Abstract
A putative ABC transporter, fit, with significant homology to several bacterial iron transporters was identified in Escherichia coli. The E. coli fit system consists of six genes designated fitA, -B, -C, -D, -E, and -R. Based on DNA sequence analysis, fit encodes an outer membrane protein (FitA), a periplasmic binding protein (FitE), two permease proteins (FitC and -D), an ATPase (FitB), and a hypothetical protein (FitR). Introduction of the E. coli fit system into E. coli strain K-12 increased intracellular iron content and transformed bacteria were more sensitive to streptonigrin, which suggested that fit transports iron in E. coli. Expression of fit was studied using a lacZ reporter assay. A functional, bidirectional promoter was identified in the intergenic region between genes fitA and fitB. The expression of the E. coli fit system was found to be induced by iron limitation and repressed when Fe2+ was added to minimal medium. Several fit mutants were created in E. coli using an in vitro transposon mutagenesis strategy. Mutations in fit did not affect bacterial growth in iron-restricted media. Using a growth promotion test, it was found that fit was not able to transport enterobactin, ferrichrome, transferrin, and lactoferrin in E. coli.
Iron is an essential microelement for bacteria (12). It is required as a cofactor for a lot of important enzymes which are involved in many fundamental cellular processes, including electron transfer, cell respiration, and superoxide metabolism. Iron is also an important factor for bacterial pathogenesis (5, 24, 36). Although the concentration of iron in the environment is sufficient to sustain the viability of microbes, most iron is present as ferric hydroxide, which is insoluble and biologically inaccessible for bacteria. In the host, iron is bound by high-affinity iron binding proteins, such as transferrin and lactoferrin (12). In human and animal body fluids, the concentration of free iron at neutral pH is estimated to be about 10−18 M, which is far too low to support bacterial growth (10, 12). However, under iron starvation, bacteria produce high-affinity iron binding molecules, such as siderophores, to scavenge iron from the environment. The iron-siderophore complexes are transported into bacteria by specific iron transport systems. Most iron transport systems consist of an outer membrane receptor, a periplasmic binding protein, an ABC transporter formed by permease, and ATPase proteins (7, 10). In Escherichia coli, there are at least nine known iron transport systems, including ferrichrome (fhu), enterobactin (fep), ferric citrate (fec), aerobactin (iut), heme (chu), rhodotorulic acid and coprogen (fhuE), salmochelin (iro), yersiniabactin, and ferrous iron (feo) transport systems (6, 7, 10, 17, 24, 25, 27, 32, 37). Besides these iron systems, other putative iron transport systems have been identified by sequence analysis of the E. coli genome (23, 35, 39).
In a previous study that employed in vivo gene expression technology, a novel gene, ivi932, was identified in a clinical E. coli isolate (18). Gene ivi932 was shown to be expressed in vivo in a mouse septicemia infection model. Furthermore, when a mutation was introduced into ivi932, the strain was attenuated about fourfold in the infection model (M. A. Khan and R. E. Isaacson, unpublished data). DNA sequence analysis suggested that ivi932 is probably related to iron transport. We report here that the open reading frames flanking ivi932 appear to encode a novel ABC iron transport system, which was termed fit (ferric or ferrous iron transport). In this report, we studied the effect of mutations in the E. coli fit system on bacterial response to iron limitation. In addition, we studied fit expression under various conditions using lacZ reporter assays. The data in this report support our hypothesis that fit encodes a novel iron transport system.
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli was grown in Difco LB broth (Becton Dickson and Company, Maryland) medium, a defined α-MEM with 10 μM FeCl3 (Invitrogen), or MM9 minimal medium (0.03% KH2PO4, 0.1% NH4Cl, 0.05% NaCl, 1 mM MgSO4, 0.3% Casamino Acids, 0.2% thiamine-HCl). Glucose was added to MM9 medium (0.2%) as a carbon source. For iron-rich medium, a solution of freshly prepared FeSO4 was added to LB medium at a final concentration of 20 μM. For iron-restricted medium, the iron chelator 2,2′-dipyridyl (DIP; Sigma) was added into LB medium at a final concentration of 200 μM.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli strains | ||
| i484 | O25:H autoagluttinating; human isolate | 18 |
| xy006 | i484, fitA::EZ-TN5 <Kan-2> | This study |
| xy005 | i484, fitB::EZ-TN5 <Kan-2> | This study |
| xy009 | i484, fepA::EZ-TN5 <DHFR> | This study |
| xy008 | i484, fepC::EZ-TN5 <DHFR> | This study |
| xy007 | i484, fhuA::EZ-TN5 <DHFR> | This study |
| oy016 | i484, lacZ::EZ-TN5 <DHFR> | This study |
| AA93 | F−araD139 ΔlacU169 rpsL150 relA1 deoC1 flbB5301 ptsF25 rbsR aroB fecB::Mud1 (Ap lac) | 26 |
| oy097 | AA93; feoB::EZ-TN5 <DHFR> | This study |
| AB1515.199 | purE42 proC14 leu-6 trpE38 thi-1 fhuA23 lacY1 (fepC::Tn5) | 6 |
| 123 | Field isolate | Pig |
| 124 | Field isolate | Pig |
| 252 | Field isolate | Pig |
| EcoR33 | Field isolate | Pig |
| 263 | Field isolate | Pig |
| 431 | Field isolate | Pig |
| 987 | Field isolate | Pig |
| 1413 | Field isolate | Pig |
| 16 | Field isolate | Human |
| 17 | Field isolate | Human |
| 18 | Field isolate | Human |
| EcoR40 | Field isolate | Human |
| EcoR60 | Field isolate | Human |
| EcoR62 | Field isolate | Human |
| EcoR8 | Field isolate | Human |
| EcoR26 | Field isolate | Human |
| EcoR42 | Field isolate | Human |
| EcoR45 | Field isolate | Human |
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdS17 (rK− mK+) deoR thi-1 supE44 λ−gyrA96 relA1 | Invitrogen |
| Plasmids | ||
| pMP220 | Contains a promoterless lacZ gene, Tetr IncP | 33 |
| pMP-fitA | fit promoter cloned into pMP220 with direction of fitA transcription | This study |
| pMP-fitB | fit promoter cloned into pMP220 with direction of fitB transcription | This study |
| pCR | Cloning plasmid pCR-XL-TOPO | Invitrogen |
| pCR63 | lacZ cloned into pCR-XL-TOPO | This study |
| pCR64 | fitR cloned into pCR-XL-TOPO | This study |
| pfit1 | Whole fit region cloned into pCR-XL-TOPO | This study |
All reagents and media were made with deionized water after passage through a Millipore reverse osmosis system (Millipore). All glassware was treated with 8 M HCl and then rinsed three times with water. When appropriate, supplements were added to media at the following concentrations: ampicillin, 100 μg/ml; tetracycline, 15 μg/ml; kanamycin, 50 μg/ml; trimethoprim, 100 μg/ml.
