Abstract
The human β-defensin 3 (hBD-3) is an inducible epithelial peptide antibiotic that has potent antistaphylococcal activity. Infection of skin epithelial cells with viable Staphylococcus aureus, a common skin pathogen, induces increased gene expression of hBD-3 and other antimicrobial peptides. The aim of this study was to identify signaling pathways and nuclear responses that contribute to the gene expression of hBD-3 in primary human keratinocytes upon contact with S. aureus. Increased hBD-3 peptide was observed by immunofluorescence microscopy in keratinocytes exposed to S. aureus and to lipoteichoic acid (LTA). Both are ligands for the cell surface Toll-like receptor 2 (TLR2), and thus the contribution of TLR2 signaling in hBD-3 expression was examined. Functional inhibition of TLR2 prior to S. aureus stimulation significantly decreased hBD-3 mRNA levels by 37%, attesting to the involvement of this surface receptor in the initial recognition and downstream signaling for hBD-3 expression. Treatment of keratinocytes with a p38 mitogen-activated protein kinase (MAPK) inhibitor prior to either S. aureus or LTA stimulation was associated with reduced hBD-3 mRNA transcripts and peptide. We also propose a role for the MAPK-regulated transcriptional activating protein 1 in S. aureus-induced hBD-3 gene expression. Combined, these studies indicate a role for TLR2 signaling and MAPK activation in the upregulation of hBD-3 and demonstrate the innate immune capacity of skin keratinocytes under conditions of S. aureus challenge to enhance the local expression of this antistaphylococcal peptide antibiotic.
Skin keratinocytes constitute a protective mechanical barrier against invading microorganisms. These epithelial cells also serve as active participants in cutaneous host defense by generating innate immune responses upon exposure to microbial pathogens that trigger inflammatory cascades. Stimulated keratinocytes also produce endogenous peptides that have direct antimicrobial activity against a broad spectrum of pathogens, including most bacteria, certain fungi, and enveloped viruses (2). The biological relevance of these peptides has been demonstrated in animal models showing that host antimicrobial peptide expression in the skin is critical to resisting infection (19). In addition, the finding of deficient antimicrobial peptide levels in the involved skin of patients with atopic dermatitis provides an explanation for the increased propensity toward Staphylococcus aureus colonization and infection in this condition (20, 21).
The β-defensins are cysteine-rich peptides of 36 to 42 amino acids in length and are stabilized by three disulfide bonds (5). The three best-characterized human β-defensins—human β-defensin (hBD-1), hBD-2, and hBD-3—have been detected in human skin and cultured keratinocytes. hBD-1 expression is primarily constitutive, whereas the expression of hBD-2 and hBD-3 is inducible by cytokines such as tumor necrosis factor alpha and interleukin-1β (IL-1β), various microorganisms, lipopolysaccharide, and other microbial products (1, 7, 8). The mechanism by which β-defensins kill or inactivate bacteria is not precisely understood but is generally thought to be a function of their pore-forming activity upon the microbial membrane (13).
S. aureus is an occasional skin flora resident, as well as a major cutaneous pathogen. Both hBD-1 and hBD-2, which display salt-sensitive antimicrobial activity against most gram-negative bacteria, are relatively inactive against S. aureus and other gram-positive organisms in vitro. However, each may have additive or synergistic antistaphylococcal activity with other antimicrobial peptides, as has been demonstrated with hBD-2 and the cathelicidin LL-37 (3, 17). On the other hand, hBD-3 exhibits potent killing activity against S. aureus and other gram-positive bacteria in addition to activity against gram-negative organisms (6, 7). Moreover, the antimicrobial action of hBD-3 is retained even at physiologic salt concentrations. hBD-3 peptide has been localized to the intercellular spaces in keratinocyte layers of the upper epidermis, where it is released from lamellar bodies (23). A keratinocyte cell line engineered to overexpress hBD-3 within epidermal sheets demonstrated significant antimicrobial activity against S. aureus (23). Thus, endogenous production of hBD-3 in the epidermis may provide an antimicrobial shield to protect cutaneous tissues from bacterial invasion against pathogens such as S. aureus.
We and others have previously demonstrated that contact of cultured epidermal keratinocytes with S. aureus triggers the upregulation of hBD-3 and other β-defensins (16, 17). Furthermore, we showed that highly purified lipoteichoic acid (LTA), a major staphylococcal cell wall constituent, was responsible at least in part for the induction of hBD-3 in skin keratinocytes. The signaling mechanisms involved in the upregulation of hBD-3 in skin epithelia upon contact with microorganisms, including S. aureus, are incompletely understood. The hBD-3 gene promoter has no discernible NF-κB binding elements but does contain transcriptional binding motifs for activator protein 1 (AP-1), interferon gamma interferon (IFN-γ) response elements, and NF-IL-6 response elements (22). IFN-γ is a potent inducer of hBD-3 in most epithelial cells, including keratinocytes. Chemical inhibitors of IFN-γ and JAK2 kinase downregulate IFN-γ-stimulated hBD-3 expression, thereby, attesting to the involvement of the JAK/STAT signaling pathway (11). In the present study, we examine the intracellular signaling pathways and nuclear responses in skin keratinocytes that contribute to gene induction of hBD-3 upon stimulation with S. aureus and its bacterial components.
MATERIALS AND METHODS
Reagents.
Pyrrolidine dithiocarbamate (PDTC) was obtained from Sigma-Aldrich (St. Louis, MO), and the inhibitors SB203580 and SP600125 were purchased from Calbiochem (San Diego, CA). PDTC was resuspended in H2O, and the other inhibitors were reconstituted in dimethyl sulfoxide (DMSO) and stored according to the manufacturer's directions. Antibodies to phospho-p38 mitogen-activated protein (MAPK) and total p38 MAP were purchased from Cell Signaling (Beverly, MA). S. aureus LTA was a generous gift of Thomas Hartung and Siegfried Morath (University of Konstanz, Konstanz, Germany) and had been purified via a butanol extraction method (18). S. aureus peptidoglycan (PGN) was obtained from Fluka. Both LTA and PGN preparations were determined to be endotoxin-free by the Limulus amebocyte assay (<3 × 10−3 endotoxin units/ml). Anti-human Toll-like receptor 2 (TLR2) monoclonal antibody (IMG-416E; Imgenex, San Diego, CA) was used in blocking experiments at 10 μg/ml for 1 h prior to the stimulation of keratinocytes.
