Abstract
Adaptive gene expression in prokaryotes is mediated by protein kinases and phosphatases. These regulatory proteins mediate phosphorylation of histidine or aspartate in two-component systems and serine/threonine or tyrosine in eukaryotic and eukaryote-like protein kinase systems. The genome sequence of Mycobacterium avium subsp. paratuberculosis, the causative agent of Johne's disease, does not possess a defined tyrosine kinase. Nevertheless, it encodes for protein tyrosine phosphatases. Here, we report that Map1985, is a functional low-molecular tyrosine phosphatase that is secreted intracellularly upon macrophage infection. This finding suggests that Map1985 might contribute to the pathogenesis of Mycobacterium avium subsp. paratuberculosis by dephosphorylating essential macrophage signaling and/or adaptor molecules.
Mycobacterium avium subsp. paratuberculosis is a parasite of animals that resides in macrophages (5). The pathogen is ubiquitously distributed in the environment (released in feces or milk) and may survive as a dormant organism for years in soil and water (24, 25). M. avium subsp. paratuberculosis is the etiological agent of Johne's disease, a chronic, infectious, and mortal enteritis in wild and domestic animals, affecting a variety of mammals, including rabbits, deer, cattle, and sheep (7, 18). Interestingly, the symptoms of this disease are very similar to those manifested in Crohn's disease, a chronic inflammatory bowel disease in humans. Due to that resemblance, it has been suggested that the pathological agent of Johne's disease might be transferred to humans through dairy products (15). Although M. avium subsp. paratuberculosis is the etiological agent fulfilling all of Koch's postulates in Johne's disease, in the case of Crohn's disease the role of this pathogen remains controversial. Nevertheless, studies have shown that M. avium subsp. paratuberculosis was detected in 50 to 90% of the clinical biopsies from Crohn's disease patients (2, 8).
Signal transduction is a ubiquitous mechanism responsible for cell adaptation to environmental changes in both prokaryotes and eukaryotes. Cellular responses are mediated by a cascade of reactions involving protein kinases, which activate protein substrates by ATP-dependent phosphorylation on specific residues such as histidine and aspartate in prokaryotes (two-component systems) and serine/threonine or tyrosine in eukaryotic or eukaryote-like protein kinases. The reverse regulation of kinases (dephosphorylation) is mediated by protein phosphatases. Protein phosphatases participate in modulating a variety of cellular events such as metabolism, gene transcription, cell cycle control, immune response, and cell growth (10, 23). In addition, protein phosphatases have also been associated with virulence contributing to the intracellular survival of pathogens. For example, the serine-threonine phosphatase YopH of Yersinia pseudotuberculosis dephosphorylates host proteins (21) and the tyrosine phosphatase SptP from Salmonella enterica serovar Typhimurium, which is translocated into the host, causes a disorganization of the actin cytoskeleton (14), while Stp, a serine-threonine phosphatase from Listeria monocytogenes dephosphorylates the host elongation factor EF-Tu (1).
According to the recently reported annotated genome of M. avium subsp. paratuberculosis, its signal transduction is regulated by 12 two-component systems based on signal-transducing histidine kinases, nine serine/threonine protein kinases, five proteins containing serine/threonine phosphatase catalytic domains, and two tyrosine phosphatases (17). Interestingly, the M. avium subsp. paratuberculosis genome, like all other deciphered counterparts in actinomycetes, lacks protein tyrosine kinases, suggesting that the presence of tyrosine phosphatases might play a role in mediating interactions with host cell substrates. Consistent with that, the closely related Mycobacterium tuberculosis strain H37Rv encodes two protein tyrosine phosphatases but no protein tyrosine kinases as well (11). One of them, the low-molecular-weight tyrosine phosphatase (LMWPTP) Mt-PtpA (Rv2234), was reported to be secreted in culture supernatant (16), to be involved in phagocytosis, and to promote actin polymerization in murine macrophages, but its secretion into the cytoplasm of those macrophages has not been demonstrated (9). Similarly, M. avium subsp. paratuberculosis also possesses the LMWPTP 1985 (Map-PtpA) (17), which shares 89% identity with its close relative Mt-PtpA from M. tuberculosis. This high homology led us to hypothesize that Map-PtpA is secreted from M. avium subsp. paratuberculosis behaving as an orthologous gene of Mt-PtpA in M. tuberculosis.
In the present study, we show that Map-PtpA is a protein phosphatase that dephosphorylates specifically tyrosine amino acids. We further demonstrate that Map-PtpA is secreted upon infection of human macrophages with M. avium subsp. paratuberculosis in a time-dependent manner.
MATERIALS AND METHODS
Strains, plasmid construction, and DNA manipulations.
