Abstract
Pseudomonas aeruginosa is an opportunistic bacterial pathogen that primarily infects immunocompromised individuals and patients with cystic fibrosis. Invasive strains of P. aeruginosa are known to induce apoptosis at a high frequency in HeLa cells and in many other cell lines, a process that is dependent on the ADP-ribosylation (ADPRT) activity of a type III secreted protein ExoS. In our previous report, it was proposed that P. aeruginosa secreting ExoS, upon infection, shuts down host cell survival signal pathways by inhibiting ERK1/2 and p38 activation, and it activates proapoptotic pathways through activation of JNK1/2, leading ultimately to cytochrome c release and activation of caspases. In this study, we demonstrate that the expression of ExoS in HeLa cells by eukaryotic expression vector effectively caused apoptosis in an ADPRT activity-dependent manner, indicating that ExoS alone is sufficient to trigger apoptotic death of host cells independent of any other bacterial factors. By expressing an EGFP-ExoS fusion protein, we were able to directly correlate the death of HeLa cells with the presence of intracellular ExoS and further proved the dependence of this process on both JNK activation and mitochondrial proapoptotic event. The cellular pathway responsible for the ExoS-induced cytotoxicity appears to be well conserved, since the expression of the ADPRT-competent ExoS also induced rapid cell death in the Drosophila melanogaster S2 cell lines. The presented study not only highlights the ability of ExoS ADPRT to modulate host cell signaling, eventually leading to apoptosis, but also establishes ExoS as a valuable tool, in principle, for the elucidation of apoptosis mechanisms.
Pseudomonas aeruginosa, an opportunistic bacterial pathogen, causes devastating human infections in patients with cystic fibrosis, severe burns, or immunosuppression (31, 52, 74). To successfully colonize and maintain its infectious cycle, P. aeruginosa orchestrates the production of a large arsenal of virulence factors in a highly regulated manner (35, 84). Some of the virulence factors are produced and injected directly into the host cell by using the cell contact-mediated type III secretion system (TTSS), which has attracted significant attention in recent years (18, 28, 44). To date, four TTSS effector molecules have been described in P. aeruginosa. Exoenzyme U (ExoU) is a necrotizing toxin with phospholipase activity (39, 81), and ExoY is an adenylate cyclase (92). The other two known effectors are ExoS and ExoT, highly homologous to each other, having a carboxy-terminal ADP-ribosyltransferase (ADPRT) domain and an amino-terminal GTPase-activating (GAP) domain (32, 53, 54, 76, 85).
The GAP activity of ExoS and ExoT targets small Rho-like GTPases, such as Rho, Rac, and Cdc42 (1, 53, 54), and has been linked to cytoskeletal rearrangements and cell rounding in vitro (32, 53). Although ExoS and ExoT share 75% amino acid identity, their ADPRT domains appear to have distinctive groups of target proteins in host cells (1, 86). The ADPRT activity of ExoS has been shown to modify Ras and several Ras-like host proteins in vivo, including RalA, Rabs, and Rac1 (5, 15, 25), which is linked to cytotoxicity toward eukaryotic cells (70); it also has non-G-protein substrates, the Ezrin/Radixin/Moesin (ERM) family of proteins (62), demonstrating another mechanism by which ExoS can modulate cytoskeleton dynamics. Glutamic acid at position 381 (E381) functions as a catalytic residue for the ExoS ADPRT domain (76), the activation of which requires a 14-3-3 family protein, termed FAS (for factor for activating ExoS), from eukaryotic host (27).
In addition, we reported that the ADPRT activity of the ExoS was required for triggering rapid apoptosis in various host cells upon infection by invasive strains of P. aeruginosa (50). In that study, the apoptotic death in infected cells was determined by several criteria, including (i) visual changes in cell morphology, (ii) the induction of chromatin condensation and nuclear marginalization, (iii) the presence of a high percentage of cells with subG1 DNA content, and (iv) the activation of caspase-3 activity. Subsequently, evidence was provided to suggest that, in infected host cells, P. aeruginosa producing ExoS not only triggers a proapoptotic pathway through JNK-mediated cytochrome c release from mitochondria but also sensitizes the host cell to proapoptotic signals by inhibiting antiapoptotic pathway(s) controlled by ERK1/2 and possibly also by p38 (48).
There are three major regulatory pathways known to regulate caspase-based death programs: the mitochondrial pathway, which involves Bcl-2 family proteins and ced-4/Apaf-1 (43, 61), the reaper-family/inhibitor of apoptosis protein (IAP) pathway (64, 99), and the death receptor pathway (68). Components of all three pathways have been documented in both mammals and insects, and perturbation of each of these pathways has been implicated in a variety of human diseases (40).
In the present study, by using multiple approaches to express ExoS with eukaryotic expression vectors, we demonstrate that ExoS alone, precisely the ADPRT of ExoS, is sufficient to trigger apoptosis in cultured host cells representing human and drosophila, independent of any other signals from the bacterial cell. Also importantly, the apoptosis induced by exogenously expressed ExoS was proven dependent on both JNK activation and mitochondrial proapoptotic event. Therefore, by extending our previous findings on P. aeruginosa-induced apoptosis in distinctive approaches, the presented data set up a foundation for us to take the advantage of a novel role ExoS may play in studying apoptosis mechanisms in eukaryotic cells.
MATERIALS AND METHODS
Materials and cell lines.
Eukaryotic cell expression vectors, pcDNA4 and pEGFP-C1, were obtained from Invitrogen (Carlsbad, CA) and BD Bioscience-Clontech (Palo Alto, CA), respectively. Reporter construct pGFP (pUF5) was kindly provided by Alfred Lewin (University of Florida College of Medicine). Transfection reagents Lipofectamine Plus and Lipofectamine 2000 were purchased from Invitrogen. Anti-green fluorescent protein (anti-GFP; 1:1,000) and anti-actin (1:500) antibodies, used at the indicated dilutions, were obtained from Santa Cruz Biotech (Santa Cruz, CA). Anti-ExoS and anti-ExoT antibodies were generated from rabbits by Lampire Biological Laboratories, Inc. (Pipersville, PA). Peroxidase-conjugated goat anti-rabbit secondary antibodies and peroxidase-conjugated goat anti-mouse antibodies were obtained from Santa Cruz Biotech and Amersham Pharmacia Biotech (Piscataway, NJ), respectively. SP600125 (SP), staurosporine (STS), cycloheximide (CHX), caspase-3 inhibitor DEVD-CHO, and pan-caspase inhibitor Boc-d-FMK were obtained from CalBiochem (San Diego, CA).
HeLa cell lines stably transfected with Bcl-xL or Neo vector alone were generously provided by Xiaodong Wang (University of Texas Southwestern Medical Center, Dallas) and grown as suggested previously (69). Other HeLa cell cultures were maintained or treated as previously described (48).
Bacterial strains and plasmids.
Strains and plasmids used in the present study are listed in Table 1. Bacterial cultures were grown in Luria-Bertani broth at 37°C. To construct pcNDA4-exoS, exoS was cloned by PCR, using PAK chromosomal DNA as a template with the forward primer 5′-CAG GAG AAG GTA CCA TCA TGG ATA TTC AAT CGC TTC AG-3′ (underlining indicates the mutated nucleotides) and the reverse primer 5′-CGT TTC GTC GCC TGG ACC TAC CTC GAC AAG AAG CA-3′. The forward primer contained the KpnI restriction site (GGTACC) upstream of the exoS initiation codon and Kozak translation initiation sequence (i.e., G/ANNATGG). The PCR product was cloned into pCR2.1-TOPO vector (Invitrogen, Inc.), the resulting plasmid (pCR 2.1 TOPO-exoS) was then digested with KpnI/EcoRI, and the exoS-containing fragment was further ligated into the same site of pcDNA4 to generate pcNDA4-exoS (pJJ0040), where exoS is under the control of cytomegalovirus (CMV) promoter. The pcNDA4-exoS was sequenced to confirm the correct coding sequence as well as the fusion junction.
TABLE 1.
List of bacterial strains and plasmids
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | F− φ 80dlacZΔ M15 endA1 recA1 hsdR17(rK− mK+) supE44 thi-1 relA1 Δ(lacZYA-argF)U169 gyrA96 deoR | 37 |
| P. aeruginosa | ||
| PAK | Laboratory strain (invasive) | D. Bradleyb |
| PAKexoS::Ω/exoT::Gem | exoS exoT double-mutant derivative of PAK | 50 |
| Plasmids | ||
| pCR2.1-TOPO | PCR cloning vector; Apr Kmr | Invitrogen |
| pcDNA4 | Constitutive mammalian expression vector with CMV promoter; Apr | Invitrogen |
| pJJ0040 | exoS of PAK cloned into pcDNA4; Apr | This study |
| pJJ0042 | exoS of pJJ0040 mutated to exoS(R146K); Apr | This study |
| pJJ0043 | exoS of pJJ0040 mutated to exoS(E381A); Apr | This study |
| pJJ0044 | exoS of pJJ0040 mutated to exoS(R146K/E381A); Apr | This study |
| pUCP18 | Broad-host-range cloning vector, IncP; Apr | 82 |
| pHW0015 | exoS derived from PAK in pUCP18; Apr | 50 |
| pExoSRK18 | exoS(R146K) in pUCP18; Apr | This study |
| pExoSEA18 | exoS(E381A) in pUCP18; Apr | This study |
| pExoSRKEA18 | exoS(R146K/E381A) in pUCP18; Apr | This study |
| pEGFP-C1 | Constitutive mammalian expression vector containing egfp(CMV promoter); Kmr | BD Clontech |
| pJJ0322 | exoS in pEGFP-C1; Kmr | This study |
| pJJ0323 | exoS(E381A) in pEGFP-C1; Kmr | This study |
| pJJ0324 | exoS(R146K/E381A) in pEGFP-C1; Kmr | This study |
| pJJ0325 | exoS(R146K) in pEGFP-C1; Kmr | This study |
| pUF5 | Reporter construct pGFP | A. Lewin |
| pcDNA4 | Constitutive mammalian expression vector with CMV promoter; Apr | Invitrogen |
| pie3 | Drosophila expression vector; Apr | Invitrogen |
| pie_exoS | exoS in pie3; Apr | This study |
| pie_exoSRK | exoS(R146K) in pie3; Apr | This study |
| pie_exoSEA | exoS(E381A) in pie3; Apr | This study |
| pie_ExoSdouble | exoS(R146K/E381A) in pie3; Apr | This study |
| pie_reaper | reaper in pie3; Apr | 49 |
| pie_Diap1 | Diap1 in pie3; Apr | 98 |
Antibiotic resistance markers: Apr, ampicillin resistance; Kmr, kanamycin resistance.
Faculty of Medicine, Memorial University of Newfoundland, Newfoundland, Canada.
Site-directed mutagenesis.
