Abstract
Helicobacter pylori is dependent upon the production of the highly abundant and active metalloenzyme urease for colonization of the human stomach. Thus, H. pylori has an absolute requirement for the transition metal nickel, a required cofactor for urease. To investigate the contribution of genes that are factors in this process, microarray analysis comparing the transcriptome of wild-type H. pylori 26695 cultured in brucella broth containing fetal calf serum (BBF) alone or supplemented with 100 μM NiCl2 suggested that HP1512 is repressed in the presence of 100 μM supplemental nickel. When measured by comparative real-time quantitative PCR (qPCR), HP1512 transcription was reduced 43-fold relative to the value for the wild type when cultured in BBF supplemented with 10 μM NiCl2. When grown in unsupplemented BBF, urease activity of an HP1512::cat mutant was significantly reduced compared to the wild type, 4.9 ± 0.5 μmol/min/mg of protein (n = 7) and 17.1 ± 4.9 μmol/min/mg of protein (n = 13), respectively (P < 0.0001). In silico analysis of the HP1511-HP1512 (HP1511-1512) intergenic region identified a putative NikR operator upstream of HP1512. Gel shift analysis with purified recombinant NikR verified nickel-dependent binding of H. pylori NikR to the HP1511-1512 intergenic region. Furthermore, comparative real-time qPCR of four nickel-related genes suggests that mutation of HP1512 results in reduced intracellular nickel concentration relative to wild-type H. pylori 26695. Taken together, these data suggest that HP1512 encodes a NikR-nickel-regulated outer membrane protein.
The neutralophilic gastric pathogen Helicobacter pylori colonizes the gastric mucosa, an environment marked by potentially rapid and drastic fluctuations in pH. The highly abundant and active urease nickel-metalloenzyme is central to the ability of H. pylori to colonize the gastric mucosa (12, 20). When left untreated, this bacterium may cause lifelong infection. Urease, which is both acid and nickel inducible in H. pylori, catalyzes the hydrolysis of urea into bicarbonate and ammonia, buffering the surrounding microenvironment and offering protection against the inhibitory effects of low pH (2, 22, 23, 27, 32, 42, 43). Thus, H. pylori has an absolute requirement for the transition metal nickel for the production of catalytically active urease.
The uptake, storage, and utilization of nickel pose significant challenges for H. pylori. On one hand, sufficient nickel is required to maintain the activities of urease and hydrogenase, both of which are required for efficient colonization (13, 31, 40). However, excess intracellular nickel concentrations can produce toxic effects resulting from the generation of free radicals (26, 29, 35). Thus, H. pylori must tightly regulate intracellular nickel concentrations.
The transport of nickel across the inner membrane, its storage in the cytoplasm, and its incorporation into active urease via accessory proteins have been well characterized (45). Nickel destined for urease crosses the inner membrane, in part, through the high-affinity nickel permease NixA (25). Once in the cytoplasm, nickel can be directed to and subsequently incorporated into the urease apoenzyme via urease accessory proteins UreE, UreF, UreG, and UreH. To safeguard against the accumulation of toxic concentrations of intracellular nickel, cytoplasmic nickel can be stored in the histidine- and cysteine-rich protein Hpn (HP1427) and possibly the Hpn-like protein HP1432 (17, 26).
Nickel metabolism is regulated by the nickel-dependent transcriptional regulatory protein NikR which can act as both a transcriptional repressor, as in the case of nixA, and a transcriptional activator, as in the case for the urease operon (15, 48). This dual role represents an expanded regulatory function of H. pylori NikR relative to its Escherichia coli homolog, which acts solely as a nickel-dependent repressor of the E. coli Nik operon (7, 41, 43, 48). To date, one seemingly critical component of a nickel transport network absent in H. pylori is a nickel-regulated outer membrane transport mechanism. Given the essential role of nickel as a micronutrient for H. pylori, it seems reasonable to hypothesize that there would be a specific mechanism for transporting nickel across the outer membrane, perhaps in a manner analogous to that described for iron acquisition.
Iron acquisition has been well characterized in H. pylori, which contains putative iron-regulated outer membrane proteins (OMPs) homologous to E. coli ferric citrate transport proteins, encoded by fecA1, fecA2, and fecA3, and homologs of the Neisseria gonorrhoeae ferric enterobactin receptor FrpB, encoded by frpB1, frpB2/3, and frpB4 (39). Interestingly, two of the putative iron-regulated OMPs, encoded by fecA3 (HP1400) and frpB4 (HP1512), are unaffected by iron concentration and by mutations in fur, which encodes the global iron regulatory protein Fur (9, 44).
During a preliminary screen for nickel-responsive genes in H. pylori 26695, HP1512 (frpB4) was identified as a gene of interest, as it was down-regulated at the transcriptional level in response to nickel supplementation. In this study, we verify and then characterize the nickel-responsive regulation of HP1512, which was originally annotated as encoding a putative iron-regulated OMP (39). The contribution of HP1512 to the synthesis of catalytically active urease was assessed by comparing urease activities of wild-type H. pylori 26695 and an isogenic HP1512::cat mutant. Finally, a role for NikR in the regulation of HP1512 was demonstrated.
MATERIALS AND METHODS
Bacterial strains and growth.
Helicobacter pylori strain 26695 was maintained on Columbia blood agar (CBA) base (Difco, Detroit, MI) supplemented with 10% defibrinated sheep blood (Hemostat Laboratories, Dixon, CA) under 7% CO2 at 37°C. Liquid H. pylori cultures were grown in brucella broth (Difco, Detroit, MI) supplemented with 10% fetal bovine serum (BBF) (Gibco, Carlsbad, CA) with shaking (200 rpm) in a hypoxic chamber (Coy Laboratories and Products Inc., Grass Lake, MI) in an atmosphere containing 10% CO2 and 5% O2. Both agar plates and BBF were supplemented with antibiotics, 20 μg/ml chloramphenicol and/or 25 μg/ml kanamycin, when necessary. Liquid cultures used for experiments were inoculated at an optical density at 600 nm (OD600) of ≈0.01 from an overnight culture, inoculated from fresh CBA plates.
H. pylori microarrays.
cDNA was synthesized from 2 μg of total RNA with SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA). Briefly, a mixture containing 2 μg total RNA and 6 μg random hexamers (Invitrogen, Carlsbad, CA) was heated at 70°C for 5 min and then cooled on ice. Synthesis was carried out overnight at 42°C in a 30-μl reaction mixture consisting of 1× synthesis buffer, 0.01 M dithiothreitol, 0.5 mM (each) deoxynucleoside triphosphates (containing a 2:3 molar ratio of aminoallyl dUTP:dTTP), 40 U RNaseOut, and 400 U SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA). The reaction was stopped by the addition of 10 μl of 0.5 M EDTA and 10 μl of 1 M NaOH, followed by a 15-min incubation at 65°C and subsequent neutralization with 25 μl of 1 M Tris (pH 7.0). Aminoallyl dUTP-labeled cDNA was purified with a Microcon YM-30 column (Millipore Corp, Billerica, MA) and dried to completion. Amine-modified cDNA was resuspended in 5 μl water and labeled with either Alexa Fluor 555 or Alexa Fluor 647 per the manufacturer's protocol (Molecular Probes, Eugene, OR). Labeled cDNA was combined, purified over a QIAquick column (QIAGEN, Valencia, CA), eluted twice with 60 μl supplied elution buffer (QIAGEN, Valencia, CA), concentrated in a Microcon YM-30 column, and dried to completion.
H. pylori whole-genome microarrays were provided by the Pathogen Functional Genomics Resource Center. The microarray consists of 2,572 70-mer oligonucleotides, printed in triplicate, representing open reading frames from Helicobacter pylori 26695 and strain J99. Labeled cDNA was resuspended in 50 μl filter-sterilized hybridization buffer (50% formamide, 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% sodium dodecyl sulfate, 0.1 M dithiothreitol, and 0.6 μg/μl salmon sperm DNA), heated to 95°C for 5 min, mixed, and heated to 95°C for an additional 5 min. Labeled probe was applied to prehybridized microarrays and placed in a hybridization chamber (Corning, Acton, MA), which was submersed in a 42°C water bath for 16 to 20 h. Arrays were scanned on a Perkin-Elmer ScanArray Express microarray scanner. Microarray analyses were conducted with the TM4 software suite (33).