DNA manipulation techniques.
General genetic techniques were performed as described previously, including genomic or plasmid DNA purification, ligation, and transformation (29). PCR amplification using Taq polymerase (Promega) or high-fidelity Pfu DNA polymerase (Stratagene) was performed according to the manufacturers' instructions. Restriction endonuclease and DNA-modifying enzymes were used according to manufacturers' protocols. E. coli DH5α cells were used as standard competent cells for DNA cloning. The oligonucleotide primers used in this study for PCR, cloning, and sequencing were commercially synthesized by Integrated DNA Technologies and are listed in Table S1 of the supplemental material. Primers used to amplify and sequence each of the fit genes were designed based on the sequence of E. coli CFT073 (39). DNA sequencing was performed by the Advanced Genetic Analysis Center at the University of Minnesota.
Cloning of the E. coli fit system.
The whole fit system was PCR amplified using the Elongase amplification system (Invitrogen). After DNA was purified using a QiaQuick gel extraction kit (QIAGEN), it was cloned into plasmid pCR-XL-TOPO (Invitrogen). Positive clones were identified by colony PCR, restriction endonuclease digestion, and sequence analysis.
Functional annotation of the E. coli fit system.
Gene prediction and annotation were performed by Open Reading Frame Finder analysis and BLAST sequence similarity searching against the GenBank database (National Center for Biotechnology Information) (40) and confirmed by analyzing with the GenQuiz server (1) and the fold recognition server GenThreader (16). Protein domains were assigned by searching against the Conserved Domain Database (CDD) (40), clusters of orthologous groups (34), and the PFAM (2) and SMART (30) databases and by sequence motif analysis (E-motif) (15). A SOSUI analysis (14) was carried out to identify transmembrane segments.
Construction of transposon-insertional mutants.
The EZ::TN transposons (Epicenter, Madison, Wis.) were used to create gene disruption mutants in E. coli i484 according to the protocols provided by the manufacturer (8, 9, 11). Briefly, the particular gene to be mutated was amplified by PCR using high-fidelity Pfu DNA polymerase. After purification of the amplified DNA, an EZ::TN transposon was inserted into the DNA fragment through in vitro transposition that employs addition of exogenous transposase. The DNA was then transformed as linear DNA by electroporation into E. coli strains containing the plasmid pKD46 (8). The lambda recombinase system carried on pKD46 promotes the homologous recombination of the linear, mutated DNA into the chromosome. Mutants were screened for antibiotic resistance carried on the inserted EZ::TN transposon and confirmed by PCR amplification and sequence analysis. Plasmid pKD46 was then eliminated from these cells by growth at 42°C.
Growth promotion test.
A plate bioassay (37) was performed to determine the substrate of the E. coli fit system. Briefly, LB agar plates were depleted of iron by adding 400 μM and 250 μM 2,2′-dipyridyl for E. coli i484 and AA93, respectively. Approximately 105 (for E. coli i484) or 107 (for E. coli AA93) CFU/ml of cells in log phase were seeded onto the plate. Discs containing 10 μl of various iron-binding or iron-containing compounds were placed on the plates. The concentrations of the compounds were as follows: 50 μM 2,3-dihydroxybenzoic acid, 10 μM ferrichrome, 10 μM heme, 10 μM heme-bovine serum albumin (BSA) complex, 10 μM lactoferrin, 25 μM transferrin, 50 μM rhodotorulic acid (RA), 50 μM desferal, iron citrate, 10 mM FeSO4, and 10 mM FeCl3. Culture supernatant from E. coli AB1515.199 grown in LB with 100 μM DIP was used as a source of enterobactin (6). Bacterial growth was monitored after 24 h of incubation at 37°C. If there was bacterial growth around the disc, it demonstrated that the bacteria could utilize the compound on the disc as an iron source.
Streptonigrin sensitivity test.
Streptonigrin sensitivity was monitored for cells grown in LB, LB supplemented with 200 μM 2,2′-dipyridyl, or α-MEM. Overnight cultures were harvested by centrifugation, washed three times, resuspended in LB or MEM, and adjusted to an A600 of 1.0. The washed cells were then inoculated into fresh medium (1:100). Cell growth was monitored by measurement of the A600 at intervals, and growth curves were drawn. To measure sensitivity to streptonigrin, three experimental groups were designed. One was supplemented with 0.4% (vol/vol) dimethyl formamide (control), and the other two groups were supplemented with 1 or 5 μg/ml streptonigrin (dissolved in dimethyl formamide).