Additional mutations of the exoS in pJJ0040, such as exoS(E381A) (ADPRT null), exoSR146K (GAP null), and exoS(R146K)/exoS(E381A) (ADPRT and GAP null double mutant), were generated by site-specific mutagenesis, resulting in pJJ0043, pJJ0042, and pJJ0044, respectively. The oligonucleotides used to generate the mutations were as follows: for exoS(R146K), 5′-GAG ATG GGG CCC TGA AAT CGC TGA GCA CC-3′, in which the Arg codon CGT was replaced with the Lys codon AAA while creating a new ApaI site; and for exoS(E381A), 5′-GAA TGA AAA AGC AAT ATT GTA TAA CAA AGA G-3′, where the Glu codon GAG was replaced with the Ala codon GCA while creating a new SspI site. The creation of new restriction sites allowed quick identification of mutant plasmids in initial screening. Base changes in the pcDNA4 constructs were verified by DNA sequencing. Subsequently, clones of the exoS(E381A) and exoS(R146K) in pUCP18 were generated from pJJ0043 and pJJ0042, respectively, by replacing the corresponding mutant fragments with that of pHW0015 (exoS clone in pUCP18 or pUCP18exoS). In this procedure, a ClaI-XbaI fragment was used to generate a pUCP18 containing exoS(E381A) (pExoSEA18), whereas XmaI was used forexoS(R146K) (pExoSRK18).
Transfection of HeLa cells.
Cells were plated on six-well plates at 1.5 × 105 cells/well in Dulbecco modified Eagle medium (DMEM) plus 10% fetal calf serum (FCS), 100 U of penicillin G sodium/ml, 100 μg of streptomycin sulfate/ml, and 2 mM l-glutamine. After 24 h, cells were transfected with the indicated constructs or vector control (2 μg/well for transfection or 6 μg/well in total for cotransfection) by using Lipofectamine 2000 reagent (Invitrogen) or Lipofectamine-Plus reagent according to the manufacturer's instructions. To select for stable transfectants, growth medium was replaced at 48 h posttransfection and then weekly for 3 weeks with fresh medium containing 600 μg of G418 (Life Technologies, Inc.)/ml.
Western blot analysis of ExoS and EGFP-ExoS.
At various times postinfection, HeLa cells were collected and suspended in protein loading buffer (62.5 mM Tris-HCl [pH 6.8], 2% [wt/vol] sodium dodecyl sulfate [SDS], 10% glycerol, 50 mM dithiothreitol, 0.1% bromophenol blue). Equal amounts of total protein samples were subjected to SDS-12% polyacrylamide gel electrophoresis under reducing conditions. After electrophoresis, the proteins were transferred to PVDF-Plus membranes (Osmonics, Inc., Minnetonka, MN) at 50 mA for 60 min using a semidry protein transfer system (Bio-Rad, Hercules, CA). Blots were blocked with 5% (wt/vol) dry milk in 1× TBS buffer (50 mM Tris-HCl, 150 mM NaCl [pH 7.4]) containing 0.5% (vol/vol) Tween 20 for 60 min and then probed with the appropriate antibodies for 1 h at room temperature or overnight at 4°C, according to suppliers' recommendations. After being washed with 1× TBS-0.1% Tween 20, the membranes were incubated with peroxidase-conjugated goat anti-mouse or goat anti-rabbit secondary antibodies for 60 min. Specific signals were developed by using the ECL-Plus system (Amersham). To ensure equal loading and even transfer of proteins, protein bands on the membrane were visualized by staining with Ponceau S (0.1% Ponceau S in 3% trichloroacetic acid [wt/vol]) and further reprobed with anti-actin antibody after treatment with stripping buffer (62.5 mM Tris-HCl [pH 6.7], 100 mM mercaptoethanol, and 2% [vol/vol] SDS) for 20 min at 60°C. The bands were quantified by using the densitometry program LabWorks (UVP, Inc., Upland, CA). All measurements of protein levels were normalized against the β-actin signal. Each Western blot experiment was conducted with two separate membranes in parallel to ensure reproducibility.
Infection of HeLa cells by P. aeruginosa.
HeLa cell monolayers were plated from suspension culture 1 day prior to infection in DMEM supplemented with 5% FCS (DMEM-5% FCS). HeLa cell monolayers (∼5 × 105 cells per well; >80% confluence) were washed with phosphate-buffered saline (PBS), mixed with bacteria (107 CFU/ml in DMEM; multiplicity of infection of 20), and incubated for 2 h at 37°C in a 5% CO2 incubator. The cells were washed with PBS to remove the nonadhering bacteria. Fresh medium, DMEM-5% FCS supplemented with 400 μg of gentamicin or amikacin/ml, was added, and the cells were incubated for an additional 3 to 24 h. As positive controls for the apoptosis, HeLa cells were incubated with 10 ng of tumor necrosis factor (TNF)/ml and 20 μg of CHX/ml or with 2 μM STS.
Caspase-3 activation analysis.
Caspase-3 activity was measured by using the caspase-3 cellular activity assay kit plus (BioMol). HeLa cells (3 × 107) infected with P. aeruginosa were washed and harvested by scraping and centrifugation (1,000 × g, 10 min) at various times postinfection. Cells were lysed with caspase-3 assay lysis buffer (BioMol) containing 0.1% Tween 20, and the cell lysates were centrifuged at 10,000 × g for 10 min. Dilutions of the cell lysates in a 96-well plate were incubated in triplicate with caspase-3 substrate DEVD-pNA, or the substrate plus caspase-3 inhibitor DEVD-CHO. Changes in the optical density at 405 nm were monitored for 2 h at 10-min intervals. Protein concentrations were determined by using the protein assay system from Bio-Rad. The specific activity is reported as picomoles of substrate cleaved/minute per microgram of protein.
Alternatively, caspase-3 activation in EGFP-ExoS-transfected HeLa cells was assayed by fluorescence-activated cell sorting (FACS) analysis. HeLa cells seeded in a six-well plate were cultivated for 1 day before transfection with EGFP-ExoS fusion constructs. The HeLa cells were collected 24 h later, and the caspase-3 activity was measured using Red-DEVD-FMK (Oncogene Research Products, San Diego, CA), which is a cell-permeable, nontoxic caspase-3 inhibitor conjugated to a sulforhodamine fluorescent marker. It binds irreversibly and selectively to activated caspase-3 at an early stage of apoptosis, and the red fluorescence label allows for the direct detection of activated caspase-3 in apoptotic cells by flow cytometry (excitation max, ∼540 nm; emission max, ∼570 nm). Flow cytometry analysis was performed using FL-2 channel on FACSort cytometer (BD Biosciences, San Jose, CA). Dot density analysis of each sample was represented as the FL-2 intensity of a single cell (x axis) and the forward light scatter of cells (y axis). As cells undergo apoptosis, their size gradually decreases, resulting in decreased forward light scatter. At the same time, leakage of cytosolic proteins in general results from compromised membrane integrity at the late stage of apoptosis. The index for a gating population with activated caspase-3 in this assay was determined by the analysis pattern for HeLa cells treated with 2 μM STS for 4 h, which served as a positive control. In all cases, 10,000 cells were analyzed by flow cytometry. The figure generation and data analysis were done by using FCS Express 2 program (De Novo Software, Ontario, Canada).
Hoechst staining of condensed chromatin.
Transfected cells were recovered by trypsinization of the cell monolayer. Cells were washed once with PBS and stained with Hoechst 33258 (Molecular Probes, Inc., Eugene, OR) at 1 mg/ml for 10 min in the dark. Chromatin condensation was examined under the fluorescence microscope by using a DAPI (4′,6′-diamidino-2-phenylindole) filter after stained cells were mounted onto slides using mounting medium (Vectashield Hard Set; Vector Laboratories, Inc., Burlingame, CA).
Cell death assay in Drosophila S2 cell line.
The coding regions for exoS, exoS(E381A), exoS(R146K), and exoS(R146K/E381A) were cut out from the respective pcDNA4 constructs and subcloned into the Pie3 vector (Invitrogen), resulting in pie_exoS, pie_exoSEA, pie_exoSRK, and pie_exoSRKEA, respectively. A cell death assay of the Drosophila S2 cell line was performed as described previously (98). Briefly, for each test, 1.0 μg of DNA was distributed into 2 wells in a 24-well plate. This included 0.2 μg of pIE-lacZ and 0.8 μg of the test DNA sample or a combination of samples in the pIE vector. Intact pIE vector was used as the vector control. At 20 h posttransfection, cells were fixed and stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)-IPTG (isopropyl-β-d-thiogalactopyranoside). Blue cells were counted to calculate the percentage of cell survival. All experiments reported here were repeated at least three times.
Statistics.
All data represent at least three independent experiments and are expressed as the means ± the standard deviations (SD) unless otherwise indicated. Differences between groups were compared by using the Student t test, for pair comparisons, to generate P values. Unless otherwise indicated, P values are indicated in all of figures and tables as follows: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
RESULTS
Transiently expressed ExoS induces apoptosis in HeLa cells.
In a previous study, we demonstrated that ExoS is the effector molecule required for the P. aeruginosa-induced rapid and high-frequency apoptosis in HeLa and other cell lines (50). Analysis of mutant forms of the ExoS further suggested that its ADPRT activity is essential for eliciting apoptosis (50). We intended to further dissect signaling pathways involving ExoS-mediated apoptosis. P. aeruginosa possesses a large array of virulence factors that can cause host cell toxicity to certain extend (2, 7, 8, 34, 47, 50, 60, 67, 75, 88, 93, 95); at the same time, it has not been demonstrated conclusively whether ExoS-dependent apoptosis induction coincides with necrosis-like cell death resulting from the diverse effects on cellular components by ExoS or whether apoptosis is secondary to that event. It is therefore important to determine whether the ExoS protein alone is sufficient to induce apoptosis in host cells independent of other bacterial virulence components. To this end, attempts were made to establish a stably transfected HeLa cell line with inducible expression of the exoS using eukaryotic expression vector T-REx expression system (Invitrogen, Inc.). In this system, the transcription of the gene of interest is blocked in the absence of an inducer, such as tetracycline or doxycycline, but is permitted or activated after the inducer is added into culture medium (33, 94). However, the desired stable cell line carrying exoS could not be generated after repetitive rounds of selection. Analysis of a few resulting colonies showed the loss of the exoS gene (data not shown). Since we were able to generate stable cell lines carrying exoS(R146K/E381A) (GAP and ADPRT null) or vector control, the failure to generate an exoS-expressing cell line is presumably due to the basal level expression of the ADPRT capable exoS, which caused effective cell death.
Therefore, a transient-expression system was adopted. HeLa cells were transiently cotransfected with wild-type or mutant forms of the exoS, driven by the CMV promoter, together with a reporter plasmid expressing GFP. GFP is unique in that its fluorophore forms spontaneously without added cofactors. As a result, the emitted fluorescence intensity provides a direct readout of GFP expression (11), which can be measured at the single-cell level without any processing steps.
