Abstract
The facultative intracellular pathogen Salmonella enterica causes a variety of diseases, including gastroenteritis and typhoid fever. Inside epithelial cells, Salmonella replicates in vacuoles, which localize in the perinuclear area in close proximity to the Golgi apparatus. Among the effector proteins translocated by the Salmonella pathogenicity island 2-encoded type III secretion system, SifA and SseG have been shown necessary but not sufficient to ensure the intracellular positioning of Salmonella vacuoles. Hence, we have investigated the involvement of other secreted effector proteins in this process. Here we show that SseF interacts functionally and physically with SseG but not SifA and is also required for the perinuclear localization of Salmonella vacuoles. The observations show that the intracellular positioning of Salmonella vacuoles is a complex phenomenon resulting from the combined action of several effector proteins.
Salmonella is a facultative intracellular pathogen that is associated with several diseases in humans. These include typhoid fever caused by Salmonella enterica serovar Typhi and gastroenteritis caused by Salmonella enterica serovar Typhimurium. After entry into host cells, Salmonella replicates within a vacuole known as the Salmonella-containing vacuole (SCV), which interacts with vesicles derived from the late endocytic pathway. Salmonella's ability to cause disease is complex and involves a multitude of virulence factors, particularly two distinct type III secretion systems (TTSS), which are important for interaction with host cells. The first TTSS is encoded by the Salmonella pathogenicity island 1 (SPI-1) and translocates proteins into the host cell necessary for bacterial invasion (28). The SPI-1 TTSS also seems to be involved in the early establishment of SCVs (24). The second TTSS (TTSS-2) is encoded by SPI-2 and translocates proteins across the vacuolar membrane that act together to enable intracellular replication as a prerequisite for systemic infection (9). Several proteins translocated by TTSS-2 have been described, but in most cases their functions remain unknown (27). The best-characterized effector protein, SifA, is encoded outside SPI-2 but is tightly regulated by the two-component regulatory system SsrAB that also regulated SPI-2 expression (3). SifA is essential for virulence in the mouse model of systemic Salmonella infection (25) and is required for the formation of Salmonella-induced filaments (Sif) (11) and maintenance of the vacuolar membrane that surrounds replicating bacteria (3). This process is mediated by interaction of SifA with SKIP, its cellular host target, which in turn down-regulates recruitment of kinesin-1 onto the vacuolar membrane (6). In epithelial cells SCVs migrate to the perinuclear region in close proximity to the Golgi network. This specific positioning involves SifA and another TTSS-2 effector, SseG (6, 23). Although translocated SseG has been shown to localize to SCV membranes (18, 23) and to target endosomal membranes and microtubules (19), its mode of action remains unclear.
As the perinuclear localization of SCVs seems to be important for intracellular replication in epithelial cells (6, 14, 20, 23), we set out to search for other TTSS-2 effectors that contribute to this positioning. We found that the SPI-2-encoded effector SseF, which displays significant amino acid sequence similarity to SseG, is also necessary for perinuclear localization of SCVs. We also show that SseF and SseG can interact with each other but not with SifA, suggesting that different mechanisms contribute to the spatial containment of SCVs in epithelial cells.
MATERIALS AND METHODS
Antibodies and reagents.
The mouse anti-Giantin antibody and monoclonal mouse anti-M45 were kindly provided by H.-P. Hauri and M. Hensel, respectively. The mouse anti-green fluorescent protein (anti-GFP; JL8; BD Biosciences), anti-c-myc 9E10, anti-hemagglutinin (anti-HA; clone 16B12; Covance), and the rat anti-HA (clone 3F10; Roche Molecular Biochemicals) antibodies were used at a dilution of 10−3. Polyclonal rabbit anti-HA was purchased from Sigma (H6908). Goat anti-mouse and anti-rabbit coupled to peroxidase (Sigma) were used at 10−4. Secondary antibodies donkey anti-rabbit or anti-mouse immunoglobulin G (IgG) conjugated to fluorescein isothiocyanate, Texas red, or cyanine 5 (Cy5) were purchased from Jackson Immunoresearch Laboratories and used at a dilution of 1:200. The goat anti-rabbit and anti-mouse IgG conjugated to Alexa Fluor 350 were from Molecular Probes and used at a dilution of 1:200. The sheep anti-human TGN46 antibody (Serotec) was used at a dilution of 1:100.
His6-SseJ was used to obtain anti-SseJ polyclonal antibodies. Recombinant His6-SseJ was produced according to standard protocols (QIAGEN) and purified by metal-chelating chromatography using Hi-trap columns according to the manufacturer's instructions (GE Healthcare). It was administered to mice by intraperitoneal injection (about 200 μg). His6-SseJ was emulsified with complete or incomplete Freund's adjuvant for initial or booster immunizations, respectively. Antibodies were checked by Western blotting using the purified antigen and total cell fractions of S. enterica serovar Typhimurium expressing SPI-2 genes under phosphate starvation at pH 7.4 as described previously (10).
Cell lines and culture conditions.
HeLa human epithelial cells were routinely grown in Dulbecco's modified Eagle's medium (Gibco-BRL) supplemented with 10% fetal calf serum (Gibco-BRL), 2 mM glutamine, and nonessential amino acids at 37°C in 5% CO2.
Bacterial stains and plasmids.