S. aureus growth conditions.
S. aureus strain DK2076, a clinical strain that is methicillin sensitive, was cultivated overnight in tryptic soy broth and then diluted into fresh TSB for logarithmic growth over ∼2 h. The culture was centrifuged, and the bacterial pellet was washed twice in phosphate-buffered saline (PBS) and then adjusted to an optical density at 600 nm of 0.40, which corresponds to ∼2 × 108 bacteria/ml. Further dilutions for keratinocyte stimulations were done using keratinocyte growth media (KGM; Clonetics, Walkersville, MD). For heat-killed preparations of S. aureus, the washed and adjusted inoculum was incubated at 68°C for 30 min. S. aureus cell-free conditioned medium was prepared by cultivating the bacteria in KGM to logarithmic growth phase, pelleting the bacteria by centrifugation, and then filtering the supernatant through a 0.2-μm-pore-size filter (Acrodisc; Gelman, Ann Arbor, MI).
Cultivation of human keratinocytes.
Pooled human neonatal epidermal keratinocytes derived from foreskins were obtained from Clonetics and propagated in KGM serum-free medium supplemented with a BulletKit (Clonetics) containing bovine pituitary extract, human epidermal growth factor, insulin, hydrocortisone, gentamicin, and amphotericin at 37°C in a CO2 incubator. Cells were passaged by trypsinization into 6-well, 24-well, or 100-mm plates. When the monolayers reached 80% confluence, the extracellular calcium concentration was increased by the addition of CaCl2 to 1.15 mM to permit differentiation. Keratinocytes were then cultivated for an additional 3 to 4 days before stimulation or infection assays.
hBD-3 immunofluorescence.
Primary human keratinocytes were seeded onto sterile four-well chamber slides (Nalge Nunc International) until near confluence and then exposed to CaCl2 1.15 mM for differentiation. After stimulation with viable S. aureus for 2 h, the monolayers were washed with PBS and then incubated with lysostaphin (Sigma) at 5 μg/ml to kill the extracellular staphylococci. The medium was replaced with fresh KGM containing lysostaphin for continued overnight incubation. Control wells were treated with KGM only. After 24 h of incubation, the monolayers were washed with PBS and then fixed for 5 min with freshly prepared 4% paraformaldehyde in PBS. The cells were permeabilized with a 10-min treatment of 0.5% Triton X-100, washed with PBS, and then blocked with 5% goat serum for 1 h. Slides were then incubated overnight at 4°C with a 1:100 dilution of polyclonal hBD-3 antibody (Orbigen, San Diego, CA) diluted in 0.1% bovine serum albumin. Controls were treated with normal rabbit serum only. After a washing step, the slides were incubated with fluorescein isothiocyanate-labeled goat anti-rabbit immunoglobulin G (IgG; Sigma) in PBS containing 1% bovine serum albumin for 1 h. After three washes with PBS (5 min each), the slides were mounted using Vectashield mounting medium with DAPI (4′,6′-diamidino-2-phenylindole; Vector Laboratories, Burlingame, CA), and fluorescent images were captured by using the Zeiss Axioplan microscope attached to a cooled charge-coupled device camera and analyzed by using MCID-M2 (version 4.0) software.
RNA isolation from stimulated keratinocytes.
Confluent keratinocyte monolayers in 24-well plates were washed with PBS and then incubated with either heat-killed or viable S. aureus suspensions as prepared above. At various time points, wells were harvested for total RNA isolation by using the Absolutely RNA Miniprep kit (Stratagene, La Jolla, CA) according to the manufacturer's directions. To examine the role of MAPKs and NF-κB transcription, cells were preincubated 60 min with the vehicle control DMSO or specific inhibitor resuspended in DMSO. The cells were next incubated with S. aureus at a multiplicity of infection (MOI) of 50 to 100, LTA, or PGN.
Real-time RT-PCR.
Total RNA (250 to 500 ng) was reverse transcribed using oligo(dT)VN23 primers (New England Biolabs, Ipswich, MA). An aliquot of the reverse transcription (RT) reaction was used as a template for real-time PCR using a SYBR Green Mastermix (ABI Biosystems, Tokyo, Japan) on an ABI Prism 7000 sodium dodecyl sulfate with SYBR green I dye as the amplicon detector and ROX as the passive reference. The gene for β-actin was amplified as an endogenous reference. Quantification was determined by using both the standard curve and the comparative ΔΔCT methods. The primers used and methods are presented in a prior investigation (17).
Electrophoretic mobility shift assay (EMSA).
Primary human keratinocytes in six-well tissue culture dishes were stimulated with viable S. aureus or heat-inactivated S. aureus at an MOI of 50 to 100 for various time intervals. After stimulation, the medium was aspirated, and ice-cold PBS was added. The cells were scraped gently and placed into 15-ml tubes for centrifugation (130 × g for 5 min 4°C). The pellet was resuspended in 1 ml of ice-cold lysis buffer (10 mM HEPES [pH 8.0], 1.5 mM MgCl2, 10 mM KCl, 1 mM dithiothreitol, protease inhibitor cocktail [Sigma catalog no. P8340]) and then incubated on ice for 15 min. Igepal-CA630 (Nonidet P-40) was added to 1%, and the solution was vortex mixed vigorously. The homogenates were centrifuged 2 to 3 min at 15,000 rpm, and the pellets were resuspended in ice-cold nuclear extraction buffer (200 mM HEPES [pH 8.0], 1.5 mM MgCl2, 25% glycerol, 420 mM NaCl, 0.2 mM EDTA, 1 mM dithiothreitol, protease inhibitor cocktail). The samples were vortexed and incubated with vigorous shaking at 4°C for 30 min. The samples were then centrifuged at 15,000 rpm for 15 min, and the collected supernatants (nuclear extracts) were stored in aliquots at −80°C until use. The protein concentration was determined by the bicinchoninic acid method (Pierce, Rockford, IL).
