Abstract
Secondary metabolites are important factors for interactions between bacteria and other organisms. Pseudomonas chlororaphis PCL1391 produces the antifungal secondary metabolite phenazine-1-carboxamide (PCN) that inhibits growth of Fusarium oxysporum f. sp. radius lycopersici the causative agent of tomato foot and root rot. Our previous work unraveled a cascade of genes regulating the PCN biosynthesis operon, phzABCDEFGH. Via a genetic screen, we identify in this study a novel TetR/AcrR regulator, named Pip (phenazine inducing protein), which is essential for PCN biosynthesis. A combination of a phenotypical characterization of a pip mutant, in trans complementation assays of various mutant strains, and electrophoretic mobility shift assays identified Pip as the fifth DNA-binding protein so far involved in regulation of PCN biosynthesis. In this regulatory pathway, Pip is positioned downstream of PsrA (Pseudomonas sigma factor regulator) and the stationary-phase sigma factor RpoS, while it is upstream of the quorum-sensing system PhzI/PhzR. These findings provide further evidence that the path leading to the expression of secondary metabolism gene clusters in Pseudomonas species is highly complex.
Among gram-negative bacteria, pseudomonads are known to produce a wide variety of secondary metabolites, such as toxins (35), rhamnolipids (25, 27), hydrogen cyanide (HCN) (28) and phenazines (6). In contrast to primary metabolites, secondary metabolites are not essential for growth and reproduction. However, many of them play an important role in interactions between Pseudomonas species and other organisms, particularly during pathogenesis and biocontrol. For example pyocyanin produced by Pseudomonas aeruginosa is suggested to be involved in lung infection of cystic fibrosis patients (19), whereas Phl (2,4-diacylphloroglucinol) and HCN produced by Pseudomonas fluorescens protect tobacco plants from black root rot (20). The elucidation of how secondary metabolism is regulated is therefore relevant for medicine, agriculture, and industry.
In most species, the GacS/GacA two-component system is a global regulator of secondary metabolism, for example, for the production of HCN and Phl (20), the production of exoprotease and phospholipase C (30), and the production of phenazine (29). After binding of an unknown signal, the membrane-associated sensor GacS activates the GacA transcriptional regulator by phosphorylation (13, 38). Direct targets of GacA are so far unknown. In addition to GacA/GacS, quorum-sensing also regulates secondary metabolism in many species. Quorum-sensing involves a LuxI homologue synthesizing N-acyl-homoserine lactone signal molecules (N-AHLs), which are able to traffic across membranes. Their extracellular concentration reflects the number of bacteria present in a (semi-) closed environment. N-AHLs bind to a LuxR homologue, thereby activating it. Activated LuxR homologues function as transcriptional regulators. Thus, N-AHLs enable bacteria to sense the density of their population and to induce specific (sets of) genes (9, 23).
Phenazine-1-carboxamide (PCN) is a secondary metabolite produced by Pseudomonas chlororaphis PCL1391, which suppresses tomato foot and root rot caused by Fusarium oxysporum f. sp. radicis lycopersici (5). PCN production and efficient root colonization for delivering PCN in the rhizosphere are crucial traits for the biocontrol ability of strain PCL1391 (4). Understanding the components regulating the synthesis of PCN is likely to give new insights in regulation of bacterial secondary metabolism in general.
Production of PCN was shown to be regulated by an intrinsic regulatory network, according to the following observations. (i) The GacS/GacA system activates a cascade of regulators upstream of the phz biosynthetic operon (6, 10). (ii) PsrA (Pseudomonas sigma factor regulator) was shown to be part of the PCN regulatory cascade. (iii) PsrA controls the production of the stationary-phase sigma factor RpoS (10). (iv) Downstream of RpoS, the LuxI homologue PhzI synthesizes N-hexanoyl-homoserine lactone (C6-HSL), the N-AHL that is supposed to bind to the LuxR homologue transcriptional regulator PhzR. Activated PhzR binds, in turn, to the lux box upstream of the phz operon, which is responsible for the synthesis of PCN at the onset of the stationary phase. Our previous results showed that a constitutively activated quorum-sensing system, PhzI/PhzR, is sufficient for synthesis of PCN when other regulators are mutated (10).