The Salmonella strains used in this work were wild-type 12023 (NTCC) and its isogenic mutant derivatives P3H6 (sifA::mTn5) (3), HH109 (ssaV::aphT) (10), HH197 (ΔsifB::cat), HH107 (sseF::apht), HH108 (sseG::apht) (16), ΔsseFsseG (this study), MvP373 (ΔsscB sseFsseG::aph) (16), ΔsopD (this study), DH217 (ΔpipB2), and DH221 (ΔpipB) (15). Bacteria were grown in Luria-Bertani (LB) medium supplemented with ampicillin (50 μg/ml), kanamycin (50 μg/ml), or chloramphenicol (30 μg/ml) as appropriate. Plasmid pFVP25.1, carrying gfpmut3A under the control of the rpsM constitutive promoter (26), was introduced into bacterial strains for fluorescence visualization. Plasmids p2888 (ProsseAsscBsseF::HA sseG::M45), p2643 (ProsseAsscBsseF::HA), and p2788 (ProsseA sseG::M45), allowing the protein secretion of epitope-tagged effector proteins under the control of the promoter sseA, were kindly provided by M. Hensel (19). For in vitro induction of SPI-2-regulated proteins, strains were grown under phosphate starvation.
Bacterial infection of HeLa cells, replication assays, and immunofluorescence.
Cells were seeded onto glass coverslips (12-mm diameter) in 10-cm dishes at a density of 106 cells per dish 24 h before infection. Bacteria were incubated overnight at 37°C with shaking, diluted 1:33 in fresh LB broth, and incubated under the same conditions for 3.5 h. The cultures were diluted in Earle's buffered salt solution, pH 7.4, and added to the cells at a multiplicity of infection of approximately 100:1. The infection was allowed to proceed for 10 min at 37°C in 5% CO2. Cells were washed three times with their growing medium containing 100 μg/ml gentamicin and incubated in this medium for 1 h, after which the gentamicin concentration was decreased to 10 μg/ml for the remainder of the experiment. For enumeration of intracellular bacteria, cells were washed three times with phosphate-buffered saline (PBS) and lysed with 0.1% Triton X-100 for 10 min, and a dilution series was plated onto LB agar plates. Plates were incubated overnight at 37°C. Colonies were counted. Each time point was performed in triplicate, and each individual experiment was performed three times or more. For immunofluorescence, coverslips were washed with PBS, fixed with 3% formaldehyde in PBS, pH 7.4, for 15 min, washed extensively with 50 mM NH4Cl in PBS, and washed twice with 0.1% saponin in PBS. Cells were incubated for 20 min with primary antibodies diluted in 19% horse serum, 0.1% saponin in PBS, washed extensively with 0.1% saponin-PBS, and incubated for 20 min with secondary antibodies. Coverslips were then washed, mounted in Mowiol, and viewed under a Zeiss 510 confocal microscope equipped with a photomultiplier. Images were processed with Zeiss LSM and Photoshop software. Quantification of the number of bacteria associated with the Golgi network was performed essentially as described previously (23).
Mouse mixed infections.
Mice were inoculated intraperitoneally with 105 CFU per mouse, as described previously (2). The spleens were aseptically removed 48 h after inoculation, and bacteria were recovered and enumerated after plating a dilution series onto LB agar and LB agar with the appropriate antibiotics. Each competitive index value is the mean of three independent mouse infections and is defined as the ratio between the mutant and wild-type strains within the output (bacteria recovered from the mouse after infection) divided by their ratios within the input (initial inoculum). Student's t test was used to analyze the competitive index, and P values of 0.05 or less are considered significant.
Recombinant DNA methods.
All open reading frames in this study were amplified from Salmonella enterica serovar Typhimurium 12023 (NTCC) using the primers given in Table 1.
TABLE 1.
Primers used in this study
| Designation | Nucleotide sequence |
|---|---|
| SseF1for | 5′-GCGAGATCTAAAATTCATATTCCGTCAGCGGC-3′ |
| SseF1rev | 5′-GCGGTCGACAGGTTTCATGGTTCTCCCC-3′ |
| SseGfor | 5′-GCGAGATCTAAACCTGTTAGCCCAAATGCTC-3′ |
| SseGrev | 5′-GCGGTCGACGATTACTCCGGCGCACG-3′ |
| SifBfor | 5′-GCGGGATCCAATTACTATCGGGAGAGG-3′ |
| SifBrev | 5′-GCGCTGCAGGGGATTGTAAATCCATAC-3′ |
| SseIfor | 5′-GCGGGATCCTTTCATATTGGAAGCGGATG-3′ |
| SseIrev | 5′-GCGGTCGACGGTGCGCTTACATTTTACCT-3′ |
| SseJfor | 5′-GCGGGATCCCCATTGAGTGTTGGACAGGG-3′ |
| SseJrev | 5′-GCGGTCGACTTTATTCAGTGGAATAATGATG-3′ |
| SseF2for | 5′-GGATCCAAAATTCATATTCCGTCAGCG-3′ |
| SseF2rev | 5′-GAATTCTCATGGTTCTCCCCGAGATG-3′ |
| Myc-rev | 5′-ATGGATCCCAGGTCCTCCTCGGAGATCAGC-3′ |
| SseF(Δ1-63)for | 5′-GCTGGATCCTTTATGCAATACACTATTCGTGC-3′ |
| SseF (Δ1-82)for | 5′-GCTGGATCCGCGGTAATTTCTGGCGGGGCAGG-3′ |
| SseF (Δ208-260)rev | 5′-TATCTAGAATAATCCAGTACCGCACCTATCGC-3′ |
| SseF (Δ208-260)for | 5′-GCTGGATCCTTTATGCAATACACTATTCGTGC-3′ |
| SseF (Δ83-145)rev | 5′-GCTCTAGACGCTGCAGCAACCGATAACCC-3′ |
| SseF (Δ83-145)for | 5′-GCTCTAGACTTAACTGCGCTAACACCCTTGC-3′ |
| SseF (83-145)for | 5′-ATGGATCCGTAATTTCTGGCGGGGCAGGATTACC-3′ |
| SseF (83-145)rev | 5′-GCTCTAGAACTTGCCCCACATTTTAAGGC-3′ |
The PCR products were subcloned into the pGemT cloning vector (Promega). The respective BglII- or BamHI- and SalI-restricted fragments of sseF and sseG were ligated into the pEGFP C1 cloning vector (Clontech) via the same restriction sites of the multiple cloning site. In order to obtain differentially tagged products for SseF and SseG, the respective BglII/Sst1 and BglII/Nco1 fragments of the pGemT constructs were subcloned into the pLitmus 29 cloning vector (New England Biolabs) restricted with BamH1/Sst1 and BamH1/Nco1. The plasmid products were digested with Sal1 and EcoR1 and subcloned into pCMVmyc and pCMVHA restricted with the same enzymes, generating the constructs pCMVmyc-sseF, pCMVHA-sseF, pCMVmyc-sseG, and pCMVHA-sseG. The latter constructs encoded the N-terminal myc- or HA-tagged proteins with a native C terminus. For generation of the N-terminal myc-tagged SifB, a construct of pQE31 (QIAGEN) obtained by insertion of the BamH1/PstI fragment of pGemT-sifB was subcloned into pLitmus 29 via BamH1 and HindIII. The KpnI/XhoI fragment of the latter construct was subcloned into a KpnI/SalI-restricted pEGFP-C1 derivate, in which gfp was replaced substituted with the myc sequence. In analogy myc-sseI was obtained by ligation of a pQE30-sseI-derived NheI/BamH1 fragment with pLitmus29 restricted with SpeI/BamH1. The SalI/EcoR1 fragment of the latter construct was inserted into the equally restricted pCMV-myc, giving rise to an N-terminal myc-tagged fusion protein. The BamHI/SalI sseJ fragment of pGemT sseJ was inserted into the equally restricted pQE30, resulting in am N-terminal His6-tagged fusion construct for expression in Escherichia coli.