Table 1 lists the strains, plasmids, and oligonucleotides used in the present study. Mycobacterium smegmatis mc2155 and Escherichia coli DH5α were utilized for protein overexpression and for genetic manipulations, respectively. M. avium subsp. paratuberculosis genomic DNA was prepared according to published protocols (3). The E. coli-Mycobacterium shuttle vector pALACE was used for the overexpression of proteins (12). The pALACE-derived plasmid pHB-1 was constructed by ligation of ptpA from M. avium subsp. paratuberculosis obtained by PCR amplification of genomic DNA using Pfu (Fermentas). The forward oligonucleotide was designed to carry the BamHI restriction site, whereas the reverse oligonucleotide contained the ClaI restriction site (Table 1). The gene was amplified by using 30 successive cycles comprising 30 s of the following stages: denaturation at 94°C, annealing at 55°C, and elongation at 72°C. The PCR-amplified product was digested with both BamHI and ClaI restriction enzymes and cloned into the linearized pALACE vector digested previously with the same set of enzymes. All of the DNA manipulations were performed according to published protocols (19). The monocyte-derived human THP-1 cells were used for macrophage infection.
TABLE 1.
Strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid or oligonucleotide | Characteristic(s) or sequence | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | F−recA1 hsdR17 thi-1 gyrA96 supE44 endA1 relA1 recA1 deoR Δ(lacZYA-argF)U169(φ80lacZΔM15) | Invitrogen |
| M. smegmatis | mc2155 | W. R. Jacobs |
| M. bovis BCG | Pasteur strain | ATCC 35374 |
| M. avium subsp. paratuberculosis | k-10 | ATCC BAA-968 |
| Plasmids | ||
| pALACE | ace promoter, hygromycin resistant | 13 |
| pHB-1 | M. avium subsp. paratuberculosis ptpA cloned into pALACE using BamHI-ClaI restriction sites | This study |
| Oligonucleotides | ||
| mapPtpA-forward | CACCGGATCCATGTCTGAACCGCTGCAC | |
| mapPtpA-reverse | ATTCAAATCGATTCACGACGATCCGTTC |
Growth conditions.
E. coli DH5α was grown in flasks using LB medium at 37°C and 250 rpm. 7H9 medium supplemented with 0.05% Tween 80 was used for culturing the mycobacterial strains. Then, 2 mg of Mycobactin J (Allied Monitor)/liter and 10% oleic acid-albumin-dextrose-catalase (OADC) were added as supplements to M. avium subsp. paratuberculosis cultures, while 1% glucose was added to M. smegmatis cultures. M. avium subsp. paratuberculosis was grown in rolling bottles at 37°C, and M. smegmatis was cultured in shaken flasks at 37°C and 250 rpm. Hygromycin (Sigma) was added as a supplement at final concentrations of 50 μg/ml for M. smegmatis and 150 μg/ml for E. coli.
THP-1 cells were cultured in RPMI 1640 (Sigma) supplemented with 1% l-glutamine (Stem Cell Technologies Vancouver, British Columbia, Canada), 10 mM HEPES (Stem Cell Technologies), 100 μg of streptomycin/ml, 100 U of penicillin/ml (Stem Cell Technologies), 0.1% amphotericin B (Fungizone; Invitrogen), and 10% fetal calf serum (Sigma). Infection of THP-1 cell with M. avium subsp. paratuberculosis was carried out in the same medium but lacking penicillin, streptomycin, and amphotericin B.
Protein overexpression.
Map-PtpA was overexpressed in M. smegmatis harboring the pHB-1 plasmid. A starter culture was initiated by picking a single colony into 7H9 medium supplemented with 0.05% Tween 80, 1% glucose, and hygromycin as described above. After 48 h, the starter was diluted 1:100 in fresh 7H9 medium and incubated under the same conditions overnight. After 24 h, bacteria were harvested and resuspended to an optical density at 600 nm of 1.0 using 7H9 medium lacking glucose to deplete the carbon source. The cells were then induced by the addition of 0.02% acetamide and incubated at 37°C overnight. Bacteria were harvested by centrifugation at 8,000 × g for 10 min and stored at −20°C until further processing. Protein purification was carried out by affinity chromatography using Ni-NTA resin (QIAGEN) and according to the instructions of the manufacturer. Briefly, frozen cells were resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole [pH 8.0]) supplemented with 1 mM phenylmethylsulfonyl fluoride and disrupted by sonication at 50 W for 20 s (three times). The lysate was centrifuged at 12,000 × g for 30 min, and the supernatant was loaded in an Ni-NTA resin preequilibrated with the same buffer. After the column was washed with washing buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole [pH 8.0]), recombinant proteins were eluted in imidazole buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole [pH 8.0]), dialyzed overnight in dialysis buffer (50 mM Tris-HCl [pH 7.5], 5% glycerol, 1 mM dithiothreitol [DTT]), divided into aliquots, and stored at −20°C.
