Abstract
Lyme disease Borrelia organisms are highly invasive spirochetes that alternate between vertebrate and arthropod hosts and that establish chronic infections and elicit inflammatory reactions in mammals. Although progress has been made in the targeted mutagenesis of individual genes in infectious Borrelia burgdorferi, the roles of the vast majority of gene products in pathogenesis remain unresolved. In this study, we examined the feasibility of using transposon mutagenesis to identify infectivity-related factors in B. burgdorferi. The transformable, infectious strain 5A18 NP1 was transformed with the spirochete-adapted Himar1 transposon delivery vector pMarGent to create a small library of 33 insertion mutants. Single mouse inoculations followed by culture of four tissue sites and serology were used to screen the mutants for infectivity phenotypes. Mutants that appeared attenuated (culture positive at some sites) or noninfectious (negative at all sites) and contained the virulence-associated plasmids lp25 and lp28-1 were examined in more extensive animal studies. Three of these mutants (including those with insertions in the putative fliG-1-encoded flagellar motor switch protein and the guaB-encoded IMP dehydrogenase) were noninfectious, whereas four clones appeared to exhibit reduced infectivity. Serological reactivity in VlsE enzyme-linked immunosorbent assays correlated with the assignment of mutants to the noninfectious or attenuated-infectivity groups. The results of this study indicate that random transposon mutagenesis of infectious B. burgdorferi is feasible and will be of value in studying the pathogenesis of Lyme disease Borrelia.
Spirochetes that cause Lyme disease (including Borrelia burgdorferi, B. garinii, and B. afzelii) are endemic to regions of the United States and Eurasia and causes a systemic illness that can result in chronic manifestations in humans, including neurological symptoms and arthritis (4, 27). These organisms have a complex life cycle that requires adaptation to disparate environments in the tick and mammalian hosts and therefore must possess a number of mechanisms involved in transmission, growth, dissemination, and persistence. To date, the characterization of potential virulence factors has focused on the use of homologous recombination for targeted insertional mutagenesis (26). Although genetic studies of Lyme disease pathogenesis have been hampered by the low transformation rate of infectious, low-passage-number B. burgdorferi strains relative to that of high-passage-number, noninfectious strains, significant progress has been made in recent years (26).
The genome of B. burgdorferi contains multiple linear and circular plasmids (7, 10); it has been shown that the linear plasmids lp25 and lp28-1 are required for full infectivity in mice (11, 16, 17, 23-25). lp25 encodes PncA, a nicotinamidase involved in NAD biosynthesis (23), and BptA, a protein essential for tick colonization and also implicated in mouse infection (25). lp28-1 contains the vls antigenic variation locus, including the gene for the variable surface lipoprotein VlsE (32). Lawrenz et al. (19) found that transformation of low-passage-number, infectious B. burgdorferi B31 containing all native plasmids with the shuttle vector pBSV2 (29) was inefficient; the only transformants isolated in that study had lost lp25. A survey of low-passage-number clones with various plasmid contents indicated that the transformation frequency with pBSV2 was low when both lp25 and lp56 were present, intermediate when either one or the other was missing, or high when both were missing (19). Examination of the open reading frames (ORFs) of these two plasmids revealed that each encoded a large predicted protein with both restriction endonuclease and DNA methyltransferase motifs. These predicted products (encoded by BBE02 on lp25 and BBQ67 on lp56) are similar to type IV restriction endonucleases (such as Eco57I) that possess both methyltransferase and endonuclease activities in one protein. The plasmid lp28-3 also encodes a putative restriction-modification protein, BBH09, that is 92% similar to BBE02 despite having a different pI. The effect of BBH09 on transformation frequency has not been examined (19).
Based on these data, it was hypothesized that BBE02 and BBQ67 interfere with transformation by cleaving incoming DNA before it can be methylated at the corresponding restriction sites. To examine this possibility, two infectious isolates containing an insertional mutation in BBE02 have been developed: 5A4 NP1, which lacks only cp9, and 5A18 NP1, which is missing lp56 and lp28-4 (15). Disruption of BBE02 increased the transformation frequency with the shuttle vector pBSV2C03::gntΔkan to intermediate levels in 5A4 NP1 and to relatively high levels in 5A18 NP1, correlating with the lack of lp56 (and hence BBQ67) in the latter clone. Both 5A4 NP1 and 5A18 NP1 retained full infectivity in mice as measured by median infectious dose following needle inoculation (15).
Transposon mutagenesis has been an important tool for the identification of potential virulence factors in other pathogenic bacteria. Two recent reports described the successful in vivo transposition of noninfectious B. burgdorferi with mariner-based transposons (21, 28). Stewart et al. (28) utilized the suicide vector pMarGent to deliver a Himar1 transposon that contains a flgBp::aacC1 cassette, thus conferring gentamicin resistance. An important feature of pMarGent is the presence of the hyperactive C9 transposase expressed from the Borrelia promoter flgBp, thus increasing expression of the transposase following transformation. The noninfectious strains A3-89 and B31-AchbC72 exhibited high transposition frequencies, correlating with the absence of lp25 and lp56 in these strains. In contrast, no mutants were obtained in the infectious B. burgdorferi strains B31-A3 and N40, most likely due to the presence of restriction-modification systems in these strains.
In another study, Morozova et al. (21) utilized the broad-host-range vector pED7, which replicates at a low copy number in B. burgdorferi, to express the C9 Himar1 transposase under control of the Borrelia flaB promoter (flaBp). B. burgdorferi B31 containing pED7 was transformed with three different suicide plasmids, containing Himar1 inverted repeat sequences, a kanamycin resistance cassette, and a colE1 origin of replication. Relatively high transposition frequencies were obtained using the noninfectious, high-passage-number B. burgdorferi strain, but transposition of infectious strains of B. burgdorferi was not reported in this analysis (21).
In the current study, we tested the hypothesis that potential infectivity-related factors could be found by transposition of the infectious B. burgdorferi clone 5A4 NP1 or 5A18 NP1 with pMarGent. Our findings indicate that transposon mutants can be obtained using the combination of 5A18 NP1 and pMarGent, and clones with reduced infectivity can be isolated by this means.
MATERIALS AND METHODS
Bacterial strains and vector.
B. burgdorferi was grown in a humidified incubator at 34°C, 3% CO2, using Barbour-Stoenner-Kelly medium without gelatin (BSK II [2]) prepared in-house. Borrelia burgdorferi B31 5A4 NP1 and 5A18 NP1 are infectious isolates derived from the B31 isolates 5A4 and 5A18, respectively. Clone 5A4 contains all plasmids, whereas 5A18 lacks plasmids lp28-4 and lp56 (24). During the course of manipulations, the 5A4 NP1 derivative had lost cp9, while 5A18 NP1 had retained its parental plasmid content. The NP1 strains contain an insertionally inactivated BBE02 allele (Kanr) that results in an increase in transformation efficiency (15). Escherichia coli GC10 (GeneChoice, Frederick, MD) was used for transposon recovery steps. Kanamycin was used at a concentration of 200 μg/ml with B. burgdorferi; gentamicin was used at a concentration of 40 μg/ml with B. burgdorferi and 7 μg/ml with E. coli. pMarGent (28) is a Himar1-based transposon vector with a ColE1 origin of replication and aacC1 (conferring gentamicin resistance) expressed from the B. burgdorferi flgB promoter (flgBp::aacC1) within the Himar1 inverted repeat sequences and the C9 variant of the Himar1 transposase with the flgB promoter in the region outside the transposable element. The plasmid replicates in E. coli but acts as a suicide vector in B. burgdorferi.