Truncated myc-sseF constructs were derived from a pRK5-myc-sseF construct, similar to pCMVmyc-sseF. A PCR product of primers SseF2for and SseF2rev was restricted with BamHI and EcoRI and ligated into pRK5-myc, resulting in an N-terminal-tagged protein. Truncated versions of pRK5myc-sseF were generated by inverse PCR using the Expand Long Template PCR system (Roche). PCR products deleted for the first 60 and 82 amino acids were generated by the primer SseF(Δ1-63)for and SseF(Δ1-82)for, respectively, and Myc-rev and with pRK5myc-sseF as template. In analogy, the C-terminal deletion (208 to 260) was obtained by SseF(Δ208-260)rev and SseF(Δ208-260) for exclusion of the 3′ end of sseF. The respective XbaI and BamHI sites were digested and ligated. Likewise, SseF(Δ83-146) was generated using the primer SseF(Δ83-145)rev and SseF(Δ83-145)for. For expression of myc::sseF(83-145), the fragment was generated by PCR using the primers SseF(83-145)for and SseF(83-145)rev and ligated into pRK5-myc via BamHI and EcoRI. All PCR products were confirmed by sequencing, and ectopically expressed proteins in HeLa cells were analyzed by Western blotting. Sequence comparison has been done with the program LALIGN at www.ch.embnet.org/.
GST pull-down.
Glutathione-S-transferase-SseF polypeptide was expressed in BL21(DE3) cells by using a pGEX-6P1 construct obtained by insertion of a BglII/Sal fragment, purified from bacterial lysates on glutathione-Sepharose 4B beads (Pharmacia Biotech) following the manufacturer's instructions and stored at −80°C in PBS-20% glycerol. Transfected HeLa cells were resuspended in lysis buffer (PBS-1% Triton X-100-EDTA-free protease inhibitor cocktail [Roche]) for 1 h at 4°C and centrifuged at 20,000 × g for 10 min. For pull-down experiments, 250 to 300 μg protein of HeLa cell extracts was incubated with 2 μg of purified protein bound to 5 μl stacked glutathione-Sepharose 4B beads and incubated for 2 h at 10°C. Beads were washed four times with lysis buffer supplemented with 0.5 M NaCl. Ten percent of the lysates and pulled-down proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting using an appropriate antibody. Western blots were developed with autoradiographic film (Super RX; Fuji) using the SuperSignal West Pico detection reagent (Pierce, Rockford, Ill.).
Coimmunoprecipitation of proteins expressed in HeLa cells.
HeLa cells were grown in culture dishes cotransfected with two plasmids by using FuGene 6 (Roche, Mannheim, Germany) according to the manufacturer's instructions. After 20 h the cells were washed with cold PBS and lysed for 20 min by treatment with 400 μl of 5% glycerol, 1% Triton X-100, and a protease inhibitor cocktail (Complete EDTA free; Roche Molecular Biochemicals). Unbroken cells and nuclei were separated by centrifugation at 1,800 × g, and the supernatant was incubated with Sepharose beads coupled to a mouse anti-myc antibody (9E10; Santa Cruz) for 2 h at 4°C. Subsequently, the beads were washed four times with PBS, 0.1% Triton X-100. The washed beads were incubated with Laemmli sample buffer at 100°C for 2 min, and proteins were analyzed by SDS-PAGE and Western blotting using appropriate antibodies without stripping the membrane.
Coimmunoprecipitation of translocated epitope-tagged proteins.
HeLa cells of three dishes of 160 cm2 were infected with the sseF sseG mutant MvP 373 carrying a plasmid allowing the secretion of both SseF-HA and SseG-M45 (p2888). For controls, plasmids allowing secretion of SseF-HA or SseG-M45 were used. At 12 h after infection, cells were washed extensively with cold PBS and lysed with 1% Triton X-100 in PBS in the presence of protease inhibitor at 4°C. Bacteria, unbroken cells, and nuclei were separated by centrifugation at 6,000 × g for 5 min at 4°C. The supernatant was subsequently incubated with rabbit anti-HA or mouse anti-M45 antibody coupled to Sepharose beads via protein G (GE Healthcare) for 3 h. The beads were washed three times with PBS-0.1% Triton X-100 and analyzed by SDS-PAGE and Western blotting using the rat anti-HA (dilution, 1:104), mouse anti-M45, and mouse anti-SseJ antibodies. The blot membranes were stripped for elimination of former antibodies with 100 mM glycine-NaOH, 150 mM NaCl, pH 2.1.