A nonradioactive EMSA method was developed by using a DNA biotinylation kit (Pierce) to label single-stranded oligonucleotide probes. The AP-1 binding motif for the hBD-3 gene was identified at position −1258 in the promoter region by using Matinspector of Genomatix suite (Pub Med accession no. NM018661). The oligonucleotide sequence of the primer containing the AP-1 site in the hBD-3 promoter region was as follows: 5′-CATCACGGTGACTTCAGCTCCCAATTG-3′. The binding reaction consisting of nuclear extract (1 to 2 μg) with labeled oligonucleotide (0.1 to 0.3 pmol) and poly(dI-dC) in 10 mM HEPES (pH 8.0) and containing 5 mM KCl, 0.2 mM EDTA, 5 mM MgCl2, and 1 mM DTT was incubated at room temperature for 20 min. For the supershift assay, 5 μl of polyclonal antibody to the fos or jun family proteins (Santa Cruz) was incubated with the nuclear extract for 30 min prior to addition of the oligonucleotide. For competition controls, a 100-fold excess of unlabeled oligonucleotide was added in addition to other binding reaction components. Protein-DNA complexes were separated from free labeled probe by electrophoresis on a 1.2% agarose gel in 40 mM Tris acetate-1 mM EDTA (pH 8.0; 1×) at 70 V over several hours. After electrophoresis, the gel was transferred overnight onto nylon membranes by using the capillary transfer method. The membrane was then UV cross-linked and processed for chemiluminescence detection by using the Phototope-Star kit (New England Biolabs).
Western blot analysis of phospho-specific MAPK activation.
For immunodetection of phosphorylated forms of p38 MAPK, stimulated keratinocytes in six-well dishes were washed with ice-cold PBS and incubated for 15 min on ice in 200 μl of lysis buffer containing 50 mM Tris-HCl (pH 7.4), 1% (vol/vol) Igepal-CA630, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate, 1 mM sodium fluoride, and protease inhibitor cocktail. The cells were then scraped from the wells into microcentrifuge tubes, suspended into an additional 100 μl of lysis buffer, and incubated on ice for 30 min. The extracts were centrifuged for 5 min, and the supernatant was retained and stored at −80°C until further use. The protein concentration of each extract was determined by the BCA method (Pierce), and 40 μg of each extract was resolved on a 10 or 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel. Separated proteins were transferred onto nitrocellulose membranes by using the Mini Trans-Blot apparatus (Bio-Rad). The membrane was blocked with 5% (wt/vol) nonfat dry milk in TBS and then incubated overnight at 4°C with phospho-specific p38 polyclonal antibody (Cellular Signaling Technology, Beverly, MA) diluted in buffer according to the manufacturer's recommendation. After being washed with Tris-buffered saline containing 0.1% Tween 20, the membrane was probed for 1 h with peroxidase-conjugated goat anti-rabbit IgG (Cellular Signaling). Chemiluminescence detection was performed with the LumiGlo substrate from Cellular Signaling. To evaluate the amount of protein loading per sample, the blots were stripped and probed with antibody recognizing the nonphosphorylated form of p38.
Statistics.
The data were collected from at least three independent experiments. Statistical significance was determined by using the paired Student t test. Differences were considered to be statistically significant for P values of <0.05.
RESULTS
S. aureus and LTA but not staphylococcal exoproducts induce hBD-3 expression in primary keratinocytes.
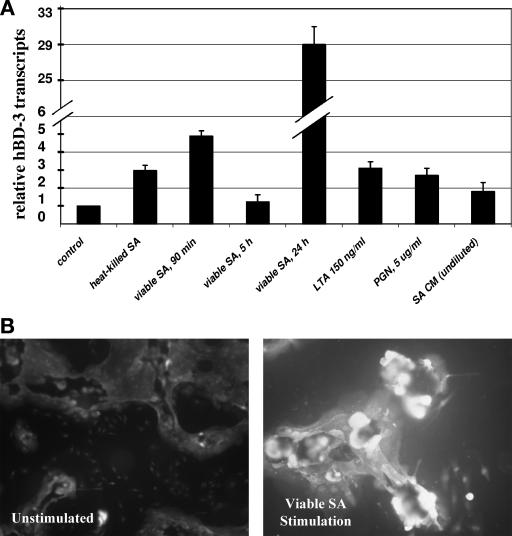
We have previously demonstrated that hBD-3 transcripts are upregulated upon contact of skin keratinocytes with viable S. aureus and LTA (16). This upregulation was most marked when viable staphylococci rather than heat-inactivated staphylococci was used as a stimulus (Fig. 1A). In the present study, we investigated whether exoproducts released by viable S. aureus organisms could induce β-defensin gene transcription. S. aureus DK2076, a methicillin-sensitive clinical isolate, was cultivated to late-logarithmic-growth phase in KGM, and the supernatant from the centrifuged culture was passed through a 0.2-μm-pore-size filter to remove bacterial cells. We then examined the effect of this cell-free conditioned S. aureus filtrate upon β-defensin gene expression in cultured keratinocytes by real-time PCR analysis. Incubation of staphylococcal conditioned medium containing bacterial exoproducts with keratinocyte cultures for 1.5- and 5-h periods did not significantly alter the hBD-3 mRNA levels from that of untreated controls (Fig. 1A). Throughout the incubation periods, the keratinocytes exhibited no signs of toxicity by light microscopy. We also evaluated the effect upon hBD-3 gene expression by PGN, another staphylococcal cell wall component. Keratinocytes stimulated with PGN at 5 μg/ml did not show a significant increase in hBD-3 mRNA levels compared to untreated controls (Fig. 1A).