Determination of protein concentration.
Protein concentration was determined by using the Bio-Rad protein assay procedure according to the manufacturer's instructions. Protein concentrations were calculated by using a calibration curve of bovine serum albumin (Sigma) as a standard protein.
Biochemical assay.
The yield of the protein purification was monitored along the purification process using the chromogenic phosphatase substrate p-nitrophenylphosphate (pNPP; Sigma). The phosphatase activity was monitored by hydrolysis of pNPP to yield p-nitrophenol, which was used at a final concentration of 1 mM. Next, 80 to 100 μl of each assayed fraction and 185 μl of pNPP substrate were added to 715 to 735 μl of reaction buffer (6.7 mM PIPES, 5 mM MgCl2, 1 mM DTT [pH 6.5]) to a final volume of 1 ml. The reaction was incubated at 37°C for 30 min, and the release of p-nitrophenol was monitored spectrophotometrically at λ = 405 nm. One unit of phosphatase activity is defined as the release of 1 μmol of p-nitrophenol per min.
The phosphatase activity on specific phosphorylated residues was determined by using the malachite green assay (13). Briefly, the malachite green solution was prepared by mixing 3 volumes of 0.045% (wt/vol) malachite green with 1 volume of 4.2% (wt/vol) ammonium molybdate tetrahydrate dissolved in 4 M HCl. The reaction mixture contained 6.7 mM PIPES (pH 6.5), 5 mM MgCl2, 1 mM DTT, and 3 μg of recombinant Map-PtpA. The reactions were carried out in a final volume of 50 μl using 96-well microplates, and three substrates—O-phosphoserine, O-phosphotyrosine, and O-phosphothreonine (Sigma)—were tested for phosphatase activity. Microplates containing the reactions were incubated at 37°C for 30 min and then stopped by the addition of 100 μl of malachite green solution, which reacts with the cleaved inorganic phosphate to give a green color. Microplates were read in a microplate reader model 550 (Bio-Rad) at 655 nm after allowing color development for 15 min. The kinetic parameters were determined by using the same substrates in concentrations varying from 0.1 to 2.5 mM. Prior to all of the phosphatase activity measurements, fractions were dialyzed extensively against 50 mM Tris-HCl (pH 7.0) in order to reduce the phosphate background, which were subtracted from the obtained readings. Sodium orthovanadate at a final concentration of 1 mM was utilized as a phosphatase inhibitor.
Time-dependent dephosphorylation of O-phosphate substrates was performed by preparing the reactions in 96-well microplates in a final volume of 50 μl as described above. The O-phospho substrates were used at a final concentration of 0.5 mM and incubated at 37°C. At selected time points, reactions were removed and stopped by addition of 100 μl of malachite green solution, and A655 measurements were obtained after color development for 15 min.
Immunoprecipitation.
Cell-free filtrate corresponding to 250 ml of culture was taken at an optical density at 600 nm of 1.0 and rocked for 2 h at room temperature after supplementation of 0.1 ml of rabbit anti-PtpA antibodies raised against mt-PtpA. Purification of the antigen-antibody complex was carried out by using the Montage antibody purification ProsepG kit (Millipore) according to the manufacturer's instructions. After elution of the complex from the column, 5 volumes of cold acetone were added to the eluate and placed at −20°C for 1 h. The precipitate was washed twice with cold acetone and resolved by sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis (SDS-15% PAGE) after the remaining acetone was dried. The resolved proteins were electroblotted onto a nitrocellulose membrane and blocked with a solution of 3% milk in phosphate-buffered saline (PBS) overnight. The membrane was then exposed to rabbit anti-Mt-PtpA antibodies diluted to 1:5,000 in 3% milk-PBS for 2 h. After three 5-min washes with 0.05% Tween 20 in PBS, the membrane was incubated with goat anti-rabbit horseradish peroxidase-conjugated antibodies and washed, and bound secondary antibodies were revealed by enhanced chemiluminescence using the Super-Signal West Pico kit (Pierce).
THP-1 cells infection.