Statistical analyses.
Statistical analyses were performed using the Student-Newman-Keuls test of the INSTAT software by GraphPad.
RESULTS
The perinuclear localization of SCVs requires multiple TTSS-2 effector proteins.
It has been shown that in epithelial cells Salmonella replication in the Golgi region requires the translocation of the TTSS-2 effectors SseG and SifA into the host cell but not SseI or SseJ (6, 23). We investigated if other TTSS-2 effectors are involved in localizing the SCV in proximity with the Golgi. To investigate this possibility, we infected HeLa cells with different mutant strains lacking functional genes encoding known translocated TTSS-2 effectors and analyzed their intracellular distribution by immunofluorescence microscopy in relation to the Golgi network. At 8 h after invasion sifB, sopD, pipB, and pipB2 mutant strains and wild-type Salmonella were found in vacuoles, the majority of which were clustered in the vicinity of the Golgi. By contrast, the majority of sseF SCVs were scattered throughout the host cytosol (Fig. 1A and B), a phenotype evocative of ssaV and sseG mutant strains (23) as well as sifA mutants that are still in a vacuole (Fig. 1A) (6). Plasmids expressing wild-type alleles of sseF or sifA were sufficient to restore the level of perinuclear localization of the corresponding mutants to that of the wild-type strain (Fig. 1A). This confirmed that the lack of perinuclear localization of sseF SCVs was specifically due to the absence of SseF. We next analyzed the contribution of SseF in the intraepithelial replication of Salmonella. Both the sseF mutant and an sseF sseG double mutant displayed a strong replication defect in HeLa cells, similar to that of ssaV (Fig. 1C) and sseG mutants (23). Altogether, these data show that deletion of sseF or sseG has a similar impact with regard to Salmonella replication and SCV localization in epithelial cells.
FIG. 1.
Identification of new effectors required for Golgi localization of SCVs in HeLa cells. (A and B) HeLa cells were infected with GFP-expressing wild-type (wt) Salmonella or mutants lacking different effectors and fixed at 8 h postinfection. (A) Cells were immunostained for Lamp1 and the Golgi marker Giantin and scored by immunofluorescence for the intracellular positioning of bacteria. Only bacteria enclosed in a Lamp1-positive compartment that were either completely or partially surrounded by the Golgi marker were counted as being Golgi associated. Bacterial clusters that were found adjacent to the Golgi but did not fulfill the above criteria were counted as nonassociated. At least 50 host cells, corresponding to more than 100 bacteria, were scored blind in each experiment. Statistical analysis for comparison of wild-type Salmonella and mutant strains indicated a significant difference for the ssaV, sseF, and sifA strains (P < 0.001), whereas no significant difference was observed in comparison of the wt to other mutants or the complemented sseF and sifA strains (P > 0.05). (B) Confocal immunofluorescence images of the dispersed distribution of the sseF mutant in contrast with the Golgi localization of the wt strain. The cell shape is marked. Bars, 10 μm. (C) Intracellular replication of Salmonella strains. Values indicate the fold increase, calculated as the ratio of intracellular bacteria between 2 and 16 h after invasion. Statistical analysis indicated a significant difference between mutant strains and wild-type Salmonella (P < 0.001), whereas no significant difference was observed between mutant strains. (D) Complementation of the intracellular positioning of the sseF and sifA mutants to the Golgi region by ectopic expression of myc-SseF and myc-SifA, respectively. (A, C, and D) Standard deviations of the means are shown and correspond to three independent experiments.
Functional domains and intracellular distribution of secreted effectors.
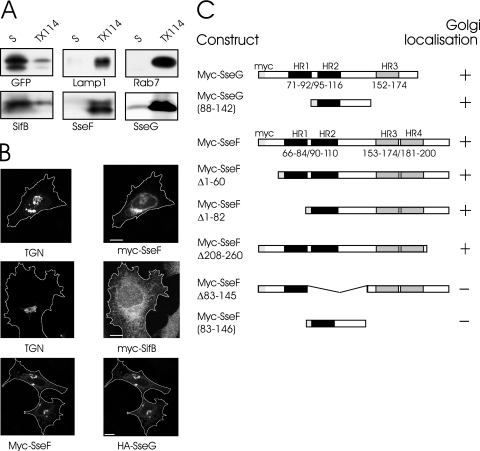
Ectopic expression of myc-SseG has been demonstrated to complement the sseG mutant for Golgi localization of the vacuole (23). Therefore, we determined the ability of ectopically expressed myc-SseF or myc-SifA to complement in trans the sseF and sifA mutant strains for their intracellular distribution in HeLa cells. Immunofluorescence microscopy analysis revealed that myc-SseF and myc-SifA were able to restore the Golgi localization of the sseF and sifA SCVs, respectively, showing that these ectopically expressed proteins are functional (Fig. 1D). Having demonstrated the efficiency of ectopically expressed SseF in complementing the mutant, we analyzed its cellular distribution in HeLa cells. We first tested the capacity of ectopically expressed effector proteins to partition between aqueous and TX-114 detergent-rich phases, as a simple approach to determine the hydrophobicity of proteins (5). GFP is a water-soluble protein and, as expected, partitioned exclusively into the aqueous phase. In contrast, the prenylated Rab7 GTPase and the integral membrane protein Lamp1 were both exclusively found in the detergent phase (Fig. 2A). SseF and SseG both possess several predicted transmembrane regions and showed an exclusive membrane distribution, whereas SifB, another TTSS-2 effector, was found in the aqueous phase. Then, we analyzed the intracellular distribution of these effectors by immunofluorescence microscopy. myc-SifB showed an even distribution throughout cells, corresponding to its cytosolic partition, while SseF exhibited a stack-like perinuclear staining (Fig. 2B). Such localization has been previously shown for myc-SseG (19, 23), which localizes to the Golgi network. Colabeling of transfected cells with TGN46, a marker for the Golgi network, and myc-tagged proteins revealed an extensive colocalization of ectopically expressed SseF with the Golgi marker (Fig. 2B). In addition, coexpressed myc-SseF and HA-SseG revealed a high degree of colocalization. Collectively, these results show that ectopically expressed SseF and SseG localized both to the Golgi network.