FIG. 1.
hBD-3 expression in human keratinocytes in response to S. aureus and staphylococcal components. (A) Real-time PCR analysis of hBD-3 transcripts after stimulation with keratinocyte growth medium (control); heat-killed S. aureus (SA) for 5 h; viable S. aureus at 90 min, 5 h, and 24 h; LTA, at 150 ng/ml for 5 h; PGN at 5 μg/ml for 5 h; and S. aureus conditioned medium (SA CM) for 5 h. (B) Immunofluorescence staining of hBD-3 in S. aureus-stimulated keratinocytes. Cells were cultured on glass chamber slides and treated with viable S. aureus or with KGM medium only (unstimulated). After 2 h of incubation, the medium was replaced with fresh KGM containing lysostaphin to kill extracellular staphylococci and allowed to incubate for 24 h before immunostaining hBD-3. Slides were fixed and reacted with polyclonal antibody against hBD-3, and then fluorescein isothiocyanate-conjugated goat anti-rabbit IgG was used in detection. Cells were photographed with a ×40 objective lens. A control using normal rabbit serum showed no immunoreactivity (data not shown).
Viable S. aureus organisms elicited higher levels of induction of hBD-3 mRNA in keratinocytes compared to that observed from stimulation with bacterial cell wall components and from stimulation with staphylococcal exoproducts that were devoid of inducing activity. We considered whether the host cell uptake of viable staphylococcal organisms was required for hBD-3 gene expression. Intracellular invasion of S. aureus in keratinocytes has previously been demonstrated (15, 16). To determine whether bacterial invasion was required for maximal S. aureus-induced hBD-3 expression, we undertook two approaches. First, we used cytochalasin D, an inhibitor of actin polymerization that has been shown to reduce S. aureus uptake in keratinocytes and in other nonprofessional phagocytic cells prior to bacterial challenge. However, our preliminary studies showed that cytochalasin D treatment of keratinocytes alone, in the absence of bacterial treatment, significantly diminished hBD-3 mRNA expression, as well as that of another defensin, hBD-2, thereby rendering further study invalid. We next examined hBD-3 mRNA induction in keratinocytes that were stimulated with the invasion-deficient S. aureus strain DU5883. This strain, which is defective in the expression of fibronectin-binding proteins A and B, was compared to its parent, the laboratory strain 8325-4. Neither the parent strain nor the mutant strain elicited any upregulation of hBD-3 mRNA (data not shown), although each was equally capable of upregulating expression of the chemokine IL-8 upon challenge with S. aureus (∼32-fold) in real-time PCR experiments.
We next examined hBD-3 peptide expression in S. aureus-stimulated keratinocytes by immunofluorescence to determine whether inducible mRNA transcripts correlated with increased defensin peptide. We have previously reported findings of elevated hBD-3 transcripts in cells 20 to 24 h after stimulation with S. aureus. Keratinocytes cultivated in chamber slides were treated with viable S. aureus for 2 h, and then the cells were washed to remove nonadherent staphylococci and treated with lysostaphin to kill adherent extracellular staphylococci. Keratinocytes harboring intracellular viable staphylococci were then incubated further for overnight cultivation prior to fixation and immunostaining with a polyclonal antibody to hBD-3. Immunofluorescence microscopy revealed basal hBD-3 peptide levels in unstimulated keratinocytes and markedly increased hBD-3 peptide expression in staphylococcus-treated cells (Fig. 1B). Keratinocytes were also treated with LTA at 5 μg/ml or with PGN at 5 μg/ml for 5 h prior to fixation and immunofluorescence staining. LTA-treated keratinocytes exhibited increased hBD-3 staining compared to untreated cells; however, no increased staining was detectable for PGN-treated cells (data not shown).
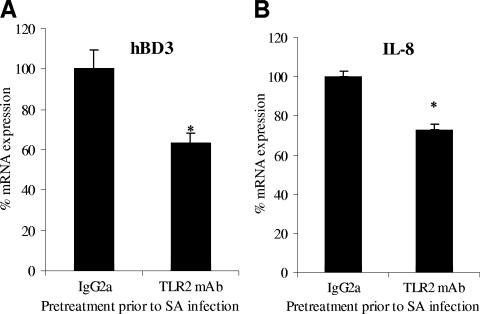
Staphylococcus-induced hBD-3 gene expression is TLR2 dependent.
We sought to identify the keratinocyte host factors involved in the initial recognition of S. aureus that would lead to increased hBD-3 transcription. In general, recognition of pathogen-associated molecular patterns by host receptors results in the activation of intracellular signaling molecules and transcription pathways, such as the MAPK family and the NF-κB and AP-1 transcription families. TLR2 is considered the major receptor for gram-positive bacteria by virtue of its capacity to recognize cell wall constituents such as LTA (24). Stimulation of cells of myeloid or epithelial origin with S. aureus organisms or with the TLR2 ligand LTA activates MAPK cascades and transcription gene programs, thereby regulating the expression of a myriad of inflammatory effector molecules such as cytokines, chemokines, and hBD-2. With the involvement of TLR2 ligands in hBD-3 gene regulation, we sought to determine whether hBD-3 gene expression was TLR2 dependent. Primary keratinocytes were preincubated for 1 h with a TLR2 neutralizing monoclonal antibody or an IgG2a isotype control antibody and then stimulated for 2 h with viable S. aureus. RNA was extracted and analyzed by real-time RT-PCR for hBD-3 transcript expression. hBD-3 mRNA levels were attenuated ca. 37% compared to that of infected keratinocytes pretreated with the isotype control antibody (Fig. 2A). To validate the functional inhibition of the TLR2 monoclonal antibody, IL-8 gene transcription was similarly analyzed since this gene is upregulated by gram-positive stimuli in part due to TLR2-initiated signaling (1, 9, 25). IL-8 gene expression was upregulated ∼33-fold by S. aureus challenge of keratinocytes and TLR2 inhibition reduced gene expression 28% (Fig. 2B).
FIG. 2.