Isolation and confirmation of pMarGent transposition mutants.
Transformation of B. burgdorferi with pMarGent was performed as described previously (28). Briefly, electrocompetent B. burgdorferi 5A4 NP1 and 5A18 NP1 were prepared from mid-to-late-log-phase cultures by multiple centrifugation/wash steps, using decreasing volumes of electroporation solution (9.3% sucrose [wt/vol], 15% glycerol). A 50-μl aliquot of cells (∼5 × 109 cells, total) was electroporated with 10 μg of pMarGent (28) using the following parameters: 2.5 kV, 25 μF, 200 Ω. Cells were allowed to recover for 24 h in BSK II without antibiotics before subsurface plating in BSK II agar plates with gentamicin and kanamycin. The presence of the pMarGent transposon in each of the recovered clones was confirmed by PCR of the aacC1 cassette region. PCR was performed using the forward primer 5185 (5′-TTA CGC AGC AGC AAC GAT GTT ACG CA-3′) and the reverse primer 5175 (5′-ACA TGC ATG CCG ATC TCG GCT TGA ACG-3′) to amplify ∼560 bp from aacC1. The reaction mixture was denatured at 94°C for 2 min, followed by five cycles of 94°C, 30 s; 51°C, 45 s; and 72°C, 45 s. Another 30 cycles of 94°C for 30 s, 60°C for 45 s, and 72°C for 45 s was followed by a final extension at 72°C for 10 min. Products were analyzed by agarose gel electrophoresis.
Identification of insertion sites.
Transposon recovery was performed as previously described (28); i.e., genomic DNA was prepared from stationary-phase B. burgdorferi, digested with HindIII, self-ligated, and chemically transformed into E. coli GC10. Plasmid DNA isolated from gentamicin-resistant colonies was sequenced using primers directed outward from the transposon and into borrelial flanking DNA (28). The sequence was used in a nucleotide BLAST search (1) at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) to determine the location of each insertion site.
Infectivity studies.
Transposon mutants were initially screened for infectivity by inoculation of one mouse per clone. For each of the 33 mutants, 1 female C3H/HeN mouse (Harlan, Indianapolis, IN) was infected with 1 × 104 log-phase B. burgdorferi organisms intradermally at the base of the tail as described previously (22). The mice were sacrificed after 2 weeks, blood was collected for enzyme-linked immunosorbent assays (ELISAs), and tissue samples from the ear, heart, urinary bladder, and tibiotarsal joint were cultured in 6 ml BSK II containing kanamycin and gentamicin. The cultures were examined for spirochetes by dark-field microscopy at 2 and 4 weeks after sacrifice. The presence of at least 1 spirochete in 10 high-power fields indicated a positive result; conversely, the absence of spirochetes in 10 high-power fields indicated a negative result. To confirm the noninfectious or attenuated infectivity phenotypes of various mutants, groups of 2 female C3H/HeN mice (Charles River Laboratories, Wilmington, MA, or Harlan, Indianapolis, IN) were inoculated intradermally with each of 3 different doses: 1 × 102, 1 × 104, or 1 × 106. Mice were sacrificed after 2 weeks and processed as described for the single mouse inoculations. Differences in culture positivity between mutants with reduced infectivity and the parental strain were examined by chi-square analysis using pooled results from 104 and 106 inocula (GraphPad Prism, version 4.03 for Windows; GraphPad Software, San Diego, California [www.graphpad.com]). Culture results for mutants were considered significantly different from those for the 5A18 NP1 parental strain at P values of <0.05.
Plasmid profiling.
The plasmid content of transposon mutants was determined by a microtiter plate-based PCR method. Template genomic DNA (50 to 100 ng per reaction) was prepared from B. burgdorferi cultures or by amplification from glycerol stocks or genomic DNA using the GenomiPhi DNA amplification system (Amersham Biosciences, Piscataway, NJ). Unique primer sets were used to amplify a product from each of the linear and circular plasmids present in B. burgdorferi (24). The reaction mixture was denatured at 94°C for 2 min, followed by 30 cycles of 94°C, 30 s; 44°C, 30 s; 72°C, 1 min; and a final extension at 72°C for 10 min. Products were analyzed by agarose gel electrophoresis.
ELISA.
Serum reactivity against the recombinant VlsE1 protein was analyzed by ELISA as previously described (18) with the following modifications. Microtiter plates were coated with 50 ng per well of recombinant VlsE1. One hundred microliters of primary sera diluted 1:200 in phosphate-buffered saline with 1% milk was added to triplicate wells. Alkaline-phosphatase-conjugated goat antimouse secondary antibody (Pierce, Rockford, IL) was diluted 1:10,000 in phosphate-buffered saline with 0.05% Tween 20. Absorbance at 405 nm was measured with a μQuant spectrophotometer (Bio-Tek Instruments, Winooski, VT). Background reactivity values (mean optical density at 405 nm [OD405] readings from control wells lacking primary antibody) were subtracted from all values on that plate. A cutoff value of 0.010 was determined by calculating 2 standard deviations above the mean reactivity of preimmune sera from 32 mice. OD readings of >0.010 were considered reactive, whereas those of ≤0.010 were deemed nonreactive. ELISAs using B. burgdorferi B31 5A4 whole-cell sonicate were performed as described above, using 600 ng protein per well as the antigen; in this case, the cutoff value was 0.014. Statistically significant differences in the mean reactivity to each mutant compared to the mean reactivity to the parental strain, 5A18 NP1, were analyzed using a two-tailed Student t test. Significance was assigned at P values of <0.05.
RESULTS
Isolation of pMarGent transposition mutants and insertion site identification.