FIG. 2.
Role of membrane-targeting domains and cellular localization of ectopically expressed effectors. (A) Protein lysates of myc-tagged proteins expressed in HeLa cells were partitioned using Triton X-114. Aliquots of each fraction were analyzed by Western blotting after SDS-PAGE. S, soluble phase; TX114, membrane-enriched phase. For controls, the partitioning of GFP, Lamp1, and Rab7 was analyzed. (B) Confocal immunofluorescence analysis of HeLa cells transfected with plasmids expressing myc-tagged effectors (two upper panels) and cotransfection of myc- and HA-tagged effectors (lower panel). The cells were fixed 24 h posttransfection and labeled using appropriate antibodies. Tagged SseF and SseG colocalized with each other as well as with the Golgi marker TGN46. Bars, 10 μm. (C) Schematic diagram showing that the region of SseF homologous to the Golgi-targeting domain of SseG is not sufficient for Golgi localization of the ectopically expressed myc-tagged protein. HR, hydrophobic region.
The region of myc-SseG required for Golgi targeting was previously identified by ectopic expression of truncated versions of the protein and consists of its second transmembrane domain with adjacent amino acids (88 to 142) (Fig. 2C) (23). Alignment of the amino acid sequences of the Golgi-targeting domain of SseG and the corresponding region of SseF (83 to 127) revealed 41.3% identity. Therefore, we investigated which region is necessary to target myc-SseF to the Golgi network. We found that neither of the hydrophilic ends nor the first of the four possible transmembrane regions of myc-SseF was necessary for its perinuclear localization (Fig. 2C). A version of myc-SseF lacking amino acid residues 83 to 145 (myc-SseFΔ83-145) was no longer able to target the Golgi network. However, ectopic expression of this domain alone (83-145) was not sufficient for Golgi localization, suggesting that additional regions encoded in the last two hydrophobic regions are involved in targeting ectopically expressed SseF to the Golgi network.
Interaction between SseF and SseG.
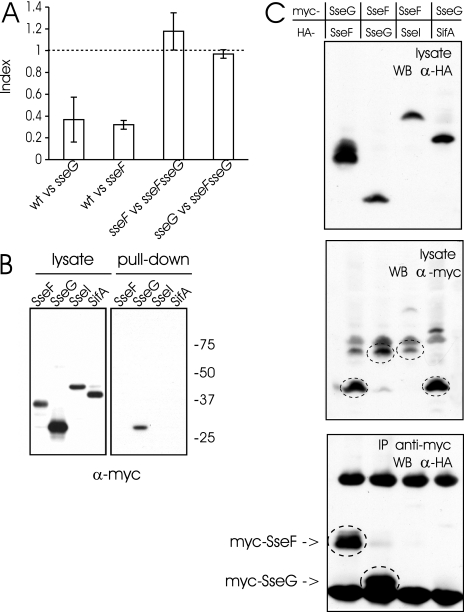
The observation that SseG, SseF, and SifA are all required for the perinuclear localization of SCVs prompted us to investigate whether they contribute to the same function in vivo. It is known that a sifA mutant is strongly attenuated in virulence in the mouse model of systemic infection (3, 25), while sseF and sseG mutant strains exhibit similar but less drastic levels of virulence attenuation in mice (16). Therefore, the competitive indices of wild type versus single mutants and of single versus double mutants (2) were determined for sseF and sseG mutants. The sseF sseG double mutant strain was not more attenuated than either single mutant (Fig. 3A). This lack of an additive effect on virulence attenuation supports the hypothesis that SseF and SseG contribute to the same Salmonella virulence function during systemic infection. This result, together with the similarities between SseF and SseG, and also their complete colocalization upon ectopic coexpression, prompted us to analyze if these TTSS-2 effectors interact. GST pull-down assays were performed. Purified GST-SseF bound to glutathione-Sepharose beads was incubated with Triton X-100 extracts of HeLa cells expressing either myc-SseF, myc-SseG, myc-SseI, or myc-SifA. Immunoblotting with an anti-myc antibody revealed the binding of myc-SseG to GST-SseF (Fig. 3B). The stability of the proposed SseF-SseG complex towards high ionic strength (0.5 M NaCl) was further confirmed with coelution analysis of E. coli-derived recombinant proteins (data not shown). This observation excludes ionic forces between SseF and SseG, suggesting that hydrophobic interactions are responsible for their binding. To analyze this complex in more detail, we performed coimmunoprecipitations using Triton X-100 extracts prepared from HeLa cells cotransfected with vectors expressing myc- and HA-tagged effectors. These experiments revealed the coimmunoprecipitation of SseF and SseG irrespective of which partner harbored the HA or the myc tag (Fig. 3C). Altogether, these experiments strongly suggest specific binding of SseF and SseG. To test whether SseF and SseG interact under physiological conditions, infection experiments were performed using Salmonella strains expressing SseF-HA and/or SseG-M45 (19). The expression of tagged effectors and of the TTSS-2 effector SseJ was controlled in extracts of bacteria grown under SPI-2-inducing conditions (Fig. 4, left panel) and in postnuclear supernatants of infected cells (Fig. 4, middle panel) using specific antibodies. We observed that translocated SseF exhibited different isoforms that differentiated from the secreted effector by their molecular weights. Whether these shifts are due to a host cell-mediated modification of SseF is unknown. Translocated SseG-M45 and SseF-HA proteins were efficiently immunoprecipitated from infected cells and coimmunoprecipitated each other (Fig. 4, right panel). SseJ could not be detected in immunoprecipitates, thus establishing the specificity of the immunoprecipitation. Collectively, these experiments demonstrate the specific direct binding of SseF and SseG upon translocation, enabling perinuclear positioning of the SCV and intracellular replication of S. enterica serovar Typhimurium.