To ensure the exoS presence in HeLa cells expressing reporter GFP, a fivefold excess of exoS-expressing plasmid (pExoS) or vector control plasmid (pcDNA4) was used over the reporter plasmid pGFP (pUF5) in all cotransfections. At 24 h after the cotransfection of vector pcDNA4 and pGFP, a significant fraction of the population was GFP positive (data not shown), and quantitative analysis indicated cotransfection efficiencies between 15 and 30%. In contrast, cotransfection with the pExoS in the equivalent population resulted in fewer and lower-intensity GFP-positive cells, suggesting that wild-type ExoS protein caused significant cytotoxicity (data not shown). Moreover, the application of cell-permeable caspase-3 inhibitor DEVD-CHO (Fig. 1) or pan-caspase inhibitor Z-VAD-FMK (data not shown) resulted in significantly increased number of GFP-positive cells after the cotransfection, suggesting that the cytotoxicity elicited by the pExoS was dependent on the activation of caspases.
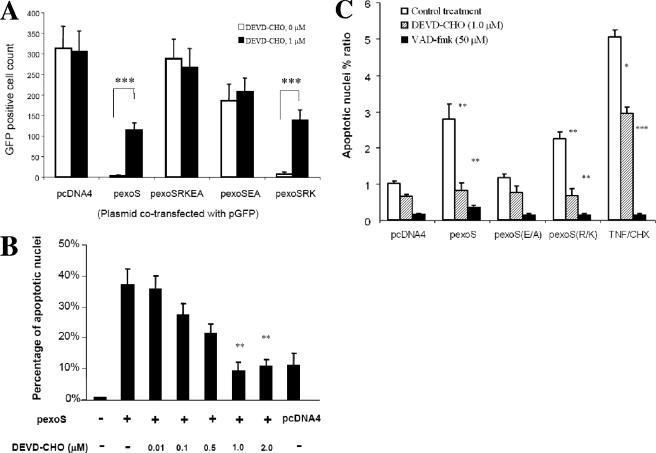
FIG. 1.
Caspase-3-dependent cytotoxicity caused by transfection of the exoS expression plasmid. (A) Disappearance of GFP signal after cotransfection. HeLa cells were cotransfected with pGFP reporter plasmid and the exoS expression plasmids as indicated, with (▪) or without (□) the presence of 1 μM DEVD-CHO, a cell-permeable caspase-3 inhibitor. HeLa cells were observed under a fluorescence microscope using GFP fluorescence at 24 h posttransfection, and GFP-positive cells were quantified from five random views. Plasmids used for cotransfection with pGFP are indicated as follows: pcDNA4, vector control; pexoS, wild-type exoS in pcDNA4(pJJ0040); pexoSEA, exoS(E381A) in pcDNA4(pJJ0043); pexoSRK, exoS(R146K) in pcDNA4(pJJ0042); and pexoSRKEA, exoS(R146K/E381A) in pcDNA4(pJJ0044). As a solvent control, HeLa cells were treated with dimethyl sulfoxide (DEVD-CHO, 0 M). The data are means ± the SD of the counts of GFP-positive cells from three replicates. P values were calculated by comparing DEVD-CHO-treated (1 μM) and untreated (0 M) groups (***, P < 0.001). (B) Nuclear condensation caused by transfection with pcDNA4-exoS. HeLa cells were transfected with pexoS, wild-type exoS in pcDNA4, or pcDNA4 alone. HeLa cells were collected at 36 h posttransfection, stained with Hoechst dye, and subjected to fluorescence microscopy. Five fields were randomly sampled from each experimental population, and all of the cells stained with Hoechst dye in each field were counted up to 500 in total. The total number of apoptotic cells with condensed or fragmented nuclei was determined in the five sampled regions and was expressed as follows: percentage of apoptosis per sample = (number of apoptotic cells/total number of cells) × 100%. During and after transfection, HeLa cells were treated with the indicated amount of DEVD-CHO, a cell-permeable specific caspase-3 inhibitor. The data are means ± the SD of the percentages from three replicates. Significant differences between certain DEVD-CHO treated and untreated groups are indicated (**, P < 0.01). (C) Apoptosis after transfection with the indicated exoS expression constructs. In each experiment, the apoptotic nucleus percent ratio was calculated by comparing the calculated percentage of apoptosis to that of the pcDNA4. During and after transfection, HeLa cells were treated without (control) or with indicated amounts of DEVD-CHO or VAD-fmk, a cell-permeable specific caspase-3 or a pan-caspase inhibitor, respectively. TNF/CHX, treatment with TNF-α and CHX was used as a positive control for apoptosis induction. The data are means ± the SD of the ratios from three replicates. Significant differences between certain DEVD-CHO- or VAD-fmk-treated and untreated control groups are indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
To quantify apoptosis events, a Hoechst staining assay was conducted to assess nuclear condensation and chromatin fragmentation (50). HeLa cells were transiently transfected with vector or pExoS before being subjected to the analysis. About 10% of cells had apoptotic nucleus changes resulting from the expression of vector control (Fig. 1B), which was considered the basal level of apoptosis caused by the transfection procedure. Remarkably, expression of the exoS resulted in more than 35% of the total cells with condensed nuclei, which was significantly reduced by the treatment of HeLa cells with the caspase-3 inhibitor DEVD-CHO in a dose-dependent manner (Fig. 1B), echoing the GFP signal rescue effect.
ADPRT activity of ExoS is sufficient, whereas GAP activity is not required, for apoptosis induction.
In contrast to observations with exoS (pJJ0040), cotransfection of the ADPRT-null mutant exoS(E381A) (pJJ0043) and pGFP showed frequencies of GFP-positive cells similar to those for the expressing vector control (data not shown). Furthermore, the GAP-null mutant exoS(R146K) (pJJ0042) caused cytotoxicity as dramatic as the wild-type exoS strain did (data not shown). Therefore, the specificity of cytotoxicity correlated with the ADPRT but not the GAP activities. Importantly, all of the GFP-positive cells cotransfected with the exoS(E381A) showed rounding morphology (data not shown), a phenomenon presumably caused by the GAP activity of the ExoS through its effect on small Rho GTPases (73). In addition, caspase inhibitors effectively reversed the cytotoxicity caused by the expression of exoS (pJJ0040) and exoS(R146K) (pJJ0042) (Fig. 1B and C), suggesting the apoptotic nature of the cytotoxicity. These observations were reproduced in different cell lines, such as 293T cells (data not shown), suggesting that this effect is not restricted to the HeLa cell line, an observation consistent with our previous findings in P. aeruginosa infection studies (50).
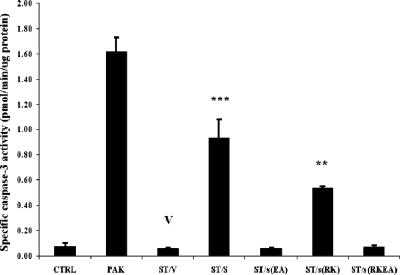
The dispensable role of ExoS GAP activity for apoptosis induction was further investigated by using a previously described bacterial infection model (50). Strain PAKexoST, defective of exoS and exoT, was complemented by the GAP-defective exoS(R146K) or other exoS clones, namely, wild-type exoS, exoS(E381A), or the exoS(R146K/E381A) double mutant (ADPRT and GAP null). As a negative control, vector pUCP18 was used. There was no noticeable difference in bacterial abilities to adhere to host cells among these strains. After infection of HeLa cells the PAKexoST/exoSR146K strain, unlike the vector control strain PAKexoST/pUCP18, caused a significant level of capase-3 activation (Fig. 2), as well as nucleus condensation (data not shown). It appeared to be slightly less effective than complementation with wild-type exoS, suggesting a minor role for the ExoS GAP activity in sensitizing HeLa cells to apoptosis induction. Finally, as expected, the strain complemented with exoS(R146K/E381A) was as “silent” as the vector control strain. In summary, transient expression of exoS is sufficient to elicit apoptosis, and this is dependent on ADPRT but not on GAP activity of the ExoS.
FIG. 2.
GAP activity of ExoS is not essential for apoptosis induction by PAK. HeLa cells were infected with the indicated PAK derivative strains (multiplicity of infection of 20) or left uninfected (CTRL). At 5 h postinfection, cells were collected, lysed, and subjected to caspase-3 assay. The exoS and exoT double mutant PAK (ST) was complemented with vector (ST/V) or with exoS (ST/S), exoS(E381A) [ST/s(EA)], exoS(R146K) [ST/s(RK)], or exoS(R146K/E381A) [ST/s(RKEA)] on the same vector. The data are means ± the SD of the ratios from four replicates. Significant differences between ST/V (V) and other PAK-derived strains are indicated (
 , P < 0.01;
, P < 0.01; 

 , P < 0.001).
, P < 0.001).
Transiently expressed EGFP-ExoS fusion protein effectively induces apoptosis in HeLa cells.
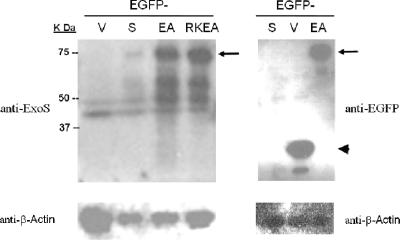
To better identify the HeLa cells expressing ExoS and to assess coincident apoptosis, wild-type exoS and its mutant derivatives were cloned into a pEGFP-C1 vector to produce the fusion proteins of EGFP-ExoS (pJJ0322), EGFP-ExoSE381A (pJJ0323), and EGFP-ExoSR146KE381A (pJJ0324). With a 15-amino-acid polypeptide as a spacer/linker, the chimeric proteins are 707 amino acids in length. The fusion constructs, as well as the pEGFP control vector, were transfected into HeLa cells, and the expression of the fusion proteins in the resulting HeLa cells was examined by Western blotting. At 12 h posttransfection, the 75-kDa full-length fusion proteins were readily recognizable by an antibody directed against either ExoS or GFP (Fig. 3). The amount of EGFP-ExoS was significantly less than that of the other two mutant fusion proteins, namely, EGFP-SE381A (EA) and ExoSR146KE381A (RKEA). Also notably, the protein levels of EGFP-ExoS started to decrease at 12 h posttransfection, whereas those of the two mutant proteins kept rising steadily until 36 h posttransfection (data not shown).
FIG. 3.
Expression of EGFP fusion proteins in HeLa cell after transient transfection. Wild type and exoS, exoS(E381A), and exoS(R146K/E381A) mutants were cloned into pEGFP-C1, with the resulting fusion constructs labeled as follows: S, wild-type ExoS; EA, ExoSE318A; and RKEA, ExoSR146KE381A double mutant. At 12 h posttransfection, the total cellular proteins were collected. The expression of EGFP or fusion proteins was determined by Western blotting with antibodies to ExoS or GFP. Arrows point to the full-length fusion protein band; the arrowhead indicates the EGFP band. Equal protein loading was controlled by Ponceau S staining of membranes and Western blotting for β-actin.