Iron content measurement.
Bacteria were grown in α-MEM supplemented with 10 μM FeCl3. When bacterial growth reached an A600 of 0.4 to 0.6, 20 ml of the culture was harvested by centrifugation at 5,000 rpm at 4°C for 15 min. The pellet was washed twice with 20 ml of ice-cold 0.1 M Tris buffer and once with metal-free double-distilled water. The pellet was resuspended in 1 ml of 37% HCl (TraceMetal grade; Fisher Scientific), lightly vortexed, and heated at 78°C for 5 min until cells were completely lysed. Nine milliliters of metal-free double-distilled water was added to the lysed cells and mixed by vortex. Samples were centrifuged at 13,000 rpm for 10 min, and the supernatant was analyzed for iron content by inductively coupled plasma atomic emission spectrometry at the Research Analytical Laboratory, University of Minnesota. One ppm yttrium was used as an internal standard. The data were normalized by bacterial dry weights, which were determined after drying cell pellets at 87°C for 18 h.
Construction of fit-lacZ transcriptional fusions.
The plasmid pMP220 (33), which contains a promoterless lacZ gene, was used to create fit-lacZ transcriptional fusion vectors. The putative fit promoter region, presumed to be located between fitA and fitB, was amplified by PCR with primers listed in Table S1 in the supplemental material. These primers contain an EcoRI site. After digestion of the PCR product with EcoRI, the fragment was cloned into plasmid pMP220. Blue colonies on LB plates containing tetracycline and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside were picked. Positive clones were identified using PCR and confirmed by DNA sequence analysis.
Assay of β-galactosidase.
Bacteria were grown at 37°C in LB broth or MM9 minimal medium. β-Galactosidase activities were measured using ortho-nitrophenyl-β-d-galactopyranoside (Sigma) as the substrate, and Miller units were calculated as described elsewhere (21). Each experiment was repeated in triplicate, and the results were analyzed using Student's t test.
Nucleotide sequence accession numbers.
GenBank accession numbers are DQ885342 to DQ885347 for fitA, fitB, fitC, fitD, fitE, and fitR, respectively.
RESULTS
Sequencing and annotation of the E. coli fit system.
In a previous study, a putative iron transport gene, ivi932, was identified in E. coli i484 by using an in vivo expression technology protocol (18). Preliminary sequence analysis showed that ivi932 was present in E. coli O157:H7 strain EDL933 and in the uropathogenic strain CFT073, but not in K-12 strain MG1655 (3, 13, 28, 39). In addition to ivi932, five additional genes are located in the vicinity of ivi932 in the genomes of E. coli EDL933 and CFT073. Based on PCR amplification, these six genes were found to be present as a gene cluster in the E. coli i484 genome. The entire gene cluster in strain i484 was amplified and sequenced. Sequence comparison demonstrated that the gene cluster in E. coli i484 was almost identical (96%) to those of the other two E. coli strains. These six genes, designated fitA, -B, -C, -D, -E, and -R, form a novel putative iron transport system which was named fit (ferric or ferrous iron transport) (Fig. 1).
FIG. 1.
Organization and orientation of the E. coli fit system. The block arrow indicates the gene transcription direction. Intergenic regions are shown as boxes. P, promoter. Arrows indicate the locations of a putative fur box and the −10 and −35 boxes of fitA and fitB, respectively.
The six open reading frames were annotated using various software packages described in Materials and Methods. fitA was deduced to encode a 78-kDa outer membrane receptor protein with homology (44%) to the outer membrane receptor of the Vibrio cholerae ferrichrome transporter FhuA. FitA has a TonB-dependent ligand-gated channel domain, which is found in the outer membrane receptors of many iron transport systems, such as FepA, FecA, FhuE, CirA, and Fiu (4, 22). fitB was deduced to encode a 30-kDa ATPase protein with homology (63%) to the ATPase of the E. coli enterobactin transporter FepC. The characteristic ATP binding domain (Walker A and B motifs) (38) was predicted in the protein. fitC and fitD were predicted to encode two permease proteins of 34 and 37 kDa, respectively. FitC had homology (55%) to Escherichia coli FepD, and FitD had homology (59%) to Yersinia enterocolitica HemU. Based on SOSUI analysis, both FitC and FitD were predicted to be strongly hydrophobic, and each protein contains nine transmembrane motifs. fitE encodes a 35-kDa periplasmic binding protein with homology (46%) to the Bacillus subtilis FhuD protein. These five components, FitA, -B, -C, -D, and -E, form a typical bacterial iron transport system. Besides these five genes, there is an extra open reading frame in the E. coli fit system, fitR, which encodes a 32-kDa hypothetical protein. The predicted FitR protein contains an RpiR domain that has been associated with other regulators and thus might be a regulatory protein (31). Between the genes fitA and fitB is a 373-bp A/T-rich (70%) region. Based on DNA sequence analysis, it is predicted to be the promoter of the fit system, containing two −35 and −10 signal sequences (Fig. 1). The −10 and −35 sequences of fitA overlap the −35 and −10 sequences of fitB, respectively, but they are oriented in opposite directions. In addition, a putative fur box has been predicted in this region.
Distribution of the fit locus in E. coli isolates.
PCR amplification was performed to detect the presence of each fit gene among 18 E. coli isolates. As shown in Table 2, when any of the fit genes was found, all the other fit genes were also detected. Of the strains investigated, fit was found only in E. coli isolates of human origin and not in pig isolates (including fecal commensal strains and enterotoxigenic strains).
TABLE 2.
