Abstract
An upshift of 10°C or more in the growth temperature of an Escherichia coli culture causes induction of extra rounds of chromosome replication. This stress replication initiates at oriC but has functional requirements different from those of cyclic replication. We named this phenomenon heat-induced replication (HIR). Analysis of HIR in bacterial strains that had complete or partial oriC deletions and were suppressed by F integration showed that no sequence outside oriC is used for HIR. Analysis of a number of oriC mutants showed that deletion of the L-13-mer, which makes oriC inactive for cyclic replication, was the only mutation studied that inactivated HIR. The requirement for this sequence was strictly correlated with Benham's theoretical stress-induced DNA duplex destabilization. oriC mutations at DnaA, FIS, or IHF binding sites showed normal HIR activation, but DnaA was required for HIR. We suggest that strand opening for HIR initiation occurs due to heat-induced destabilization of the L-13-mer, and the stable oligomeric DnaA-single-stranded oriC complex might be required only to load the replicative helicase DnaB.
The initiation of chromosome replication is the central event in the bacterial cell cycle and the step that controls the frequency with which chromosomes are replicated. Initiation is a precisely timed event that occurs once during the cell cycle. In Escherichia coli replication initiates at a unique site, oriC, located at min 84.5 on the genetic map. The minimal sequence that promotes replication was found to be 258 bp long (25). The left side of oriC is an AT-rich region consisting of an AT cluster and three similar sequences, each 13 nucleotides long and each starting with GATC (1, 5). This AT-rich region provides the helical instability of oriC. Apparently, it is the AT richness that is important in this region, and except for the R-13-mer, the sequence can be replaced by different AT-rich sequences with similar properties (5, 6, 21). Within oriC there are eight 9-mer DnaA binding sites; five of these sites can bind either DnaA-ADP or DnaA-ATP with equal affinity (R boxes) (27), but the remaining three sites preferentially bind DnaA-ATP (I boxes) (23, 26). The minimal oriC sequence also contains 11 GATC sites specific for Dam methyltransferase that must be methylated for optimal origin function. In addition, there are binding sites for the DNA bending protein IHF, which assists in oriC unwinding, and the DNA bending protein Fis. There are also binding sites for IciA, ROB, SeqA, and Cnu/Hha among other proteins (18, 25).
Initiation of replication begins with the binding to oriC and cooperative interaction of 20 to 30 monomers of DnaA-ATP and bending of the DNA that promotes the unwinding of the AT-rich region (31). This strand opening also requires transcriptional activation (2, 29, 34) and is enhanced by a certain DNA topology (9, 30). DnaA-ATP then binds to the single-stranded DNA, providing the initial stabilization of the single-stranded state. The stable oligomeric DnaA-oriC complex is required for the interaction of DnaA with the two replicative helicase DnaB-DnaC hexamers and their loading into the unwound DNA (7, 28, 33). The binding of DnaB to the single-stranded DNA seems to facilitate dissociation of DnaC from the complexes. Subsequent loading of primase permits the recruitment of the rest of the proteins for replisome assembly.