No gentamicin-resistant colonies were obtained in two experiments when 5A4 NP1 was transformed with either pMarGent (28) or the shuttle vector pBSV2-G, which also carries the gentamicin resistance cassette flgBp::aacC1 (8). This result indicates that transformation with pMarGent (or pBSV2-G) is still an inefficient process if BBE02 is disrupted but lp56 (and hence BBQ67) is still present. In contrast, transformation of 5A18 NP1 with pMarGent resulted in 4 gentamicin-resistant colonies in the first transformation and 29 such colonies in the second experiment. The 33 gentamicin-resistant clones were designated MG001 to MG033. To confirm the presence of the gentamicin resistance cassette in the clones, an aliquot of liquid culture or isolated DNA was used as a template in PCR to amplify a region of aacC1. Bands of the predicted size were found in all clones (data not shown). The transposon recovery procedure described by Stewart et al. (28) was used to determine the insertion site of each of the mutants. The precise insertion sites were successfully identified in 31 of the 33 mutants; 6 were located in the chromosome, 12 in the linear plasmids, and 13 in the circular plasmids. Sequencing revealed that each of the 31 transposon insertions occurred at a TA dinucleotide Himar1 recognition site (data not shown), indicating that each mutant was the result of transposition and not an illegitimate recombination event. Overall, 12 (39%) of the insertions mapped to locations with an identified database match, 10 (∼32%) mapped to hypothetical genes, 1 (∼3%) was found in a conserved hypothetical gene, and 8 (∼26%) of the insertions were found in intergenic regions (Table 1). Two transposon mutants remain uncharacterized (MG001 and MG013), despite repeated efforts and modifications to the procedure, including the use of inverse PCR (12, 21) and attempts to rescue the insertion site region by using different restriction enzymes.
TABLE 1.
Insertion sites of pMarGent mutants (grouped by infectivity phenotype)
| Infectivitya | Clone | Replicon/positionb | Insertion site(s)c | Description |
|---|---|---|---|---|
| + | MG004 | Chrom./108542 | BB0110 (108307-109671) | Hypothetical protein |
| + | MG005 | cp32-6/24624 | BBM36 (24619-25044) | Borrelia plasmid protein B (bppB) (putative) |
| + | MG006 | lp36/17582 | BBK25 (17565-16966) promoter | Transposase-like protein (putative), authentic frameshift |
| + | MG007 | Chrom./534145 | Intergenic; BB0524 (534301-535115) and BB0523 (533980-534072) | BB0524 possible myoinositol-1 (or 4)- monophosphatase (putative); BB0523 is a hypothetical protein |
| + | MG010 | cp32-3/27222 | BBS41 (26708-27298) | Outer surface protein G (erpG) |
| + | MG011 | lp28-3/4210 | BBH09 (7728-3892) | Hypothetical protein |
| + | MG012 | lp28-1/4515 | BBF10 (4488-4985) | Hypothetical protein |
| + | MG016 | cp32-8/25599 | BBL38 (25951-25175) | Borrelia plasmid protein C (bppC) (putative) |
| + | MG017 | lp38/33790 | Intergenic; BBJ45 (32498-33220) and BBJ46 (34668-34372) | BBJ45 and BBJ46 are hypothetical proteins |
| + | MG018 | cp32-4/24618 | BBR37 (24210-24635) | Borrelia plasmid protein B (bppB) (putative) |
| + | MG020 | cp26/23390 | BBB28 (23255-24499) | Hypothetical protein |
| + | MG025 | lp21/7829 | Intergenic; BBU05 (2868-3656) and BBU06 (14633-15235) | Within central 63-bp repeat region of lp21 |
| + | MG026 | lp36/23615 | BBK37 (23953-22910) | Homolog of immunogenic protein P37 |
| + | MG027 | cp32-4/7055 | BBR11 (6711-7823) | Hypothetical protein |
| + | MG028 | cp32-4/17887 | BBR29 (18664-17573) | Conserved hypothetical protein |
| + | MG029 | lp28-1/5088 | Intergenic; BBF10 (4488-4985) and BBF11 (5435-5542) | BBF10 and BBF11 are hypothetical proteins |
| + | MG030 | cp26/25676 | BBB29 (24825-26453) | Phosphotransferase system enzyme, maltose- and glucose-specific IIABC component (malX) |
| + | MG032 | lp17/9413 | Intergenic; BBD14 (8269-9381) and BBD15 (10015-9593) | BBD14 is a conserved hypothetical protein and BBD15 is a hypothetical protein |
| +/− (lp28-1−) | MG008 | Chrom./660583 | BB0629 (661524-659644) | Phosphotransferase system enzyme II (fruA-2) |
| +/− (lp28-1−) | MG015 | lp38/25996 | BBJ34 (26407-25337) | Hypothetical protein |
| +/− (lp28-1−) | MG021 | lp28-2/20360 | BBG24 (22310-19620) | Hypothetical protein |
| +/− (lp28-1−) | MG022 | lp25/21344 | Intergenic; BBE29 (19697-20886) and BBE30 (21558-21704) | BBE29, putative adenine-specific methyltransferase, authentic frameshift; BBE30, hypothetical protein |
| +/− (lp28-1−) | MG023 | lp21/6922 | Intergenic; BBU05 (2868-3656) and BBU06 (14633-15235) | Within central 63-bp repeat region of lp21 |
| +/−(lp28-1−) | MG031 | cp26/12701 | BBB16 (12014-13606) | Oligopeptide ABC transporter, periplasmic oligopeptide-binding protein (oppAIV) |
| +/− (lp28-1+) | MG009 | Chrom./786112 | BB0743 (785057-786754) | Hypothetical protein |
| +/− (lp28-1+) | MG013 | Not determined | ||
| +/− (lp28-1+) | MG014 | Chrom./841814 | BB0797 (839244-841832) | DNA mismatch repair protein (mutS) |
| +/− (lp28-1+) | MG019 | cp9/5623 | BBC08 (5534-5980) | Conserved hypothetical protein |
| +/− (lp28-1+) | MG033 | cp32-7/22694 | BBO35 (22755-22627) | Hypothetical protein |
| − | MG001 | Not determined | ||
| − | MG002 | cp26/14424 | BBB17 (15107-13893) | IMP dehydrogenase (guaB) |
| − (lp25−) | MG003 | cp26/11461 | Intergenic between BBB14 (11417-10920) and BBB15 (11636-11740) | BBB14 and BBB15 are hypothetical proteins |
| − | MG024 | Chrom./226987 | BB0221 (227836-226610) | Flagellar motor switch protein (fliG-1) |
+, culture positive at all sites; +/−, culture positive at some sites; −, culture negative at all sites.
Chrom., chromosome.
Position of gene given in parentheses. Reverse coordinates indicate that the gene is encoded on the negative strand.
Infectivity studies.
Each of the mutants was screened initially for infectivity using single mouse inoculations with a relatively large dose of 104 organisms/mouse. The transposon mutant clones were divided into three tentative infectivity phenotypes, depending on the pattern of spirochete isolation from the four tissues examined. Mutants isolated from all four tissues were considered to be fully infectious and were not examined further in this study. Transposon mutants isolated from one to three tissues were provisionally designated attenuated, and mutants not isolated from any of the four tissues were tentatively considered noninfectious. Of the 33 mutants tested using single mouse inoculations, 18 (55%) were fully infectious, 11 (33%) appeared to be attenuated, and 4 (12%) were considered noninfectious.