S. aureus-induced hBD-3 gene expression is TLR2 dependent. Cultured human keratinocytes were preincubated with 10 μg of anti-TLR2 or an IgG2a isotype control antibody/ml for 1 h prior to and during challenge with viable S. aureus (SA). After bacterial stimulation for 2 h, RNA was harvested for the cDNA preparation, which was then used in quantitative real-time PCR for analysis of hBD-3 (A) and IL-8 (B) expression relative to β-actin expression. The data shown are representative of two independent experiments performed in quadruplicate and are expressed relative to the values measured in the IgG2a-treated controls. *, P < 0.05.
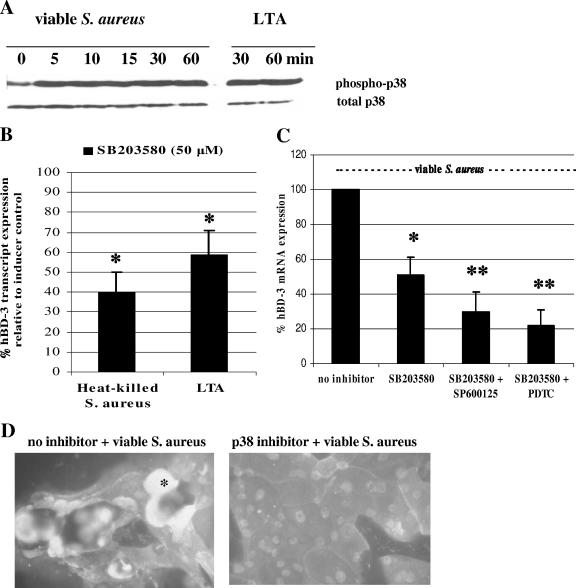
Host p38 MAPK activity contributes to enhanced hBD-3 gene transcription and peptide expression by S. aureus.
The p38 and other intracellular MAPK pathways have been implicated in the regulation of another β-defensin, hBD-2, in a number of bacterial stimulation models (4, 12). To examine the overall level of p38 activation (phosphorylation) in keratinocytes challenged with S. aureus, the cells were lysed at various time points after exposure to viable S. aureus, and the lysate was used for detection of the phosphorylated form of p38 by immunoblotting. At baseline, unstimulated cells contained a discernible level of activated p38; however, the level of phosphorylated p38 rapidly increased within 5 min of exposure to S. aureus and remained elevated for at least 60 min (Fig. 3A). In addition, treatment of keratinocytes with 5 μg of LTA/ml increased p38 activation by 30 min, and this level of activation persisted for at least 60 min (Fig. 3A).
FIG. 3.
Role of p38 activation in S. aureus-induced hBD-3 gene upregulation. (A) Activation of p38 MAPK in epidermal keratinocytes was examined in cell lysates collected at various times after stimulation with either viable S. aureus (MOI of ∼50) or LTA (5 μg/ml) and screened by Western blotting for phosphorylated (phospho-) p38 and total p38. (B) Effect of p38 inhibition upon hBD-3 mRNA levels in stimulated keratinocytes as determined by real-time RT-PCR analysis. Keratinocytes were pretreated with the specific p38 inhibitor SB203580 (50 μM) for 60 min prior to and during stimulation with heat-killed S. aureus or LTA at 5 μg/ml. After 5 h of stimulation, RNA was harvested for RT, and the cDNA was used in real-time PCR analysis to quantitate hBD-3 transcripts using the β-actin gene as the internal control. Values are means and standard deviations of at least three independent determinations and are expressed as relative hBD-3 mRNA transcripts normalized to the housekeeping β-actin gene. (C) Keratinocytes were pretreated with SB203580 (50 μM) alone or in combination with the JNK inhibitor SP600125 (1 μM) or the NF-κB inhibitor PDTC (50 μM) for 60 min prior to and during stimulation with viable S. aureus. Total RNA was harvested for RT and real-time PCR analysis. Values are the means and standard deviations of at least three independent determinations and are expressed as the percentage of the S. aureus treatment control without the inhibitor, considered to be 100%. *, P < 0.05; **, P < 0.02 (compared to conditions without inhibitor). (D) Immunofluorescence staining of hBD-3 in S. aureus-stimulated keratinocytes pretreated with the p38 inhibitor SB203580 (50 μM) for 60 min prior to and during stimulation with viable S. aureus (*, hBD-3 signal).
The contribution of p38 MAPK activation by S. aureus in the enhanced gene expression of hBD-3 was investigated by using the selective inhibitor SB203580, which specifically inhibits phosphorylation by downstream kinases of p38 isoforms α and β. The levels of hBD-3 mRNA in cells treated with SB203580 in the vehicle DMSO were not different from that of untreated cells. Pretreatment of cells with SB203580 (50 μm) for 1 h prior to challenge with heat-killed S. aureus, viable S. aureus, or LTA significantly attenuated hBD-3 mRNA levels (Fig. 3B and C). The reduction in mRNA transcripts by the p38 inhibitor correlated with reduced hBD-3 peptide immunostaining in cultured keratinocytes (Fig. 3D). Although the c-Jun-NH2-terminal kinase (JNK) inhibitor, SP600125, did not effectively lower the induction of hBD-3 mRNA levels by S. aureus, when combined with the p38 inhibitor, the two markedly abrogated S. aureus-induced hBD-3 transcripts by 71% (Fig. 3C). In summary, these experiments highlight a role for the p38 signaling pathway and, to a lesser extent, JNK, in the inducible hBD-3 gene regulation during S. aureus infection.
Pretreatment of keratinocytes with a combination of the p38 inhibitor SB203580 and the NF-κB inhibitor PDTC resulted in an additive inhibition of hBD-3 mRNA levels, whereas PDTC pretreatment alone prior to S. aureus challenge did not significantly alter relative hBD-3 mRNA levels (Fig. 3C). The additive inhibition of hBD-3 transcripts with the use of p38 and NF-κB inhibitors was unexpected since the hBD-3 promoter region has no discernible NF-κB binding sites (22). Using the Matinspector software program of Genomatix Suite, we also were unable to detect NF-κB motifs.