THP-1 cells (2 × 105 cells/well in 24-well plate) were differentiated into macrophages on tissue culture-treated coverslips (Fisher Scientific) by overnight incubation with 20 ng of phorbol myristate acetate/ml. Before infection, M. avium subsp. paratuberculosis cells were washed with 7H9 medium and labeled with 10 μg of Alexa 350 carboxylic acid succinimidyl ester (Molecular Probes) dissolved in 1 ml of fresh 7H9 medium for 1 h by rocking at room temperature. Killed M. avium subsp. paratuberculosis cells were used as a negative control and were prepared by supplementing 20 μg of gentamicin (Gibco)/ml overnight at 37°C with gentle rocking and labeled with Alexa 350 as described above. After labeling, live and killed cells were washed extensively with Hanks buffer (Sigma) and opsonized for 30 min at 37°C with 10% human AB+ serum. Prior to the infection, macrophages were gently washed three times with Hank's buffer. Macrophage infection was carried out at a multiplicity of infection of 5:1 (bacteria:THP-1 cells) and incubated at 37°C and 5% CO2 for 4 h. Then, noninternalized bacilli were removed by several washes with Hanks buffer. Plates were returned to the incubator, and coverslips were examined at 24, 48, and 72 h postinfection. All of the experiments were performed in triplicates.
THP-1 cell lysate immunoprecipitation.
THP-1 cells were infected with opsonized M. avium subsp. paratuberculosis cells at a multiplicity of infection of 10:1 and, after 4 h, noninternalized bacteria were washed away with Hanks balanced salt solution. After 72 h, infected cells were scraped from the plates, combined, centrifuged (3,000 rpm for 5 min), and lysed by using a solution containing 10 mM NaCl and 20 mM Tris-HCl (pH 7.5). Macrophage debris and bacteria were separated from the lysate by centrifugation (12,000 rpm for 30 min). The cell-free lysate was filtered through a 0.22-μm-pore-size filter, exposed to the primary antibody, purified by using a ProsepG kit, and resolved as described above.
Immunostaining and fluorescence microscopy.
Infected macrophages were fixed with 2.5% p-formaldehyde for 30 min at room temperature. Sodium borohydrate (0.05% [wt/vol] in PBS) was utilized as a quencher after the cells were washed with warmed Hanks buffer. After 15 min of incubation, the quencher was washed away with Hanks buffer, and the cells were permeabilized and analyzed by fluorescence microscopy according to described previously protocols (20).
RESULTS
Map-PtpA is conserved among actinomycetes.
LMWPTPs are ubiquitously distributed among prokaryotes and eukaryotes. They are characterized by the presence of the consensus CX5R in the active site; nevertheless, the overall homology is divergent (6). BLAST analysis of Map-PtpA revealed a homology of 89% with its counterparts in both M. bovis (Mb2258) and M. tuberculosis (Rv2232) and 71% homology to the PtpA from M. smegmatis. Interestingly, the most prominent difference of LMWPTPA from M. smegmatis and other mycobacterial LMWPTPs is the presence of an additional region at the N terminus region comprising 13 residues in M. smegmatis (Fig. 1). In addition, other LMWPTPs in gram-positive bacteria have shown a relatively high homology to Map-PtpA as well. For example, an identity of 60% was observed with Nocardia farcinica and an identity of 51% was observed with Corynebacterium species (Table 2).
FIG. 1.
Map-PtpA alignment to mycobacterial LMWPTPs. Mycobacterial LMWPTPs were aligned to Map-PtpA based on an E score value of <1e−60. The LMWPTPs from the following strains were aligned (accession numbers): M. avium subsp. paratuberculosis (NP-960919), M. tuberculosis (NP-216750), M. bovis (NP-855907), and M. smegmatis (MSMEG4314). The conserved catalytic site is marked by a box.
TABLE 2.
Homologies of selected LMWPTPs corresponding to high-G-C content bacteria
| Strain | Accession no. | % Identity | % Similarity | E-valuea |
|---|---|---|---|---|
| Mycobacterium bovis | NP-855907 | 81 | 89 | 6e−76 |
| Mycobacterium tuberculosis | NP-216750 | 81 | 89 | 6e−76 |
| Mycobacterium smegmatis | MSMEG4314 | 73 | 80 | 2e−61 |
| Nocardia farcinica | BAD56477.1 | 60 | 66 | 5e−44 |
| Corynebacterium diphtheriae | CAE50212.1 | 51 | 68 | 2e−38 |
| Corynebacterium glutamicum | CAF20582.1 | 51 | 64 | 1e−36 |
| Streptomyces avermitilis | BAC71984.1 | 49 | 60 | 4e−32 |
| Streptomyces coelicolor | CAB46959.1 | 47 | 55 | 3e−29 |
Obtained from BLAST analysis.
Expression and purification of Map-PtpA.