Distribution of the E. coli fit system among E. coli clinical isolates
| E. coli strain source and type | fit system |
|---|---|
| Pig commensal strains | |
| 123 | − |
| 124 | − |
| 252 | − |
| EcoR33 | − |
| Pig enterotoxigenic strains | |
| 263 | − |
| 431 | − |
| 987 | − |
| 1413 | − |
| Human extraintestinal pathogenic strains | |
| 16 | + |
| 17 | − |
| 18 | + |
| EcoR40 | − |
| EcoR60 | + |
| EcoR62 | + |
| Human commensal strains | |
| EcoR8 | − |
| EcoR26 | − |
| EcoR42 | + |
| EcoR45 | − |
Intracellular iron content measurement.
To determine if the E. coli fit system is involved in iron transport, the fit system was introduced into E. coli K-12 strain AA93 and we then asked if fit contributed to intracellular iron accumulation in this strain. The intracellular iron content was measured indirectly using streptonigrin as a probe. Streptonigrin interacts with intracellular iron to form reactive oxygen species that lead to DNA damage and eventual cell death (41). Thus, sensitivity to streptonigrin can be used as a measure of the intracellular iron content in strains AA93/pCR and AA93/pfit1. It was expected that if fit mediated iron accumulation, AA93/pfit1 would be more sensitive to streptonigrin than AA93/pCR. As shown in Fig. 2A, there was no difference between these two strains when cells were grown in LB containing no streptonigrin. However, in LB supplemented with 1 μg/ml streptonigrin (dissolved in dimethyl formamide), AA93/pCR grew very well while the growth of AA93/pfit1 was inhibited, which indicated that AA93/pfit1 was more sensitive to streptonigrin. Similar phenomena were observed when both strains were grown in α-MEM (Fig. 2B) or in iron-restricted medium (Fig. 2C). The E. coli feo system is a major ferrous iron transporter (17). To decrease background iron uptake in AA93, we created a feoB knockout (oy097) and then introduced the fit system into this strain. Strain oy097/pCR grew well in LB medium with 5 μg/ml streptonigrin, while the growth of oy097/pfit1 was completely inhibited (Fig. 2D). These results showed that oy097/pfit1 was more sensitive to streptonigrin than oy097/pCR.
FIG. 2.
Effect of streptonigrin on growth of E. coli AA93 derivatives. Growth of strain AA93 with plasmids in LB (A), α-MEM (B), or LB with 200 μM dipyridyl (C) and growth of strain AA93/feoB with plasmids in LB (D) were measured. Open symbols, strains carrying pfit1; filled symbols, strains carrying pCR; squares, growth in medium without streptonigrin; triangles, growth in medium with 1 μg/ml streptonigrin; diamonds, growth in medium with 5 μg/ml. All experiments were performed at least three times to assess reproducibility. The figure presents the results of one typical experiment.
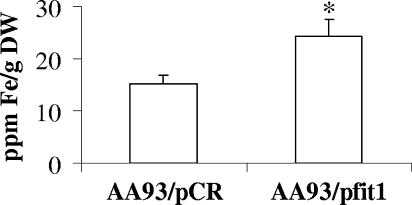
To confirm the above results, the intracellular iron content was measured directly from lysed cells using inductively coupled plasma atomic emission spectrometry. As shown in Fig. 3, there is a significantly higher iron content in E. coli AA93/pfit1 (25 ppm iron/g [dry weight]) than in AA93/pCR (15 ppm iron/g [dry weight]).
FIG. 3.
Measurement of the iron content of the E. coli AA93 derivatives by inductively coupled plasma atomic emission spectrometry. Cells were grown in α-MEM supplemented with 10 μM FeCl3. The data were calculated from three independent replicates. Error bars indicate standard deviations. The asterisk indicates statistical significance using Student's t test (P < 0.05).
Regulation of expression of the E. coli fit system.
To study the regulation of fit expression, a lacZ reporter assay was developed by insertion of the predicted fit promoter into the lacZ reporter plasmid pMP220 in two directions. Transcriptional fusion plasmids pMP-fitA and pMP-fitB were used to measure the expression of fitA and fitB, respectively. We first asked whether β-galactosidase was produced from strains containing either of the fusion plasmids. As shown in Table 3, β-galactosidase activity was not detected in cells containing pMP220 while pMP-fitA and pMP-fitB expressed β-galactosidase, although fitB was expressed at a much higher level (about 36-fold) than fitA. When cells were grown in MM9 minimal medium, expression of fitA and fitB was induced 2.8- and 4.9-fold, respectively, compared with gene expression levels in cells grown in LB medium. These results indicated that there is a bidirectional promoter in the intergenic region between fitA and fitB.
TABLE 3.
Expression of β-galactosidase from E. coli oy016 derivatives grown in LB or MM9 medium
| E. coli derivative | β-Galactosidase activity (Miller units, mean ± SD)
|
|
|---|---|---|
| LB | MM9-glucose | |
| oy016 | 0.0 | 0.0 ± 0.3 |
| pMP220 | 2.4 ± 0.3 | 0.75 ± 0.1 |
| pMP-fitA | 4.2 ± 0.3 | 11.7 ± 1.2 |
| pMP-fitB | 153 ± 12 | 759 ± 82 |
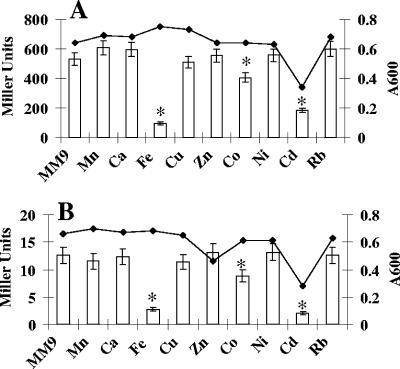
To determine if fit expression was affected by bacterial growth phase or iron depletion, β-galactosidase activity was measured for E. coli oy016 carrying pMP-fitA or pMP-fitB. Cells were first grown in LB for 3 h. The culture was split in two groups. DIP (200 μM) was added to one group. The cells were further incubated, and samples were collected at intervals. As shown in Fig. 4, there was no significant difference among the β-galactosidase levels at different cell growth phases. When iron was depleted from the medium by addition of DIP, expression of fitA and fitB was increased about four- and fivefold, respectively. This increase was observed at all time points after the first 3-hour period. To determine if metal ions other than iron affected expression of fit, β-galactosidase activities were measured from bacteria grown in MM9-glucose medium supplemented with different metal ions. Our results showed that pMP-fitB expressed 531 units of β-galactosidase when cells were grown in this minimal medium without adding any metal ion (Fig. 5A). Addition of Fe2+, Co2+, or Cd2+ resulted in significant reduction of fitB expression. Similar trends were observed for pMP-fitA (Fig. 5B). These results demonstrated that both fitA and fitB are significantly repressed by Fe2+, Co2+, and Cd2+.