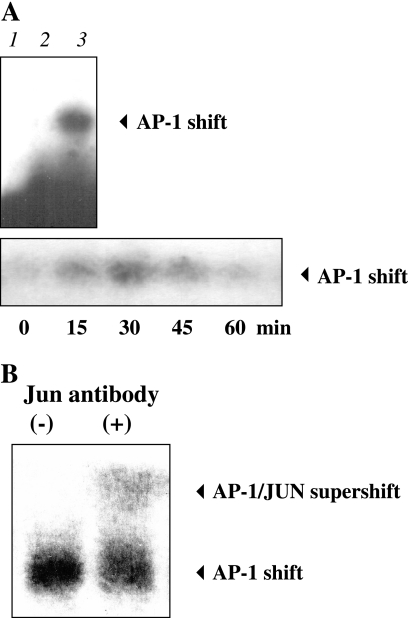
AP-1 transcription factors are involved in the induction of hBD-3 gene transcription in keratinocytes stimulated with S. aureus.
MAPK signaling pathways, notably JNK and p38, are known to regulate the transcription factor AP-1 by increasing transcription of AP-1 family proteins and altering their phosphorylation (27). AP-1 regulates gene activity in response to a plethora of stimuli, including cytokines, growth factors, and bacterial and viral infections. Therefore, for the putative AP-1 site at the hBD3-promoter, we investigated S. aureus induction of AP-1 nuclear translocation by EMSA analysis. The AP-1 binding motif for the hBD-3 gene was identified at position −1258 in the promoter region. Nuclear extracts from S. aureus-treated keratinocytes were prepared and analyzed by EMSA using biotinylated AP-1 oligonucleotides that contained the putative AP-1 binding site of the hBD-3 promoter or the AP-1 consensus sequence. AP-1 binding was observed in the nuclear extracts of keratinocytes challenged with live S. aureus for 30 min, and the specificity of binding was demonstrated using a 100-fold excess of unlabeled probe (Fig. 4A, upper panel). A time course analysis revealed that with either labeled probe there was a low level of constitutive AP-1 binding in unstimulated keratinocytes, and there was maximal AP-1 binding at 30 min after S. aureus stimulation (Fig. 4A, lower panel). Supershift analysis using polyclonal antibody to either jun or fos family proteins demonstrated additional shifting of the bands, indicating the presence of both jun and fos family proteins in the protein-DNA complex (Fig. 4B [shown for jun only]).
FIG. 4.
Activation of the transcriptional factor activator protein (AP-1) at hBD-3-specific promoter sites in S. aureus-treated keratinocytes as determined by EMSA. Keratinocytes were treated with viable S. aureus and then lysed at various time points for nuclear extract preparation for biotinylated EMSA analysis using oligonucleotide probes based on the AP-1 motifs in the hBD-3 promoter. (A, upper panel) EMSA showing free probe control (lane 1), 100-fold excess unlabeled probe control (lane 2), and AP-1 binding after challenge with S. aureus for 30 min (lane 3). (A, lower panel) Time course of AP-1 binding upon S. aureus stimulation. (B) Supershift of AP-1 complexes binding to the hBD-3 gene promoter is shown in nuclear extracts from S. aureus-treated keratinocytes using polyclonal antibodies to jun family proteins (second lane).
DISCUSSION
S. aureus and, to a lesser extent, its cell wall component LTA strongly induce keratinocyte expression of hBD-3, a potent antistaphylococcal peptide. Although viable staphylococcal organisms were more potent in hBD-3 induction than heat-killed staphylococci and LTA, we did not observe any inducible hBD-3 expression from the contact of keratinocytes with conditioned medium containing secreted bacterial exoproducts. The observed differences in hBD-3 gene inducibility between viable and heat-killed staphylococci may be explained by heat-associated alterations in bacterial cell surface ligands that may diminish host signaling or host cell-pathogen interactions that only intact and viable staphylococci can efficiently elicit.
We provide evidence that S. aureus-induced hBD-3 gene expression in cultured keratinocytes is subject to multiple regulatory pathways that are TLR2 dependent and involve activation of the p38 MAPK signaling pathway, as well as the AP-1 gene transcription network. Epithelial cells express TLR receptors, including TLR2, and demonstrate TLR2-dependent activation and nuclear translocation of NF-κB for the upregulation of IL-8 and another β-defensin, hBD-2, in response to TLR2 ligands (9). In the present study, we have demonstrated that functional inhibition of TLR2 in keratinocytes significantly reduces hBD-3 gene expression in response to challenge with S. aureus.
Mediators of downstream signaling pathways of TLRs following pathogen recognition include MAPKs. In our cell culture model, p38 MAPK is rapidly phosphorylated in staphylococcus-infected and LTA-treated skin keratinocytes. We provide evidence that the p38 MAPK pathway participates in S. aureus-induced hBD-3 gene transcription. SB203580, a well-characterized chemical inhibitor of p38, attenuated hBD-3 expression as induced by staphylococcal organisms. This response to the p38 inhibitor was observed with stimulation with either heat-inactivated or with viable staphylococci. The JNK MAPK pathway is also a participant since stronger inhibition of hBD-3 mRNA levels was observed with treatment with both JNK and p38 inhibitors prior to S. aureus challenge. The equally strong inhibition of hBD-3 mRNA levels observed with both the NF-κB inhibitor PDTC and the p38 inhibitor was unexpected since NF-κB binding elements are not apparent within the immediate vicinity of the hBD-3 promoter. The possibility exists for unrecognized NF-κB binding motifs located elsewhere near the hBD-3 gene that may control gene expression by mechanisms dependent upon p38 phosphorylation.
Although NF-κB binding motifs are not apparent within the immediate hBD-3 promoter region, we were able to identify a putative motif for transcription factor AP-1 binding. The involvement of the MAPK family in hBD-3 gene transcription supports a role for AP-1 since members of the MAPK family regulate AP-1 activation. We found that contact of S. aureus with keratinocytes led to the nuclear translocation of AP-1 at a specific site within the hBD-3 promoter and that both fos and jun family proteins were involved, supporting a role for AP-1 in mediating hBD-3 gene transcription.