The noninfectious mutants and attenuated mutants were analyzed for the presence of the vlsE-encoding plasmid lp28-1. PCR revealed that 6 of the 11 attenuated mutants had lost lp28-1, a plasmid required for full infectivity of B. burgdorferi (16, 24, 32). B. burgdorferi strains lacking lp28-1 exhibit reduced infectivity in mice; typically, mice inoculated with lp28-1− clones are culture positive in only a limited number of cultures from joint and other tissues at 2 weeks postinoculation and are consistently culture negative at 4 weeks after inoculation (16, 24, 32). Further plasmid profiling demonstrated that the noninfectious clone MG003 is missing lp25, a plasmid required for infectivity of mice (11, 16, 17, 23-25). Its ability to grow in BSK II containing kanamycin was therefore likely due to a spontaneous mutation, since the kanamycin resistance cassette on lp25 was no longer present. Therefore, the lp28-1− attenuated clones MG008, MG015, MG021, MG022, MG023, and MG031 and the lp25− clone MG003 were excluded from further infectivity studies, since their attenuation appeared to be the result of plasmid loss and not transposition. Clone MG033 could not be consistently recovered from frozen stocks and was also excluded from this study.
Seven transposon mutants containing lp28-1 and lp25 had reduced infectivity in the initial screen. To confirm the attenuated and noninfectious phenotypes of these mutants, two replicate studies were performed using intradermal inoculation of 106, 104, or 102 organisms and 2 mice per inoculum. After 2 weeks, the mice were sacrificed and tissue from the ear, heart, bladder, and joint was cultured in BSK II. The plasmid content of these clones was also determined, as described below. For each of the seven mutants, chi-square analyses indicated that the number of culture-positive organ sites from mice inoculated with 104 or 106 organisms was significantly lower than that with the parental strain, 5A18 NP1 (P < 0.05).
The combined results of these experiments are shown in Table 2. The parental clone, 5A18 NP1, was isolated from 16/16 sites at an inoculum of 106 organisms and 15/16 sites at an inoculum of 1 × 104 organisms; however, positive cultures were not obtained from the mice inoculated with 102 bacteria. A median infectious dose of 83 organisms was previously obtained for 5A18 NP1 (15); a higher dose was required for infection in the current study, perhaps due to plasmid loss within the population (9).
TABLE 2.
Infectivities of noninfectious and lp28-1+ attenuated mutantsa
| Strain and inoculum | No. of tissue samples infected/no. tested
|
No. of mice infected/no. tested | No. of mice reactive/no. testedc | ||||
|---|---|---|---|---|---|---|---|
| Ear | Bladder | Heart | Joint | All sitesb | |||
| 5A18 NP1 (parental strain) | |||||||
| 102 | 0/4 | 0/4 | 0/4 | 0/4 | 0/16 | 0/4 | 0/4 |
| 104 | 3/4 | 4/4 | 4/4 | 4/4 | 15/16 | 4/4 | 4/4 |
| 106 | 4/4 | 4/4 | 4/4 | 4/4 | 16/16 | 4/4 | 4/4 |
| Single mouse (104) | 1/1 | 1/1 | 1/1 | 1/1 | 4/4 | 1/1 | 1/1 |
| Total | 8/13 | 9/13 | 9/13 | 9/13 | 35/52 | 9/13 | 9/13 |
| MG001 (insertion site ND; no plasmids lost) | |||||||
| 102 | 0/4 | 0/4 | 0/4 | 0/4 | 0/16 | 0/4 | 0/4 |
| 104 | 0/4 | 0/4 | 0/4 | 0/4 | 0/16 | 0/4 | 1/4 |
| 106 | 0/4 | 0/4 | 0/4 | 0/4 | 0/16 | 0/4 | 2/4 |
| Single mouse (104) | 0/1 | 0/1 | 0/1 | 0/1 | 0/4 | 0/1 | 1/1 |
| Total | 0/13 | 0/13 | 0/13 | 0/13 | 0/52 | 0/13 | 4/13 |
| MG002 (guaB; cp9− cp32-3−) | |||||||
| 102 | 0/4 | 0/4 | 0/4 | 0/4 | 0/16 | 0/4 | 0/4 |
| 104 | 0/4 | 0/4 | 0/4 | 0/4 | 0/16 | 0/4 | 0/4 |
| 106 | 0/4 | 0/4 | 0/4 | 0/4 | 0/16 | 0/4 | 1/4 |
| Single mouse (104) | 0/1 | 0/1 | 0/1 | 0/1 | 0/4 | 0/1 | 0/1 |
| Total | 0/13 | 0/13 | 0/13 | 0/13 | 0/52 | 0/13 | 1/13 |
| MG024 (fliG-1; no plasmids lost) | |||||||
| 102 | 0/4 | 0/4 | 0/4 | 0/4 | 0/16 | 0/4 | 0/4 |
| 104 | 0/4 | 0/4 | 0/4 | 0/4 | 0/16 | 0/4 | 0/4 |
| 106 | 0/4 | 0/4 | 0/4 | 0/4 | 0/16 | 0/4 | 0/4 |
| Single mouse (104) | 0/1 | 0/1 | 0/1 | 0/1 | 0/4 | 0/1 | 0/1 |
| Total | 0/13 | 0/13 | 0/13 | 0/13 | 0/52 | 0/13 | 0/13 |
| MG009 (BB0743; cp9−) | |||||||
| 102 | 0/4 | 1/4 | 1/4 | 2/4 | 4/16 | 2/4 | 2/4 |
| 104 | 1/4 | 4/4 | 4/4 | 3/4 | 12/16 | 4/4 | 4/4 |
| 106 | 0/4 | 4/4 | 3/4 | 4/4 | 11/16 | 4/4 | 4/4 |
| Single mouse (104) | 0/1 | 1/1 | 1/1 | 1/1 | 3/4 | 1/1 | 1/1 |
| Total | 1/13 | 10/13 | 9/13 | 10/13 | 30/52 | 11/13 | 11/13 |
| MG013 (insertion site ND; cp9−) | |||||||
| 102 | 0/4 | 1/4 | 0/4 | 1/4 | 2/16 | 1/4 | 0/4 |
| 104 | 0/4 | 2/4 | 1/4 | 3/4 | 6/16 | 3/4 | 4/4 |
| 106 | 2/4 | 4/4 | 0/4 | 4/4 | 10/16 | 4/4 | 4/4 |
| Single mouse (104) | 0/1 | 1/1 | 0/1 | 1/1 | 2/4 | 1/1 | 1/1 |
| Total | 2/13 | 8/13 | 1/13 | 9/13 | 20/52 | 9/13 | 9/13 |
| MG014 (mutS; cp9−, cp32-6−) | |||||||
| 102 | 0/4 | 0/4 | 0/4 | 0/4 | 0/16 | 0/4 | 0/4 |
| 104 | 2/4 | 3/4 | 2/4 | 3/4 | 10/16 | 3/4 | 3/4 |
| 106 | 1/4 | 4/4 | 3/4 | 4/4 | 12/16 | 4/4 | 4/4 |
| Single mouse (104) | 0/1 | 1/1 | 1/1 | 1/1 | 3/4 | 1/1 | 1/1 |
| Total | 3/13 | 8/13 | 6/13 | 8/13 | 25/52 | 8/13 | 8/13 |
| MG019 (BBC08; lp38−) | |||||||
| 102 | 0/4 | 0/4 | 0/4 | 0/4 | 0/16 | 0/4 | 0/4 |
| 104 | 0/4 | 1/4 | 0/4 | 1/4 | 2/16 | 1/4 | 0/4 |
| 106 | 0/4 | 2/4 | 1/4 | 0/4 | 3/16 | 2/4 | 2/4 |
| Single mouse (104) | 0/1 | 0/1 | 1/1 | 1/1 | 2/4 | 1/1 | 1/1 |
| Total | 0/13 | 3/13 | 2/13 | 2/13 | 7/52 | 4/13 | 3/13 |
Transposon insertion sites and plasmids missing (in addition to lp28-4 and lp56, which are absent in the parental strain 5A18 NP1) are indicated for each clone. Single mouse, results from single mouse inoculation study (see Table 1). ND, not determined.