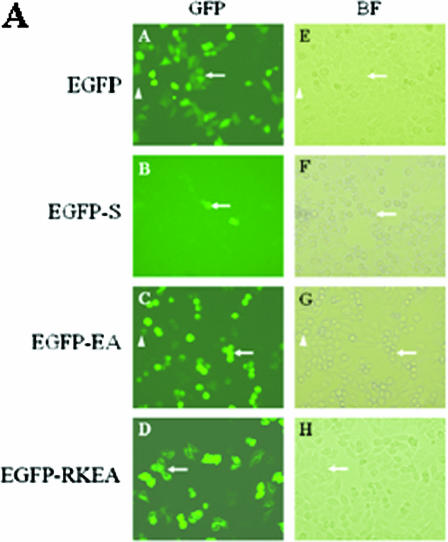
Next, we evaluated cell morphology and GFP fluorescence in HeLa cells transfected with the chimera constructs. Upon transient expression of EGFP-ExoS, very few GFP-positive cells were detectable throughout the 48 h of observation (Fig. 4A and data not shown). Within the same time frame, a large fraction of the cells gradually developed apoptotic morphology, such as membrane blebbing and nuclear condensation. In contrast, HeLa cells transfected with the other three constructs (EGFP, EGFP-ExoSE381A, or EGFP-ExoSR146KE381A) all resulted in high proportions of GFP-positive population, with significantly higher GFP intensity (Fig. 4A). These results strongly indicated that expression of the EGFP-ExoS caused substantial cytotoxicity in HeLa cells. These results were also consistent with the evaluation of the GFP signal in HeLa cells using flow cytometry analysis, which measures the GFP fluorescence intensity of each cell. Transfection with fusion constructs containing exoS, exoS(E381A), and exoS(R146K/E381A) resulted in 11.1, 74.7, and 73.3%, respectively, of total cells marked as GFP positive 24 h after transfection (Fig. 4B). In the GFP-signal-versus-cell-number histogram, these egfp-exoS-transfected cells showed substantially lower GFP values than those in the other two cases (Fig. 4B). Remarkably, all EGFP-ExoSEA-expressing cells, but not EGFP-ExoSRKEA-expressing cells, exhibited rounding morphology, suggesting an intact GAP activity in the fusion protein (Fig. 4C). Taken together, the majority of EGFP-ExoS-expressing cells died and lost the GFP signal rapidly, whereas most of the EGFP-ExoSE381A- and EGFP-ExoSR146KE381A-expressing cells stayed alive and maintained strong GFP signal over the time.
FIG. 4.
(A) Transient expression of EGFP-ExoS in HeLa cells. HeLa cells were transfected with EGFP fusion protein constructs as indicated. HeLa cells were observed under a microscope using GFP fluorescence (A to D) or a bright-field (BF) filter (E to H) at 16 h posttransfection. A and E, EGFP vector control; B and F, EGFP-S, wild-type exoS in fusion construct; C and G, EGFP-EA, exoS(E381A) in fusion construct; D and H, EGFP-RKEA, exoS(R146K) in fusion construct. In each pair of images, the arrows refer to the same cells with detectable GFP. (B) FACS analysis of GFP signals in HeLa cells transiently transfected with fusion constructs. HeLa cells were collected at 24 h posttransfection and subjected to flow cytometry analysis using FL-1 channel on a FACSort cytometer. Each color-coded line on the histogram represents a collection of cells that were transfected with an indicated construct, with the GFP intensity of each cell on the x axis and counts of the cells with the same intensity on the y axis. A threshold intensity for a GFP-positive cell (GFP+ in the figure) was set based on a vector control, and the percentages for GFP-positive cells, of the total, are presented on the right. CTRL means no plasmid DNA was added for transfection. The results are from a single experiment that is representative of three replicated experiments. (C) Cytosolic localization of fusion proteins in HeLa cells after transient transfection with fusion protein expression plasmids. HeLa cells were transfected with EGFP fusion protein constructs as indicated. The resulting HeLa cells were fixed for 16 h after transfection and then stained with DAPI and observed under a phase-contrast DMIRB inverted fluorescence microscope (Leica, Germany) using a GFP fluorescence (a and b) or UV (DAPI, c and d) filter; merged images of GFP and DAPI are also shown (e and f). Panels a, c, and e show EGFP-EA [exoS(E381A)] and panels b, c, and f show EGFP-RKEA [exoS(R146K/E381A)] in fusion constructs.
Furthermore, caspase-3 activation was examined in the transfected HeLa cells using Red-DEVD-FMK. EGFP-ExoS expression led to gradually enhanced caspase-3 activation, resulting in activated caspase-3 in 53.6% of the total cells or nearly 80% of the GFP-positive subpopulation by 24 h posttransfection, which was comparable to the 60.0% of HeLa cells at 3 h after STS treatment (2 μM) (Fig. 5A and Table 2). STS treatment served as a positive control for the apoptosis induction. In contrast, at 24 h posttransfection, the expression of EGFP, EGFP-ExoSEA, and EGFP-ExoSRKEA resulted in caspase-3 activation in 13.8, 11.6, and 7.0% of total cells or 9.1, 9.2, and 4.1% among EGFP-positive populations, respectively (Fig. 5A and Table 2). Combining the data obtained at all three time points posttransfection, in GFP-positive HeLa cells, the caspase-3 activity was consistently and significantly higher for transfection with EGFP-ExoS than with EGFP (Table 2). Importantly, treatment of HeLa cells with Boc-d-FMK, a cell-permeable broad-spectrum caspase inhibitor (22, 66), significantly reduced apoptosis elicited by the EGFP-ExoS, as judged by caspase-3 activity assay (Fig. 5B) and Hoechst staining assay (data not shown). In contrast, glycine, a necrosis inhibitor (51), failed to reverse the caspase-3 activation (data not shown), indicating an apoptosis-specific phenomenon that is not secondary to possible necrosis.
FIG. 5.
(A) Caspase-3 activation in EGFP-ExoS transfected HeLa cells. HeLa cells were transfected with EGFP fusion protein constructs as indicated. The resulting HeLa cells were collected 24 h after transfection, and a caspase-3 activation assay was performed using Red-DEVD-FMK as described in Materials and Methods. Flow cytometry analysis was performed using a FL-2 channel on a FACSort cytometer. Dot density analysis of each sample is represented as the FL-2 intensity of a single cell (x axis) and the forward light scatter of the cells (y axis). The index for the gating population with activated caspase-3 in this assay was determined from the analysis pattern for HeLa cells treated with 2 μM STS for 4 h, which served as a positive control (subpanel B). Remaining subpanels: A, CTRL, no DNA was added for the transfection procedure; C, EGFP-S, wild-type exoS in fusion construct; D, EGFP-EA, exoS(E381A) in fusion construct; E, EGFP, vector control; F, EGFP-RKEA, exoS(R146K/E381A) in fusion construct. (B) Caspase-3 activation in total transfected HeLa cells is reduced by pan-caspase inhibitor. Caspase-3 activation in HeLa cells was assayed 24 h after transfection with the indicated constructs or 3 h after STS treatment. HeLa cells were treated with Boc-d-fmk at 80 μM immediately after transfection. The data are means ± the SD from three replicates from one transfection experiment. Significant differences between Boc-d-fmk-treated and untreated (CTRL) groups are shown (** P < 0.01).
TABLE 2.
Caspase-3 is activated in EGFP-ExoS-transfected HeLa cellsa
| Time posttransfection (h) | Fraction | Mean content (%) ± SDb
|
|||
|---|---|---|---|---|---|
| EGFP | S | EA | RKEA | ||
| 8 | Total | 3.8 ± 1.9 | 4.8 ± 1.6 | 3.7 ± 1.3 | 3.8 ± 1.5 |
| GFP+ | 4.0 ± 0.9 | 26.6 ± 4.7* | 4.1 ± 0.6 | 3.6 ± 0.8 | |
| 12 | Total | 4.9 ± 2.8 | 10.9 ± 2.7 | 4.5 ± 1.4 | 3.6 ± 1.0 |
| GFP+ | 5.7 ± 0.9 | 34.3 ± 3.6** | 5.1 ± 0.7 | 2.9 ± 0.5 | |
| 24 | Total | 13.8 ± 3.7 | 53.6 ± 10.8** | 11.6 ± 2.1 | 7.0 ± 1.4 |
| GFP+ | 9.1 ± 1.9 | 75.2 ± 6.9*** | 9.2 ± 0.6 | 4.1 ± 0.5 | |
HeLa cells were transfected with EGFP fusion protein constructs as indicated. The resulting HeLa cells were collected at the indicated times after transfection, and a caspase-3 activation assay was performed using Red-DEVD-FMK as described in Materials and Methods. Flow cytometry analysis was performed using FL-1 and FL-2 channels on a FACSort cytometer. Cells with activated caspase-3 were determined based on the FL-2 channel reading as described in Fig. 5. Cells with EGFP (GFP+) were determined based on the FL-1 channel reading as described in Fig. 4. Detection through both channels was adjusted based on signal-positive control (i.e., either without Red-DEVE-FMK staining, for FL-2 negative, or without EGFP transfection, for FL-1 negative, respectively) in order to minimize false positives. The percentages of cells with caspase-3 activation in transfected HeLa cells are shown as in total HeLa cells (Total) or in cells with detectable GFP signal (GFP+). EGFP, vector control; S, wild-type exoS in fusion construct; EA, exoS(E381A) in fusion construct; RKEA, exoS(R146K/E381A) in fusion construct. Data are means from three replicates. Significant differences between EGFP- and EGFP-ExoS-transfected groups are indicated as follows: *, P < 0.05; **, P < 0.01, and ***, P < 0.001.
JNK signaling and the mitochondrion proapoptotic pathway were both required in EGFP-ExoS-induced apoptosis.
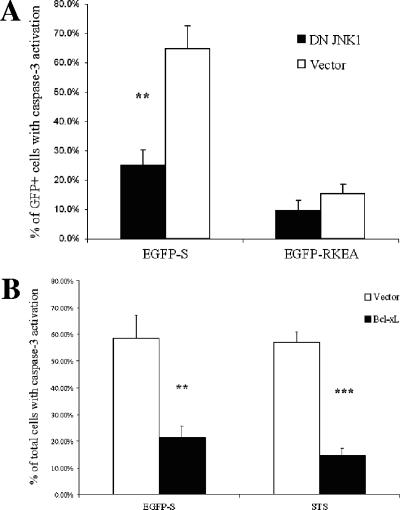
Previously, we demonstrated that P. aeruginosa secreting ExoS triggers a proapoptotic pathway through JNK-mediated cytochrome c release from mitochondria (48). Here, we used the HeLa cells overexpressing DN JNK1 (48) to test the role of JNK in the EGFP-ExoS-induced apoptosis. Upon transient transfection with EGFP-ExoS, HeLa/DN JNK1 cells were much less sensitive to apoptosis induction than the HeLa/pcDNA3 (vector control) (Fig. 6A). Furthermore, overexpression of the antiapoptotic mitochondrial protein Bcl-xL in HeLa cells (65, 69) significantly reduced capase-3 activation after transfection with EGFP-ExoS (Fig. 6B). As a control in this experiment (Fig. 6B), STS-triggered apoptosis was also blocked by the overexpression of Bcl-xL in a similar fashion (87). In agreement with this, HeLa/Bcl-xL cells exhibited significantly reduced sensitivity to apoptosis induction by PAK infection compared to the HeLa/Neo control cell line, although there is no difference in terms of ExoS secretion efficiency for the involved cell lines (data not shown). These results demonstrate that both JNK activation and cytochrome c release from mitochondria play important roles in promoting apoptosis in the current model.