Construction of H. pylori mutants.
HP1512 was PCR amplified from H. pylori 26695 chromosomal DNA using primers HP1512.1 and HP1512.2 (Table 1) with the Expand Taq Long Template PCR system (Roche Diagnostic Corp., Indianapolis, IN). The resulting amplicon was ligated into Invitrogen pCR2.1 vector and transformed into electrocompetent E. coli TOP10 cells (Invitrogen Corporation, Carlsbad, CA). Transformants were screened for an insert via PCR and then verified by restriction digestion. H. pylori HP1512 was inactivated by inserting a chloramphenicol acetyltransferase (cat) cassette (46) into a unique StuI restriction site within HP1512. pCR2.1 plasmid containing the HP1512::cat construct was electroporated (800 Ω, 2.5 kV, 25 μF) into H. pylori, recovered on nonselective blood agar plates for 17 to 24 h, and then transferred to BBF plates supplemented with chloramphenicol (20 μg/ml). Transformants were screened via PCR to verify incorporation of HP1512::cat into the H. pylori chromosome. Mutations in strains other than strain 26695 were generated using the H. pylori 26695 HP1512::cat construct resulting in mutants with a disrupted copy of H. pylori 26695 HP1512 in place of their respective wild-type HP1512 homolog. Immediately upstream of HP1512 is a 327-bp hypothetical protein (HP1511). Downstream, HP1513 encodes a putative selenocysteine synthase (SelA) protein. Both HP1511 and HP1513 are transcribed in the same direction as HP1512; however, on the basis of the operon prediction tools on The Institute for Genomic Research (TIGR) website, these genes do not form an operon. H. pylori strain 26695 HP1512::cat nixA::aphA3 double mutants were generated using electrocompetent H. pylori 26695 nixA::aphA3 as recipient cells. Construction of the H. pylori nixA mutant has been previously described (4, 30).
TABLE 1.
Oligonucleotide primers used in this study
| Locus (gene) | Direction of primera | Primerb | Sequence (5′→3′) |
|---|---|---|---|
| HP1512 (frpB4) | F | HP1512.1 | GTTTGTCTCTTGTATTGTCG |
| R | HP1512.2 | CCACATATTGCTTCTTAAAG | |
| HP1338 (nikR) | F | NikR-1 | ACTAGTCCATGGATACACCCAATAAAGAC |
| R | N4_NotI | GCGGCCGCCGCTCAACAGTACCCTATCTATCCC | |
| HP1511-1512 intergenic region | F | IG1511-12-1 | TCACGAACAGTGCTTGATTCAG |
| R | HP1511-12-2 | CGATACGAAATTGATTTCTAAGC | |
| HP1512 (frpB4) | F | HP1512RTF | TCTAGCTGCAACGACTGGGAATGT |
| R | HP1512RTR | AGCGCGAGAGCCAGGTTAGGATA | |
| HP1400 (fecA3) | F | HP1400RTF | TTGAGCGATCGCATTGAAGCTTGG |
| R | HP1400RTR | AAGCCAATGTTGGTTGAGGGCATC | |
| HP1010 (ppk) | F | HP1010RTF | GCGCGTTAGTCGTTTATGGCGTTT |
| R | HP1010RTR | AGCGCTCAAAGGGTTGTAATTGCC | |
| HP1077 (nixA) | F | HP1077RTF | CGTTTGATGCGGATCACATCGCTT |
| R | HP1077RTR | TCTTCTAGCATCGGCGTGTGTTCT | |
| HP1338 (nikR) | F | HP1338RTF | CGCCGTGCTTGTGGTGATTTATGA |
| R | HP1338RTR | CCATGTGAATGTGCGTGGTGCATA | |
| HP0073 (ureA) | F | HP0073RTF | CTGAATTGATGCAAGAAGGGCGCA |
| R | HP0073RTR | CCACTTCATGGATCATGCTTGCCA |
F, forward; R, reverse.
Primers HP1512.1 and HP1512.2 were used for construction of the HP1512::cat mutants. Primers NikR-1 and N4_NotI were used for expression of recombinant NikR in pET30a. Primers used for real-time PCR are designated with RTF (real-time forward) and RTR (real-time reverse) in the primer name. HP1010 (ppk) served as a housekeeping gene for the quantitative real-time PCR experiments.
Whole-cell lysate preparation for urease activity.
H. pylori strains and their isogenic mutants were cultured in 5 to 7 ml of BBF medium alone or supplemented with different NiCl2 concentrations in a 50-ml conical tube. Cultures were incubated for 18 to 21 h with shaking (200 rpm) and harvested by centrifugation at 4,000 × g, for 20 min at 4°C. Cells were washed once in 20 ml of 50 mM ice-cold HEPES buffer (pH 7.5), once with 1 ml of 50 mM ice-cold HEPES buffer (pH 7.5), and resuspended in 150 μl of 50 mM HEPES buffer (pH 7.5). Cells were lysed in a chilled water bath sonicator (Sonicator 3000; Misonix, Farmingdale, NY) with approximately three 30-s pulses (with equivalent “pulse off” times). Sonicated lysate was centrifuged (12,000 × g for 12 min at 4°C), and the supernatant was transferred to chilled microcentrifuge tubes and placed on ice.
Total protein concentration was determined with the bicinchoninic acid assay (BCA protein assay; Pierce Chemical Company, Rockford, IL) using the 30-minute microtiter plate protocol. Urease activity was measured using the phenol-hypochlorite assay (47) modified for a 96-well microtiter plate by incubating 10 μl of protein lysate (0.1 μg/μl) with 90 μl urease buffer (50 mM HEPES, pH 7.5, 25 mM urea) on ice, followed by 5-, 12-, and 20-min incubations at 37°C. After incubation, 10 μl of the urease reaction mixture was mixed with phenol-sodium nitroprusside and alkaline hypochlorite (47), 150 μl each, and incubated for an additional 30 min at 37°C. Absorbance (OD595) was measured with a Universal microplate reader ELx800 (Bio-Tek Instruments, Inc., Winooski, VT). A standard NH4Cl concentration curve was generated for each assay. Urease activity was expressed as micromoles of urea hydrolyzed per minute per milligram of total protein.
RNA isolation and comparative, quantitative real-time PCR.
Total RNA was isolated from 18-hour cultures using a combination of TRIzol reagent (Invitrogen, Carlsbad, CA) followed by QIAGEN RNeasy column purification (QIAGEN, Valencia, CA). Cells were harvested by centrifugation for 1 min at 13,000 × g at room temperature, cell pellets were resuspended in 500 μl TRIzol at 65°C, and the suspensions were incubated at room temperature for 5 min. After incubation, the TRIzol solution was extracted twice with 100 μl chloroform. Supernatant was removed, 250 μl of 100% ethanol was added and mixed by pipetting, and the solution was applied to an RNeasy column. The manufacturer's protocols were followed once the solution was applied to the RNeasy column, and RNA was eluted in 50 μl RNase-free water. DNA was removed from RNA preparations by DNase I digestion with 40 U RNase-free DNase I for 30 min at 37°C followed by a second RNeasy (QIAGEN, Valencia, CA) column purification. Total RNA was eluted in 30 μl RNase-free water, quantified on a NanoDrop (NanoDrop, Wilmington, DE) spectrophotometer and visualized on an ethidium bromide-stained agarose gel.
Total RNA served as a template for cDNA synthesis using the SuperScript II first strand synthesis kit (Invitrogen, Carlsbad, CA). Synthesis reactions were carried out following manufacturer's protocol, starting with 2 μg total RNA and 500 ng random hexamers per 40-μl reaction mixture. cDNA was purified on a QIAGEN QIAquick (QIAGEN, Valencia, CA) column per the manufacturer's protocol and eluted in 30 μl supplied elution buffer. Purified cDNA was quantified on a Nanodrop (Wilmington, DE) spectrophotometer and diluted to 6 ng/μl. RNA transcripts were quantified on a Stratagene MX3000P real-time PCR machine using Stratagene's Brilliant SYBR green QPCR master mix (Stratagene, La Jolla, CA) in 25-μl reaction mixtures containing 30 ng of total cDNA. Optimal primer concentrations were determined empirically. Table 1 lists the genes of interest and their respective primer sequences. For each experiment, the transcript level was normalized to the level of HP1010 (ppk, polyphosphate kinase), and changes were determined relative to an experiment-specific calibrator using the Stratagene MXPro v 3.00 software package (Stratagene, La Jolla, CA).