χ2 analysis found the combined 104 and 106 inoculum data for each of the mutants to be statistically different from those for the 5A18 NP1 control (P < 0.05).
Reactive mice are those for which a significant serological response was demonstrated in a VlsE ELISA.
The clones MG001, MG002, and MG024 were noninfectious at the three different inocula tested, as predicted from the results of the single mouse inoculations for these clones (Table 2). The transposon insertion site for MG002 is within the cp26 gene for the IMP dehydrogenase GuaB, whereas the insertion point in MG024 is within the chromosomal gene encoding the predicted flagellar motor switch protein FliG-1 (Table 1). The location of the transposon mutation in MG001 has not been determined.
Most of the clones tentatively identified as attenuated during single mouse inoculations exhibited reduced infectivity in the multiple-dose experiments (Table 2). Clone MG009 (with a transposon insertion in the hypothetical gene BB0743) was able to colonize tissues at doses as low as 102 organisms, indicative of full infectivity; however, only 1/12 ear cultures were positive. Clone MG013 (transposon location not determined) had an interesting pattern in that only 1/12 of the heart cultures and 2/12 of the ear cultures were positive, whereas the bladder and joint cultures were positive in 7/12 and 8/12 specimens, respectively. The mutS (BB0797) mutant MG014 appeared to have a reduced infection rate, most notably in the ear tissue specimens, where only 3/12 cultures were positive. Clone MG019 (containing an insertion in the cp9 conserved hypothetical gene BBC08) was the most attenuated, with only 5 of 48 cultures being positive. Because cp9 (and hence BBC08) is not required for infectivity (24), the reduced infectivity of MG019 may be related to the absence of the plasmid lp38 in this clone rather than the transposon insertion.
Antibody responses to VlsE.
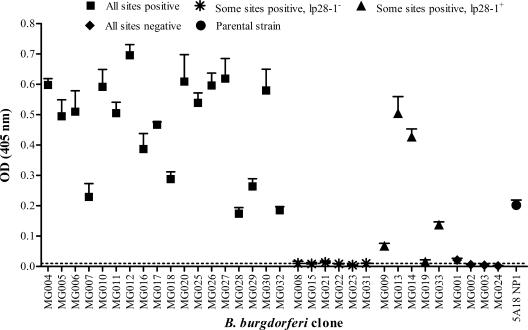
Serum antibody reactivity to VlsE was also examined in the single mouse inoculation study (Fig. 1). Sera from mice infected with mutants culture positive at all sites were consistently reactive. Sera from mice inoculated with the noninfectious mutants MG002, MG003, and MG024 were nonreactive, while the reactivity of sera from the mouse inoculated with the noninfectious mutant MG001 was only slightly greater than the cutoff value (Fig. 1). Sera from five out of six mice inoculated with lp28-1− attenuated mutants were nonreactive (since vlsE is encoded on lp28-1), and the response for the sixth mouse was very close to the cutoff value (Fig. 1). Sera of mice infected with lp28-1+ attenuated mutants displayed various degrees of reactivity toward VlsE.
FIG. 1.
Serological reactivities to VlsE for individual mice inoculated with pMarGent transposon mutants MG001 through MG033. As a screening assay, each transposon mutant was inoculated into one mouse (two mice were used for the parental strain, 5A18 NP1), and the mice were sacrificed after 2 weeks. Serum from each mouse was assayed using a VlsE-based ELISA. Each data point represents the mean for triplicate samples plus standard deviation. The isolates are arranged according to the infectivity results obtained from the individual mouse inoculation screening.
Sera from all mice in the multiple-dose experiments were also tested for serological reactivity to VlsE (Fig. 2). Reactivity of mice inoculated with one of the three noninfectious mutants (MG001, MG002, or MG024) was low; 3/12, 1/12, and 0/12 mice, respectively, were reactive by our criteria (Fig. 2A; Table 2), but the reactive serum samples all yielded low OD405s (0.011 to 0.047). The weak serological response identified for some of the mice infected with 104 or 106 MG001 organisms or 106 MG002 organisms most likely reflects a response to VlsE present in the initial inoculum. Anti-VlsE reactivity for the attenuated mutants was similar to that for the 5A18 NP1 control, except that 10/12 mice inoculated with MG019 were nonreactive.
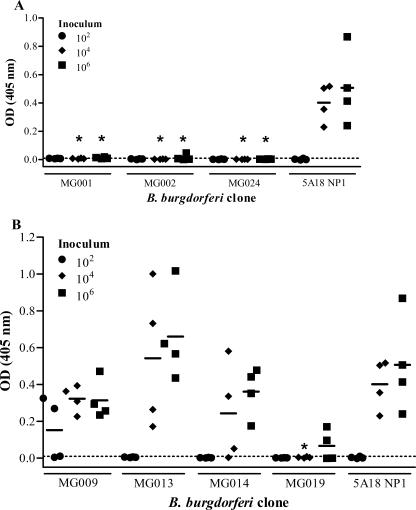
FIG. 2.
Antibody responses of mice inoculated with lp25+ lp28-1+ attenuated and noninfectious pMarGent mutants from two independent multiple-dose experiments. In each experiment, groups of two mice each were inoculated with 1 × 102, 1 × 104, or 1 × 106 spirochetes. Mice were sacrificed after 2 weeks, and serum reactivity was measured using serum from each mouse in a VlsE ELISA. Each symbol represents the mean for triplicate samples from one mouse. Each horizontal bar indicates the mean reactivity for four mice inoculated with a given dose. The dotted line indicates the cutoff value for reactivity (OD405 = 0.010), defined as 2 standard deviations above the mean VlsE reactivity for preimmune sera from 32 mice. An asterisk indicates that the reactivity for that inoculum was statistically different from the reactivity observed for the corresponding inoculum of 5A18 NP1 (P < 0.05). The transposon mutants are subdivided into those that were identified as being (A) noninfectious or (B) attenuated in the single mouse inoculation study.
Overall, the occurrence of anti-VlsE antibodies correlated well with culture positivity (Fig. 2; Table 2), with concordance of these results occurring with 90/96 mice (94%). In a few instances, anti-VlsE reactivity was obtained in the absence of culture positivity (4/96 mice) and vice versa (2/96 mice).