FIG. 3.
Interaction of SseF and SseG. (A) Competitive index analysis of Salmonella mutant strains. The competitive indices of wild type (wt) versus sseG (n = 3) and of wt versus sseF (n = 6) are not significantly different (P = 0.71) but are significantly (P = 0.0012) or very significantly (P = 0.0006) lower than 1, respectively. The competitive indices of sseG versus sseF sseG (n = 3; P = 0.30) and sseF versus sseF sseG (n = 4; P = 0.12) are not significantly different from 1. These data indicate that the two genes are functionally linked. (B) GST-SseF bound to beads was incubated with extracts of HeLa cells expressing myc-tagged Salmonella effectors. Total lysates and proteins bound to washed beads were analyzed by Western blotting using an anti-myc antibody. (C) HeLa cells were cotransfected with plasmids coding for myc- and HA-tagged effector proteins as indicated, lysed, and immunoprecipitated (IP) using an anti-myc antibody coupled to Sepharose beads. HA-tagged proteins present in lysates (upper panels) and coimmunoprecipitated with myc-tagged effectors (lower panel) were analyzed by Western blotting (WB). The lysate membrane was reanalyzed for myc-tagged proteins (encircled bands, middle panel). Controls indicated the effective binding of myc-tagged proteins to the beads (data not shown). The top and bottom bands seen after anti-myc immunoprecipitation (lower panel) correspond to mouse IgG heavy and light chains.
FIG. 4.
Interaction of translocated SseF and SseG. HeLa cells were infected with an S. enterica serovar Typhimurium sseF sseG double mutant harboring plasmid p2888 (sseF-HA and sseG-M45) (C). As controls, double mutants harboring plasmids p2643 (sseF-HA) (A) or p2788 (sseG-M45) (B) were used. The tagged proteins were detected in extracts of bacteria grown under phosphate starvation, inducing ssrAB-regulated proteins in vitro (left panel). SseG-M45 and SseF-HA were detected in the postnuclear supernatant (PNS) of HeLa cells infected for 12 h (middle panel). The signal for SseF-HA was intensified by successive incubations of the blotting membrane with a rabbit anti-rat and a goat anti-rabbit antibody, both coupled to peroxidase. Tagged proteins were subsequently immunoprecipitated (IP) from the PNS using anti-HA or anti-M45 antibodies coupled to the Sepharose beads. Immunoprecipitated proteins were analyzed by Western blotting (WB, right panel). The fractions of translocated SseF detected in the coimmunoprecipitation or PNS differ in electrophoretic mobility (apparent molecular masses, 33.7, 32.5, and 30.5 kDa) compared to SseF-HA secreted in vitro (apparent molecular mass, 31.6 kDa). Expression and secretion of SseJ were controlled using Salmonella wild type, the double mutant strain expressing sseF-HA and sseG-M45, and an sseJ mutant. The absence of translocated SseJ with sseF-HA and sseG-M45 was used to check the specificity of the immunoprecipitations.
DISCUSSION
Following entry into host epithelial cells, Salmonella replicates in vacuoles that are juxtanuclear and particularly concentrated near the Golgi. This localization requires a functional TTSS-2 and more specifically the translocated effector proteins SseG (23) and SifA (6). The present study shows that a third TTSS-2 effector, SseF, contributes in this process and that SseF and SseG are physically and functionally linked. In the mouse model of systemic infection, sseF and sseG mutant strains have comparable levels of virulence attenuation. The double mutant is no more attenuated than either single mutant, indicating that these TTSS-2 effectors contribute to the same function in vivo. In contrast, the sseJ sifA double mutant displays a higher attenuation than the respective single mutants, suggesting a different mode of action (22). Interestingly, both SseF and SseG are targeted to the Golgi compartment upon ectopic expression and interact, as demonstrated by their coprecipitation. In addition, we have shown that in the physiological context of Salmonella-infected cells, translocated SseF and SseG interact specifically. Molecular analysis further indicates that the region encompassing the last three hydrophobic regions of SseF is necessary for targeting the effector protein to the Golgi.
SifA, SseF, and SseG have been shown to be required for the formation of Sifs (13, 25). Following their translocation from intravacuolar bacteria, the three effectors are found mainly on SCV and Sif membranes, while SseG is also found on endosomal compartments (18, 23). These TTSS-2 effectors localize to the Golgi apparatus when they are expressed from a eukaryotic expression vector in uninfected cells. This difference raises the question of whether their Golgi localization, although useful for biochemical analysis, is relevant in physiological terms. It is likely that in the absence of SCVs, the lipid composition of the Golgi, or a Golgi-associated protein, may provide a signal for targeting and subsequent insertion of these effectors into Golgi membranes. Interestingly, it was recently shown that the N-terminal WEK(I/M)xxFF translocation motifs of SifA, SopD2, and SspH2 (21) are sufficient to localize GFP to the Golgi apparatus (7). Consistent with that, the solubility of SseG was found to be elevated in 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS)-containing buffers, in contrast to those with other detergents (e.g., Triton X-100 or dodecylmaltoside) (data not shown). CHAPS resembles cholesterol, a major component of Golgi membranes that accumulates in SCVs in a TTSS-2-dependent manner (8). In infected cells, effectors are likely to be inserted into the SCV membrane immediately upon translocation, thereby preventing cytoplasmic diffusion and insertion in the Golgi membranes. Upon insertion, effectors diffuse laterally through vacuolar and Sif membranes. Nonetheless, ectopically expressed effectors were found to rescue the intracellular dispersed distribution of their respective mutants (Fig. 1) (23). This result implies that ectopically expressed effectors remain functional, even though we were unable to detect SseG or SseF on SCVs (unpublished results). How ectopically expressed SseG is capable of rescuing the juxtanuclear localization defect of the sseG mutant is unknown. In the case of SifA, ectopically expressed protein was found to localize to SCV membranes and Sifs (3).