FIG. 6.
Caspase-3 activation in EGFP-ExoS transfected HeLa cells is dependent on JNK1 activation and cytochrome c release. (A and B) HeLa/pcDNA3 (vector) or HeLa/DN JNK1 cells (A) and HeLa/Neo vector (vector) or HeLa/Bcl-xL cells (B) were transfected with the EGFP-ExoS construct and subjected to caspase-3 activation assay with Red-DEVD-FMK as described above. As a control for apoptosis induction, STS (2 μM) was used to treat the indicated cells for 3 h. The data are means ± the SD from three replicates from one transfection experiment. Significant differences between HeLa/pcDNA3 (vector) or HeLa/DN JNK1 cells (A) or between HeLa/Neo vector (vector) and HeLa/Bcl-xL cells (B) are indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
The cellular pathway(s) mediating ExoS-induced cytotoxicity is evolutionarily conserved.
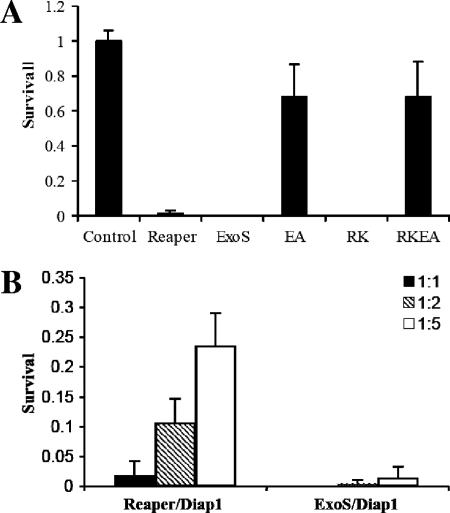
To test whether the molecular mechanism of ExoS-induced cell death is conserved, we expressed exoS, exoS(E381A), exoS(R146K), and exoS(R146K/E381A) in Drosophila S2 cells. Expression of ExoS using the Pie3 insect virus promoter induced rapid cell death within 20 h after the transfection (Fig. 7A), displaying an efficiency similar to that induced by the expression of reaper, a proapoptotic gene in Drosophila (99). This result is not a total surprise. Previous studies have shown that P. aeruginosa infects Drosophila larvae and adults and that the TTSS is responsible for killing the organisms (3, 19, 24). We demonstrated here that the intracellular expression of ExoS alone is sufficient to induce apoptosis in Drosophila cells. Similar to its effect on mammalian cells, ExoS-induced cytotoxicity in insect cells mainly depends on its ADPRT activity. The ADPRT-null mutation (E381A) abolished ExoS-induced cell death, whereas the GAP-null mutation (R146K) had little effect (Fig. 7A). The IAP and IAP-antagonist (Reaper) pathway play important roles in regulating cell death during insect development; IAP-antagonist, thus far, is the most potent proapoptotic regulator in insect cells (10). However, whereas the coexpression of DIAP1 (for Drosophila inhibitor of apoptosis protein 1) significantly suppressed IAP-antagonist-induced cell death, it had little effect on the ExoS-induced cell death (Fig. 7B), suggesting that either ExoS-induced cytotoxicity is not mediated through the IAP pathway or the direct target of ExoS is located lower in the signal transduction cascade than the level of IAP function in Drosophila cells.
FIG. 7.
(A) ExoS induced cell death in Drosophila S2 cells. Similar to its effect on mammalian cells, the cytotoxicity of ExoS in Drosophila S2 cells is dependent on its ADP-ribosylating activity (EA, ExosE381A; RK, ExoSR146K; RKEA, ExoSR146K/E381A). Wild type and ExoSRK killed essentially 100% transfected cells, whereas the ExoSEA and ExoSRKEA double mutants have dramatically reduced cytotoxic activity. (B) While increasing the level of dIAP1 significantly suppressed reaper-induced cell death, it had little effect on ExoS-induced cytotoxicity. The colors of the bar reflect the relative ratio between proapoptotic gene constructs and Diap1. For both panels A and B, the data are represented as averages (n = 5 and 4, respectively), and error bars indicate the SD.
Collectively, ExoS, either by itself or in the form of a chimeric protein, efficiently induces apoptosis in HeLa cells, as well as Drosophila S2 cells, independent of other factors from P. aeruginosa. This induction correlates directly with its functional ADPRT domain but not the GAP domain. Also importantly, the apoptotic signal transduction events that are involved in the current ExoS transfection model, such as the essential roles played by JNKs, mitochondria, and caspases, mirror the previously described pathway wherein apoptosis was triggered upon infection by ExoS-secreting P. aeruginosa (48).
DISCUSSION
ExoS, along with other P. aeruginosa type III secretion effectors, has been shown to be an important virulence determinant in various animal infection models (42, 57-59, 83). In addition, most cystic fibrosis (CF) patients harbor antibodies to ExoS (4). On the other hand, clinical isolates from CF patients have also been shown to predominantly lack the ability to secrete type III effector molecules, including ExoS (20, 80). Indeed, ExoS plays an important role in the initial colonization and persistence in the CF lung but not during the chronic stage of the disease (59, 83). It was further suggested that the CF strains accumulate a number of mutations to ensure the reduced production of type III effectors and therefore bacterial toxicity to the host (59). Nonetheless, it remains a partially elucidated picture how the “on and off” switch of ExoS secretion in susceptible human host correlates with the progression of the P. aeruginosa infection. In recent years, increasing numbers of studies, both in vitro or in vivo, describe apoptosis induction by P. aeruginosa (17, 23, 36, 38, 46, 77, 89); some of the studies identified a variety of apoptosis-inducing factors from the bacterium (2, 7, 8, 34, 47, 50, 88). Bruno et al. (7) and we (50) first reported ExoS as one of these factors. The present study stems from our previous observations, directly correlating the apoptosis induction with the ADPRT-competent ExoS independent of any other bacterial factor.
Our attempts to generate stable HeLa cell lines with inducible expression of wild-type ExoS were not successful. The failure is likely due to a leakiness of the control of transcription in the selected system, a finding similar to those observed by others (94). Indeed, trace amounts of ExoS were detected in transfectants even in the absence of inducer. However, an elevated expression of exoS was achieved in the transfectants that were transiently transfected using the same system, and the resulting expression of ExoS, which was confirmed by Western blotting (data not shown), led to a substantial increase in apoptotic cells, as assessed by both caspase-3 activation and nucleus condensation, whereas that of ExoSE381A failed to do so.
A GFP reporter, carried either by pUF5 or pEGFP-C1, was used as an indicator of successful transfection in analysis of ExoS-mediated apoptosis in present study. The total GFP signal intensity in the cell is affected by the rate of de novo protein synthesis and the stability of the protein, both of which are likely affected during the ExoS-induced apoptosis process. This is evident in the cotransfection experiment, in which, although strong blockage of caspase-3 activation and nuclei condensation were achieved upon application of a caspase inhibitor (Fig. 1), the GFP signal was only partially restored in the transfected population (Fig. 1 and data not shown), suggesting that exogenously expressed ExoS affects protein synthesis and/or protein stability in the cell in addition to triggering apoptosis. Apoptosis-related inhibition of translation has been shown in an array of cell lines, including HeLa cells (12, 63). In particular, caspase-3-independent degradation of PABP [for poly(A)-binding protein] has been reported (63). Nevertheless, the effect on translation should not be the direct cause of the efficient apoptosis induction demonstrated in our study, since the inhibition of translation in HeLa cells by CHX alone did not lead to rapid apoptosis (data not shown; see also reference 6); it is rather the result of rapid self-deterioration of apoptotic cells. Moreover, this effect should not be significantly contributed by the GAP activity of the ExoS since transfection featuring ExoSE381A led to most, if not all, of the resulting cells being EGFP positive (Fig. 4B), indicating maintenance of the cellular protein level, despite the dramatic alteration of the cellular morphology resulting from the intact GAP activity (25, 32, 54).
Interestingly, in the experiments with EGFP chimeric proteins, an extensive degree of degradation was observed for all derivatives of full-length chimeric proteins (Fig. 3), suggesting that the degradation of EGFP fusion protein is at least partially triggered by an ExoS-specific proteasome pathway. It is worth noting that, when treated with Boc-d-FMK, a general caspase inhibitor, EGFP-ExoS-transfected HeLa cells were only marginally (although noticeably) rescued for the GFP signal (data not shown), whereas cells that were cotransfected with GFP and ExoS (i.e., the two proteins were expressed independently) displayed much more significant restoration of the signal (Fig. 1), suggesting a possible ExoS-associated degradation. Type III toxins secreted by Salmonella enterica reportedly undergo proteasome-dependent protein degradation, which plays an essential role in the temporal regulation of their function (56). It will be interesting to evaluate and confirm the possible degradation of ExoS in future studies.
After injection into the host cell cytosol through the TTSS of P. aeruginosa, ExoS has been shown to localize to the perinuclear region within host cells due to the function of a membrane localization domain localized between amino acid residues 51 and 72 (55, 71, 72). The EGFP-transfected HeLa cells, as previously well-documented (79), display both cytoplasmic and nuclear fluorescence, whereas in their EGFP-ExoSE381A and EGFP-ExoSR146KE381A counterparts, the fluorescence appeared to be excluded from nuclei (Fig. 4C), which closely resembles the pattern of perinuclear localization of the ExoS mediated by the membrane localization domain (55, 97). Therefore, the subcellular distribution pattern and GAP domain of the chimeric proteins remained intact, and the cell death data presented above also suggested that the ADPRT domain in EGFP-ExoS retained its catalytic function and targeting specificity as well.
Furthermore, it has been documented that cells in different phases of the cell cycle have different sensitivities to apoptosis induction (21). Therefore, it is conceivable that in the EGFP-ExoS transfection experiments, the variation of EGFP intensity in each cell may reflect a cell cycle-dependent sensitivity to ExoS-induced apoptosis. In those less-sensitive cells, the proapoptotic signal needs to overcome a higher threshold in order to push the balance toward apoptosis. As a result, more EGFP-ExoS fusion proteins are accumulated before apoptosis is triggered in these cells, and thus the cells displayed a higher intensity of GFP. In the EGFP-ExoS-transfected HeLa cells, nearly 54% of the total population was apoptotic (Fig. 5A), which correlates with a consistent >60% frequency of transfection. However, only 11% of the total cells displayed detectable EGFP; thus, we believe that the majority of the transfected, yet EGFP signal-negative HeLa cells underwent apoptosis. Considering ca. 60% of the STS-treated HeLa cells were apoptotic, 80% of the GFP-positive subpopulation undergoing apoptosis (Fig. 5C) is highly significant for EGFP-ExoS-transfected cells. These data strongly suggested that the appearance of GFP signal in these cells is an indication of their temporal insensitivity to apoptosis induction, possibly due to a higher tolerance at a certain cell cycle state. Studies are under way to test this possibility.