In addition to cyclic replication during exponential growth, there are stress conditions that can induce extra chromosome replication. An increase in the environmental temperature of 10°C or more induces the initiation of extra rounds of replication (4, 13). We named this phenomenon heat-induced replication (HIR). This stress replication initiates at oriC, is not a heat shock response, and, in contrast to cyclic replication, requires neither RNA polymerase activity nor protein synthesis (4). Other physiological changes, such as DNA damage and a change in the growth medium, can induce replication by one of two ways, termed inducible stable DNA replication and constitutive stable DNA replication (19). HIR exhibits some homology with both types of stable DNA replication. The three replication modes do not require protein synthesis. In contrast to constitutive stable DNA replication, HIR requires active RNase H; in contrast to inducible stable DNA replication, HIR is independent of SOS induction; and, as we showed in this work, HIR initiates only in oriC and requires DnaA.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All of the strains used are derivatives of E. coli K-12 and are listed in Table 1. Bacteria were incubated at 30°C in M9 minimal medium containing 0.4% glucose, 20 μg/ml required amino acids, 2 μg/ml thiamine, and 0.1% Casamino Acids. Antibiotics, including 20 μg/ml ampicillin, 50 μg/ml kanamycin, and 15 μg/ml tetracycline, were added when appropriate.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant genotype | Other characteristics | Reference or source |
|---|---|---|---|
| CM1565 | ilv192 asnB32 relA1 spoT1 thi-1 lys | 35 | |
| CM1588 | CM1565 zif::pCM197 | F′ ilv+ | 35 |
| CM1671 | CM1588 ΔoriC1071 | Deletion from gidB to asnA | 35 |
| CM1793 | CM1565 zij::pKN500 | R1 derivative, asnA+ | 35 |
| CM1843 | CM1793 ΔoriC1071 | Deletion from gidB to asnA | 35 |
| CM735 | metE46 trp3 his4 thi1 | 8 | |
| CM735-intFs-B | CM735 ΔoriC::pAE15 | Nucleotides 23 to 38 deleted (L-13-mer), integratively suppressed by mini-F | 8 |
| ER | asnA asnB thi1 | 20 | |
| LK211 | ER ΔoriC::pKN1562 | Nucleotides 23 to 38 deleted (L-13-mer), integratively suppressed by R1 derivative | 20 |
| WM2482 | ilvG | MG1655 | 36 |
| WM2759 | WM2482 oriC160 | Δ275-352 | 36 |
| WM2762 | WM2482 oriC13 | R2 scrambled | 36 |
| WM2763 | WM2482 oriC14 | R3 scrambled | 36 |
| WM2764 | WM2482 oriC15 | R4 scrambled | 36 |
| WM2765 | WM2482 oriC21 | R1 at R3 position | 36 |
| WM2766 | WM2482 oriC131 | Mutant FIS site | 36 |
| WM2767 | WM2482 oriC132 | Mutant IHF site | 36 |
| WM2768 | WM2482 oriC136 | R4 inverted | 36 |
| WM2844 | WM2482 oriC17 | M scrambled | 36 |
| WM2845 | WM2482 oriC162 | +14 bp between R3 and R4 | 36 |
| JK607 | thyA arg his thi | Our lab | |
| JK876 | JK607 dnaA46 | dnaA46 | Our lab |
DNA synthesis and number of origins.
DNA synthesis was determined by growing cells in minimal medium containing 1 μg/ml thymidine and 1 μCi/ml [methyl-3H]thymidine (20 Ci/mmol; ICN) in the presence of 1.5 mM uridine. The isotope was added to both the overnight and exponentially growing cultures. Trichloroacetic acid-insoluble material from 0.2-ml aliquots was assayed using a Beckman LS3801 scintillation counter. Rifampin (150 μg/ml) was added to exponentially growing cultures to inhibit initiation of replication, and replication runout (ΔG at 30°C) was determined by measuring the [3H]thymidine that was incorporated over 4 h. All measurements were made relative to the radioactive count at the time that rifampin was added, which was always around 5,000 dpm. The accumulated DNA synthesis was proportional to the number of active replication forks, as in these growth conditions bacteria had overlapping replication cycles. The number of overlapping replication cycles (n) was determined from the formula ΔG = 2n × n × ln2/(2n − 1) − 1, as described by Sueoka and Yoshikawa (32).
Measurement of HIR.
HIR was determined by shifting a culture from 30 to 41°C and adding 150 μg/ml rifampin. Replication runout (ΔG at 41°C) was determined by measuring the [3H]thymidine that accumulated over 4 h. The number of induced origins was determined as described by Jiménez-Sánchez and Guzmán (17). Briefly, if i is the frequency of reinitiated origins after any inductive treatment resulting in complete chromosome replication and n is the number of replication cycles per chromosome in the exponentially growing culture at 30°C, the final amount of chromosomes is 2n(i + 1) and the replication runout is ΔG = 2n × (i + 1) × n × ln2/(2n − 1) − 1.
SIDD.
The probability that superhelically stressed DNA underwent strand separation events was analyzed by introducing DNA sequences into Benham's stress-induced DNA duplex destabilization (SIDD) program (http://genomics.ucdavis.edu/benham/sidd/index.php) and determining the probability profile using near-neighbor computing (3).