FIG. 4.
Expression of fit was induced by iron depletion. E. coli oy016 cells carrying pMP-fitB (A) or pMP-fitA (B) were first grown in LB medium for 3 h. Then the culture was split into two groups. Into one group, DIP was added at a final concentration of 200 μM, and into the other group no DIP was added. Cells were then allowed to grow, samples were taken at various time points, and β-galactosidase activities were measured as described elsewhere (21). Open bars, cells grown in LB; filled bars, cells grown in LB with DIP. Values are the means from three independent experiments. Error bars indicate standard deviations. Asterisks indicate statistical significance using Student's t test (P < 0.05).
FIG. 5.
Expression of fit was repressed by iron. E. coli oy016 cells carrying pMP-fitB (A) and pMP-fitA (B) were grown in MM9 medium with various metal ions. A 100 μM concentration each of Mn2+, Ca2+, Fe2+, Cu2+, Ni2+, and Rb+, 50 μM each of Zn2+ and Co2+, and 10 μM Cd2+ were used in the experiment. Cells were harvested at mid-log phase, and β-galactosidase activities were measured as described previously (21). Error bars indicate standard deviations (n = 3). Asterisks indicate statistical significance using Student's t test (P < 0.05).
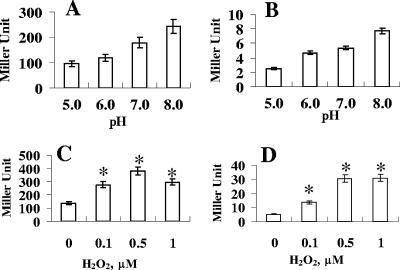
Previous studies reported that some E. coli iron transporters are regulated by oxidative stress and pH (19, 42). To measure the pH effect on fit expression, LB medium was buffered by the addition of 100 mM homopiperazine-N,N′-bis(2-ethanesulfonic acid), and the pH was adjusted to 5.0, 6.0, 7.0, or 8.0. When cells were grown in LB of pH 5.0, 6.0, 7.0, or 8.0, expression levels of fitB and fitA were increased with increasing pH (Fig. 6A and B). To measure the effect of H2O2 on fit expression, mid-log-phase cells grown in LB medium were split into four groups. One group was used as the control group. The other three groups were exposed to 0.1, 0.5, or 1.0 μM H2O2 for 10 min. Our results showed that expression of fitB was increased about 2.0-, 2.8-, and 2.2-fold when cells were exposed to 0.1, 0.5, or 1.0 μM H2O2, respectively (Fig. 6C). This result indicated that fitB was induced significantly by H2O2. Similar results were obtained for pMP-fitA (Fig. 6D).
FIG. 6.
Effects of pH and H2O2 on fit expression, based on β-galactosidase activities of oy016 cells carrying pMP-fitB (A and C) and pMP-fitA (B and D). (A and B) Cells were grown in LB at various pHs. (C and D) Cells were grown in LB, and when bacterial growth reached an A600 of 0.6, various amounts of H2O2 were added to the medium. After 10 min, cells were collected and β-galactosidase activities were measured as described elsewhere (21). Values are the means from three independent experiments. Error bars indicate standard deviations (n = 3). Asterisks indicate statistical significance using Student's t test (P < 0.05).
Bacterial growth in iron-restricted medium.
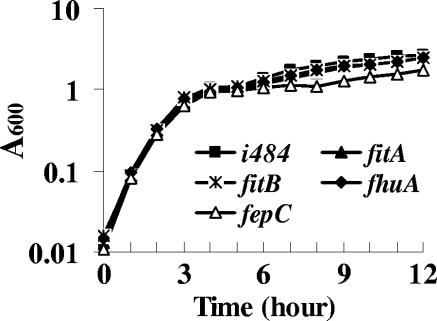
To determine if fit is involved in iron transport in E. coli, the growth of fit mutants in LB containing 200 μM DIP was compared with that of the parental strain, E. coli i484. As shown in Fig. 7, mutations in fitA or fitB did not affect bacterial growth. In this report, the growth of a fepC (enterobactin transport) mutant and fhuA (ferrichrome transport) mutant in the iron-restricted medium also was studied. Results showed that when fepC was mutated, bacterial growth in this medium was significantly inhibited compared to the parental strain, while a mutation in fhuA did not affect bacterial growth.
FIG. 7.
Bacterial growth in iron-restricted medium. LB medium was made iron depleted by adding DIP to a final concentration of 200 μM. Bacteria were grown at 37°C with shaking, and the A600 was measured during growth. All tests were repeated three times, and the results of one typical experiment are presented here.
Growth promotion test.