Sera from mice receiving the noninfectious mutant MG001, MG002, or MG024 did not produce a strong anti-VlsE response in the single mouse inoculation study or the multiple-dose study (Fig. 1 and 2), yet a robust response against a B. burgdorferi whole-cell lysate was measured in mice inoculated with 104 or 106 organisms (data not shown). Seroconversion with high inocula of noninfectious (i.e., heat-inactivated) B. burgdorferi has been observed previously (3), indicating the activation of antibody responses by Borrelia components present in the inoculum. Therefore, a whole-cell-based ELISA was not a useful tool for screening infectivity phenotypes in our analysis.
Plasmid profiling.
The plasmid content of the noninfectious mutants MG009, MG013, MG014, and MG019 and the noninfectious mutants MG001, MG002, and MG024 was determined by PCR (Table 2). No plasmids previously identified as having roles in infectivity were absent from the transposon mutants. However, MG014 is lacking cp32-6 and MG019 is missing lp38; both plasmids have undefined roles in the infectious process (24).
In vitro growth analysis.
The lp28-1+ attenuated mutants MG009, MG013, MG014, and MG019 and the noninfectious mutants MG001, MG002, and MG024 were further characterized by comparing in vitro growth rates to that of the parental strain, 5A18 NP1. Triplicate cultures of each clone were analyzed daily by dark-field microscopy. The growth rates of every mutant were comparable to the rate of the parental strain (data not shown).
During the enumeration of organisms for the in vitro growth analysis, no mutants appeared to have a defect in motility, and only one mutant exhibited morphological differences from the parental strain; MG019 cells appeared to be shorter than those of the parental strain, although quantitative measurements have not as yet been performed.
DISCUSSION
We have demonstrated the successful use of in vivo transposon mutagenesis to identify potential virulence determinants in a low-passage-number, infectious isolate of B. burgdorferi. Previous studies on in vivo transposition of B. burgdorferi had been successful only with high-passage-number, noninfectious isolates (21, 28). The work presented here thus indicates the feasibility of large-scale analyses of genes involved in the infectious processes of B. burgdorferi using transposon mutagenesis techniques.
The transposon delivery vector pMarGent (28) was transformed into the infectious, transformable strains 5A4 NP1 and 5A18 NP1 (15). A small pool of gentamicin-resistant clones was isolated from transformations with 5A18 NP1, but no clones were isolated using 5A4 NP1. This difference is likely due to the presence of lp56 (and hence BBQ67, encoding a putative restriction-modification protein) in 5A4 NP1 and the absence of this plasmid in 5A18 NP1. In addition, the suicide vector pMarGent used in the current studies may possess different transformation properties relative to the initially tested shuttle vector pBSV2C03::gntΔkan (15). Although it cannot be stated definitively that transposition occurred stochastically within this small initial library of transposon mutants, it is evident that the insertion sites are distributed throughout the chromosome and plasmids (Table 1). Published reports on Himar1-based transposition for B. burgdorferi (21, 28), Leptospira interrogans (5), and Leptospira biflexa (20) also support this observation.
Six of the transposon insertion sites mapped to the chromosome, whereas 13 mutations were in the circular plasmids and 12 were in the linear plasmids. The relatively low number of insertions mapping to the chromosome (representing ∼60% of the genomic DNA) is likely attributable to a higher proportion of essential genes required for in vitro multiplication in the chromosome and the resulting failure to recover such mutants. Although this bias could potentially result from differences in copy numbers between the chromosome and the plasmids, available data indicate that the plasmids of B. burgdorferi are maintained at a copy number of approximately one per chromosome (6, 13). Differences in the density of the Himar1 insertion site TA could also potentially contribute to this bias. However, there is essentially no difference between the G+C content of the chromosome (28.6%) and that of the plasmids (27.6%) (7, 10).
Each of the mutants was inoculated into an individual mouse to serve as an initial screen for defects in infectivity introduced by random insertional mutagenesis. A total of 18 mutants appeared to remain fully infectious for mice, 11 mutants exhibited an attenuated phenotype in vivo, and 4 mutants were tentatively considered noninfectious. Based on the observation that some clones had lost plasmids related to infectivity, six attenuated clones lacking lp28-1 (MG008, MG015, MG021, MG022, MG023, and MG031) and one noninfectious clone lacking lp25 (MG003) were not characterized further in this study. The noninfectious mutant MG033 was not pursued further in this study due to difficulties in reliably culturing this clone from frozen stocks.
To examine the validity of the single mouse inoculation results, lp28-1+ lp25+ mutants that appeared to be noninfectious (MG001, MG002, and MG024) or attenuated (MG009, MG013, MG014, and MG019) were each inoculated into six mice using different doses. Again, spirochete outgrowth was used to determine the infectivity phenotype of each of the mutants (Table 2). The phenotypes of these mutants in relation to the transposon insertion site are discussed below, with the understanding that conclusive evidence for the roles of each of the affected genes will require more-detailed analyses, including gene complementation and other studies.
The mutants MG001, MG002, and MG024 were noninfectious in their respective single mouse inoculation and multiple-inoculation studies. Of particular interest are the two noninfectious mutants with known insertion sites, MG002 and MG024. MG002 contains a transposon insertion in guaB (BBB17), which encodes an IMP dehydrogenase and is involved in the interconversion of inosine and guanine. The insertion site is at nucleotide (nt) 683 of the guaB ORF and thus is predicted to result in a product truncated at 227 amino acids, representing 52% of the predicted protein sequence. guaB is the downstream gene in a bicistronic operon with guaA (encoding GMP synthase), so its disruption should not have a polar effect. guaB had been inactivated previously in a noninfectious B. burgdorferi strain, and this mutant was capable of survival and growth in vitro (30). The clone MG002 also lacks plasmids cp9 and cp32-3 (Table 2), but earlier studies indicated that these plasmids are not required for mouse infection (9, 24). It is possible, but unlikely, that plasmids cp9 and cp32-3 encode redundant functions, so that infectivity is reduced when both are absent.
The noninfectious transposon mutant MG024 contains an insertion in fliG-1 (BB0221), encoding a homolog of a protein involved in the flagellar motor switch, i.e., the change in direction of flagellar rotation. The transposon insertion in this case occurred at nt 777 of the fliG-1 ORF, resulting in expression of a 259-amino-acid fragment (71%) of the predicted gene product. fliG-1 may be part of a large and diverse operon, including genes that encode a putative glucose-6-phosphate-1-dehydrogenase (BB0222), an alanyl t-RNA synthetase (BB0220), and a putative zinc permease (BB0219). Thus, disruption of BB0221 could have polar effects on the downstream genes, BB0220 and BB0219. A B. burgdorferi paralog exists (fliG-2; BB0290), but it is only moderately similar to fliG-1 (26% identical, 52% similar) and is located in a large flagellar operon. Most bacteria contain only one copy of fliG; spirochetes, on the other hand, contain anywhere from two to four paralogs of fliG. E. coli defective in fliG do not produce flagella and are consequently not motile (14). However, MG024 is motile in liquid culture and does not exhibit any in vitro growth defect compared to the parental strain (data not shown). It is unknown what role the spirochetal paralogs of fliG fulfill, but they are likely to be involved in their invasive nature and ability to disseminate and colonize many different tissue types. The plasmid content of the fliG-1 mutant MG024 and that of the 5A18 NP1 parental strain are identical.