Our data add to an emerging framework for understanding S. aureus-induced β-defensin expression in keratinocytes as part of cutaneous host immunity. Both LTA, which is anchored in the membrane, and PGN, which comprises the polymeric cell wall structure, are considered critical ligands for host cell recognition of S. aureus by TLR2 Recognition of staphylococcal components by TLR2 transduces signals that alter the expression of a myriad of proinflammatory mediators through activation of gene transcription factors such as NF-κB. Staphylococcal and other microbial products also can rapidly activate MAPK cascades, which in turn can modulate nuclear responses of transcriptional pathways.
Certainly, the involvement of TLR2 in hBD-3 gene upregulation by staphylococci does not preclude the involvement of other pattern recognition receptors. Internalized staphylococci may activate nucleotide-binding oligomerization domain 2 (NOD-2), an intracellular family of proteins that act as sensing receptors for muramyl dipeptide, the basic subunit of PGN (10, 14). As an intracellular PAMP receptor, NOD-2 mediates NF-κB and MAPK activation, which leads to the induction of α-defensins in macrophages. There is also evidence that NOD-2 may be involved in the induction of another antimicrobial peptide, hBD-2, in keratinocytes (26). Nod proteins have also been shown to be induced in murine osteoblasts in response to bacterial challenge with S. aureus (14). We did not observe significant hBD03 mRNA induction or AP-1 nuclear translocation using PGN as a stimulus; however, this does not exclude a NOD-2-activating role from PGN of intact staphylococci from an intracellular site. Since viable staphylococcal organisms were a more potent stimulus for hBD-3 upregulation than cell wall components, bacterial invasion of metabolically active staphylococci may be required to evoke this response.
Of the human β-defensins thus far characterized, hBD-3 has potent antimicrobial activity in vitro against S. aureus, including strains of methicillin-resistant S. aureus, and is bactericidal at low micromolar concentrations (7). Regulation of the expression of hBD-3 and perhaps other antimicrobial peptides by pathogens such as S. aureus may be important in maintaining or augmenting the endogenous antimicrobial barrier in the skin in response to bacterial invasion. Elucidation of the host signaling pathways that contribute to β-defensin expression may aid in the understanding of host susceptibility to staphylococcal infections during various disease states, e.g., atopic dermatitis, and in identifying therapeutic targets for enhancing innate defenses within the skin.
Acknowledgments
We gratefully acknowledge the gift of purified LTA from Thomas Hartung and Siegfried Morath of the University of Konstanz, Konstanz, Germany.
This study was supported by a VA Merit Award from the Department of Veterans Affairs and grant RO3 AI053122 from the National Institute of Allergy and Infectious Diseases to B.E.M.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 5 September 2006.
REFERENCES
- 1.Becker, M. N., G. Diamond, M. W. Verghese, and S. H. Randell. 2000. CD14-dependent lipopolysaccharide-induced beta-defensin-2 expression in human tracheobronchial epithelium. J. Biol. Chem. 275:29731-29736. [DOI] [PubMed] [Google Scholar]
- 2.Braff, M. H., A. Bardan, V. Nizet, and R. L. Gallo. 2005. Cutaneous defense mechanisms by antimicrobial peptides. J. Investig. Dermatol. 125:9-13. [DOI] [PubMed] [Google Scholar]
- 3.Chen, X., F. Niyonsaba, H. Ushio, D. Okuda, I. Nagaoka, S. Ikeda, K. Okumura, and H. Ogawa. 2005. Synergistic effect of antibacterial agents human β-defensins, cathelicidin LL-37 and lysozyme against Staphylococcus aureus and Escherichia coli. J. Dermatol. Sci. 40:123-132. [DOI] [PubMed] [Google Scholar]
- 4.Chung, W. O., and B. A. Dale. 2004. Innate immune response of oral and foreskin keratinocytes: utilization of different signaling pathways by various bacterial species. Infect. Immun. 72:352-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ganz, T. 2003. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3:710-720. [DOI] [PubMed] [Google Scholar]
- 6.Garcia, J.-R., F. Jaumann, S. Schulz, A. Krause, J. Rodriguez-Jimenez, U. Forssmann, K. Adermann, E. Kluver, C. Vogelmeier, D. Becker, R. Hedrich, W.-G. Forssmann, and R. Bals. 2001. Identification of a novel, multifunctional beta-defensin (human β-defensin 3) with specific antimicrobial activity. Cell Tissue Res. 306:257-264. [DOI] [PubMed] [Google Scholar]
- 7.Harder, J., J. Bartels, E. Christophers, and J.-M. Schroder. 2001. Isolation and characterization of human β-defensin-3, a novel human inducible peptide antibiotic. J. Biol. Chem. 276:5707-5713. [DOI] [PubMed] [Google Scholar]
- 8.Harder, J., U. Meyer-Hoffert, L. M. Teran, L. Schwichtenberg, J. Bartels, S. Maune, and J.-M. Schroder. 2000. Mucoid Pseudomonas aeruginosa, TNF-α, and IL-1β, but not IL-6, induce human β-defensin-2 in respiratory epithelia. Am. J. Respir. Cell Mol. Biol. 22:714-721. [DOI] [PubMed] [Google Scholar]
- 9.Hertz, C. J., Q. Wu, E. M. Porter, Y. J. Zhang, K. H. Weismuller, P. J. Godowski, T. Ganz, S. H. Randell, and R. L. Modlin. 2003. Activation of Toll-like receptor 2 on human tracheobronchial epithelial cells induces the antimicrobial peptide human beta defensin-2. J. Immunol. 171:6820-6826. [DOI] [PubMed] [Google Scholar]