It was hypothesized that if the E. coli fit system were responsible for transporting iron compounds, the ability of bacteria to transport this substrate would be attenuated or lost by creating mutations in fit. To address this question, a growth promotion test was performed. As shown in Table 4, the parental strain E. coli i484 was able to utilize all the compounds, with the exception of transferrin and lactoferrin, as external iron sources. When the fep system was mutated (strains xy008 and xy009), the mutants lost their abilities to utilize enterobactin (the growth supernatant of E. coli strain AB1515.199 under iron limitation). It also was found that the fhuA mutant (xy007) lost its ability to utilize ferrichrome. When fitA or fitB was mutated, bacteria were still able to utilize enterobactin, ferrichrome, heme, heme-BSA, RA, desferal, FeSO4, or FeCl3 as the only iron source to support their growth. This test was also performed in E. coli AA93, and similar results were found (Table 4). It was found that AA93/pfit1 did not utilize heme, heme-BSA, transferrin, lactoferrin, desferal, or iron citrate. These data suggested that the E. coli fit system does not transport enterobactin, ferrichrome, transferrin, lactoferrin, heme, or iron citrate.
TABLE 4.
Growth promotion test of E. coli strainsa
| Substrate | Growth of strain
|
|||||||
|---|---|---|---|---|---|---|---|---|
| i484 | fitA-(xy006) | fitB-(xy005) | fhuA-(xy007) | fepC-(xy008) | fepA-(xy009) | AA93/ pfit1 | AA93/ pCR | |
| Si484 | + | + | + | + | + | + | + | + |
| SAB1515 | + | + | + | + | − | − | + | + |
| Heme | + | + | + | ND | ND | ND | − | − |
| Heme-BSA | + | + | + | ND | ND | ND | − | − |
| DHBA | + | + | + | ND | ND | ND | − | − |
| Lactoferrin | − | − | − | ND | ND | ND | − | − |
| Transferrin | − | − | − | ND | ND | ND | − | − |
| Desferal | + | + | + | ND | ND | ND | − | − |
| RA | + | + | + | ND | ND | ND | + | + |
| Ferrichrome | + | + | + | − | ND | ND | + | + |
| Iron citrate | + | + | + | ND | ND | ND | − | − |
| FeSO4 | + | + | + | + | + | + | + | + |
| FeCl3 | + | + | + | + | + | + | + | + |
| dH2O | − | − | − | − | − | − | − | − |
Concentrations of iron compounds used are reported in Materials and Methods. Si484 and SAB1515 represent supernatants of E. coli i484 and AB1515.199 cells, respectively, grown in iron-restricted media. Heme-BSA was made by mixing 100 μM heme and 100 μM BSA at a 1:1 molar ratio. Iron citrate was made by mixing fresh FeSO4 with sodium citrate at a molar ratio of 1:1,000. +, positive; −, negative; ND, not determined; DHBA, dihydroxybenzoic acid; dH2O, distilled water.
DISCUSSION
Iron is an essential element for almost all bacteria. In the host, iron is nearly unavailable for bacteria due to the binding of transferrin and lactoferrin, etc. To support their growth, pathogenic bacteria have developed elaborate mechanisms to overcome this iron deficiency. One of these mechanisms involves the production of siderophores, which are low-molecular-weight chelators that have high affinities for iron (10, 22). Once siderophores bind iron, the siderophore-iron complexes are transported into cells by specific transport systems.
In this paper, we described the discovery of a new E. coli iron transport system. The fit operon was discovered by using an in vivo expression technique coupled with directed DNA sequencing in an E. coli strain that causes human septicemia. Based on DNA sequence analysis, the likely functions of the genes in this cluster include a TonB-dependent outer membrane receptor, a periplasmic binding protein, two permease proteins, and an ATPase. These components form a typical iron transport system in gram-negative bacteria (7). However, unlike most iron transport systems, the fit operon contains a sixth gene that encodes a hypothetical protein. Based on CDD analysis, the predicted protein FitR has an RpiR domain, which suggested that it might be a regulator.
To determine if mutations in the fit operon affected bacterial growth under iron limitation, a collection of fit mutants were created. Under in vitro iron-limiting conditions, no significant differences in growth characteristics between the parental strain and the fit mutants were found. It is likely that this is because E. coli strain i484 encodes other iron transport systems, including fep, fhu, fec, iro, fhuE, feo, heme, aerobactin, and yersiniabactin transport systems (their presence in E. coli i484 was confirmed by PCR amplification [data not shown]). These iron transporters probably abrogate the effects of fit mutation on iron uptake.
E. coli i484 produces four kinds of siderophores (enterobactin, aerobactin, salmochelin, and yersiniabactin) under iron starvation (data not shown), and it possesses multiple iron transport systems. An E. coli K-12 strain, AA93, does not produce enterobactin and salmochelin due to an aroB mutation (26). Genes for synthesis of aerobactin and yersiniabactin were not detected in this strain (data not shown). AA93 also is a fec deletion mutant, and the fit system is not present in its genome. Thus, fit-mediated iron uptake in E. coli i484 is likely to be masked due to the redundancy of iron transport systems. Therefore, to study the contribution of the fit system to iron uptake, the whole fit system was cloned and introduced into E. coli AA93. The relative intracellular iron content was determined indirectly by measuring sensitivity to streptonigrin. Streptonigrin is an antibiotic whose toxicity is dependent on intracellular iron. Increased sensitivity to this drug is a sign of increased availability of intracellular free iron. Our results showed that introduction of the E. coli fit system into strain AA93 resulted in the strain being much more sensitive to streptonigrin, which suggested that there is more intracellular iron in AA93/pfit1 than in AA93/pCR. The plasmid pCR is a high-copy-number cloning vector. Thus, we assume that pfit1 was present in high copy numbers in E. coli AA93. The presence of a high-copy-number plasmid in bacteria may cause metabolic burden effects, which induce a significant shift in the normal metabolism and a reduced bacterial growth. Thus, the accumulation of iron observed in E. coli AA93/pfit1 possibly is an indirect effect which is mediated through the metabolic burden effects due to the presence of the high-copy-number plasmid. To address this question, a fitR clone, pCR64, and a lacZ clone, pCR63, were used as controls in this test. It was found that E. coli AA93/pCR64 and AA93/pCR63 had growth curves similar to that of AA93/pfit1, while AA93/pfit1 was more sensitive to streptonigrin than AA93/pCR64 and AA93/pCR63 (data not shown). This suggested that the iron accumulation effect in AA93/pfit1 was not due to an increased burden from the presence of a high-copy-number plasmid. To confirm the results from the streptonigrin sensitivity test, inductively coupled plasma atomic emission spectrometry was performed to directly measure the iron content within the cells. Our data showed that E. coli AA93/pfit1 had significantly higher iron content than AA93/pCR. These results suggested that the E. coli fit system transports iron when it is expressed from the plasmid pfit1.