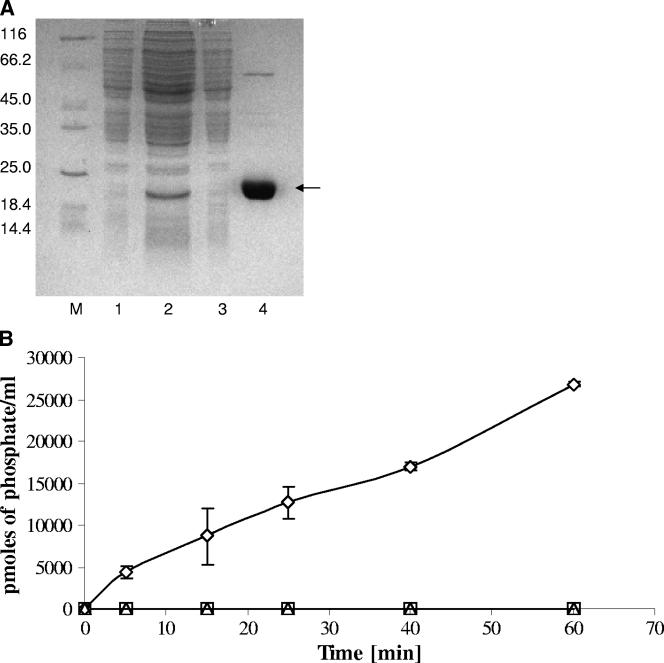
The map1985 gene was cloned and expressed in M. smegmatis as described in Materials and Methods. As shown in Table 3, a 115-fold purification of Map-PtpA was accomplished by using Ni-NTA affinity chromatography, giving a yield of 97%. The specific activity of the purified enzyme in the elution fraction reached a value of 153 U/mg of protein, in contrast to 1.33 U/mg of protein measured in the starting material. Aliquots taken during the purification process were resolved by SDS-12% PAGE, and the results showed a single protein band matching the molecular weight of PtpA (Fig. 2A, lane 4).
TABLE 3.
Purification of recombinant Map-PtpA
| Purification step (fraction) | Vol (ml) | Total activity (U)a | Total protein (mg) | Sp act (U/mg of protein) | Yield (%) | Fold increase |
|---|---|---|---|---|---|---|
| Starting material | 10 | 23.6 | 1.77 | 1.33 | 100 | 1 |
| Washing with 20 mM imidazole | 3 | 1.65 | 0.45 | 1.22 | 6.99 | 0.91 |
| Elution with 250 mM imidazole | 0.8 | 22.9 | 0.15 | 153 | 97 | 115 |
The phosphatase activity was measured by using pNPP as a substrate. One unit of phosphatase activity is defined as the formation of 1 μmol of p-nitrophenol per min.
FIG. 2.
(A) Purification of the overexpressed Mab-PtpA by metal-chelation affinity chromatography. Soluble recombinant Mab-PtpA was prepared after the induction of M. smegmatis harboring the pHB-1 plasmid. Proteins were purified by using Ni-NTA resin. Aliquots of column fractions were resolved by SDS-12% PAGE and stained with Coomassie blue. Lanes: M, molecular mass marker, 1, uninduced cells (4.3 μg); 2, start material (26.6 μg); 3, wash with 20 mM imidazole (6.7 μg); 4, elution with 250 mM imidazole (8.7 μg). The protein amounts loaded in each fraction are indicated in parentheses. The molecular mass markers are indicated on the left. (B) Time-dependent dephosphorylation of O-phospho substrates. Dephosphorylation of O-phosphorylated amino acids was assayed in a time-dependent manner. The reactions were tracked by the malachite green assay using 500 μM O-phosphotyrosine (⋄), O-phosphothreonine (□), and O-phosphoserine (▵). The data represent the means of three independent experiments, and the error bars indicate the standard deviations.
Map1985 is a protein tyrosine phosphatase.
In order to determine the specificity of the phosphatase activity, recombinant Map-PtpA was assayed utilizing the malachite green assay as described in Materials and Methods. Three O-phospho residue substrates—O-phosphoserine, O-phosphotyrosine, and O-phosphothreonine—were tested. The results showed that Map-PtpA phosphatase activity was limited to O-phosphotyrosine, and phosphatase activity using O-phosphoserine and O-phosphothreonine as substrates was undetectable. The calculated kinetic parameters for O-phosphotyrosine phosphatase activity were a Vmax of 1.716 pmol of released phosphate/min and a Km of 0.4047 mM, values similar to those reported for Mt-PtpA (13). It is noteworthy that the phosphatase activity was abolished completely when sodium orthovanadate, a specific phosphatase inhibitor, was added to the reaction.
A time-dependent dephosphorylation using all three O-phosphate substrates was carried out. The results showed that only the dephosphorylation of the O-phosphotyrosine substrate increased concomitantly in a time-dependent manner. A release of 27,000 pmol of phosphate was observed after a reaction time of 1 h (Fig. 2B).