The apoptosis induction, as a result of the expression of either ExoS alone or EGFP-ExoS, appeared to be attenuated compared to that resulting from P. aeruginosa infection (48, 50). The rate of apoptosis elicitation was slower, and the intensity was reduced in terms of caspase-3 activation. In contrast to the direct injection of ExoS into HeLa cells by the bacteria at a fast and uniform rate, the uptake of exogenous DNA and its expression are much more time and energy consuming, which might be responsible for the reduced tempo and intensity of the apoptosis induction.
It remains obscure how the ADPRT activity of ExoS leads to apoptosis in host cells. The ADPRT of ExoS shares a similar motif with the arginine-specific mono-ADP-ribosyltransferase of eukaryotic cells (96). Endogenous mono-ADPRT of proteins has not been associated with the apoptotic process. However, pretreatment with a potent inhibitor of arginine-specific mono-ADPRT has been demonstrated to suppress TNF-induced DNA fragmentation in U937 cells in a dose-dependent manner (90). In addition, cytosolic mono-ADPRT activity is elevated in cells undergoing apoptosis (91). These data support the hypothesis that mono-ADPRT may function in apoptotic signal transduction. On the other hand, it has also been speculated that the hyperactivity of ADPRT may deplete the cellular levels of NAD, and subsequently ATP, which ultimately causes cell death due to impaired energy metabolism (9).
A great deal of progress has been made in defining the biological targets of ExoS in host cells. ExoS recognizes a wide variety of genetically or structurally diverse proteins for ADPRT, including the monomeric GTPases, vimentin, and other undefined host proteins (6, 13, 62). Most dominant substrates are Ras proteins, especially H-Ras, N-Ras, and K-Ras, three proteins that are ubiquitously expressed in mammalian cells (26, 41). Ras and several related proteins, including Rab3, Rab4, Rab5, Ral, Rap1A, and Rap2, were identified as targets of ExoS (15, 16, 26, 41). The ADPRT of these components by ExoS disrupted signaling by inhibiting guanine nucleotide exchange factor-catalyzed nucleotide exchange, which uncoupled respective signal transduction (5, 29, 30, 78). Ras and Ral, both important in the proliferation and survival of the host cell and ADP-ribosylated by ExoS, represent the antiapoptosis barrier that ExoS ADPRT will have to overcome to induce apoptosis. Most recently, Jansson et al. (45) reported that when Yersinia pseudotuberculosis TTSS was used as a tool to deliver ExoS proteins into fibroblast and kidney cells, activated Ras was shown to protect infected cells against ExoS-induced apoptosis, regardless of whether activated Ras was modified by ExoS. We have previously drawn a direct link between ExoS ADPRT domain and its ability to trigger the proapoptotic signaling through a JNK/mitochondrion/caspase-3 pathway that is facilitated by the inhibition of the Erk survival pathway (48, 50). Differential sensitivities of the two signaling pathways to ExoS might explain the cell type-dependent sensitivity to the killing by P. aeruginosa (14). Two critical questions remain unanswered. (i) Is the ExoS-mediated blockage of Ras signaling sufficient to trigger host cell apoptosis? (ii) If it is sufficient, how does the disruption of Ras signaling warrant JNK activation and the downstream proapoptosis pathway? If not, what else is involved? We should be able to better define the unique involvement of ExoS ADPRT in apoptosis induction by comparing ExoS with other known endogenous mono-ADPRTs with regard to intracellular processing, cofactor binding, and in vivo substrate targets.
Even though the nature of the cellular pathway(s) mediating ExoS induces cell death remains elusive, it is clear that the responsible pathway(s) is genetically conserved. We have shown that ExoS, when expressed, induces rapid cell death in Drosophila S2 cells. Similar to its effect on mammalian systems, the cytotoxic activity of ExoS is mainly dependent on the ADPRT activity. Intriguingly, identification of the putative target(s) of ExoS in S2 cells may very likely provide a great deal of information that helps elucidate its role in apoptosis induction in mammalian systems. These findings paved the way for using the powerful genetic system offered by the fruit fly to conduct systematic searches for the cellular targets mediating ExoS-induced cytotoxicity.
Overall, by using two approaches to express ExoS in HeLa cells, we demonstrate that ExoS alone is sufficient to mediate apoptosis in host cells. Furthermore, the ADPRT of ExoS is the key factor for initiating the proapoptotic signaling pathways in host cells. Therefore, TTSS secreted ExoS serves as an effective apoptosis-inducing agent for P. aeruginosa to manipulate the fate of host cell and to promote its prominent infection cycle in susceptible host. Finally, with ongoing study to fully elucidate the cellular mechanism for its action, ExoS presents a great deal of potential as an apoptosis-inducing molecule that may be tested as a novel cancer therapeutic agent in the future.
Acknowledgments
We thank Alfred Lewin for the gift of pUF5, Xiaodong Wang for the HeLa-Bcl-xL cell lines, Lung-Ji Chang for critical support, Neal Benson and Bhavna Bhardwaj (ICBR, University of Florida) for assistance on flow cytometry, and Carl P. Santos for technical assistance.
This study was supported by a Research Scholar Award from the American Cancer Society (to S.J.) and a National Institutes of Health grant (to L.Z.).
Editor: J. B. Bliska
Footnotes
Published ahead of print on 11 September 2006.
REFERENCES
- 1.Aktories, K., and J. T. Barbieri. 2005. Bacterial cytotoxins: targeting eukaryotic switches. Nat. Rev. Microbiol. 3:397-410. [DOI] [PubMed] [Google Scholar]
- 2.Allen, L., D. H. Dockrell, T. Pattery, D. G. Lee, P. Cornelis, P. G. Hellewell, and M. K. Whyte. 2005. Pyocyanin production by Pseudomonas aeruginosa induces neutrophil apoptosis and impairs neutrophil-mediated host defenses in vivo. J. Immunol. 174:3643-3649. [DOI] [PubMed] [Google Scholar]
- 3.Avet-Rochex, A., E. Bergeret, I. Attree, M. Meister, and M. O. Fauvarque. 2005. Suppression of Drosophila cellular immunity by directed expression of the ExoS toxin GAP domain of Pseudomonas aeruginosa. Cell Microbiol. 7:799-810. [DOI] [PubMed] [Google Scholar]
- 4.Banwart, B., M. L. Splaingard, P. M. Farrell, M. J. Rock, P. L. Havens, J. Moss, M. E. Ehrmantraut, D. W. Frank, and J. T. Barbieri. 2002. Children with cystic fibrosis produce an immune response against exoenzyme S, a type III cytotoxin of Pseudomonas aeruginosa. J. Infect. Dis. 185:269-270. [DOI] [PubMed] [Google Scholar]
- 5.Barbieri, A. M., Q. Sha, P. Bette-Bobillo, P. D. Stahl, and M. Vidal. 2001. ADP-Ribosylation of Rab5 by ExoS of Pseudomonas aeruginosa affects endocytosis. Infect. Immun. 69:5329-5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbieri, J. T., and J. Sun. 2004. Pseudomonas aeruginosa ExoS and ExoT. Rev. Physiol. Biochem. Pharmacol. 152:79-92. [DOI] [PubMed] [Google Scholar]
- 7.Bruno, T. F., D. E. Woods, and C. H. Mody. 2000. Exoenzyme S from Pseudomonas aeruginosa induces apoptosis in T lymphocytes. J. Leukoc. Biol. 67:808-816. [DOI] [PubMed] [Google Scholar]
- 8.Buommino, E., F. Morelli, S. Metafora, F. Rossano, B. Perfetto, A. Baroni, and M. A. Tufano. 1999. Porin from Pseudomonas aeruginosa induces apoptosis in an epithelial cell line derived from rat seminal vesicles. Infect. Immun. 67:4794-4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carson, D. A., S. Seto, D. B. Wasson, and C. J. Carrera. 1986. DNA strand breaks, NAD metabolism, and programmed cell death. Exp. Cell Res. 164:273-281. [DOI] [PubMed] [Google Scholar]
- 10.Cashio, P., T. V. Lee, and A. Bergmann. 2005. Genetic control of programmed cell death in Drosophila melanogaster. Semin. Cell Dev. Biol. 16:225-235. [DOI] [PubMed] [Google Scholar]
- 11.Cheng, L., J. Fu, A. Tsukamoto, and R. G. Hawley. 1996. Use of green fluorescent protein variants to monitor gene transfer and expression in mammalian cells. Nat. Biotechnol. 14:606-609. [DOI] [PubMed] [Google Scholar]
- 12.Clemens, M. J., M. Bushell, I. W. Jeffrey, V. M. Pain, and S. J. Morley. 2000. Translation initiation factor modifications and the regulation of protein synthesis in apoptotic cells. Cell Death Differ. 7:603-615. [DOI] [PubMed] [Google Scholar]
- 13.Coburn, J., S. T. Dillon, B. H. Iglewski, and D. M. Gill. 1989. Exoenzyme S of Pseudomonas aeruginosa ADP-ribosylates the intermediate filament protein vimentin. Infect. Immun. 57:996-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coburn, J., and D. W. Frank. 1999. Macrophages and epithelial cells respond differently to the Pseudomonas aeruginosa type III secretion system. Infect. Immun. 67:3151-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coburn, J., and D. M. Gill. 1991. ADP-ribosylation of p21ras and related proteins by Pseudomonas aeruginosa exoenzyme S. Infect. Immun. 59:4259-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coburn, J., R. T. Wyatt, B. H. Iglewski, and D. M. Gill. 1989. Several GTP-binding proteins, including p21c-H-ras, are preferred substrates of Pseudomonas aeruginosa exoenzyme S. J. Biol. Chem. 264:9004-9008. [PubMed] [Google Scholar]