To identify the substrate of the E. coli fit system, a growth promotion test was performed using various iron compounds. BLAST and CDD analyses suggested that fit was most closely homologous to ferrichrome (fhu) or enterobactin (fep) iron transport systems. Our results showed that when fepC (encoding the ATPase of the fep system), or fhuA (encoding the receptor of the fhu system) was mutated, these mutants lost their abilities to utilize enterobactin or ferrichrome, respectively, as iron sources. These results demonstrated that, contrary to our expectation, enterobactin and ferrichrome are not the substrates of the E. coli fit system. We also studied if fit transported ferric citrate, since FitC and FitD had FecCD domains, which are conserved in the permease proteins of the ferric citrate transport (fec) system (26). We found that strain AA93/pfit1 did not use ferric citrate as the only iron source, which suggested that fit did not transport ferric citrate.
Gene regulation information can provide clues to the function of the fit system. In this study, a lacZ reporter assay was employed to study the expression of fit. Sequence data suggested that the intergenic region between fitA and fitB contained a putative bidirectional promoter for the E. coli fit system. The experimental results indicated that this was true, since both pMP-fitA and pMP-fitB expressed β-galactosidase. Our data showed that the β-galactosidase activity from the fitA promoter was much lower than that of the fitB promoter, which suggested that fitB was expressed at a higher level than fitA. However, the responses of fitA to iron depletion, pH, or H2O2 were of a magnitude similar to those of fitB. Both genes were regulated at similar levels under these conditions.
Of those well-characterized E. coli iron transporters mentioned above, all are regulated by iron (4, 20). Since fit encodes a putative iron transport system, we hypothesized that fit was regulated by iron. Our data demonstrated that the expression of fit was induced by depletion of iron and repressed by addition of exogenous iron. This result serves as strong evidence that fit is involved in E. coli iron metabolism.
We further examined if other environmental condition regulators were involved in the regulation of fit expression. For example, E. coli responds to pH changes encountered in the digestive tract by regulating different sets of genes. At low pH, for example, acid consumption, proton export, and oxidative stress responses are induced. At high pH, proton import is induced while the energy-expensive systems of flagellar biosynthesis and chemotaxis are repressed (19). Some iron transporters are found to be regulated by pH. For example, fecAB and fhuD are induced at high pH (19). Similarly, we found that the E. coli fit system also was induced at high pH (8.0). In this study, we also found that fit was induced by hydrogen peroxide. Since sequence analysis indicated that the fit promoter region lacks an OxyR binding site, currently the mechanism of this induction is unknown.
We have described here a novel E. coli iron transport system, fit, which has homology to many bacterial iron transporters. Our data showed that the fit system contributes to iron accumulation in E. coli AA93. We also found that the expression of the E. coli fit system was repressed by addition of exogenous iron and derepressed by depletion of iron. Based on these experimental data, combined with the sequence information, we believe that the E. coli fit system is involved in iron acquisition.
Supplementary Material
Acknowledgments
We thank James R. Johnson, University of Minnesota, for wild-type human isolates of E. coli. Sandra K. Armstrong, University of Minnesota, provided the plasmid pMP220. Volkmar Braun, University of Tubingen, provided E. coli strain AA93. Charles Earhart, University of Texas, provided E. coli strain AB1515.199.
This work was supported in part by a grant from the USDA-NRI, 2002-35201-12542.
Editor: F. C. Fang
Footnotes
Published ahead of print on 18 September 2006.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Andrade, M. A., N. P. Brown, C. Leroy, S. Hoersch, A. de Daruvar, C. Reich, A. Franchini, J. Tamames, A. Valencia, C. Ouzounis, and C. Sander. 1999. Automated genome sequence analysis and annotation. Bioinformatics 15:391-412. [DOI] [PubMed] [Google Scholar]
- 2.Bateman, A., E. Birney, R. Durbin, S. R. Eddy, K. L. Howe, and E. L. Sonnhammer. 2000. The Pfam protein families database. Nucleic Acids Res. 28:263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 4.Braun, V., and M. Braun. 2002. Iron transport and signaling in Escherichia coli. FEBS Lett. 529:78-85. [DOI] [PubMed] [Google Scholar]
- 5.Calderwood, S. B., and J. J. Mekalanos. 1987. Iron regulation of Shiga-like toxin expression in Escherichia coli is mediated by the fur locus. J. Bacteriol. 169:4759-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chenault, S. S., and C. F. Earhart. 1991. Organization of genes encoding membrane proteins of the Escherichia coli ferrienterobactin permease. Mol. Microbiol. 5:1405-1413. [DOI] [PubMed] [Google Scholar]
- 7.Clarke, T. E., L. W. Tari, and H. J. Vogel. 2001. Structural biology of bacterial iron uptake systems. Curr. Top. Med. Chem. 1:7-30. [DOI] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faraldo-Gomez, J. D., and M. S. Sansom. 2003. Acquisition of siderophores in gram-negative bacteria. Nat. Rev. Mol. Cell Biol. 4:105-116. [DOI] [PubMed] [Google Scholar]
- 11.Goryshin, I. Y., and W. S. Reznikoff. 1998. Tn5 in vitro transposition. J. Biol. Chem. 273:7367-7374. [DOI] [PubMed] [Google Scholar]
- 12.Griffiths, E. 1999. Iron in biological systems, p. 1-26. In J. Bullen and E. Griffiths (ed.), Iron and infection. Wiley, Chichester, United Kingdom.