Cloning, expression, and purification of H. pylori 26695 NikR.
H. pylori 26695 nikR (HP1338) was amplified via PCR with cloned Pfu polymerase (Stratagene, La Jolla, CA) from chromosomal DNA using primers NikR-1 and N4-NotI (primers modified from reference 9) (Table 1). The resulting amplicon was digested with NcoI and NotI and ligated into pET30a (Novagen, Madison, WI) digested with the same enzymes. This construct expresses recombinant H. pylori NikR (HpNikR) with a N-terminal His tag and S tag and no C-terminal fusions. Ligated plasmid was electroporated into Invitrogen TOP10 cells (Invitrogen Technologies, Carlsbad, CA) and plated on kanamycin, screened via PCR with T7 primers for the presence of nikR, and purified using a QIAprep Spin Miniprep kit (QIAGEN, Valencia, CA). The nikR sequence was verified by DNA sequencing, and the pET30a_nikR plasmid construct was electroporated into E. coli BL21(DE3)pLysS (Novagen, Madison, WI) and plated on Luria agar containing 20 μg/ml chloramphenicol and 25 μg/ml kanamycin.
Recombinant H. pylori nikR was expressed in Luria broth (1 liter) containing chloramphenicol (20 μg/ml) and kanamycin (25 μg/ml) inoculated at a dilution of 1:100 from an overnight culture of E. coli BL21(DE3)pLysS containing pET30_nikR. Expression cultures were incubated at 37°C with shaking (200 rpm) until they reached an OD600 of ≈0.4 at which time isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.4 mM. Upon induction, the culture was shifted to room temperature and incubated, with shaking, for an additional 5 h. After incubation, cells were harvested by centrifugation at 12,000 × g for 20 min at 4°C and washed once in 50 ml ice-cold HEPES, and the pellets were frozen at −80°C. Frozen cells were thawed at room temperature and resuspended in 20 ml of lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, 50 μM NiCl2; pH 8.0) containing 10 U benzonase (Sigma-Aldrich, St. Louis, MO). The resuspended pellet was pulled through a 16-gauge syringe several times and then passaged twice through a French pressure cell at 20,000 lb/in2. Cellular debris was pelleted by centrifugation at 10,000 × g for 25 min at 4°C, and the supernatant was transferred to a clean tube.
Recombinant protein was purified by Ni-nitrilotriacetic acid (Ni-NTA) affinity purification under native conditions by adding 5.0 ml of 50% Ni-NTA agarose (QIAGEN, Valencia, CA) per liter of starting culture followed by gentle shaking for 60 min at 4°C. The Ni-NTA slurry was applied to a column and sequentially washed with 20 ml lysis buffer followed by 4- to 10-ml sequential washes with lysis buffer containing 56 mM, 96 mM, 128 mM, and 164 mM imidazole. Bound recombinant NikR was eluted in 20 ml elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole; pH 8.0) and collected in approximately 0.5-ml fractions. Elution fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis under denaturing conditions, followed by staining with Coomassie blue R-250. Fractions containing the recombinant protein, with no contaminating bands, were pooled and concentrated with a Centricon YM-10 column (Millipore, Billerica, MA). Following the manufacturer's protocol, tagged protein was subjected to recombinant enterokinase digestion/capture (Novagen, Madison, WI), which removed both fusion tags, leaving a single N-terminal alanine on the recombinant HpNikR protein.
Gel shift assay.
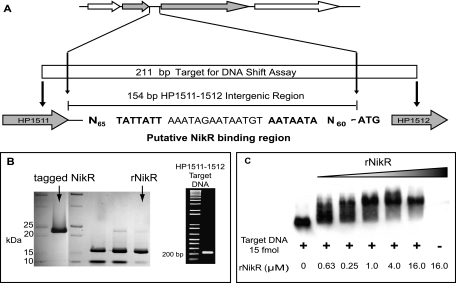
Target DNA from the HP1511-HP1512 (HP1511-1512) intergenic region was amplified from H. pylori 26695 chromosomal DNA via PCR with cloned Pfu polymerase (Stratagene, La Jolla, CA) using primers HP1511-12-1 and HP1511-12-2 (Table 1). The approximately 200-bp amplicon was gel isolated, labeled with digoxigenin (DIG) according to the manufacturer's protocol (Roche Diagnostics, Mannheim, Germany), and used in gel shift assays with the recombinant H. pylori NikR. Binding reaction mixtures consisted of approximately 15 fmol of DIG-labeled target DNA and fourfold dilutions of recombinant NikR protein ranging from 16 μM to 63 nM. Reactions were conducted in 1× binding buffer (20 mM Tris, pH 8.0, 100 mM KCl, 3 mM MgCl2, 0.1% Nonidet P-40, 5% glycerol, and 100 μM NiCl2) for 30 min at room temperature in a total volume of 20 μl. The entire binding reaction mixture was electrophoresed on a 7% nondenaturing polyacrylamide gel in a Tris-borate buffer (25 mM Tris, 300 mM borate; pH 7.6). Nickel was added to the polyacrylamide gel and running buffer to a final concentration of 100 μM NiCl2. The gel was prerun for 30 min at 100 V prior to loading; once the gel was loaded, it was run at ∼32 mA, while migration was monitored with a bromophenol blue marker. Proteins and DNA were electroblotted onto a positively charged Hybond-N+ nylon membrane (GE Healthcare) for 30 min at 400 mA in 0.5× Tris-borate-EDTA. DIG-labeled DNA was UV cross-linked to the membrane and detected according to the manufacturer's protocol (Roche Diagnostics, Mannheim, Germany).
RESULTS
Effect of nickel supplementation on HP1512.
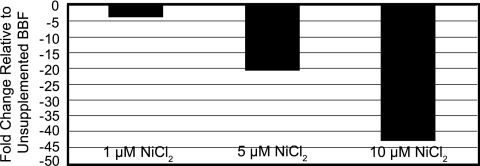
Using whole-genome microarrays, we identified Helicobacter pylori 26695 HP1512 as a potential nickel-responsive gene of interest. This gene was down-regulated when H. pylori 26695 was cultured in the presence of 100 μM NiCl2. To verify our initial findings and measure the effects of nickel supplementation on HP1512 transcription, wild-type H. pylori 26695 was cultured in the presence of supplemental nickel (0 to 10 μM), and the level of transcripts was determined by comparative qPCR. The level of transcripts in the presence of nickel was determined relative to that of HP1512 transcript in unsupplemented BBF (0 μM supplemental nickel). HP1512 transcript decreased in a stepwise fashion as NiCl2 concentration was increased (Fig. 1). With 10 μM supplemental NiCl2, the level of HP1512 transcripts was decreased by 43-fold (n = 3) relative to the level measured in unsupplemented BBF. These data support our original findings that HP1512 transcript is significantly down-regulated in the presence of nickel.
FIG. 1.
HP1512 transcript is significantly down-regulated in the presence of nickel as measured by real-time qPCR. H. pylori 26695 was grown in BBF supplemented with 0, 1, 5 or 10 μM NiCl2. The transcript level was determined relative to the transcript levels from cultures with no additional nickel added. The values (changes [n-fold]) represent the averages from three independent assays. As the nickel concentration increased, the HP1512 transcript level decreased in a stepwise fashion relative to the HP1512 transcript level for cells grown in unsupplemented BBF. With the addition of 10 μM NiCl2, the HP1512 transcript level decreased approximately 43-fold.
Urease activity of H. pylori HP1512::cat mutant.