- 17.Coopersmith, C. M., P. E. Stromberg, W. M. Dunne, C. G. Davis, D. M. Amiot II, T. G. Buchman, I. E. Karl, and R. S. Hotchkiss. 2002. Inhibition of intestinal epithelial apoptosis and survival in a murine model of pneumonia-induced sepsis. JAMA 287:1716-1721. [DOI] [PubMed] [Google Scholar]
- 18.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 19.D'Argenio, D. A., L. A. Gallagher, C. A. Berg, and C. Manoil. 2001. Drosophila as a model host for Pseudomonas aeruginosa infection. J. Bacteriol. 183:1466-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dacheux, D., I. Attree, and B. Toussaint. 2001. Expression of ExsA in trans confers type III secretion system-dependent cytotoxicity on noncytotoxic Pseudomonas aeruginosa cystic fibrosis isolates. Infect. Immun. 69:538-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Danial, N. N., and S. J. Korsmeyer. 2004. Cell death: critical control points. Cell 116:205-219. [DOI] [PubMed] [Google Scholar]
- 22.Deas, O., C. Dumont, M. MacFarlane, M. Rouleau, C. Hebib, F. Harper, F. Hirsch, B. Charpentier, G. M. Cohen, and A. Senik. 1998. Caspase-independent cell death induced by anti-CD2 or staurosporine in activated human peripheral T lymphocytes. J. Immunol. 161:3375-3383. [PubMed] [Google Scholar]
- 23.Epelman, S., G. G. Neely, L. L. Ma, M. Gjomarkaj, E. Pace, M. Melis, D. E. Woods, and C. H. Mody. 2002. Distinct fates of monocytes and T cells directly activated by Pseudomonas aeruginosa exoenzyme S. J. Leukoc. Biol. 71:458-468. [PubMed] [Google Scholar]
- 24.Fauvarque, M. O., E. Bergeret, J. Chabert, D. Dacheux, M. Satre, and I. Attree. 2002. Role and activation of type III secretion system genes in Pseudomonas aeruginosa-induced Drosophila killing. Microb. Pathog. 32:287-295. [DOI] [PubMed] [Google Scholar]
- 25.Fraylick, J. E., M. J. Riese, T. S. Vincent, J. T. Barbieri, and J. C. Olson. 2002. ADP-ribosylation and functional effects of Pseudomonas exoenzyme S on cellular RalA. Biochemistry 41:9680-9687. [DOI] [PubMed] [Google Scholar]
- 26.Fraylick, J. E., E. A. Rucks, D. M. Greene, T. S. Vincent, and J. C. Olson. 2002. Eukaryotic cell determination of ExoS ADP-ribosyltransferase substrate specificity. Biochem. Biophys. Res. Commun. 291:91-100. [DOI] [PubMed] [Google Scholar]
- 27.Fu, H., J. Coburn, and R. J. Collier. 1993. The eukaryotic host factor that activates exoenzyme S of Pseudomonas aeruginosa is a member of the 14-3-3 protein family. Proc. Natl. Acad. Sci. USA 90:2320-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galan, J. E., and A. Collmer. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284:1322-1328. [DOI] [PubMed] [Google Scholar]
- 29.Ganesan, A. K., D. W. Frank, R. P. Misra, G. Schmidt, and J. T. Barbieri. 1998. Pseudomonas aeruginosa exoenzyme S ADP-ribosylates Ras at multiple sites. J. Biol. Chem. 273:7332-7337. [DOI] [PubMed] [Google Scholar]
- 30.Ganesan, A. K., T. S. Vincent, J. C. Olson, and J. T. Barbieri. 1999. Pseudomonas aeruginosa exoenzyme S disrupts Ras-mediated signal transduction by inhibiting guanine nucleotide exchange factor-catalyzed nucleotide exchange. J. Biol. Chem. 274:21823-21829. [DOI] [PubMed] [Google Scholar]
- 31.Garau, J., and L. Gomez. 2003. Pseudomonas aeruginosa pneumonia. Curr. Opin. Infect. Dis. 16:135-143. [DOI] [PubMed] [Google Scholar]
- 32.Goehring, U.-M., G. Schmidt, K. J. Pederson, K. Aktories, and J. T. Barbieri. 1999. The N-terminal domain of Pseudomonas aeruginosa exoenzyme S is a GTPase-activating protein for Rho GTPases. J. Biol. Chem. 274:36369-36372. [DOI] [PubMed] [Google Scholar]
- 33.Gossen, M., S. Freundlieb, G. Bender, G. Muller, W. Hillen, and H. Bujard. 1995. Transcriptional activation by tetracyclines in mammalian cells. Science 268:1766-1769. [DOI] [PubMed] [Google Scholar]
- 34.Goto, M., T. Yamada, K. Kimbara, J. Horner, M. Newcomb, T. K. D. Gupta, and A. M. Chakrabarty. 2003. Induction of apoptosis in macrophages by Pseudomonas aeruginosa azurin: tumour-suppressor protein p53 and reactive oxygen species, but not redox activity, as critical elements in cytotoxicity. Mol. Microbiol. 47:549-559. [DOI] [PubMed] [Google Scholar]
- 35.Govan, J. R., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grassme, H., S. Kirschnek, J. Riethmueller, A. Riehle, G. von Kurthy, F. Lang, M. Weller, and E. Gulbins. 2000. CD95/CD95 ligand interactions on epithelial cells in host defense to Pseudomonas aeruginosa. Science 290:527-530. [DOI] [PubMed] [Google Scholar]
- 37.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 38.Hauser, A. R., and J. N. Engel. 1999. Pseudomonas aeruginosa induces type-III-secretion-mediated apoptosis of macrophages and epithelial cells. Infect. Immun. 67:5530-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hauser, A. R., P. J. Kang, and J. N. Engel. 1998. PepA, a secreted protein of Pseudomonas aeruginosa, is necessary for cytotoxicity and virulence. Mol. Microbiol. 27:807-818. [DOI] [PubMed] [Google Scholar]
- 40.Hay, B. A., J. R. Huh, and M. Guo. 2004. The genetics of cell death: approaches, insights and opportunities in Drosophila. Nat. Rev. Genet. 5:911-922. [DOI] [PubMed] [Google Scholar]
- 41.Henriksson, M. L., C. Sundin, A. L. Jansson, A. Forsberg, R. H. Palmer, and B. Hallberg. 2002. Exoenzyme S shows selective ADP-ribosylation and GTPase-activating protein (GAP) activities toward small GTPases in vivo. Biochem. J. 367:617-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holder, I. A., A. N. Neely, and D. W. Frank. 2001. Type III secretion/intoxication system important in virulence of Pseudomonas aeruginosa infections in burns. Burns 27:129-130. [DOI] [PubMed] [Google Scholar]
- 43.Horvitz, H. R. 1999. Genetic control of programmed cell death in the nematode Caenorhabditis elegans. Cancer Res. 59:1701s-1706s. [PubMed] [Google Scholar]
- 44.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jansson, A. L., L. Yasmin, P. Warne, J. Downward, R. H. Palmer, and B. Hallberg. 2006. Exoenzyme S of Pseudomonas aeruginosa is not able to induce apoptosis when cells express activated proteins, such as Ras or protein kinase B/Akt. Cell Microbiol. 8:815-822. [DOI] [PubMed] [Google Scholar]
- 46.Jendrossek, V., S. Fillon, C. Belka, I. Muller, B. Puttkammer, and F. Lang. 2003. Apoptotic response of Chang cells to infection with Pseudomonas aeruginosa strains PAK and PAO-I: molecular ordering of the apoptosis signaling cascade and role of type IV pili. Infect. Immun. 71:2665-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jenkins, C. E., A. Swiatoniowski, A. C. Issekutz, and T.-J. Lin. 2004. Pseudomonas aeruginosa exotoxin A induces human mast cell apoptosis by a caspase-8 and -3 dependent mechanism. J. Biol. Chem. 279:37201-37207. [DOI] [PubMed] [Google Scholar]
- 48.Jia, J., M. Alaoui-El-Azher, M. Chow, T. C. Chambers, H. Baker, and S. Jin. 2003. c-Jun NH2-terminal kinase-mediated signaling is essential for Pseudomonas aeruginosa ExoS-induced apoptosis. Infect. Immun. 71:3361-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones, G., D. Jones, L. Zhou, H. Steller, and Y. Chu. 2000. Deterin, a new inhibitor of apoptosis from Drosophila melanogaster. J. Biol. Chem. 275:22157-22165. [DOI] [PubMed] [Google Scholar]
- 50.Kaufman, M. R., J. Jia, L. Zeng, U. Ha, M. Chow, and S. Jin. 2000. Pseudomonas aeruginosa mediated apoptosis requires the ADP-ribosylating activity of ExoS. Microbiology 146(Pt. 10):2531-2541. [DOI] [PubMed] [Google Scholar]
- 51.Kim, J. S., L. He, T. Qian, and J. J. Lemasters. 2003. Role of the mitochondrial permeability transition in apoptotic and necrotic death after ischemia/reperfusion injury to hepatocytes. Curr. Mol. Med. 3:527-535. [DOI] [PubMed] [Google Scholar]
- 52.Kipnis, E., T. Sawa, and J. Wiener-Kronish. 2006. Targeting mechanisms of Pseudomonas aeruginosa pathogenesis. Med. Mal. Infect. 36:78-91. [DOI] [PubMed] [Google Scholar]
- 53.Krall, R., G. Schmidt, K. Aktories, and J. T. Barbieri. 2000. Pseudomonas aeruginosa ExoT is a rho GTPase-activating protein. Infect. Immun. 68:6066-6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krall, R., J. Sun, K. J. Pederson, and J. T. Barbieri. 2002. In vivo rho GTPase-activating protein activity of Pseudomonas aeruginosa cytotoxin ExoS. Infect. Immun. 70:360-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krall, R., Y. Zhang, and J. T. Barbieri. 2004. Intracellular membrane localization of Pseudomonas ExoS and Yersinia YopE in mammalian cells. J. Biol. Chem. 279:2747-2753. [DOI] [PubMed] [Google Scholar]
- 56.Kubori, T., and J. E. Galan. 2003. Temporal regulation of salmonella virulence effector function by proteasome-dependent protein degradation. Cell 115:333-342. [DOI] [PubMed] [Google Scholar]
- 57.Kudoh, I., J. P. Wiener-Kronish, S. Hashimoto, J. F. Pittet, and D. Frank. 1994. Exoproduct secretions of Pseudomonas aeruginosa strains influence severity of alveolar epithelial injury. Am. J. Physiol. 267:L551-L556. [DOI] [PubMed] [Google Scholar]
- 58.Lee, E. J., B. A. Cowell, D. J. Evans, and S. M. Fleiszig. 2003. Contribution of ExsA-regulated factors to corneal infection by cytotoxic and invasive Pseudomonas aeruginosa in a murine scarification model. Investig. Ophthalmol. Vis. Sci. 44:3892-3898. [DOI] [PubMed] [Google Scholar]
- 59.Lee, V. T., R. S. Smith, B. Tummler, and S. Lory. 2005. Activities of Pseudomonas aeruginosa effectors secreted by the type III secretion system in vitro and during infection. Infect. Immun. 73:1695-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li, L., D. Hooi, S. R. Chhabra, D. Pritchard, and P. E. Shaw. 2004. Bacterial N-acylhomoserine lactone-induced apoptosis in breast carcinoma cells correlated with down-modulation of STAT3. Oncogene 23:4894-4902. [DOI] [PubMed] [Google Scholar]