The defect in the Golgi localization of various TTSS-2 mutants correlates with an attenuated virulence in the mouse model of infection and, for sseF and sseG mutants, with a defect of replication in HeLa cells. Perturbation of Golgi function by treatment of infected cells with brefeldin A inhibits bacterial replication (23), and post-Golgi transport is redirected in Salmonella-infected cells (17). Therefore, the presence of a functional exocytic pathway appears to be an important aspect of bacterial virulence, although the physiological consequences of the proximity of SCVs to the Golgi apparatus remain unknown. The attenuated virulence in mice of the sifA mutant is not directly linked to the mislocalization of the SCV but to the release of bacteria into the macrophage cell cytoplasm, which is toxic to Salmonella (4). This is not the case for sseF and sseG mutants, which remain in a vacuole. Therefore, how can we explain the inhibition of replication of these mutant strains? The Golgi area is a region of intense vesicular traffic at the crossroads of the exocytic and endocytic pathways. Microtubules and their associated motor proteins play a fundamental role in mediating vesicular transport as well as determining the steady-state localization of organelles and vesicles along these pathways. The requirement for intact microtubule (11) and Golgi networks in the intracellular replication of Salmonella suggests that this pathogen might induce fusion of vesicles to the SCV for nutrition using further TTSS-2-dependent effectors than those already described.
The intracellular positioning of SCVs and the regulation of SCV membrane dynamics involve the action of TTSS-2 and both plus-end- and minus-end-directed microtubule-associated molecular motors (12). We recently demonstrated that PipB2 mediates the recruitment of kinesin-1 on the SCV (15) while SifA favors the release of this motor, as shown by the accumulation of kinesin-1 on sifA mutant vacuolar membranes. Interestingly, dynein seems also to be involved in these processes, since inhibition of its activity prevents loss of membrane from vacuoles enclosing sifA mutant bacteria (12) and the capacity of intracellular Salmonella to replicate (14, 20). The scattered localization of sseF and sseG mutants may indicate that plus-end-directed motors are predominantly associated with their vacuoles. SseF and SseG may, like SifA, be involved the inhibition of a plus-end motor or, alternatively, required for the recruitment of a minus-end motor. Consistent with this hypothesis, dynein accumulates in the vicinity of bacterial microcolonies (12), and sseF mutants have been found reduced in dynein recruitment to the SCV (1).
Our data suggest that the SseG and SseF interact to form a functional complex, though immunohistochemical analysis revealed only partial colocalization of simultaneously translocated SseF-HA and SseG-M45 (19, 18). However, since the wild-type strain and not the sseF sseG double mutant was used in that study, the difference may be due to the formation of mixed complexes of tagged and untagged SseF and SseG. The identification of host target molecules of these TTSS-2 effectors will further increase our understanding of the mechanisms that govern the localization of Salmonella in the Golgi area and the contribution of each effector to the intracellular replication of this important pathogen.
Acknowledgments
We are grateful to Silke Stender and Michael Hensel for critical review of the manuscript and to Rolf Hilgenfeld for discussion.
E.B. and T.H. were recipients of fellowships from the French Ministry of Research, FRM (E.B.) and ARC (T.H.). This work was supported by grants from Deutsche Forschungsgemeinschaft (De 788/1-1) to J.D. and from Ministerium für Wissenschaft, Wirtschaft und Verkehr to N.P., by grants from the Medical Research Council and Wellcome Trust (United Kingdom) to D.W.H., by institutional grants from the CNRS and INSERM, and through the Microban EU network no. MRTN-CT-2003-504227.
Editor: F. C. Fang
Footnotes
Published ahead of print on 2 October 2006.
