein. Infect. Immun. 63:2213-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fiocca, R., O. Luinetti, L. Villani, A. M. Chiaravalli, C. Capella, and E. Solcia. 1994. Epithelial cytotoxicity, immune responses, and inflammatory components of Helicobacter pylori gastritis. Scand. J. Gastroenterol. Suppl. 205:11-21. [PubMed] [Google Scholar]
- 17.Grant, R. A., D. J. Filman, S. E. Finkel, R. Kolter, and J. M. Hogle. 1998. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat. Struct. Biol. 5:294-303. [DOI] [PubMed] [Google Scholar]
- 18.Gupta, S., and D. Chatterji. 2003. Bimodal protection of DNA by Mycobacterium smegmatis DNA-binding protein from stationary phase cells. J. Biol. Chem. 278:5235-5241. [DOI] [PubMed] [Google Scholar]
- 19.Harris, A. G., F. E. Hinds, A. G. Beckhouse, T. Kolesnikow, and S. L. Hazell. 2002. Resistance to hydrogen peroxide in Helicobacter pylori: role of catalase (KatA) and Fur, and functional analysis of a novel gene product designated ‘KatA-associated protein’, KapA (HP0874). Microbiology 148:3813-3825. [DOI] [PubMed] [Google Scholar]
- 20.Harris, A. G., J. E. Wilson, S. J. Danon, M. F. Dixon, K. Donegan, and S. L. Hazell. 2003. Catalase (KatA) and KatA-associated protein (KapA) are essential to persistent colonization in the Helicobacter pylori SS1 mouse model. Microbiology 149:665-672. [DOI] [PubMed] [Google Scholar]
- 21.Hong, Y., G. Wang, and R. J. Maier. 2006. Helicobacter hepaticus Dps protein plays an important role in protecting DNA from oxidative damage. Free Radic. Res. 40:597-605. [DOI] [PubMed] [Google Scholar]
- 22.Ishikawa, T., Y. Mizunoe, S. Kawabata, A. Takade, M. Harada, S. N. Wai, and S. Yoshida. 2003. The iron-binding protein Dps confers hydrogen peroxide stress resistance to Campylobacter jejuni. J. Bacteriol. 185:1010-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loughlin, M. F., F. M. Barnard, D. Jenkins, G. J. Sharples, and P. J. Jenks. 2003. Helicobacter pylori mutants defective in RuvC Holliday junction resolvase display reduced macrophage survival and spontaneous clearance from the murine gastric mucosa. Infect. Immun. 71:2022-2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montecucco, C., and M. de Bernard. 2003. Molecular and cellular mechanisms of action of the vacuolating cytotoxin (VacA) and neutrophil-activating protein (HP-NAP) virulence factors of Helicobacter pylori. Microbes Infect. 5:715-721. [DOI] [PubMed] [Google Scholar]
- 25.Namavar, F., M. Sparrius, E. C. Veerman, B. J. Appelmelk, and C. M. Vandenbroucke-Grauls. 1998. Neutrophil-activating protein mediates adhesion of Helicobacter pylori to sulfated carbohydrates on high-molecular-weight salivary mucin. Infect. Immun. 66:444-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nardone, G., A. Rocco, and P. Malfertheiner. 2004. Review article: Helicobacter pylori and molecular events in precancerous gastric lesions. Aliment Pharmacol. Ther. 20:261-270. [DOI] [PubMed] [Google Scholar]
- 27.Nishioka, H., I. Baesso, G. Semenzato, L. Trentin, R. Rappuoli, G. Del Giudice, and C. Montecucco. 2003. The neutrophil-activating protein of Helicobacter pylori (HP-NAP) activates the MAPK pathway in human neutrophils. Eur. J. Immunol. 33:840-849. [DOI] [PubMed] [Google Scholar]
- 28.Olczak, A. A., J. W. Olson, and R. J. Maier. 2002. Oxidative-stress resistance mutants of Helicobacter pylori. J. Bacteriol. 184:3186-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olczak, A. A., R. W. Seyler, Jr., J. W. Olson, and R. J. Maier. 2003. Association of Helicobacter pylori antioxidant activities with host colonization proficiency. Infect. Immun. 71:580-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olczak, A. A., G. Wang, and R. J. Maier. 2005. Up-expression of NapA and other oxidative stress proteins is a compensatory response to loss of major Helicobacter pylori stress resistance factors. Free Radic. Res. 39:1173-1182. [DOI] [PubMed] [Google Scholar]
- 31.Ramarao, N., S. D. Gray-Owen, and T. F. Meyer. 2000. Helicobacter pylori induces but survives the extracellular release of oxygen radicals from professional phagocytes using its catalase activity. Mol. Microbiol. 38:103-113. [DOI] [PubMed] [Google Scholar]