RESULTS
oriC is the only initiation site for HIR.
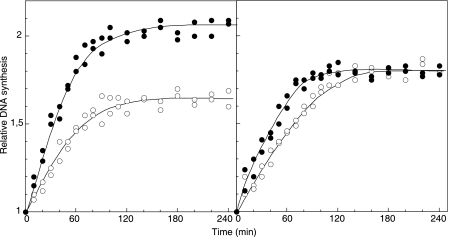
To study the strict requirement of oriC for HIR, we used strain CM1671, in which the complete oriC sequence from gidB to asnA was deleted. Inhibition of replication was suppressed by F plasmid integrated at min 85. Replication runout after inhibition of initiation with rifampin was used to determine the relative numbers of origins before and after the temperature shift from 30°C to 41°C in the control strain and the strain with oriC deleted (Fig. 1). We found HIR in about 24% of the origins present in parental strain CM1565 and in strain CM1588 with F integrated but not in strain CM1671 with oriC deleted (Table 2). Thus, HIR is initiated neither outside oriC nor in the F replication origins.
FIG. 1.
Replication runout of strains CM1588 (left panel) and CM1671 (right panel) after addition of rifampin at 30°C (○) or together with a shift to 41°C (•).
TABLE 2.
HIR of a strain with oriC deleted
| Strain | Characteristics | ΔG at 30°C | ΔG at 41°C | HIR (%) |
|---|---|---|---|---|
| CM1565 | oriC+ | 64 | 103 | 23.8 |
| CM1588 | oriC+zif::F | 65 | 105 | 24.2 |
| CM1671 | ΔoriC1071 zif::F | 80 | 80 | 0 |
Functional oriC mutations allow HIR.
In order to detect the specific oriC sequence required for HIR, a number of oriC mutant strains were used. These strains included strains with scrambled, shifted, and inverted DnaA boxes, altered IHF and FIS binding sites, deletion of nucleotides 275 to 352, and insertion of 14 nucleotides between R3 and R4. All the mutations tested induced HIR after the temperature upshift (Table 3). Some of the mutations tested affected the synchrony of cyclic initiation to different extents, but others did not affect the synchrony of cyclic initiation (36). All mutant strains induced about the same amount of HIR, and there was no detectable correlation with the asynchrony phenotype.
TABLE 3.
HIR of oriC mutant strains
| Strain | Characteristics | Asynchrony phenotypea | HIR (%) |
|---|---|---|---|
| WM2482 | MG1655 | None | 19.5 |
| WM2759 | Δ275-352 | None | 22.8 |
| WM2762 | R2 scrambled | Low | 19.2 |
| WM2763 | R3 scrambled | None | 20.0 |
| WM2764 | R4 scrambled | High | 20.0 |
| WM2765 | R1 at R3 position | None | 19.0 |
| WM2766 | Mutant FIS site | None | 22.0 |
| WM2767 | Mutant IHF site | Low | 19.5 |
| WM2768 | R4 inverted | None | 23.4 |
| WM2844 | M scrambled | Low | 20.4 |
| WM2845 | +14 bp between R3 and R4 | High | 10.1 |
| CM735 intFs-B | L-13-mer deleted, integratively suppressed by mini-F | High | 0 |
Induction of HIR requires the L-13-mer of oriC.
Deletion of the L-13-mer made oriC inactive and could be rescued only by integrative suppression. To test the induction of HIR in a mutant bacterium, we used strain CM735 intFs-B, in which the L-13-mer was replaced by a mini-F plasmid (8). No HIR was detected in this strain (Table 3). This shows that L-13-mer is a sequence that is required for HIR.
HIR can be induced in other replicons.
Above we show that HIR is specific for oriC and is not induced from the F plasmid. It might seem that this stress replication is specific for the E. coli oriC. To check for this specificity, we studied HIR in a strain with all of oriC deleted, CM1843, and in a strain with L-13-mer deleted, LK211, both suppressed by the integration of an R1 derivative plasmid. When these R1-suppressed strains were shifted from 30°C to 41°C, they exhibited high levels of HIR induction (Table 4). This indicates that a replicon other than oriC can perform HIR.