- 61.Liu, X., C. N. Kim, J. Yang, R. Jemmerson, and X. Wang. 1996. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell 86:147-157. [DOI] [PubMed] [Google Scholar]
- 62.Maresso, A. W., M. R. Baldwin, and J. T. Barbieri. 2004. Ezrin/radixin/moesin proteins are high-affinity targets for ADP-ribosylation by Pseudomonas aeruginosa ExoS. J. Biol. Chem. 279:38402-38408. [DOI] [PubMed] [Google Scholar]
- 63.Marissen, W. E., D. Triyoso, P. Younan, and R. E. Lloyd. 2004. Degradation of poly(A)-binding protein in apoptotic cells and linkage to translation regulation. Apoptosis 9:67-75. [DOI] [PubMed] [Google Scholar]
- 64.Martin, S. J. 2002. Destabilizing influences in apoptosis: sowing the seeds of IAP destruction. Cell 109:793-796. [DOI] [PubMed] [Google Scholar]
- 65.Martinou, J. C., and D. R. Green. 2001. Breaking the mitochondrial barrier. Nat. Rev. Mol. Cell. Biol. 2:63-67. [DOI] [PubMed] [Google Scholar]
- 66.Maurer, B. J., L. S. Metelitsa, R. C. Seeger, M. C. Cabot, and C. P. Reynolds. 1999. Increase of ceramide and induction of mixed apoptosis/necrosis by N-(4-hydroxyphenyl)-retinamide in neuroblastoma cell lines. J. Natl. Cancer Inst. 91:1138-1146. [DOI] [PubMed] [Google Scholar]
- 67.Morimoto, H., and B. Bonavida. 1992. Diphtheria toxin- and Pseudomonas A toxin-mediated apoptosis: ADP-ribosylation of elongation factor-2 is required for DNA fragmentation and cell lysis and synergy with tumor necrosis factor-alpha. J. Immunol. 149:2089-2094. [PubMed] [Google Scholar]
- 68.Nagata, S. 1999. Fas ligand-induced apoptosis. Annu. Rev. Genet. 33:29-55. [DOI] [PubMed] [Google Scholar]
- 69.Nijhawan, D., M. Fang, E. Traer, Q. Zhong, W. Gao, F. Du, and X. Wang. 2003. Elimination of Mcl-1 is required for the initiation of apoptosis following ultraviolet irradiation. Genes Dev. 17:1475-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pederson, K. J., and J. T. Barbieri. 1998. Intracellular expression of the ADP-ribosyltransferase domain of Pseudomonas exoenzyme S is cytotoxic to eukaryotic cells. Mol. Microbiol. 30:751-759. [DOI] [PubMed] [Google Scholar]
- 71.Pederson, K. J., R. Krall, M. J. Riese, and J. T. Barbieri. 2002. Intracellular localization modulates targeting of ExoS, a type III cytotoxin, to eukaryotic signalling proteins. Mol. Microbiol. 46:1381-1390. [DOI] [PubMed] [Google Scholar]
- 72.Pederson, K. J., S. Pal, A. J. Vallis, D. W. Frank, and J. T. Barbieri. 2000. Intracellular localization and processing of Pseudomonas aeruginosa ExoS in eukaryotic cells. Mol. Microbiol. 37:287-299. [DOI] [PubMed] [Google Scholar]
- 73.Pederson, K. J., A. J. Vallis, K. Aktories, D. W. Frank, and J. T. Barbieri. 1999. The amino-terminal domain of Pseudomonas aeruginosa ExoS disrupts actin filaments via small-molecular-weight GTP-binding proteins. Mol. Microbiol. 32:393-401. [DOI] [PubMed] [Google Scholar]
- 74.Pirnay, J.-P., D. De Vos, C. Cochez, F. Bilocq, J. Pirson, M. Struelens, L. Duinslaeger, P. Cornelis, M. Zizi, and A. Vanderkelen. 2003. Molecular epidemiology of Pseudomonas aeruginosa colonization in a burn unit: persistence of a multidrug-resistant clone and a silver sulfadiazine-resistant clone. J. Clin. Microbiol. 41:1192-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Punj, V., S. Bhattacharyya, D. Saint-Dic, C. Vasu, E. A. Cunningham, J. Graves, T. Yamada, A. I. Constantinou, K. Christov, B. White, G. Li, D. Majumdar, A. M. Chakrabarty, and T. K. Das Gupta. 2004. Bacterial cupredoxin azurin as an inducer of apoptosis and regression in human breast cancer. Oncogene 23:2367-2378. [DOI] [PubMed] [Google Scholar]
- 76.Radke, J., K. J. Pederson, and J. T. Barbieri. 1999. Pseudomonas aeruginosa exoenzyme S is a biglutamic acid ADP-ribosyltransferase. Infect. Immun. 67:1508-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rajan, S., G. Cacalano, R. Bryan, A. J. Ratner, C. U. Sontich, A. van Heerckeren, P. Davis, and A. Prince. 2000. Pseudomonas aeruginosa induction of apoptosis in respiratory epithelial cells: analysis of the effects of cystic fibrosis transmembrane conductance regulator dysfunction and bacterial virulence factors. Am. J. Respir. Cell Mol. Biol. 23:304-312. [DOI] [PubMed] [Google Scholar]
- 78.Riese, M. J., A. Wittinghofer, and J. T. Barbieri. 2001. ADP-ribosylation of Arg41 of Rap by ExoS inhibits the ability of Rap to interact with its guanine nucleotide exchange factor, C3G. Biochemistry 40:3289-3294. [DOI] [PubMed] [Google Scholar]
- 79.Rizzuto, R., M. Brini, P. Pizzo, M. Murgia, and T. Pozzan. 1995. Chimeric green fluorescent protein as a tool for visualizing subcellular organelles in living cells. Curr. Biol. 5:635-642. [DOI] [PubMed] [Google Scholar]
- 80.Roy-Burman, A., R. H. Savel, S. Racine, B. L. Swanson, N. S. Revadigar, J. Fujimoto, T. Sawa, D. W. Frank, and J. P. Wiener-Kronish. 2001. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J. Infect. Dis. 183:1767-1774. [DOI] [PubMed] [Google Scholar]
- 81.Sato, H., D. W. Frank, C. J. Hillard, J. B. Feix, R. R. Pankhaniya, K. Moriyama, V. Finck-Barbancon, A. Buchaklian, M. Lei, R. M. Long, J. Wiener-Kronish, and T. Sawa. 2003. The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J. 22:2959-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schweizer, H. P. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97:109-121. [DOI] [PubMed] [Google Scholar]
- 83.Shaver, C. M., and A. R. Hauser. 2004. Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. Infect. Immun. 72:6969-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Smith, R. S., and B. H. Iglewski. 2003. P. aeruginosa quorum-sensing systems and virulence. Curr. Opin. Microbiol. 6:56-60. [DOI] [PubMed] [Google Scholar]
- 85.Sun, J., and J. T. Barbieri. 2003. Pseudomonas aeruginosa ExoT ADP-ribosylates CT10 regulator of kinase (Crk) proteins. J. Biol. Chem. 278:32794-32800. [DOI] [PubMed] [Google Scholar]
- 86.Sun, J., A. W. Maresso, J.-J. P. Kim, and J. T. Barbieri. 2004. How bacterial ADP-ribosylating toxins recognize substrates. Nat. Struct. Mol. Biol. 11:868-876. [DOI] [PubMed] [Google Scholar]
- 87.Tang, D., J. M. Lahti, and V. J. Kidd. 2000. Caspase-8 activation and Bid cleavage contribute to MCF7 cellular execution in a caspase-3-dependent manner during staurosporine-mediated apoptosis. J. Biol. Chem. 275:9303-9307. [DOI] [PubMed] [Google Scholar]
- 88.Tateda, K., Y. Ishii, M. Horikawa, T. Matsumoto, S. Miyairi, J. C. Pechere, T. J. Standiford, M. Ishiguro, and K. Yamaguchi. 2003. The Pseudomonas aeruginosa autoinducer N-3-oxododecanoyl homoserine lactone accelerates apoptosis in macrophages and neutrophils. Infect. Immun. 71:5785-5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Watt, A. P., J. Courtney, J. Moore, M. Ennis, and J. S. Elborn. 2005. Neutrophil cell death, activation, and bacterial infection in cystic fibrosis. Thorax 60:659-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wright, S. C., P. Kumar, A. W. Tam, N. Shen, M. Varma, and J. W. Larrick. 1992. Apoptosis and DNA fragmentation precede TNF-induced cytolysis in U937 cells. J. Cell Biochem. 48:344-355. [DOI] [PubMed] [Google Scholar]
- 91.Wright, S. C., Q. S. Wei, D. H. Kinder, and J. W. Larrick. 1996. Biochemical pathways of apoptosis: nicotinamide adenine dinucleotide-deficient cells are resistant to tumor necrosis factor or ultraviolet light activation of the 24-kD apoptotic protease and DNA fragmentation. J. Exp. Med. 183:463-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yahr, T. L., A. J. Vallis, M. K. Hancock, J. T. Barbieri, and D. W. Frank. 1998. ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc. Natl. Acad. Sci. USA 95:13899-13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yamada, T., M. Goto, V. Punj, O. Zaborina, K. Kimbara, T. K. Das Gupta, and A. M. Chakrabarty. 2002. The bacterial redox protein azurin induces apoptosis in J774 macrophages through complex formation and stabilization of the tumor suppressor protein p53. Infect. Immun. 70:7054-7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yao, F., and E. Eriksson. 1999. A novel tetracycline-inducible viral replication switch. Hum. Gene Ther. 10:419-427. [DOI] [PubMed] [Google Scholar]
- 95.Zhang, J., H. Takayama, T. Matsuba, R. Jiang, and Y. Tanaka. 2003. Induction of apoptosis in macrophage cell line, J774, by the cell-free supernatant from Pseudomonas aeruginosa. Microbiol. Immunol. 47:199-206. [DOI] [PubMed] [Google Scholar]
- 96.Zhang, L., H. Wang, S. C. Masters, B. Wang, J. T. Barbieri, and H. Fu. 1999. Residues of 14-3-3 zeta required for activation of exoenzyme S of Pseudomonas aeruginosa. Biochemistry 38:12159-12164. [DOI] [PubMed] [Google Scholar]
- 97.Zhang, Y., and J. T. Barbieri. 2005. A leucine-rich motif targets Pseudomonas aeruginosa ExoS within mammalian cells. Infect. Immun. 73:7938-7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhou, L., G. Jiang, G. Chan, C. P. Santos, D. W. Severson, and L. Xiao. 2005. Michelob_x is the missing inhibitor of apoptosis protein antagonist in mosquito genomes. EMBO Rep. 6:769-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhou, L., and H. Steller. 2003. Distinct pathways mediate UV-induced apoptosis in Drosophila embryo. Dev. Cell 4:599-605. [DOI] [PubMed] [Google Scholar]