- 32.Rautelin, H., B. Blomberg, H. Fredlund, G. Jarnerot, and D. Danielsson. 1993. Incidence of Helicobacter pylori strains activating neutrophils in patients with peptic ulcer disease. Gut 34:599-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Satin, B., G. Del Giudice, V. Della Bianca, S. Dusi, C. Laudanna, F. Tonello, D. Kelleher, R. Rappuoli, C. Montecucco, and F. Rossi. 2000. The neutrophil-activating protein (HP-NAP) of Helicobacter pylori is a protective antigen and a major virulence factor. J. Exp. Med. 191:1467-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seyler, R. W., Jr., J. W. Olson, and R. J. Maier. 2001. Superoxide dismutase-deficient mutants of Helicobacter pylori are hypersensitive to oxidative stress and defective in host colonization. Infect. Immun. 69:4034-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teneberg, S., H. Miller-Podraza, H. C. Lampert, D. J. Evans, Jr., D. G. Evans, D. Danielsson, and K. A. Karlsson. 1997. Carbohydrate binding specificity of the neutrophil-activating protein of Helicobacter pylori. J. Biol. Chem. 272:19067-19071. [DOI] [PubMed] [Google Scholar]
- 36.Thoreson, A. C., A. Hamlet, J. Celik, M. Bystrom, S. Nystrom, L. Olbe, and A. M. Svennerholm. 2000. Differences in surface-exposed antigen expression between Helicobacter pylori strains isolated from duodenal ulcer patients and from asymptomatic subjects. J. Clin. Microbiol. 38:3436-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tonello, F., W. G. Dundon, B. Satin, M. Molinari, G. Tognon, G. Grandi, G. Del Giudice, R. Rappuoli, and C. Montecucco. 1999. The Helicobacter pylori neutrophil-activating protein is an iron-binding protein with dodecameric structure. Mol. Microbiol. 34:238-246. [DOI] [PubMed] [Google Scholar]
- 38.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waidner, B., S. Greiner, S. Odenbreit, H. Kavermann, J. Velayudhan, F. Stahler, J. Guhl, E. Bisse, A. H. van Vliet, S. C. Andrews, J. G. Kusters, D. J. Kelly, R. Haas, M. Kist, and S. Bereswill. 2002. Essential role of ferritin Pfr in Helicobacter pylori iron metabolism and gastric colonization. Infect. Immun. 70:3923-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, G., P. Alamuri, M. Z. Humayun, D. E. Taylor, and R. J. Maier. 2005. The Helicobacter pylori MutS protein confers protection from oxidative DNA damage. Mol. Microbiol. 58:166-176. [DOI] [PubMed] [Google Scholar]
- 41.Wang, G., R. C. Conover, A. A. Olczak, P. Alamuri, M. K. Johnson, and R. J. Maier. 2005. Oxidative stress defense mechanisms to counter iron-promoted DNA damage in Helicobacter pylori. Free Radic. Res. 39:1183-1191. [DOI] [PubMed] [Google Scholar]
- 42.Wang, G., Y. Hong, M. K. Johnson, and R. J. Maier. Lipid peroxidation as a source of oxidative damage in Helicobacter pylori: protective roles of peroxiredoxins. Biochim. Biophys. Acta, in press. [DOI] [PubMed]
- 43.Wang, G., and R. J. Maier. 2004. An NADPH quinone reductase of Helicobacter pylori plays an important role in oxidative stress resistance and host colonization. Infect. Immun. 72:1391-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, G., A. A. Olczak, J. P. Walton, and R. J. Maier. 2005. Contribution of the Helicobacter pylori thiol peroxidase bacterioferritin comigratory protein to oxidative stress resistance and host colonization. Infect. Immun. 73:378-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiedenheft, B., J. Mosolf, D. Willits, M. Yeager, K. A. Dryden, M. Young, and T. Douglas. 2005. An archaeal antioxidant: characterization of a Dps-like protein from Sulfolobus solfataricus. Proc. Natl. Acad. Sci. USA 102:10551-10556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamamoto, Y., K. Fukui, N. Koujin, H. Ohya, K. Kimura, and Y. Kamio. 2004. Regulation of the intracellular free iron pool by Dpr provides oxygen tolerance to Streptococcus mutans. J. Bacteriol. 186:5997-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamamoto, Y., L. B. Poole, R. R. Hantgan, and Y. Kamio. 2002. An iron-binding protein, Dpr, from Streptococcus mutans prevents iron-dependent hydroxyl radical formation in vitro. J. Bacteriol. 184:2931-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zanotti, G., E. Papinutto, W. Dundon, R. Battistutta, M. Seveso, G. Giudice, R. Rappuoli, and C. Montecucco. 2002. Structure of the neutrophil-activating protein from Helicobacter pylori. J. Mol. Biol. 323:125-130. [DOI] [PubMed] [Google Scholar]
- 49.Zhao, G., P. Ceci, A. Ilari, L. Giangiacomo, T. M. Laue, E. Chiancone, and N. D. Chasteen. 2002. Iron and hydrogen peroxide detoxification properties of DNA-binding protein from starved cells. A ferritin-like DNA-binding protein of Escherichia coli. J. Biol. Chem. 277:27689-27696. [DOI] [PubMed] [Google Scholar]
- 50.Zirkle, R. E., and N. R. Krieg. 1996. Development of a method based on alkaline gel electrophoresis for estimation of oxidative damage to DNA in Escherichia coli. J. Appl. Bacteriol. 81:133-138. [DOI] [PubMed] [Google Scholar]