- 10.Inohara, N., Y. Ogura, A. Fontalba, O. Gutierrez, F. Pons, J. Crespo, K. Fukase, S. Inamura, S. Kusumoto, M. Hashimoto, S. J. Foster, A. P. Moran, J. L. Fernandez-Luna, and G. Nunez. 2003. Host recognition of bacterial muramyl dipeptide mediated through NOD2: implications for Crohn's disease. J. Biol. Chem. 278:5509-5512. [DOI] [PubMed] [Google Scholar]
- 11.Joly, S., C. C. Organ, G. K. Johnson, J. P. B. McCray, and J. M. Guthmiller. 2005. Correlation between β-defensin expression and induction profiles in gingival keratinocytes. Mol. Immunol. 42:1073-1084. [DOI] [PubMed] [Google Scholar]
- 12.Krisanaprakornkit, S., J. R. Kimball, A. Weinberg, R. P. Darveau, B. W. Bainbridge, and B. A. Dale. 2000. Inducible expression of human beta-defensin 2 by Fusobacterium nucleatum in oral epithelial cells: multiple signaling pathways and role of commensal bacteria in innate immunity and the epithelial barrier. Infect. Immun. 68:2907-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lehrer, R. I., A. Barton, K. A. Daher, S. S. Harwig, T. Ganz, and M. E. Selsted. 1989. Interaction of human defensins with Escherichia coli: mechanism of bactericidal activity. J. Clin. Investig. 84:553-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marriott, I., D. M. Rati, S. H. McCall, and S. L. Tranguch. 2005. Induction of Nod1 and Nod2 intracellular pattern recognition receptors in murine osteoblasts following bacterial challenge. Infect. Immun. 73:2967-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mempel, M., C. Schnopp, M. Hojka, H. Fesq, S. Weidinger, M. Schaller, H. C. Korting, J. Ring, and D. Abeck. 2002. Invasion of human keratinocytes by Staphylococcus aureus and intracellular bacterial persistence represent haemolysin-independent virulence mechanisms that are followed by features of necrotic and apoptotic keratinocyte cell death. Br. J. Dermatol. 146:943-951. [DOI] [PubMed] [Google Scholar]
- 16.Menzies, B. E., and A. Kenoyer. 2005. Staphylococcus aureus infection of epidermal keratinocytes promotes expression of innate antimicrobial peptides. Infect. Immun. 73:5241-5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Midorikawa, K., K. Ouhara, H. Komatsuzawa, T. Kawai, S. Yamada, T. Fujiwara, K. Yamazaki, K. Sayama, M. A. Taubman, H. Kurihara, K. Hashimoto, and M. Sugai. 2003. Staphylococcus aureus susceptibility to innate antimicrobial peptides, beta-defensins and CAP18, expressed by human keratinocytes. Infect. Immun. 71:3730-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morath, S., A. Geyer, and T. Hartung. 2001. Structure-function relationship of cytokine induction by lipoteichoic acid from Staphylococcus aureus. J. Exp. Med. 193:393-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nizet, V., T. Ohtake, X. Lauth, J. Trowbridge, J. Rudisill, R. A. Dorschner, V. Pestonjamasp, J. Piraino, K. Huttner, and R. L. Gallo. 2001. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature 414:454-457. [DOI] [PubMed] [Google Scholar]
- 20.Nomura, I., E. Goleva, M. D. Howell, Q. A. Hamid, P. Y. Ong, C. F. Hall, M. A. Darst, B. Gao, M. Boguniewicz, J. B. Travers, and D. Y. M. Leung. 2003. Cytokine milieu of atopic dermatitis, as compared to psoriasis, skin prevents induction of innate immune response genes. J. Immunol. 171:3262-3269. [DOI] [PubMed] [Google Scholar]
- 21.Ong, P. Y., T. Ohtake, C. Brandt, I. Strickland, M. Boguniewicz, T. Ganz, R. L. Gallo, and D. Y. Leung. 2002. Endogenous antimicrobial peptides and skin infections in atopic dermatitis. N. Engl. J. Med. 347:1151-1160. [DOI] [PubMed] [Google Scholar]
- 22.Peng Jia, H., B. C. Schutte, A. Schudy, R. Linzmeier, J. M. Guthmiller, G. K. Johnson, B. F. Tack, J. P. Mitros, A. Rosenthal, T. Ganz, and P. B. McCrayJr. 2001. Discovery of new human β-defensins using a genomics-based approach. Gene 263:211. [DOI] [PubMed] [Google Scholar]
- 23.Sawamura, D., M. Goto, A. Shibaki, M. Akiyama, J. R. McMillan, Y. Abiko, and H. Shimizu. 2005. Beta defensin-3 engineered epidermis shows highly protective effect for bacterial infection. Gene Ther. 12:857-861. [DOI] [PubMed]
- 24.Schroder, N. W. J., S. Morath, C. Alexander, L. Hamann, T. Hartung, U. Zahringer, U. B. Gobel, J. R. Weber, and R. R. Schumann. 2003. Lipoteichoic acid (LTA) of Streptococcus pneumoniae and Staphylococcus aureus activates immune cells via Toll-like receptor (TLR)-2, lipopolysaccharide-binding protein (LBP), and CD14, whereas TLR-4 and MD-2 are not involved. J. Biol. Chem. 278:15587-15594. [DOI] [PubMed] [Google Scholar]
- 25.Vora, P., A. Youdim, L. S. Thomas, M. Fukata, S. Y. Tesfay, K. Lukasek, K. S. Michelsen, A. Wada, T. Hirayama, M. Arditi, and M. T. Abreu. 2004. Beta-defensin-2 expression is regulated by TLR signaling in intestinal epithelial cells. J. Immunol. 173:5398-5405. [DOI] [PubMed] [Google Scholar]
- 26.Voss, E., J. Wehkamp, K. Wehkamp, E. F. Stange, J. M. Schroder, and J. Harder. 2006. NOD2/CARD15 mediates induction of the antimicrobial peptide human beta-defensin-2. J. Biol. Chem. 281:2005-2011. [DOI] [PubMed] [Google Scholar]
- 27.Whitmarsh, A. J., and R. J. Davis. 1996. Transcription factor AP-1 regulation by mitogen-activated protein kinase signal transduction pathways. J. Mol. Med. 74:589-607. [DOI] [PubMed] [Google Scholar]