- 13.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 14.Hirokawa, T., S. Boon-Chieng, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378-379. [DOI] [PubMed] [Google Scholar]
- 15.Huang, J. Y., and D. L. Brutlag. 2001. The EMOTIF database. Nucleic Acids Res. 29:202-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones, D. T. 1999. GenTHREADER: an efficient and reliable protein fold recognition method for genomic sequences. J. Mol. Biol. 287:797-815. [DOI] [PubMed] [Google Scholar]
- 17.Kammler, M., C. Schon, and K. Hantke. 1993. Characterization of the ferrous iron uptake system of Escherichia coli. J. Bacteriol. 175:6212-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan, M. A., and R. E. Isaacson. 2002. Identification of Escherichia coli genes that are specifically expressed in a murine model of septicemic infection. Infect. Immun. 70:3404-3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maurer, L. M., E. Yohannes, S. S. Bondurant, M. Radmacher, and J. L. Slonczewski. 2005. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J. Bacteriol. 187:304-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McHugh, J. P., F. Rodriguez-Quinones, H. Abdul-Tehrani, D. A. Svistunenko, R. K. Poole, C. E. Cooper, and S. C. Andrews. 2003. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J. Biol. Chem. 278:29478-29486. [DOI] [PubMed] [Google Scholar]
- 21.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Moeck, G. S., and J. W. Coulton. 1998. TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport. Mol. Microbiol. 28:675-681. [DOI] [PubMed] [Google Scholar]
- 23.Mori, H., K. Isono, T. Horiuchi, and T. Miki. 2000. Functional genomics of Escherichia coli in Japan. Res. Microbiol. 151:121-128. [DOI] [PubMed] [Google Scholar]
- 24.Negre, V. L., S. Bonacorsi, S. Schubert, P. Bidet, X. Nassif, and E. Bingen. 2004. The siderophore receptor IroN, but not the high-pathogenicity island or the hemin receptor ChuA, contributes to the bacteremic step of Escherichia coli neonatal meningitis. Infect. Immun. 72:1216-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neilands, J. B. 1992. Mechanism and regulation of synthesis of aerobactin in Escherichia coli K-12 (pColV-K30). Can. J. Microbiol. 38:728-733. [DOI] [PubMed] [Google Scholar]
- 26.Ochs, M., S. Veitinger, I. Kim, D. Welz, A. Angerer, and V. Braun. 1995. Regulation of citrate dependent iron transport system in Escherichia coli: FecR is required for transcriptional activation by FecI. Mol. Microbiol. 15:119-132. [DOI] [PubMed] [Google Scholar]
- 27.Ozenberger, B. A., M. S. Nahlik, and M. A. McIntosh. 1987. Genetic organization of multiple fep genes encoding ferric enterobactin transport functions in Escherichia coli. J. Bacteriol. 169:3638-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perna, N. T., G. Plunkett, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sorensen, K. I., and B. Hove-Jensen. 1996. Ribose catabolism of Escherichia coli: characterization of the rpiB gene encoding ribose phosphate isomerase B and of the rpiR gene, which is involved in regulation of rpiB expression. J. Bacteriol. 178:1003-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sorsa, L. J., S. Dufke, J. Heesemann, and S. Schubert. 2003. Characterization of an iroBCDEN gene cluster on a transmissible plasmid of uropathogenic Escherichia coli: evidence for horizontal transfer of a chromosomal virulence factor. Infect. Immun. 71:3285-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spaniak, H. P., R. J. H. Okker, C. A. Wijffelman, E. Pees, and B. J. J. Lugtenberg. 1987. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol. Biol. 9:27-39. [DOI] [PubMed] [Google Scholar]
- 34.Tatusov, R. L., D. A. Natale, I. V. Garkavtsev, T. A. Tatusova, U. T. Shankavaram, B. S. Rao, B. Kiryutin, M. Y. Galperin, N. D. Fedorova, and E. V. Koonin. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29:22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas, G. H. 1999. Completing the E. coli proteome: a database of gene products characterised since the completion of the genome sequence. Bioinformatics 15:860-861. [DOI] [PubMed] [Google Scholar]
- 36.Torres, A. G., P. Redford, R. A. Welch, and S. M. Payne. 2001. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect. Immun. 69:6179-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Torres, A. G., and S. M. Payne. 1997. Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 23:825-833. [DOI] [PubMed] [Google Scholar]
- 38.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wheeler, D. L., D. M. Church, A. E. Lash, D. D. Leipe, T. L. Madden, J. U. Pontius, G. D. Schuler, L. M. Schriml, T. A. Tatusova, L. Wagner, and B. A. Rapp. 2001. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 29:11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yeowell, H. N., and J. R. White. 1982. Iron requirement in the bactericidal mechanism of streptonigrin. Antimicrob. Agents Chemother. 22:961-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J Bacteriol. 183:4562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.