Map-PtpA is secreted in vitro and in vivo.
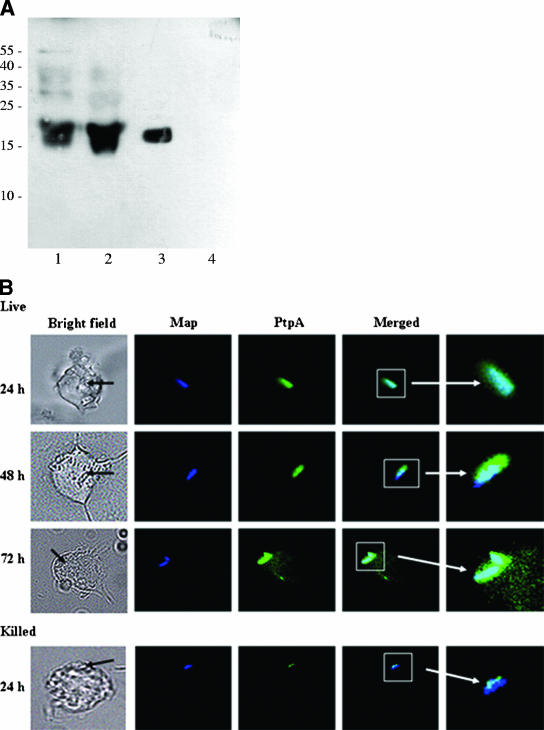
According to the results shown in Fig. 3A, a band migrating close to the molecular weight of the recombinant Map-PtpA (lane 3) was detected by immunostaining. This band was observed in both M. avium subsp. paratuberculosis cell culture filtrate (lane 1) and in the lysate corresponding to M. avium subsp. paratuberculosis-infected THP-1 cells (lane 2), whereas in noninfected THP-1 cells (lane 4) this band was not observed. In order to show that Map-PtpA is secreted from the pathogen and is localized in the cytoplasm of macrophages, its secretion was monitored by immunostaining and fluorescence microscopy. Killed bacteria were used as a negative control. The confocal images showed that macrophages could engulf both live and killed M. avium subsp. paratuberculosis cells, as shown by the blue fluorescence inside macrophages corresponding to the labeled bacilli (Fig. 3B). Killed bacterium-containing phagosomes were not observed after 24 h, suggesting that these phagosomes have fused to the lysosomes and the engulfed bacteria were digested and processed (data not shown). However, phagosomes containing live M. avium subsp. paratuberculosis were visible up to 72 h postinfection. The presence of Map-PtpA was detected inside macrophages by the formation of a green halo surrounding these phagosomes, which was observed in all macrophages that contained live M. avium subsp. paratuberculosis, whereas it was not observed in macrophages infected with killed bacteria (Fig. 3B). Interestingly, the Map-PtpA green halo grows concomitantly in a time-dependent manner (48 and 72 h postinfection), suggesting a probable link to the progress of the macrophage infection. Therefore, Map-PtpA appears to be continually secreted inside the phagosome and diffuses from this vesicle into the cytoplasm of macrophages (green color in Fig. 3B).
FIG. 3.
(A) Immunoblotting of Mab-PtpA. Samples obtained after immunoprecipitation were resolved by SDS-15% PAGE, electroblotted onto a nitrocellulose membrane, and exposed to rabbit anti-mt-PtpA antibodies as described in Materials and Methods. Lanes: 1, immunoprecipitate of cell-free filtrate; 2, immunoprecipitate of M. avium subsp. paratuberculosis-infected THP-1 lysate; 3, recombinant Mab-PtpA; 4, THP-1 cell lysate. (B) Fluorescence microscopy of infected macrophages. Macrophages were infected with fluor-labeled bacteria. Samples were obtained at 24, 48, and 72 h postinfection; processed; and permeabilized with primary and secondary antibodies as described in Materials and Methods. Fluorescence microscopy was used for image processing. Black arrows indicate phagosomes within macrophages.
DISCUSSION
The ability of mycobacterial pathogens to survive and multiply in macrophages indicated an extraordinary adaptability based on an orchestrated modulation of intracellular activation pathways in the host. This capability has evolved in order to evade the mechanisms leading to the delivery of engulfed pathogen to the proper organelles required for its destruction.
Signal transduction pathways play a crucial role in different aspects of cell adaptation, including cell division, metabolite transport regulation, and pathogenesis. In mycobacteria, one of the signal transduction mechanisms is driven by eukaryote-like kinases that require the complementary work of phosphatases (4). The existence of tyrosine phosphatases and the absence of tyrosine kinases in some pathogenic mycobacteria suggest that tyrosine phosphatases may mediate interaction with host substrates.