TABLE 4.
HIR at R1 replicon
| Strain | Characteristics | ΔG at 30°C | ΔG at 41°C | HIR (%) |
|---|---|---|---|---|
| CM1793 | oriC+zij::R1 | 60 | 95 | 21.8 |
| CM1843 | ΔoriC1071 zij::R1 | 60 | 100 | 25.0 |
| ER | oriC+ | 75 | 115 | 22.8 |
| LK211 | L-13-mer deleted, integratively suppressed by R1 | 81 | 120 | 21.5 |
Expected SIDD sites in oriC suggest that there is local DNA denaturation of L-13-mer for HIR initiation.
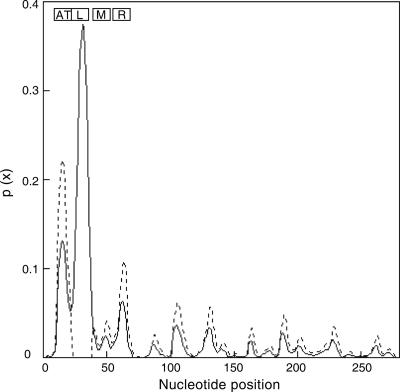
Local denaturation is a fundamental process required for initiation of replication, as well as other biological processes, such as transcription. We studied the SIDD of the wild-type oriC sequence and of mutants with mutations that did or did not affect HIR induction. The analysis of wild-type oriC showed that there was a high local probability of stress-inducible denaturation for a site located between positions 25 and 36 containing the L-13-mer (Fig. 2). This SIDD analysis performed with the mutant sequences, which conferred an HIR-inducible origin, revealed the same denaturation profile with the same local stress-destabilized site as the wild type. Deletion of the 16 bases from positions 23 to 38 eliminated the high probability of a stress-destabilized site, as expected. These results indicate that the heat-induced destabilization of the L-13-mer is an essential determinant for HIR initiation.
FIG. 2.
Benham's probability of stress-induced DNA duplex destabilization in wild-type oriC (solid line) or in the sequence with L-13-mer deleted (dashed line). The nucleotide positions are the positions described by Gille and Messer (11). The boxes show the positions of the AT cluster and the L-, M-, and R-13-mers.
HIR requires DnaA protein.
RNA polymerase activity is required at the initiation step of chromosome replication for local DNA destabilization, facilitating opening of the chromosome by DnaA (2, 5, 29). In previous work we showed that HIR does not require RNA polymerase activity (4). In this study we found that mutations in single DnaA binding sequences that allow oriC function also allow HIR. Consequently, we tested whether the DnaA protein was also required for HIR initiation. We used a dnaA46 mutant strain, which was thermosensitive at the initiation of chromosome replication due to the failure of DnaA46 in ATP binding and its thermolabile interaction with other proteins (14, 15, 16, 24). Table 5 shows that the presence of the mutant DnaA protein prevented HIR induction. This dependence of HIR on DnaA must be oriC specific, since R1 suppressed strain LK211, which did not require DnaA for initiation of replication, exhibited similar HIR values in a dnaA wild-type background and in a dnaA null mutant strain (data not shown).
TABLE 5.
DnaA requirement for HIR
| Strain | Characteristics | ΔG at 30°C | ΔG at 41°C | HIR (%) |
|---|---|---|---|---|
| JK607 | Wild type | 42 | 70 | 19.7 |
| JK876 | dnaA46 | 45 | 35 | 0 |
DISCUSSION
Thermal stress induces in all kinds of cells a number of new functions that counteract the adverse consequences of a sudden increase in the environmental temperature. The main physiological switch in E. coli is induction of the synthesis of σ32, which promotes expression of the heat shock genes (12). We reported previously that another consequence of thermal stress is induction of unscheduled replication, which we named HIR (4, 13). This stress replication was not a heat shock response, as it was not affected either by an rpoH deletion or by RNA or protein synthesis inhibition, and by using marker frequency analysis we determined that it initiates in oriC.