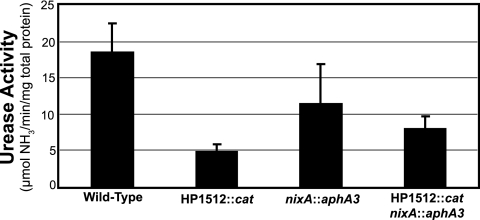
To determine the contribution of HP1512 (frpB4) to the production of catalytically active urease, the urease activities of wild-type H. pylori strain 26695 and an isogenic HP1512::cat mutant were compared following culture in unsupplemented BBF (Fig. 2). Urease activity for the HP1512::cat mutant is presented as the average urease activity from three independent colonies selected from a plate of transformants. When cultured in unsupplemented BBF, urease activity of the H. pylori 26695 HP1512::cat mutant was significantly reduced compared to the wild type, 4.9 ± 0.5 μmol/min/mg protein (n = 7) and 17.1 ± 4.9 μmol/min/mg protein (n = 13), respectively (P < 0.0001). The same trend was observed in three additional H. pylori strains (UMAB41, 43504, and SS1; data not shown).
FIG. 2.
Urease activity of wild-type H. pylori 26695, nixA::aphA3 mutant, HP1512::cat mutant, and nixA::aphA3 HP1512::cat double mutant strains. In addition to an HP1512::cat mutant, a nixA::aphA3 mutant and a nixA::aphA3 HP1512::cat double mutant were constructed. Urease activity was measured via the phenol-hypochlorite urease assay after approximately 18 h of growth in unsupplemented BBF. The urease activities of the HP1512::cat (n = 13) and nixA::aphA3 (n = 3) single mutants were significantly less than that of the wild type (n = 7) (P < 0.001 and P < 0.01, respectively). The urease activity of the H. pylori 26695 HP1512::cat nixA::aphA3 (n = 3) double mutant was significantly less (P < 0.001) than the wild type but not significantly different that that of each single mutant.
To further explore the contribution of HP1512 to urease activity in H. pylori 26695, a nixA::aphA3 HP1512::cat double mutant was constructed. Urease activity of the nixA::aphA3 HP1512::cat double mutant was significantly less than that of the wild type (P < 0.001), as was the nixA::aphA3 mutant (P < 0.01) (Fig. 2). Urease activity of the double HP1512::cat nixA::aphA3 mutant was not significantly different from that of the nixA::aphA3 single mutant (Fig. 2). However, urease activity of the HP1512::cat single mutant was significantly less than both the nixA::aphA3 mutant (P < 0.05) and the double mutant (P < 0.01) (Fig. 2). These data suggest that HP1512 and nixA do not act synergistically; that is, the urease activity of the double mutant is not less than each individual mutation, consistent with a model to be presented below.
Growth, MIC, and MBC of H. pylori 26695 HP1512::cat mutant.
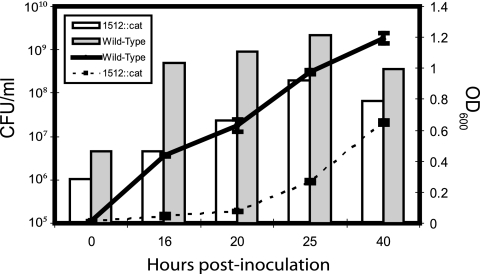
Insertional inactivation of HP1512 resulted in a reduced growth rate of the HP1512 mutant relative to wild-type H. pylori 26695 (Fig. 3). However, the MIC and minimum bactericidal concentration (MBC) to NiCl2 of the HP1512 mutant were similar to those of the wild type (750 μM and 1,750 μM NiCl2, respectively), which is not surprising, since cytoplasmic nickel concentrations are dictated by NixA-mediated transport across the inner membrane. Thus, while the ability of the mutant to transport nickel across the outer membrane may be impaired, transport across the inner membrane can be increased to compensate. Comparison of nixA transcript abundance in the HP1512::cat mutant relative to the wild type reflects this up-regulation. At all NiCl2 concentrations, the level of nixA transcripts is increased in the HP1512 mutant relative to the wild type (Fig. 4). Thus, the cytoplasmic nickel concentration, while reduced (as evidenced by decreased urease activity in the HP1512::cat mutant), is not low enough to abrogate the toxic effects of nickel accumulation. A slight difference in MIC and MBC between the wild type and the HP1512::cat mutant cannot be ruled out, but it was not evident in our assay.
FIG. 3.
Growth curve of the HP1512::cat (1512::cat) mutant compared to the wild type. Insertional inactivation of the HP1512 gene resulted in reduced growth relative to the wild-type H. pylori 26695 strain cultured in unsupplemented BBF. The optical density at 600 nm (indicated by the lines) was monitored in three independent cultures. At each time point, the number of CFU per milliliter was determined by combining the independent cultures in equal volumes and plating for viable colonies; thus, each bar represents the CFU/ml from a single pooled culture.
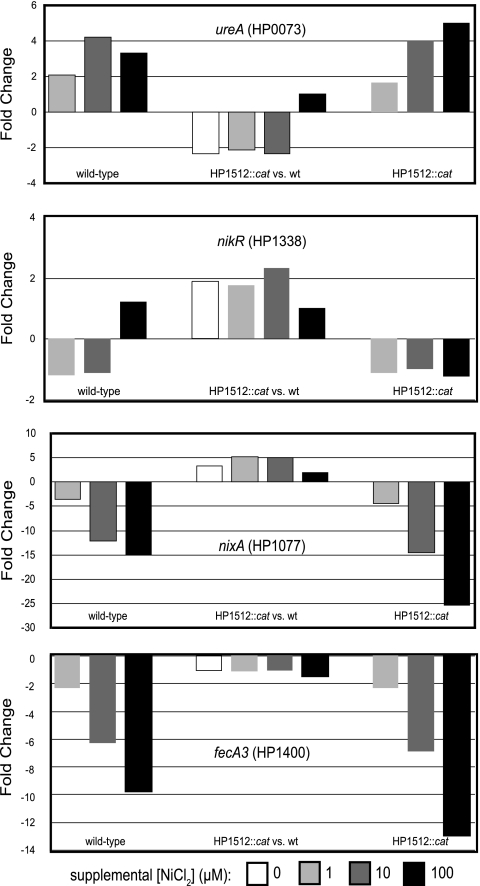
FIG. 4.
Effects of nickel supplementation on the transcription of selected genes in wild-type H. pylori 26695 and the HP1512::cat mutant. The levels of transcripts of four nickel-related genes were quantified after 18 h of growth in BBF supplemented with three different concentrations of NiCl2 (1, 5, and 10 μM). HP1010, encoding polyphosphate kinase (ppk), served as a housekeeping gene control. For each transcript measured, three comparisons were made. First, for each gene investigated, its response to nickel supplementation was determined by comparing the relative levels of transcripts in wild-type H. pylori 26695 grown in 1, 5, and 10 μM supplemental NiCl2 to the transcript level of cells grown in unsupplemented BBF (left three bars). Second, to determine whether mutation of HP1512 affected the ability of H. pylori to respond to nickel supplementation, comparisons were made between transcript levels in the HP1512 mutant with nickel supplementation of 1, 5, and 10 μM NiCl2 relative to that in cells grown in unsupplemented BBF (right three bars). Finally, at each supplemental nickel concentration (0, 1, 5, 10 μM), the relative transcript level in the HP1512 mutant was compared to that of wild-type (wt) H. pylori 26695 to measure the effect of HP1512 mutation on nickel responsiveness (middle four bars). The values (changes [n-fold]) are reported as the averages from three independent assays.
Effects of nickel concentration on transcription of select ion-related genes.
In addition to measuring the effects of nickel supplementation on HP1512 transcription, the levels of four additional nickel-related gene transcripts were compared in the wild-type H. pylori 26695 strain and the HP1512::cat mutant (Fig. 4). These nickel-related genes were largely chosen on the basis of previous work; however, our microarray analysis identified ureA as significantly up-regulated in the presence of 100 μM supplemental NiCl2. nixA, ureA, and nikR have been shown to respond to the nickel concentration at the transcriptional level (1, 9, 10, 15, 42, 43, 48). fecA3 (HP1400) was chosen as a gene of interest on the basis of its pattern of iron-responsive gene expression; it displayed a pattern of expression identical to that of HP1512 (44).