REFERENCES
- 1.Abrahams, G. L., and M. Hensel. 2006. Manipulating cellular transport and immune responses: dynamic interactions between intracellular Salmonella enterica and its host cells. Cell Microbiol. 8:728-737. [DOI] [PubMed] [Google Scholar]
- 2.Beuzon, C. R., and D. W. Holden. 2001. Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect. 3:1345-1352. [DOI] [PubMed] [Google Scholar]
- 3.Beuzon, C. R., S. Meresse, K. E. Unsworth, J. Ruiz-Albert, S. Garvis, S. R. Waterman, T. A. Ryder, E. Boucrot, and D. W. Holden. 2000. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 19:3235-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beuzon, C. R., S. P. Salcedo, and D. W. Holden. 2002. Growth and killing of a Salmonella enterica serovar Typhimurium sifA mutant strain in the cytosol of different host cell lines. Microbiology 148:2705-2715. [DOI] [PubMed] [Google Scholar]
- 5.Bordier, C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256:1604-1607. [PubMed] [Google Scholar]
- 6.Boucrot, E., T. Henry, J. P. Borg, J. P. Gorvel, and S. Meresse. 2005. The intracellular fate of salmonella depends on the recruitment of kinesin. Science 308:1174-1178. [DOI] [PubMed] [Google Scholar]
- 7.Brown, N. F., J. Szeto, X. Jiang, B. K. Coombes, B. B. Finlay, and J. H. Brumell. 2006. Mutational analysis of Salmonella translocated effector members SifA and SopD2 reveals domains implicated in translocation, subcellular localization and function. Microbiology 152:2323-2343. [DOI] [PubMed] [Google Scholar]
- 8.Catron, D. M., M. D. Sylvester, Y. Lange, M. Kadekoppala, B. D. Jones, D. M. Monack, S. Falkow, and K. Haldar. 2002. The Salmonella-containing vacuole is a major site of intracellular cholesterol accumulation and recruits the GPI-anchored protein CD55. Cell Microbiol. 4:315-328. [DOI] [PubMed] [Google Scholar]
- 9.Chan, K., C. C. Kim, and S. Falkow. 2005. Microarray-based detection of Salmonella enterica serovar Typhimurium transposon mutants that cannot survive in macrophages and mice. Infect. Immun. 73:5438-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deiwick, J., T. Nikolaus, S. Erdogan, and M. Hensel. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31:1759-1773. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-del Portillo, F., M. B. Zwick, K. Y. Leung, and B. B. Finlay. 1993. Salmonella induces the formation of filamentous structures containing lysosomal membrane glycoproteins in epithelial cells. Proc. Natl. Acad. Sci. USA. 90:10544-10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guignot, J., E. Caron, C. Beuzon, C. Bucci, J. Kagan, C. Roy, and D. W. Holden. 2004. Microtubule motors control membrane dynamics of Salmonella-containing vacuoles. J. Cell Sci. 117:1033-1045. [DOI] [PubMed] [Google Scholar]
- 13.Guy, R. L., L. A. Gonias, and M. A. Stein. 2000. Aggregation of host endosomes by salmonella requires SPI2 translocation of SseFG and involves SpvR and the fms-aroE intragenic region. Mol. Microbiol. 37:1417-1435. [DOI] [PubMed] [Google Scholar]
- 14.Harrison, R. E., J. H. Brumell, A. Khandani, C. Bucci, C. C. Scott, X. Jiang, B. B. Finlay, and S. Grinstein. 2004. Salmonella impairs RILP recruitment to Rab7 during maturation of invasion vacuoles. Mol. Biol. Cell 15:3146-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henry, T., C. Couillault, P. Rockenfeller, E. Boucrot, A. Dumont, N. Schroeder, A. Hermant, L. A. Knodler, P. Lecine, O. Steele-Mortimer, J.-P. Borg, J.-P. Gorvel, and S. Meresse. 2006. The Salmonella effector protein PipB2 is a linker for kinesin-1. Proc. Natl. Acad. Sci. USA 103:13497-13502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hensel, M., J. E. Shea, S. R. Waterman, R. Mundy, T. Nikolaus, G. Banks, A. Vazquez-Torres, C. Gleeson, F. C. Fang, and D. W. Holden. 1998. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 30:163-174. [DOI] [PubMed] [Google Scholar]
- 17.Kuhle, V., G. L. Abrahams, and M. Hensel. 2006. Intracellular Salmonella enterica redirect exocytic transport processes in a Salmonella pathogenicity island 2-dependent manner. Traffic 7:716-730. [DOI] [PubMed] [Google Scholar]
- 18.Kuhle, V., and M. Hensel. 2002. SseF and SseG are translocated effectors of the type III secretion system of Salmonella pathogenicity island 2 that modulate aggregation of endosomal compartments. Cell Microbiol. 4:813-824. [DOI] [PubMed] [Google Scholar]
- 19.Kuhle, V., D. Jackel, and M. Hensel. 2004. Effector proteins encoded by Salmonella pathogenicity island 2 interfere with the microtubule cytoskeleton after translocation into host cells. Traffic 5:356-370. [DOI] [PubMed] [Google Scholar]
- 20.Marsman, M., I. Jordens, C. Kuijl, L. Janssen, and J. Neefjes. 2004. Dynein-mediated vesicle transport controls intracellular Salmonella replication. Mol. Biol. Cell 15:2954-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miao, E. A., and S. I. Miller. 2000. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc. Natl. Acad. Sci. USA. 97:7539-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruiz-Albert, J., X. J. Yu, C. R. Beuzon, A. N. Blakey, E. E. Galyov, and D. W. Holden. 2002. Complementary activities of SseJ and SifA regulate dynamics of the Salmonella typhimurium vacuolar membrane. Mol. Microbiol. 44:645-661. [DOI] [PubMed] [Google Scholar]
- 23.Salcedo, S. P., and D. W. Holden. 2003. SseG, a virulence protein that targets Salmonella to the Golgi network. EMBO J. 22:5003-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steele-Mortimer, O., J. H. Brumell, L. A. Knodler, S. Meresse, A. Lopez, and B. B. Finlay. 2002. The invasion-associated type III secretion system of Salmonella enterica serovar Typhimurium is necessary for intracellular proliferation and vacuole biogenesis in epithelial cells. Cell Microbiol. 4:43-54. [DOI] [PubMed] [Google Scholar]
- 25.Stein, M. A., K. Y. Leung, M. Zwick, F. Garcia-del Portillo, and B. B. Finlay. 1996. Identification of a Salmonella virulence gene required for formation of filamentous structures containing lysosomal membrane glycoproteins within epithelial cells. Mol. Microbiol. 20:151-164. [DOI] [PubMed] [Google Scholar]
- 26.Valdivia, R. H., and S. Falkow. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277:2007-2011. [DOI] [PubMed] [Google Scholar]
- 27.Waterman, S. R., and D. W. Holden. 2003. Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell Microbiol. 5:501-511. [DOI] [PubMed] [Google Scholar]
- 28.Zhou, D., and J. Galan. 2001. Salmonella entry into host cells: the work in concert of type III secreted effector proteins. Microbes Infect. 3:1293-1298. [DOI] [PubMed] [Google Scholar]