The attenuation pattern observed as a result of the single mouse inoculation of MG009 was reproduced in the multiple-dose experiment. Organisms were not recovered from the ear or heart after inoculation of a single mouse with this mutant, indicating that this clone possesses a defect that negatively impacts its ability to colonize or persist in the tissues in the pinna (outer ear). Spirochetes were isolated from only 1/12 ear cultures after 4 weeks but from 26 of 36 reisolation attempts from other sites. MG009 contains an insertion in BB0743, a hypothetical gene located on the chromosome. The gene product of BB0743 contains WD40-like repeat sequences and is part of COG1520, but the functional role of predicted proteins containing this motif is unknown. BB0743 is likely to be cotranscribed with the upstream gene BB0742, encoding an ABC transporter ATP-binding protein. The downstream gene, BB0744, appears to be transcribed separately, given the presence of a 105-nt intergenic region between BB0743 and BB0744. Therefore, the transposon insertion in MG009 is unlikely to have polar effects. Only cp9 was missing in this clone relative to the parental strain. Previous studies have demonstrated that the absence of cp9 does not have a detectable effect on mouse infection. The tissue distribution observed with MG009 suggests that BB0743 may play a role in dissemination or tissue tropism.
The tissue localization patterns observed with MG013 were also consistent between the single- and multiple-dose animal studies. This mutant failed to colonize the ear (2/12 positive cultures) and the heart (1/12 positive cultures) effectively. The transposon insertion site in MG013 has not as of yet been identified, but the transposon insertion appears to prevent successful colonization of the ear and heart. Only cp9 was lost from MG013 relative to the parental strain, 5A18 NP1. Pending identification of the transposon insertion site and the relationship between the MG013 genotype and its tissue localization pattern will be characterized further.
The clone MG014, which contains an insertion 18 nucleotides from the 3′ end of a putative mutS gene (BB0797), appears to be attenuated with regard to infection of the ear (3/12 positive cultures in the multiple-inoculum experiments) and the heart (5/12 positive cultures). It is not clear how a DNA mismatch repair protein could influence dissemination and tissue localization. However, DNA repair pathways could play a role in counteracting damage induced by host defense mechanisms, including reactive oxygen species (31). In MG014, the insertion site is at the very end of the mutS ORF (nt 2570, six codons from the 3′ end). BB0797 appears to be in an operon with BB0798, encoding a predicted protein weakly homologous to competence protein F; members of this gene family are thought to have amidophosphotransferase activity. Other downstream genes are also potentially part of the same operon. Thus, the main effect of the transposon insertion in MG014 may be in reducing the expression of this downstream gene.
The infectivity results for MG019 clearly show a marked attenuation in mice. Organisms were found only in the heart and joint of the mouse used for the single inoculation and in only 5/48 tissues in the multiple dose study. MG019 contains an insertion less than 100 nucleotides from the 5′ end of the monocistronic, conserved hypothetical gene BBC08 in cp9. Previous work has demonstrated that cp9 has no discernible effect on mouse infectivity (24). Interestingly, MG019 is missing lp38, a plasmid that has no defined role in infectivity or pathogenesis. Given the negative correlation between cp9 and infectivity and the loss of lp38, these data strongly suggest that lp38 encodes one or more factor(s) essential for full infectivity in the mouse. Until MG019 can be further characterized, the implication of lp38 in infectivity remains speculative.
Data from VlsE ELISAs performed with sera from transposon mutant-infected mice revealed a pattern of reactivity that generally paralleled the infectivity phenotype determined by culture (Fig. 1 and 2; Table 2). Overall, mice inoculated with the noninfectious mutants MG001, MG002, and MG024 or the attenuated mutants MG009, MG013, MG014, and MG019 generated similar patterns of reactivity using sera from single mouse inoculations and the multiple-dose studies. In the multiple-dose experiments, culture positivity and VlsE reactivity correlated for 90/96 (94%) of the mice. There were four instances where VlsE reactivity was observed in the absence of culture positivity and only two instances of culture positivity in the absence of a serological response. These data demonstrate that anti-VlsE serologic reactivity is a strong indicator of infection and could be used effectively as a screening assay for the identification of noninfectious clones.
In this work, we demonstrated the feasibility of performing transposon mutagenesis with infectious B. burgdorferi using Himar1 derivatives. In recent studies, more than 2,000 additional transposon mutant clones have been isolated, indicating that saturating mutagenesis of the genome is possible (T. Lin, L. Gao, and S. J. Norris, unpublished results). While this report presents an initial survey of putative virulence factors, additional work is needed to more fully understand each of the attenuated and noninfectious mutants. Complementation of the disrupted genes must be performed to provide definitive evidence for the infectivity phenotypes observed; this analysis would also reveal the importance of polar transcriptional effects in the observed phenotype. Quantitative PCR and histopathological analyses would also reveal the effects of each mutation on spirochete burden and tissue pathology. As shown in this and previous studies, the determination of plasmid content is also an important factor that requires careful attention when performing infectivity analysis. High-throughput methods for transformation, mutant recovery, insertion site identification, plasmid content analysis, and infectivity screening are under development and may permit the use of more extensive transposon insertion libraries for the systematic identification of genes required for the mammalian and tick phases of the infectious cycle of Lyme disease Borrelia.
Acknowledgments
We thank Jerrilyn Howell, Mary Hubbard, Melanie McGill, Tao Lin, and Lihui Gao for helpful suggestions and assistance with the animal studies. We also thank Kit Tilly for invaluable advice regarding this project and Jerrilyn Howell for performing the plasmid content analysis.
This work was supported by NIH grants R01 AI59048 (S.J.N.) and T32 AI055449 (S.J.N. and D.J.B.) and by the Intramural Research Program of the NIH, NIAID (P.A.R. and P.E.S.).