Here we found that HIR initiates in oriC and only in this site, as indicated by the following findings. First, a bacterial strain in which oriC was completely deleted and which replicated from an integrated F plasmid did not induce HIR. Second, deletion of the L-13-mer of oriC and insertion of a mini-F plasmid at this site, which made the chromosome replicate from the integrated plasmid replicon, did not induce HIR. These results show that the L-13-mer sequence is required for HIR and that no other sequence outside oriC is used for this stress-induced initiation of replication.
To look for other specific sequence requirements in oriC for HIR, we tested a number of mutants with altered DnaA, Fis, or IHF binding sites or with deletions and insertions that altered oriC functionality, such as the synchrony of the initiation step, but did not inhibit the initiation activity. All these mutant strains induced HIR, and no effect of the asynchrony phenotype on the number of stress-induced origins was observed.
DNA strand opening is the first step required for initiation of replication. The required oriC opening for the initiation of cyclic replication is accomplished by binding of the DnaA protein to the eight DnaA boxes in oriC with the aid of RNA polymerase and other proteins. Structural distortion of DNA is a prerequisite for the second function of DnaA in the initiation process, loading of the replicative helicase DnaB. Using chemically modified single-stranded DNA, Gille and Messer (11) obtained the first in vivo evidence that the region containing the three 13-mers in the left part of oriC is the first sequence that destabilizes during unwinding of oriC, and they supported the proposal that unwinding is initiated at the R-13-mer (5, 6). Our results obtained with deletion mutants show that HIR requires the L-13-mer, and in the absence of this sequence no other site is used to initiate this stress replication. The study of the SIDD of oriC by computer simulation as described by Fye and Benham (10) showed that there is a highly local stress-inducible denaturation site consisting of 12 nucleotides in the L-13-mer. In the absence of this sequence, no other SIDD site appears. This analysis, although not probative, correlates with our results for inhibition of HIR in the absence of L-13-mer. Consequently, we propose that HIR initiates by denaturation of the L-13-mer. This local denaturation could promote the unwinding of the nearby AT-rich region, assisted by the transient changes in DNA topology due to the thermal stress (22). We also found similar SIDD sequences in the oriF sequences of mini-F and in oriR1 of mini-R1 (data not shown), although an integrated F plasmid showed no detectable HIR. RepE initiator protein-mediated incC-oriF DNA looping negatively regulates F plasmid replication by inhibiting the formation of an open complex at oriF sequences in vitro (37). We suggest that the oriF SIDD sequence is not available for duplex destabilization as a result of steric hindrance, due to RepE-oriF complex formation, and therefore is not available for HIR.
Thermal DNA unwinding might supersede the requirement for the melting action of DnaA assisted by RNA polymerase. In previous work we showed that this stress replication does not require RNA polymerase for initiation (4). In the present work we found that DnaA activity is required for HIR in oriC. Consequently, we suggest that the initial local denaturation induced by the heat stress is located at the L-13-mer and that a functional oriC is required for HIR initiation. In this initiation step the DnaA protein might not be required for strand opening, as strand opening does not require the RNA polymerase destabilization function, but DnaA might be required for other initiation functionality, such as DnaB loading.
Acknowledgments
We are grateful to A. Løbner-Olesen, K. Nordström, and J. Zyskind for providing bacterial strains, to A. Løbner-Olesen and D. Chattoraj for critically reading the manuscript, and to Encarna Ferrera for technical help.
This work was supported by grant BMC2002-00830 from the Ministerio de Educación y Ciencia, Spain. R.G.-S. acknowledges a studentship from FPI, Ministerio de Educación y Ciencia, Spain.
Footnotes
Published ahead of print on 15 September 2006.