Here we showed that Map1985 is a phosphatase that could dephosphorylate specifically phosphorylated tyrosine amino acids in a time-dependent manner. This observation was expected since the protein shared a high homology with other LMPTPs. The kinetic parameters of Map-PtpA were also similar to those reported for others LMWPTPs (13).
The presence of Map-PtpA in culture filtrates, as well as in infected macrophage lysates, indicates that the protein is secreted in both in vivo and in vitro growth conditions. The in vitro results are consistent with previous studies, which have shown that Mt-PtpA from M. tuberculosis is secreted (16) and that the expression of a gfp reporter under the control of the mt-ptpA promoter was induced (13).
In the present study, in order to demonstrate that Map-PtpA is produced and secreted in vivo, we utilized two complementary approaches: immunoprecipitation with anti-Mt-PtpA antibodies and in situ visualization of in vivo secretion by fluorescence microscopy. In the first approach, the protein was extracted from lysates of M. avium subsp. paratuberculosis-infected macrophages by immunoprecipitation and a band migrating at a molecular weight corresponding to Map-PtpA was observed after immunoblotting with specific antibodies. Consistent with that finding, proteins obtained by immunoprecipitation of cell-free filtrate showed a band migrating at the same position as well. In the second approach, a visualization of Map-PtpA secretion from the pathogen within the macrophage was determined by fluorescence microscopy.
Further analyses of the live bacterium-infected macrophages showed that the secretion of Map-PtpA commences in the phagosome and is diffused into the host cytoplasm, based on the green intensity of the fluorescein isothiocyanate emission (anti-PtpA antibodies). The appearance of a green halo surrounding the phagosome grows concomitantly with the infection time course, indicating that Map-PtpA is continually secreted from the pathogen. Although the phagosome membrane might serve as an obstacle for the proper diffusion and spreading of Map-PtpA in the host cytoplasm, it was reported that mycobacterial proteins secreted from phagosomes with a cutoff of 70 kDa may freely transit to the macrophage cytoplasm (22).
In conclusion, the secretion of Map-PtpA inside macrophage cytoplasm suggests that this protein might mediate the interaction with host cell substrate(s), interfering in pathways necessary for killing engulfed bacteria. This interference, in part, might be associated with the capacity of this pathogen to survive in macrophages. Efforts to identify the host substrates are under way to elucidate the host signal transduction pathway that is blocked by the activity of Map-PtpA to enable the survival of the pathogen.
Acknowledgments
We thank K. Papavinasasundaram for helpful discussions.
This study was supported by the British Columbia Lung Association. H.B. is a postdoctoral fellow of Michael Smith Foundation for Health Research.
Editor: W. A. Petri, Jr.
Footnotes
Published ahead of print on 18 September 2006.
REFERENCES
- 1.Archambaud, C., E. Gouin, J. Pizarro-Cerda, P. Cossart, and O. Dussurget. 2005. Translation elongation factor EF-Tu is a target for Stp, a serine-threonine phosphatase involved in virulence of Listeria monocytogenes. Mol. Microbiol. 56:383-396. [DOI] [PubMed] [Google Scholar]
- 2.Autschbach, F., S. Eisold, U. Hinz, S. Zinser, M. Linnebacher, T. Giese, T. Loffler, M. W. Buchler, and J. Schmidt. 2005. High prevalence of Mycobacterium avium subspecies paratuberculosis IS900 DNA in gut tissues from individuals with Crohn's disease. Gut 54:944-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Av-Gay, Y., and J. Davies. 1997. Components of eukaryotic-like protein signaling pathways in Mycobacterium tuberculosis. Microb. Comp. Genomics 2:63-73. [Google Scholar]
- 4.Av-Gay, Y., and M. Everett. 2000. The eukaryotic-like Ser/Thr protein kinases of Mycobacterium tuberculosis. Trends Microbiol. 8:238-244. [DOI] [PubMed] [Google Scholar]
- 5.Bannantine, J. P., and J. R. Stabel. 2002. Killing of Mycobacterium avium subspecies paratuberculosis within macrophages. BMC Microbiol. 2:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barford, D., A. K. Das, and M. P. Egloff. 1998. The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu. Rev. Biophys. Biomol. Struct. 27:133-164. [DOI] [PubMed] [Google Scholar]