For each transcript measured, three comparisons were made. First, for each gene investigated, its response to nickel supplementation was determined by comparing the relative levels of transcripts in wild-type H. pylori 26695 grown in 1, 5, and 10 μM supplemental NiCl2 to the transcript level in unsupplemented BBF (Fig. 4, left three bars). Second, to determine whether mutation of HP1512 affected the ability of H. pylori to respond to nickel supplementation, comparisons were made between transcript abundance in the HP1512::cat mutant with nickel supplementation of 1, 5, and 10 μM NiCl2 relative to unsupplemented BBF (Fig. 4, right three bars). Finally, at each nickel concentration, relative transcript abundance in the HP1512::cat mutant was compared to that of wild-type H. pylori 26695 to measure the effect of HP1512 mutation on nickel responsiveness (Fig. 4, middle four bars).
Transcription of ureA (HP0073), encoding the UreA urease structural subunit, was increased in both the wild type and the HP1512::cat mutant in response to nickel supplementation. Thus, mutation of HP1512 does not affect H. pylori's ability to regulate ureA transcript in response to nickel. When comparing ureA transcription in the HP1512::cat mutant relative to wild-type H. pylori 26695, ureA transcription is down-regulated approximately twofold at supplemental NiCl2 concentrations of 0, 1, and 5 μM, and only at 10 μM NiCl2 does the ureA transcript in the mutant approach wild-type levels. Assuming that the ureA transcript is an indirect indicator of cytoplasmic nickel availability, decreased ureA transcript abundance suggests reduced cytoplasmic nickel concentrations in the HP1512::cat mutant.
Transcription of nixA (HP1077), which encodes the high-affinity nickel transport protein NixA (25), has been shown to be repressed in response to nickel supplementation (15, 48). In both wild-type H. pylori 26695 and the HP1512::cat mutant, nickel supplementation resulted in an incremental and dramatic repression of nixA transcription relative to growth in unsupplemented BBF, 19-fold and 25-fold at 10 μM NiCl2, respectively (Fig. 4). Comparison of the level of nixA transcript in the HP1512 mutant relative to the wild type revealed increased transcription in the mutant relative to the wild type at each NiCl2 concentration tested (0, 1, 5, and 10 μM). Taken as an indirect indicator of cytoplasmic nickel concentration, the nixA data suggest reduced cytoplasmic nickel concentrations in the HP1512 mutant relative to the wild type. Thus, the transcriptional responses of both ureA and nixA are in agreement and indicative of reduced cytoplasmic nickel concentrations in the HP1512 mutant.
H. pylori FecA3 (encoded by HP1400) is annotated as a putative iron(III) dicitrate transport protein; however, fecA3 transcription is unaffected by iron supplementation and fur mutation (44). In fact, fecA3 presents the same transcriptional profile in response to iron as does HP1512 (44), making it an interesting candidate as an additional nickel transport protein. Transcription of fecA3 was repressed in a nickel-dependent manner in both the wild type (9-fold at 10 μM NiCl2) and an HP1512 mutant (13-fold at 10 μM NiCl2) relative to their transcription in unsupplemented BBF (Fig. 4).
Finally, transcription of HP1338, encoding the transcriptional regulator NikR, was measured. Nickel supplementation up to 10 μM NiCl2 had no measurable affect on HP1512 transcription in the wild type, HP1512 mutant, or in the mutant relative to the wild type. NikR has been demonstrated to act as a transcriptional repressor of its own transcription in vitro; however, the binding of NikR to its promoter was seen only at high nickel concentrations and thus may not act as an autorepressor at the nickel concentrations used in this study (1, 10).
Mobility shift DNA-binding assay.
Studies mapping the NikR operators for ureA, exbB, nikR, and fur proposed a putative NikR operator consensus sequence (10). In silico analyses of the 154-bp intergenic region between HP1511 and HP1512 identified a sequence that perfectly matched the putative NikR consensus sequence (Fig. 5A). The putative NikR operator consists of 28 bp, spanning a region from −88 bp to −60 bp upstream of the HP1512 start codon. The ability of NikR to bind the HP1511-1512 intergenic region was demonstrated using purified recombinant H. pylori NikR (Fig. 5B) in an in vitro mobility shift DNA-binding assay (Fig. 5C). In the presence of 100 μM NiCl2, recombinant HpNikR at >1 μM was able to shift a PCR amplicon containing the putative NikR operator consensus sequence (Fig. 5C).
FIG. 5.
NikR binds the HP1511-1512 intergenic region. (A) In silico analysis identified a putative NikR consensus sequence (10) upstream of HP1512 within the 154-bp HP1511-1512 intergenic region (proposed operator regions in bold). (B) The nikR sequence was cloned into pET30a_nikR plasmid and expressed in E. coli BL21(DE3)pLysS. Recombinant protein was purified by Ni-NTA affinity purification under native conditions (tagged NikR) followed by recombinant enterokinase digestion/capture to remove fusion tags. The resulting recombinant H. pylori NikR (rNikR) protein (∼17 kDa) contains a single N-terminal alanine from the expression vector. Target DNA, encompassing the HP1511-1512 intergenic region, was amplified from H. pylori 26695 chromosomal DNA via PCR, labeled with digoxigenin (DIG), and used in gel shift assays with the recombinant H. pylori NikR. Binding reaction mixtures consisted of approximately 15 fmol of DIG-labeled target DNA and fourfold dilutions of recombinant NikR protein ranging from 16 μM to 63 nM. Binding reaction mixtures were electrophoresed on 7% nondenaturing polyacrylamide gels; nickel was added to the gel and running buffer to a final concentration of 100 μM NiCl2. (C) Proteins and DNA were electroblotted onto a positively charged nylon membrane and visualized via chemiluminescence detection with alkaline phosphate and disodium 3-(4-methoxyspiro[1,2-dioxetane-3,2′-(5′-chloro)tricyclo]3.3.1.1(3,7)]decan]-4-yl) phenyl phosphate (CSPD).
DISCUSSION
In this study, we demonstrate that the Helicobacter pylori 26695 outer membrane protein HP1512 (encoded by frpB4) is regulated by nickel. HP1512 is repressed at the transcriptional level by NikR in the presence of nickel. Insertional inactivation of HP1512 produces a significant reduction in urease activity relative to the wild-type level, suggesting that HP1512 is responsible for the transport of nickel across the outer membrane. Furthermore, transcriptional profiling of select nickel-related genes suggests that inactivation of HP1512 reduces intracellular nickel concentrations. Identification of an outer membrane nickel transport protein adds an additional component to the NikR regulatory network, while providing a mechanism for nickel transport into the periplasm for its subsequent transport, via NixA, into the cytoplasm where is it incorporated into the urease apoenzyme.
H. pylori is unique in its ability to colonize and persist in the harsh environment of the stomach, where it colonizes the gastric mucosa close to the epithelial cell surface (5, 24, 38). Within the gastric mucosa, a pH gradient is established ranging from slightly acidic (pH of ∼6.5) at the epithelial surface to highly acidic (pH of ∼2.0) in the gastric lumen (34). By sensing and responding to this pH gradient, H. pylori is able to maintain its spatial orientation within the gastric mucosa and colonize the less acidic region close to the gastric epithelium (34). While capable of surviving exposure to pH 1.0 for several hours (36, 37), localization to the epithelial surface generally protects against prolonged exposure to the harsh conditions of the gastric lumen. While controversial, the mucosal region colonized by H. pylori is thought to be approximately pH 5.0, with occasional bouts of lower pH at regions of tissue damage. Thus, acid shock is likely the most significant environmental stress encountered by Helicobacter pylori.
The 1.1-MDa urease nickel-metalloenzyme (19) is the cornerstone of H. pylori's ability to respond to acid shock. The urease enzyme is both highly active and abundant, comprising up to 6% of total cellular protein (12, 20). In the cytoplasm, urease catalyzes the conversion of urea to ammonia and bicarbonate, which then buffer the surrounding microenvironment (22, 23, 27, 36). The urease operon is present as a single copy and consists of the structural subunit genes, ureA and ureB, and the urease accessory protein genes encoded by ureIEFGH (39). The urease operon is arranged into two transcriptional units, ureAB and ureIEFGH (2). Urease transcription, as well as its subsequent ability to respond to pH, is mediated through at least two regulatory systems, the HP0166-HP0165 (ArsRS) two-component system and the nickel-responsive transcriptional regulator NikR (15, 32, 43).