REFERENCES
- 1.Asai, T., M. Takanami, and M. Imai. 1990. The AT richness and gid transcription determine the left border of the replication origin of the E. coli chromosome. EMBO J. 9:4065-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, T. A., and A. Kornberg. 1988. Transcriptional activation of initiation of replication from the E. coli chromosomal origins: an RNA-DNA hybrid near oriC. Cell 55:113-123. [DOI] [PubMed] [Google Scholar]
- 3.Bi, C.-P., and C. J. Benham. 2004. WebSIDD: server for prediction of the stress-induced duplex destabilized sites in superhelical DNA. Bioinformatics 20:1477-1479. [DOI] [PubMed] [Google Scholar]
- 4.Botello, E., and A. Jiménez-Sánchez. 1997. A temperature upshift induces initiation of replication at oriC on the Escherichia coli chromosome. Mol. Microbiol. 26:133-144. [DOI] [PubMed] [Google Scholar]
- 5.Bramhill, D., and A. Kornberg. 1988. Duplex opening by DnaA protein at novel sequences in initiation of replication at the origin of E. coli chromosome. Cell 52:743-755. [DOI] [PubMed] [Google Scholar]
- 6.Bramhill, D., and A. Kornberg. 1988. A model for initiation at origins of DNA replication. Cell 54:915-918. [DOI] [PubMed] [Google Scholar]
- 7.Carr, K. M., and J. M. Kaguni. 1996. The A184V missense mutation of the dnaA5 and dnaA46 alleles confers a defect in ATP binding and thermolability in initiation of Escherichia coli DNA replication. Mol. Microbiol. 20:1307-1318. [DOI] [PubMed] [Google Scholar]
- 8.Eliasson, A., R. Bernander, and K. Nordstrom. 1996. Random initiation of replication of plasmids P1 and F (oriS) when integrated into the Escherichia coli chromosome. Mol. Microbiol. 20:1025-1032. [DOI] [PubMed] [Google Scholar]
- 9.Filutowicz, M., and J. Jonczyk. 1981. Essential role of the gyrB gene product in the transcriptional event coupled to DnaA-dependent initiation of E. coli chromosome replication. Mol. Gen. Genet. 183:134-138. [DOI] [PubMed] [Google Scholar]
- 10.Fye, R. M., and C. J. Benham. 1999. Exact method for numerically analyzing a model of local denaturation in superhelically stressed DNA. Phys. Rev. 59:2408-2426. [Google Scholar]
- 11.Gille, H., and W. Messer. 1991. Localized DNA melting and structural perturbations in the origin of replication, oriC, of Escherichia coli in vitro and in vivo. EMBO J. 10:1579-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gross, C. A. 1996. Function and regulation of the heat shock proteins, p. 1382-1399. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 13.Guzmán, E. C., A. Jiménez-Sánchez, E. Orr, and R. H. Pritchard. 1988. Heat stress in the presence of low RNA polymerase activity increases chromosome copy number in E. coli. Mol. Gen. Genet. 212:203-206. [DOI] [PubMed] [Google Scholar]
- 14.Hansen, F. G., S. Koefoed, and T. Atlung. 1992. Cloning and nucleotide sequence determination of twelve mutant dnaA genes of Escherichia coli. Mol. Gen. Genet. 234:14-21. [DOI] [PubMed] [Google Scholar]
- 15.Hansen, F. G., and K. V. Rasmussen. 1977. Regulation of the dnaA product in Escherichia coli. Mol. Gen. Genet. 155:219-225. [DOI] [PubMed] [Google Scholar]
- 16.Hwang, D. S., and J. M. Kaguni. 1988. Interaction of dnaA46 protein with a stimulatory protein in replication from the Escherichia coli chromosomal origin. J. Biol. Chem. 263:10633-10640. [PubMed] [Google Scholar]
- 17.Jiménez-Sánchez, A., and E. C. Guzmán. 1988. Direct procedure for the determination of the number of replication forks and the reinitiation fraction in bacteria. Comput. Appl. Biosci. 4:431-433. [DOI] [PubMed] [Google Scholar]
- 18.Kim, M. S., S.-H. Bae, S. H. Yun, H. J. Lee, S. C. Ji, J. H. Lee, P. Srivastava, S.-H. Lee, H. Chae, Y. Lee, B.-S. Choi, D. Chattoraj, and H. M. Lim. 2005. Cnu, a novel oriC-binding protein of Escherichia coli. J. Bacteriol. 187:6998-7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kogoma, T. 1997. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol. Mol. Biol. Rev. 61:212-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koppes, L., and K. Nordstrom. 1986. Insertion of an R1 plasmid into the origin of replication of the E. coli chromosome: random timing of replication of the hybrid chromosome. Cell 44:117-124. [DOI] [PubMed] [Google Scholar]