Editor: V. J. DiRita
Footnotes
Published ahead of print on 2 October 2006.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Barbour, A. G. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J. Biol. Med. 57:521-525. [PMC free article] [PubMed] [Google Scholar]
- 3.Barthold, S. W., E. Fikrig, L. K. Bockenstedt, and D. H. Persing. 1995. Circumvention of outer surface protein A immunity by host-adapted Borrelia burgdorferi. Infect. Immun. 63:2255-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benach, J. L., E. M. Bosler, J. P. Hanrahan, J. L. Coleman, G. S. Habicht, T. F. Bast, D. J. Cameron, J. L. Ziegler, A. G. Barbour, W. Burgdorfer, R. Edelman, and R. A. Kaslow. 1983. Spirochetes isolated from the blood of two patients with Lyme disease. N. Engl. J. Med. 308:740-742. [DOI] [PubMed] [Google Scholar]
- 5.Bourhy, P., H. Louvel, I. Saint Girons, and M. Picardeau. 2005. Random insertional mutagenesis of Leptospira interrogans, the agent of leptospirosis, using a mariner transposon. J. Bacteriol. 187:3255-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casjens, S., and W. M. Huang. 1993. Linear chromosomal physical and genetic map of Borrelia burgdorferi, the Lyme disease agent. Mol. Microbiol. 8:967-980. [DOI] [PubMed] [Google Scholar]
- 7.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. M. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 8.Elias, A. F., J. L. Bono, J. J. Kupko III, P. E. Stewart, J. G. Krum, and P. A. Rosa. 2003. New antibiotic resistance cassettes suitable for genetic studies in Borrelia burgdorferi. J. Mol. Microbiol. Biotechnol. 6:29-40. [DOI] [PubMed] [Google Scholar]
- 9.Elias, A. F., P. E. Stewart, D. Grimm, M. J. Caimano, C. H. Eggers, K. Tilly, J. L. Bono, D. R. Akins, J. D. Radolf, T. G. Schwan, and P. Rosa. 2002. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect. Immun. 70:2139-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidman, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fujii, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 11.Grimm, D., C. H. Eggers, M. J. Caimano, K. Tilly, P. E. Stewart, A. F. Elias, J. D. Radolf, and P. A. Rosa. 2004. Experimental assessment of the roles of linear plasmids lp25 and lp28-1 of Borrelia burgdorferi throughout the infectious cycle. Infect. Immun. 72:5938-5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartl, D. L., and H. Ochman. 1996. Inverse polymerase chain reaction. Methods Mol. Biol. 58:293-301. [DOI] [PubMed] [Google Scholar]
- 13.Hinnebusch, J., and A. G. Barbour. 1992. Linear- and circular-plasmid copy numbers in Borrelia burgdorferi. J. Bacteriol. 174:5251-5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Irikura, V. M., M. Kihara, S. Yamaguchi, H. Sockett, and R. M. Macnab. 1993. Salmonella typhimurium fliG and fliN mutations causing defects in assembly, rotation, and switching of the flagellar motor. J. Bacteriol. 175:802-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawabata, H., S. J. Norris, and H. Watanabe. 2004. BBE02 disruption mutants of Borrelia burgdorferi B31 have a highly transformable, infectious phenotype. Infect. Immun. 72:7147-7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Labandeira-Rey, M., E. Baker, and J. Skare. 2001. Decreased infectivity in Borrelia burgdorferi strain B31 is associated with loss of linear plasmid 25 or 28-1. Infect. Immun. 69:446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Labandeira-Rey, M., J. Seshu, and J. T. Skare. 2003. The absence of linear plasmid 25 or 28-1 of Borrelia burgdorferi dramatically alters the kinetics of experimental infection via distinct mechanisms. Infect. Immun. 71:4608-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrenz, M. B., J. M. Hardham, R. T. Owens, J. Nowakowski, A. C. Steere, G. P. Wormser, and S. J. Norris. 1999. Human antibody responses to VlsE antigenic variation protein of Borrelia burgdorferi. J. Clin. Microbiol. 37:3997-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawrenz, M. B., H. Kawabata, J. E. Purser, and S. J. Norris. 2002. Decreased electroporation efficiency in Borrelia burgdorferi containing linear plasmids lp25 and lp56: impact on transformation of infectious B. burgdorferi. Infect. Immun. 70:4798-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Louvel, H., I. Saint Girons, and M. Picardeau. 2005. Isolation and characterization of FecA- and FeoB-mediated iron acquisition systems of the spirochete Leptospira biflexa by random insertional mutagenesis. J. Bacteriol. 187:3249-3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morozova, O. V., L. P. Dubytska, L. B. Ivanova, C. X. Moreno, A. V. Bryksin, M. L. Sartakova, E. Y. Dobrikova, H. P. Godfrey, and F. C. Cabello. 2005. Genetic and physiological characterization of 23S rRNA and ftsJ mutants of Borrelia burgdorferi isolated by mariner transposition. Gene 357:63-72. [DOI] [PubMed] [Google Scholar]
- 22.Norris, S. J., J. K. Howell, S. A. Garza, M. S. Ferdows, and A. G. Barbour. 1995. High- and low-infectivity phenotypes of clonal populations of in vitro-cultured Borrelia burgdorferi. Infect. Immun. 63:2206-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purser, J. E., M. B. Lawrenz, M. J. Caimano, J. K. Howell, J. D. Radolf, and S. J. Norris. 2003. A plasmid-encoded nicotinamidase (PncA) is essential for infectivity of Borrelia burgdorferi in a mammalian host. Mol. Microbiol. 48:753-764. [DOI] [PubMed] [Google Scholar]
- 24.Purser, J. E., and S. J. Norris. 2000. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc. Natl. Acad. Sci. USA 97:13865-13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Revel, A. T., J. S. Blevins, C. Almazan, L. Neil, K. M. Kocan, J. de la Fuente, K. E. Hagman, and M. V. Norgard. 2005. bptA (bbe16) is essential for the persistence of the Lyme disease spirochete, Borrelia burgdorferi, in its natural tick vector. Proc. Natl. Acad. Sci. USA 102:6972-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosa, P. A., K. Tilly, and P. E. Stewart. 2005. The burgeoning molecular genetics of the Lyme disease spirochaete. Nat. Rev. Microbiol. 3:129-143. [DOI] [PubMed] [Google Scholar]
- 27.Steere, A. C., R. L. Grodzicki, A. N. Kornblatt, J. E. Craft, A. G. Barbour, W. Burgdorfer, G. P. Schmid, E. Johnson, and S. E. Malawista. 1983. The spirochetal etiology of Lyme disease. N. Engl. J. Med. 308:733-740. [DOI] [PubMed] [Google Scholar]
- 28.Stewart, P. E., J. Hoff, E. Fischer, J. G. Krum, and P. A. Rosa. 2004. Genome-wide transposon mutagenesis of Borrelia burgdorferi for identification of phenotypic mutants. Appl. Environ. Microbiol. 70:5973-5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stewart, P. E., R. Thalken, J. L. Bono, and P. Rosa. 2001. Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Mol. Microbiol. 39:714-721. [DOI] [PubMed] [Google Scholar]
- 30.Tilly, K., L. Lubke, and P. Rosa. 1998. Characterization of circular plasmid dimers in Borrelia burgdorferi. J. Bacteriol. 180:5676-5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, G., P. Alamuri, M. Z. Humayun, D. E. Taylor, and R. J. Maier. 2005. The Helicobacter pylori MutS protein confers protection from oxidative DNA damage. Mol. Microbiol. 58:166-176. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, J. R., J. M. Hardham, A. G. Barbour, and S. J. Norris. 1997. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell 89:275-285. [DOI] [PubMed] [Google Scholar]