NikR is a member of the ribbon-helix-helix family of transcriptional regulators which acts as a nickel-dependent transcriptional repressor in E. coli (8). E. coli NikR is responsible for regulation of nickel uptake through a nickel-specific ATP-dependent transport system encoded by the nikABCDE operon (11, 28, 49). When the nickel concentration in the cytoplasm is elevated, the C-terminal region of the tetrameric NikR protein bind Ni2+, which allows the N-terminal DNA-binding domain to interact with the nikABCDE operator (7, 8, 11, 49). Thus, in response to high intracellular nickel concentrations, the nik operon is repressed with the net result being decreased nickel import (11, 49).
In comparison, the NikR homolog in H. pylori can act as both a transcriptional activator and repressor (15, 43). In its role as an activator, nickel-bound H. pylori NikR binds an operator region in the ureA promoter, inducing transcription of the urease operon (10, 15, 42, 43). The role of NikR as a transcriptional activator appears to be unique to H. pylori, and it has been hypothesized that the positional relationship between the NikR binding region and the promoter determines whether NikR acts as a repressor or activator (15). Consistent with our results (Fig. 3), H. pylori not only increases urease transcription in response to increased intracellular nickel availability but also decreases the further transport of nickel into the cytoplasm by repressing transcription of nixA, which encodes the high-affinity nickel permease NixA (15, 48). The net result of these actions, coupled with the demonstrated nickel storage capacity of the cytoplasmic histidine-and cysteine-rich protein Hpn (HP1427) (16, 17, 26), is an overall reduction in intracellular nickel concentration.
Much of what is currently known about the H. pylori NikR regulatory network has been determined through the use of whole-genome microarray comparisons of wild-type H. pylori to isogenic nikR mutants (9). nikR mutants are more susceptible to higher nickel concentrations (9, 43) and are attenuated in the mouse model (5). Mutation of nikR affected expression of genes related to iron transport and metabolism, chemotaxis, hydrogenase, motility, and respiration (5, 9, 41). However, one potential component of H. pylori's nickel metabolism network noticeably missing from previous work is a nickel-regulated OMP. Given the role of urease as a virulence factor and its dependence upon nickel for catalytic activity, it seems reasonable to hypothesize that H. pylori would utilize specific nickel-regulated OMPs in much the same manner as those previously characterized for iron transport.
H. pylori contains six putative iron-regulated OMPs (3, 39). These OMPs can be divided into two groups on the basis of homology. Three are homologous to E. coli ferric citrate transport proteins, encoded by fecA1 (HP0686), fecA2 (HP0807), and fecA3 (HP1400), and the remaining are homologs of the Neisseria gonorrhoeae ferric enterobactin receptor FrpB, encoded by frpB1 (HP0876), frpB2/3 (HP0915/0916), and frpB4 (HP1400) (39). While conducting a survey of nickel-responsive genes, HP1512 (frpB4) was identified as a gene of interest. The presence of HP1512 in membrane preparations has been documented in studies specifically targeting H. pylori OMPs (3, 6, 14). Previous work characterizing the iron transport and storage genes of H. pylori 26695 demonstrated that while two of the frpB alleles, frpB1 (HP0876) and frpB2/3 (HP0915/0916), were iron responsive, frpB4 (HP1512) was not responsive to either the iron concentration or fur mutation (14, 44). Furthermore, HP1512 transcription was derepressed in a nikR mutant, a response attributed to indirect regulation via Fur (9). Consistent with studies at the transcriptional level, proteomic analysis of a H. pylori fur mutant did not reveal an effect of fur mutation on the expression of HP1512 at the protein level (21).
If HP1512 were in fact an outer membrane nickel transport protein, it follows that inactivation by allelic exchange mutagenesis would result in decreased intracellular nickel concentrations. Reduced intracellular nickel concentrations should be reflected both in urease activity and in transcription of nickel-responsive genes. Thus, one might expect that relative to the wild type, the HP1512 mutant would display reduced urease activity, decreased ureA transcript abundance, and increased nixA transcript abundance. While these indicators are indirect estimates of nickel concentration, they provide an assessment of intracellular nickel availability.
Reduction, but not complete abolition, of urease activity in both nixA and HP1512 mutants suggests a role for alternative nickel transport mechanisms acting across both the outer and inner membranes. One possible candidate as an additional outer membrane nickel transport protein is FecA3 (encoded by HP1400). HP1400 has been identified in H. pylori outer membrane preparations (14), and while annotated as an iron-responsive OMP, HP1400 transcription is unaffected by the iron concentration and fur mutation in a fashion similar to that of HP1512 (14, 44). Comparative qPCR of HP1400 transcript levels in the wild-type H. pylori 26695 demonstrates that HP1400 transcription is reduced in the presence of nickel (Fig. 4). While the HP1400 transcript decreases significantly and in a stepwise fashion as the nickel concentration is increased, the repression is not as dramatic as that measured for HP1512 (10-fold versus 43-fold at 10 μM NiCl2, respectively). Our observations are in agreement with those of Ernst et al. (14), who independently demonstrated NikR-mediated transcriptional repression of HP1400 and then mapped the NikR operator located upstream of HP1400.
The role of H. pylori NikR in the regulation of nickel homeostasis, coupled with the demonstrated regulation of HP1512 at the transcriptional level, suggests that NikR may directly regulate HP1512 transcription. The NikR regulatory network and mechanisms of regulation by NikR have been studied extensively in H. pylori (1, 9, 10, 15, 41, 48). While there does not appear to be a strong NikR consensus sequence in H. pylori, alignment of several mapped NikR binding regions led to the identification of a putative consensus sequence (10). In silico analysis of the 154-bp HP1511-HP1512 intergenic region identified a perfect match to the putative NikR binding region 60 bp upstream of the HP1512 start codon (Fig. 5A). DNA shift analysis (Fig. 5C) demonstrated that recombinant H. pylori NikR is capable of binding and subsequently shifting target DNA containing the HP1511-1512 intergenic region. These findings provide evidence for a direct role of NikR in the nickel-dependent repression of HP1512 transcription. In an independent study, Ernst et al. (14) demonstrated nickel-dependent NikR transcriptional repression and mapped the NikR binding region of HP1512. The spatial relationship among the NikR operator region and the promoters of nixA, HP1400, and HP1512 are consistent with NikR's role as a transcriptional repressor (14, 15). Furthermore, in a nikR mutant, both HP1512 and HP1400 are transcribed constitutively, regardless of the nickel concentration (14).
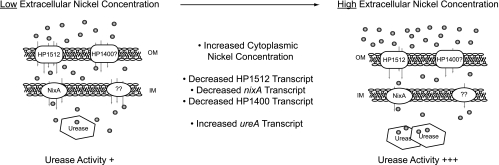
The addition of HP1512 to the nickel regulatory network of H. pylori allows for the development of a model to explain nickel trafficking from the extracellular environment to the cytoplasm (Fig. 6). Basal levels of nixA, HP1512, and urease expression can be adjusted to adapt to fluctuating nickel concentrations. Intracellular nickel concentrations modulate the expression of nickel uptake and utilization mechanisms. Thus, as cytoplasmic nickel concentrations increase, nickel ions associate with cytoplasmic NikR, which confers DNA binding capabilities to NikR. Nickel-bound NikR is able to repress transcription of nixA, HP1512, and as recently demonstrated (14), HP1400, resulting in decreased nickel uptake capacity. While the capacity to import nickel into the cytoplasm is reduced, the transcription of urease subunits is increased, resulting in the ability to utilize accumulating cytoplasmic nickel.
FIG. 6.