- 21.Kowalski, D., and M. J. Eddy. 1989. The DNA unwinding element: a novel, cis-acting component that facilitates opening of the Escherichia coli replication origin. EMBO J. 8:4335-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.López-García, P., and P. Forterre. 2000. DNA topology and the thermal stress response, a tale from mesophiles and hyperthermophiles. BioEssays 22:738-746. [DOI] [PubMed] [Google Scholar]
- 23.McGarry, K. C., V. T. Ryan, J. E. Grimwade, and A. C. Leonard. 2004. Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by the initiator DnaA-ATP. Proc. Natl. Acad. Sci. USA 101:2811-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Messer, W., F. Blaesing, J. Majka, J. Nardmann, S. Schaper, A. Schmidt, H. Seitz, C. Speck, D. Tüngler, G. Wegrzyn, C. Weigel, M. Welzeck, and J. Zakrzewska-Czerwinska. 1999. Functional domains of DnaA proteins. Biochimie 81:819-825. [DOI] [PubMed] [Google Scholar]
- 25.Messer, W., and C. Weigel. 1996. Initiation of chromosome replication, p 1579-1601. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 26.Ryan, V. T., J. E. Grimwade, C. J. Nievera, and A. C. Leonard. 2002. IHF and HU stimulate assembly of pre-replication complexes at Escherichia coli oriC by two different mechanisms. Mol. Microbiol. 46:113-124. [DOI] [PubMed] [Google Scholar]
- 27.Schaper, S., and W. Messer. 1995. Interaction of the initiator protein DnaA of Escherichia coli with its DNA target. J. Biol. Chem. 270:17622-17626. [DOI] [PubMed] [Google Scholar]
- 28.Seitz, H., C. Weigel, and W. Messer. 2000. The interaction domains of the DnaA and DnaB replication proteins of Escherichia coli. Mol. Microbiol. 37:1270-1279. [DOI] [PubMed] [Google Scholar]
- 29.Skarstad, K., T. A. Baker, and A. Kornberg. 1990. Strand separation required for initiation of replication at the chromosomal origin of E. coli is facilitated by a distant RNA-DNA hybrid. EMBO J. 9:2341-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smelkova, N., and K. J. Marians. 2001. Timely release of both replication forks from oriC requires modulation of origin topology. J. Biol. Chem. 276:39186-39191. [DOI] [PubMed] [Google Scholar]
- 31.Speck, C., and W. Messer. 2001. Mechanism of origin unwinding: sequential binding of DnaA to double- and single-stranded DNA. EMBO J. 20:1469-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sueoka, N., and H. Yoshikawa. 1965. The chromosome of Bacillus subtilis. I. Theory of marker frequency analysis. Genetics 52:747-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sutton, M. D., K. V. Carr, M. Vicente, and J. M. Kaguni. 1998. Escherichia coli DnaA protein: the N-terminal domain and loading of DnaB helicase at the E. coli chromosomal origin. J. Biol. Chem. 273:34255-34262. [DOI] [PubMed] [Google Scholar]
- 34.van der Ende, A., T. A. Baker, T. Ogawa, and A. Kornberg. 1985. Initiation of enzymatic replication at the origin of the Escherichia coli chromosome: primase as the sole priming enzyme. Proc. Natl. Acad. Sci. USA 82:3954-3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.von Meyenburg, K., and F. G. Hansen. 1980. The origin of replication, oriC, of the Escherichia coli chromosome: genes near oriC and construction of oriC deletion mutations, p 137-159. In B. Alberts (ed.), Mechanistic studies of DNA replication and genetic recombination. Academic Press Inc., New York, N.Y.
- 36.Weigel, C., W. Messer, S. Preiss, M. Welzeck, Morigen, and E. Boye. 2001. The sequence requirements for a functional Escherichia coli replication origin are different for the chromosome and a minichromosome. Mol. Microbiol. 40:498-507. [DOI] [PubMed] [Google Scholar]
- 37.Zzaman, S., and D. Bastia. 2005. Oligomeric initiator protein-mediated DNA looping negatively regulates plasmid replication in vitro by preventing origin melting. Mol. Cell 20:833-843. [DOI] [PubMed] [Google Scholar]