- 7.Beard, P. M., M. J. Daniels, D. Henderson, A. Pirie, K. Rudge, D. Buxton, S. Rhind, A. Greig, M. R. Hutchings, I. McKendrick, K. Stevenson, and J. M. Sharp. 2001. Paratuberculosis infection of nonruminant wildlife in Scotland. J. Clin. Microbiol. 39:1517-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bull, T. J., E. J. McMinn, K. Sidi-Boumedine, A. Skull, D. Durkin, P. Neild, G. Rhodes, R. Pickup, and J. Hermon-Taylor. 2003. Detection and verification of Mycobacterium avium subsp. paratuberculosis in fresh ileocolonic mucosal biopsy specimens from individuals with and without Crohn's disease. J. Clin. Microbiol. 41:2915-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castandet, J., J. F. Prost, P. Peyron, C. Astarie-Dequeker, E. Anes, A. J. Cozzone, G. Griffiths, and I. Maridonneau-Parini. 2005. Tyrosine phosphatase MptpA of Mycobacterium tuberculosis inhibits phagocytosis and increases actin polymerization in macrophages. Res. Microbiol. 156:1005-1013. [DOI] [PubMed] [Google Scholar]
- 10.Charbonneau, H., and N. K. Tonks. 1992. 1002 protein phosphatases? Annu. Rev. Cell Biol. 8:463-493. [DOI] [PubMed] [Google Scholar]
- 11.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 12.Cowley, S., M. Ko, N. Pick, R. Chow, K. J. Downing, B. G. Gordhan, J. C. Betts, V. Mizrahi, D. A. Smith, R. W. Stokes, and Y. Av-Gay. 2004. The Mycobacterium tuberculosis protein serine/threonine kinase PknG is linked to cellular glutamate/glutamine levels and is important for growth in vivo. Mol. Microbiol. 52:1691-1702. [DOI] [PubMed] [Google Scholar]
- 13.Cowley, S. C., R. Babakaiff, and Y. Av-Gay. 2002. Expression and localization of the Mycobacterium tuberculosis protein tyrosine phosphatase PtpA. Res. Microbiol. 153:233-241. [DOI] [PubMed] [Google Scholar]
- 14.Fu, Y., and J. E. Galan. 1998. The Salmonella typhimurium tyrosine phosphatase SptP is translocated into host cells and disrupts the actin cytoskeleton. Mol. Microbiol. 27:359-368. [DOI] [PubMed] [Google Scholar]
- 15.Grant, I. R., E. I. Hitchings, A. McCartney, F. Ferguson, and M. T. Rowe. 2002. Effect of commercial-scale high-temperature, short-time pasteurization on the viability of Mycobacterium paratuberculosis in naturally infected cows' milk. Appl. Environ. Microbiol. 68:602-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koul, A., A. Choidas, M. Treder, A. K. Tyagi, K. Drlica, Y. Singh, and A. Ullrich. 2000. Cloning and characterization of secretory tyrosine phosphatases of Mycobacterium tuberculosis. J. Bacteriol. 182:5425-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, L., J. P. Bannantine, Q. Zhang, A. Amonsin, B. J. May, D. Alt, N. Banerji, S. Kanjilal, and V. Kapur. 2005. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc. Natl. Acad. Sci. USA 102:12344-12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raizman, E. A., S. J. Wells, P. A. Jordan, G. D. DelGiudice, and R. R. Bey. 2005. Mycobacterium avium subsp. paratuberculosis from free-ranging deer and rabbits surrounding Minnesota dairy herds. Can. J. Vet. Res. 69:32-38. [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Sendide, K., A. E. Deghmane, J. M. Reyrat, A. Talal, and Z. Hmama. 2004. Mycobacterium bovis BCG urease attenuates major histocompatibility complex class II trafficking to the macrophage cell surface. Infect. Immun. 72:4200-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sory, M. P., A. Boland, I. Lambermont, and G. R. Cornelis. 1995. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc. Natl. Acad. Sci. USA 92:11998-12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teitelbaum, R., M. Cammer, M. L. Maitland, N. E. Freitag, J. Condeelis, and B. R. Bloom. 1999. Mycobacterial infection of macrophages results in membrane-permeable phagosomes. Proc. Natl. Acad. Sci. USA 96:15190-15195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walton, K. M., and J. E. Dixon. 1993. Protein tyrosine phosphatases. Annu. Rev. Biochem. 62:101-120. [DOI] [PubMed] [Google Scholar]
- 24.Whittington, R. J., I. B. Marsh, and L. A. Reddacliff. 2005. Survival of Mycobacterium avium subsp. paratuberculosis in dam water and sediment. Appl. Environ. Microbiol. 71:5304-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whittington, R. J., D. J. Marshall, P. J. Nicholls, I. B. Marsh, and L. A. Reddacliff. 2004. Survival and dormancy of Mycobacterium avium subsp. paratuberculosis in the environment. Appl. Environ. Microbiol. 70:2989-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]