Regulation of nickel transport across the H. pylori inner and outer membranes. The addition of HP1512 to the nickel regulatory network of H. pylori allows for the development of a model to explain nickel trafficking from the extracellular environment to the cytoplasm. Basal levels of nixA, HP1512, and urease expression can be adjusted to adapt to fluctuating nickel concentrations. As cytoplasmic nickel concentrations increase, nickel ions associate with cytoplasmic NikR, which confers DNA binding capabilities to NikR. Nickel-bound NikR is able to repress transcription of nixA, HP1512, and HP1400, resulting in decreased nickel uptake capacity. While the capacity to import nickel into the cytoplasm is reduced, the transcription of urease subunits is increased, resulting in the ability to utilize accumulating cytoplasmic nickel.
There is no difference in nickel tolerance between the wild type and HP1512::cat mutant. It might be expected that a mutant exhibiting deficient nickel import would have a higher nickel tolerance; however, differences in nickel tolerance are ultimately regulated at the inner membrane. The comparative qPCR analysis of nixA transcripts in the wild type versus HP1512::cat mutant reveals increased nixA transcription in the HP1512 mutant relative to the wild type. Up-regulation of nixA suggests that the intracellular nickel concentration is lower in the HP1512::cat mutant than in the wild type and that the cell responds by increasing the capacity to transport nickel into the cytoplasm; that is, it increases transcription of the high-affinity nickel transport protein NixA. Compensation for decreased nickel availability by increasing nixA transcription could be masking the effect of mutating HP1512 in regard to nickel tolerance. However, a slight difference in nickel tolerance cannot be ruled out, as testing the nickel concentrations between those used in our assay may result in a reduced susceptibility in the HP1512 mutant relative to the wild type.
While beyond the scope of this study, it is at least interesting to speculate that frpB4-like OMPs may be unique to gastric helicobacters. Urease is essential for colonization of the gastric mucosa; thus, it would not be surprising if gastric helicobacters, with an absolute requirement for nickel, have evolved dedicated outer membrane nickel transport mechanisms. In both sequenced H. pylori strains, 26695 and J99, there are three copies each of fecA and frpB homologs. Alignment of the H. pylori frpB sequences, along with frpB homologs of Neisseria meningitidis, Brucella melitensis, Helicobacter hepaticus, and Vibrio parahaemolyticus followed by neighbor-joining analysis produced a distinct H. pylori clade. Within the H. pylori clade, sequences were grouped by frpB allele number, i.e., frpB1 alleles were more closely related to one another than to frpB2 alleles. Genes demonstrated to be iron responsive, H. pylori 26695 frpB1 and frpB2/3 and H. pylori J99 frpB1 and frpB2, were more closely related to one another than to the H. pylori 26695 frpB4 and H. pylori J99 frpB3 cluster. The H. hepaticus frpB homolog falls outside the H. pylori frpB cluster.
In conclusion, we have demonstrated that the OMPs HP1512 (FrpB4) and HP1400 (FecA3) are regulated at the transcriptional level in response to nickel. Insertional inactivation of HP1512 results in reduced urease activity in all H. pylori strains tested, and for strain 26695, the reduction in urease activity is statistically significant (P < 0.001). Transcriptional profiling of select nickel-responsive genes suggests that the cytoplasmic nickel concentration is reduced in the HP1512 mutant relative to the wild type; further studies with 63Ni could conclusively verify decreased nickel transport in the HP1512 mutant. DNA shift analysis suggests that the nickel-responsive transcriptional regulatory protein NikR binds the region immediately upstream of HP1512, likely serving as a transcriptional repressor in the presence of nickel. This work has expanded the regulatory network of NikR, which regulates transcription of genes responsible for nickel transport across the inner membrane and the ability to utilize nickel once inside the cytoplasm via up-regulation of the urease operon, to include transport across the outer membrane.
Acknowledgments
We thank Arnoud van Vliet for critically reading the manuscript.
This work was funded in part by Public Health Service grant AI25567 from the National Institutes of Health.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 9 October 2006.
REFERENCES
- 1.Abraham, L. O., Y. Li, and D. B. Zamble. 2006. The metal- and DNA-binding activities of Helicobacter pylori NikR. J. Inorg. Biochem. 100:1005-1014. [DOI] [PubMed] [Google Scholar]
- 2.Akada, J. K., M. Shirai, H. Takeuchi, M. Tsuda, and T. Nakazawa. 2000. Identification of the urease operon in Helicobacter pylori and its control by mRNA decay in response to pH. Mol. Microbiol. 36:1071-1084. [DOI] [PubMed] [Google Scholar]
- 3.Alm, R. A., J. Bina, B. M. Andrews, P. Doig, R. E. Hancock, and T. J. Trust. 2000. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect. Immun. 68:4155-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauerfeind, P., R. M. Garner, and L. T. Mobley. 1996. Allelic exchange mutagenesis of nixA in Helicobacter pylori results in reduced nickel transport and urease activity. Infect. Immun. 64:2877-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bury-Mone, S., J. M. Thiberge, M. Contreras, A. Maitournam, A. Labigne, and H. De Reuse. 2004. Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori. Mol. Microbiol. 53:623-638. [DOI] [PubMed] [Google Scholar]
- 6.Carlsohn, E., J. Nystrom, I. Bolin, C. L. Nilsson, and A. M. Svennerholm. 2006. HpaA is essential for Helicobacter pylori colonization in mice. Infect. Immun. 74:920-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chivers, P. T., and R. T. Sauer. 2002. NikR repressor: high-affinity nickel binding to the C-terminal domain regulates binding to operator DNA. Chem. Biol. 9:1141-1148. [DOI] [PubMed] [Google Scholar]
- 8.Chivers, P. T., and R. T. Sauer. 2000. Regulation of high affinity nickel uptake in bacteria. Ni2+-dependent interaction of NikR with wild-type and mutant operator sites. J. Biol. Chem. 275:19735-19741. [DOI] [PubMed] [Google Scholar]
- 9.Contreras, M., J. M. Thiberge, M. A. Mandrand-Berthelot, and A. Labigne. 2003. Characterization of the roles of NikR, a nickel-responsive pleiotropic autoregulator of Helicobacter pylori. Mol. Microbiol. 49:947-963. [DOI] [PubMed] [Google Scholar]
- 10.Delany, I., R. Ieva, A. Soragni, M. Hilleringmann, R. Rappuoli, and V. Scarlato. 2005. In vitro analysis of protein-operator interactions of the NikR and Fur metal-responsive regulators of coregulated genes in Helicobacter pylori. J. Bacteriol. 187:7703-7715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Pina, K., V. Desjardin, M. A. Mandrand-Berthelot, G. Giordano, and L. F. Wu. 1999. Isolation and characterization of the nikR gene encoding a nickel-responsive regulator in Escherichia coli. J. Bacteriol. 181:670-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunn, B. E., G. P. Campbell, G. I. Perez-Perez, and M. J. Blaser. 1990. Purification and characterization of urease from Helicobacter pylori. J. Biol. Chem. 265:9464-9469. [PubMed] [Google Scholar]
- 13.Eaton, K. A., C. L. Brooks, D. R. Morgan, and S. Krakowka. 1991. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect. Immun. 59:2470-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ernst, F. D., J. Stoof, W. M. Horrevoets, E. J. Kuipers, J. G. Kusters, and A. H. M. van Vliet. 2006. NikR mediates nickel-responsive transcriptional repression of the Helicobacter pylori outer membrane proteins FecA3 (HP1400) and FrpB4 (HP1512). Infect. Immun. 74:6821-6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ernst, F. D., E. J. Kuipers, A. Heijens, R. Sarwari, J. Stoof, C. W. Penn, J. G. Kusters, and A. H. van Vliet. 2005. The nickel-responsive regulator NikR controls activation and repression of gene transcription in Helicobacter pylori. Infect. Immun. 73:7252-7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ge, R., R. M. Watt, X. Sun, J. A. Tanner, Q. Y. He, J. D. Huang, and H. Sun. 2006. Expression and characterization of a histidine-rich protein, Hpn: potential for nickel storage in Helicobacter pylori. Biochem. J. 393:285-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilbert, J. V., J. Ramakrishna, F. W. Sunderman, Jr., A. Wright, and A. G. Plaut. 1995. Protein Hpn: cloning and characterization of a histidine-rich metal-binding polypeptide in Helicobacter pylori and Helicobacter mustelae. Infect. Immun. 63:2682-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reference deleted.
- 19.Ha, N. C., S. T. Oh, J. Y. Sung, K. A. Cha, M. H. Lee, and B. H. Oh. 2001. Supramolecular assembly and acid resistance of Helicobacter pylori urease. Nat. Struct. Biol. 8:505-509. [DOI] [PubMed] [Google Scholar]
- 20.Hu, L. T., and H. L. Mobley. 1990. Purification and N-terminal analysis of urease from Helicobacter pylori. Infect. Immun. 58:992-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, H. W., Y. H. Choe, D. K. Kim, S. Y. Jung, and N. G. Lee. 2004. Proteomic analysis of a ferric uptake regulator mutant of Helicobacter pylori: regulation of Helicobacter pylori gene expression by ferric uptake regulator and iron. Proteomics 4:2014-2027. [DOI] [PubMed] [Google Scholar]
- 22.Marcus, E. A., A. P. Moshfegh, G. Sachs, and D. R. Scott. 2005. The periplasmic α-carbonic anhydrase activity of Helicobacter pylori is essential for acid acclimation. J. Bacteriol. 187:729-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marshall, B. J., L. J. Barrett, C. Prakash, R. W. McCallum, and R. L. Guerrant. 1990. Urea protects Helicobacter (Campylobacter) pylori from the bactericidal effect of acid. Gastroenterology 99:697-702. [DOI] [PubMed] [Google Scholar]
- 24.Merrell, D. S., and A. Camilli. 2002. Acid tolerance of gastrointestinal pathogens. Curr. Opin. Microbiol. 5:51-55. [DOI] [PubMed] [Google Scholar]
- 25.Mobley, H. L., R. M. Garner, and P. Bauerfeind. 1995. Helicobacter pylori nickel-transport gene nixA: synthesis of catalytically active urease in Escherichia coli independent of growth conditions. Mol. Microbiol. 16:97-109. [DOI] [PubMed] [Google Scholar]
- 26.Mobley, H. L., R. M. Garner, G. R. Chippendale, J. V. Gilbert, A. V. Kane, and A. G. Plaut. 1999. Role of Hpn and NixA of Helicobacter pylori in susceptibility and resistance to bismuth and other metal ions. Helicobacter 4:162-169. [DOI] [PubMed] [Google Scholar]
- 27.Mobley, H. L., M. D. Island, and R. P. Hausinger. 1995. Molecular biology of microbial ureases. Microbiol. Rev. 59:451-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navarro, C., L. F. Wu, and M. A. Mandrand-Berthelot. 1993. The nik operon of Escherichia coli encodes a periplasmic binding-protein-dependent transport system for nickel. Mol. Microbiol. 9:1181-1191. [DOI] [PubMed] [Google Scholar]
- 29.Nies, D. H. 2003. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27:313-339. [DOI] [PubMed] [Google Scholar]
- 30.Nolan, K. J., D. J. McGee, H. M. Mitchell, T. Kolesnikow, J. M. Harro, J. O'Rourke, J. E. Wilson, S. J. Danon, N. D. Moss, H. L. Mobley, and A. Lee. 2002. In vivo behavior of a Helicobacter pylori SSI nixA mutant with reduced urease activity. Infect. Immun. 70:685-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olson, J. W., and R. J. Maier. 2002. Molecular hydrogen as an energy source for Helicobacter pylori. Science 298:1788-1790. [DOI] [PubMed] [Google Scholar]
- 32.Pflock, M., S. Kennard, I. Delany, V. Scarlato, and D. Beier. 2005. Acid-induced activation of the urease promoters is mediated directly by the ArsRS two-component system of Helicobacter pylori. Infect. Immun. 73:6437-6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saeed, A. I., V. Sharov, J. White, J. Li, W. Liang, N. Bhagabati, J. Braisted, M. Klapa, T. Currier, M. Thiagarajan, A. Sturn, M. Snuffin, A. Rezantsev, D. Popov, A. Ryltsov, E. Kostukovich, I. Borisovsky, Z. Liu, A. Vinsavich, V. Trush, and J. Quackenbush. 2003. TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34:374-378. [DOI] [PubMed] [Google Scholar]
- 34.Schreiber, S., M. Konradt, C. Groll, P. Scheid, G. Hanauer, H. O. Werling, C. Josenhans, and S. Suerbaum. 2004. The spatial orientation of Helicobacter pylori in the gastric mucus. Proc. Natl. Acad. Sci. USA 101:5024-5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silver, S. 1996. Bacterial resistances to toxic metal ions—a review. Gene 179:9-19. [DOI] [PubMed] [Google Scholar]
- 36.Stingl, K., K. Altendorf, and E. P. Bakker. 2002. Acid survival of Helicobacter pylori: how does urease activity trigger cytoplasmic pH homeostasis? Trends Microbiol. 10:70-74. [DOI] [PubMed] [Google Scholar]
- 37.Stingl, K., E.-M. Uhlemann, G. Deckers-Hebestreit, R. Schmid, E. P. Bakker, and K. Altendorf. 2001. Prolonged survival and cytoplasmic pH homeostasis of Helicobacter pylori at pH 1. Infect. Immun. 69:1178-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomsen, L. L., J. B. Gavin, and C. Tasman-Jones. 1990. Relation of Helicobacter pylori to the human gastric mucosa in chronic gastritis of the antrum. Gut 31:1230-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 40.Tsuda, M., M. Karita, M. G. Morshed, K. Okita, and T. Nakazawa. 1994. A urease-negative mutant of Helicobacter pylori constructed by allelic exchange mutagenesis lacks the ability to colonize the nude mouse stomach. Infect. Immun. 62:3586-3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Vliet, A. H., F. D. Ernst, and J. G. Kusters. 2004. NikR-mediated regulation of Helicobacter pylori acid adaptation. Trends Microbiol. 12:489-494. [DOI] [PubMed] [Google Scholar]
- 42.van Vliet, A. H., E. J. Kuipers, B. Waidner, B. J. Davies, N. de Vries, C. W. Penn, C. M. Vandenbroucke-Grauls, M. Kist, S. Bereswill, and J. G. Kusters. 2001. Nickel-responsive induction of urease expression in Helicobacter pylori is mediated at the transcriptional level. Infect. Immun. 69:4891-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Vliet, A. H., S. W. Poppelaars, B. J. Davies, J. Stoof, S. Bereswill, M. Kist, C. W. Penn, E. J. Kuipers, and J. G. Kusters. 2002. NikR mediates nickel-responsive transcriptional induction of urease expression in Helicobacter pylori. Infect. Immun. 70:2846-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Vliet, A. H., J. Stoof, R. Vlasblom, S. A. Wainwright, N. J. Hughes, D. J. Kelly, S. Bereswill, J. J. Bijlsma, T. Hoogenboezem, C. M. Vandenbroucke-Grauls, M. Kist, E. J. Kuipers, and J. G. Kusters. 2002. The role of the ferric uptake regulator (Fur) in regulation of Helicobacter pylori iron uptake. Helicobacter 7:237-244. [DOI] [PubMed] [Google Scholar]
- 45.Voland, P., D. L. Weeks, E. A. Marcus, C. Prinz, G. Sachs, and D. Scott. 2003. Interactions among the seven Helicobacter pylori proteins encoded by the urease gene cluster. Am. J. Physiol. Gastrointest. Liver Physiol. 284:G96-G106. [DOI] [PubMed] [Google Scholar]
- 46.Wang, Y., and D. E. Taylor. 1990. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene 94:23-28. [DOI] [PubMed] [Google Scholar]
- 47.Weatherburn, M. W. 1967. Phenol-hypochlorite reaction for determination of ammonia. Anal. Chem. 39:971-974. [Google Scholar]
- 48.Wolfram, L., E. Haas, and P. Bauerfeind. 2006. Nickel represses the synthesis of the nickel permease NixA of Helicobacter pylori. J. Bacteriol. 188:1245-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu, L. F., and M. A. Mandrand-Berthelot. 1986. Genetic and physiological characterization of new Escherichia coli mutants impaired in hydrogenase activity. Biochimie 68:167-179. [DOI] [PubMed] [Google Scholar]