Here we describe the identification of pip (phenazine inducing protein), a novel gene that is involved in controlling PCN synthesis. Our results show that Pip, a putative transcriptional regulator, acts downstream of PsrA and RpoS and stimulates the expression of the phz operon via the quorum-sensing system PhzI/PhzR.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Pseudomonas strains were cultured at 28°C in liquid MVB1 (34), LC (10), or King's medium B (16) and shaken at 195 rpm on a Janke and Kunkel shaker KS501D (IKA Labortechnik, Staufen, Germany). Escherichia coli strains were grown at 37°C in LC medium under vigorous aeration. Media were solidified with 1.8% Bacto agar (Difco, Detroit, MI). When appropriate, growth medium was supplemented with kanamycin (50 μg/ml), carbenicillin (200 μg/ml), gentamicin (30 μg/ml), 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (40 μg/ml), or hexanoyl-homoserine lactone (C6-HSL) (5 μM) (Fluka, Sigma-Aldrich, Zwijndrecht, The Netherlands). To follow growth, the absorbance of liquid cultures diluted 10-fold was measured at 620 nm.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Characteristics and descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| P. chlororaphis | ||
| PCL1391 | wt; producing phenazine-1-carboxamide; biocontrol strain of tomato foot and root rot caused by F. oxysporum f. sp. radicis-lycopersici | 5 |
| PCL1103 | phzI; derivative of PCL1391 in which a promoterless Tn5luxAB has been inserted in phzI; Kmr | 6 |
| PCL1104 | phzR; derivative of PCL1391 in which a promoterless Tn5luxAB has been inserted in phzR; Kmr | 6 |
| PCL1111 | psrA; derivative of PCL1391 in which a promoterless Tn5luxAB has been inserted in psrA; Kmr | 7 |
| PCL1114 | pip; derivative of PCL1391 in which a promoterless Tn5luxAB has been inserted in pip; Kmr | This study |
| PCL1123 | gacS; derivative of PCL1391 in which a promoterless Tn5luxAB has been inserted in gacS; Kmr | 7 |
| PCL1954 | rpoSSHR; derivative of PCL1391; rpoS::pMP7418; Kmr | 10 |
| PCL1955 | rpoSSHR; Ptac rpoS; derivative of PCL1955 containing pMP7420; Kmr,Gmr | 10 |
| PCL1962 | psrA (empty vector); derivative of PCL1111 containing pBBR1-MCS5; Kmr Gmr | 10 |
| PCL2001 | phzR (empty vector); derivative of PCL1104 containing pBBR1-MCS5; Kmr Gmr | 10 |
| PCL2008 | pipSHR; derivative of PCL1391; pip::pMP7451; Kmr | This study |
| PCL2011 | pip (empty vector); derivative of PCL1114 containing pBBR1-MCS5; Kmr Gmr | This study |
| PCL2012 | pip Ptac pip; derivative of PCL1114 containing pMP7455; Kmr Gmr | This study |
| PCL2013 | pip Ptac phzR; derivative of PCL1114 containing pMP7447; Kmr Gmr | This study |
| PCL2019 | wt; Ptac pip; derivative of PCL1391 containing pMP7455; Gmr | This study |
| PCL2036 | pip Ptac rpoS; derivative of PCL1114 containing pMP7420; Kmr Gmr | This study |
| PCL2038 | psrA Ptac pip; derivative of PCL1111 containing pMP7455; Kmr Gmr | This study |
| PCL2040 | rpoSSHR Ptac pip; derivative of PCL1954 containing pMP7455; Kmr Gmr | This study |
| PCL2082 | phzI (empty vector); derivative of PCL1103 containing pBBR1MCS-5; Kmr Gmr | This study |
| PCL2083 | phzI Ptac phzR; derivative of PCL1103 containing pMP7447; Kmr Gmr | This study |
| PCL2085 | pip Ppip pip; derivative of PCL1114 containing pMP7487; Kmr Gmr | This study |
| PCL2086 | phzR Ppip pip; derivative of PCL1104 containing pMP7487; Kmr Gmr | This study |
| PCL2087 | psrA Ppip pip; derivative of PCL1111 containing pMP7487; Kmr Gmr | This study |
| PCL2089 | wt; Ppip pip; derivative of PCL1391 containing pMP7487; Gmr | This study |
| C. violaceum CV026 | Double mini-Tn5 mutant from C. violaceum ATCC 31532; AHL biosensor | 24 |
| Escherichia coli DH5α | λ− φ80dlacZΔM15Δ(lacZYA-argF)U169recA1 endA1hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | 12 |
| Plasmids | ||
| pRL1063a | Harboring promoterless Tn5luxAB transposon; Kmr | 36 |
| pRK2013 | Helper plasmid for triparental mating | 8 |
| pIC20H | General purpose cloning vector; Cbr | 22 |
| pGEM-T easy | Plasmid designed for direct ligation of PCR fragments | Promega |
| pBBR1MCS-5 | Empty vector; general purpose cloning vector; Gmr | 17 |
| pMP5285 | Suicide vector for Pseudomonas spp.; used for homologous recombination; Kmr Cbr | 18 |
| pMP7420 | pBBR1MCS-5 containing the rpoS gene of PCL1391 downstream of the Ptac promoter, obtained by EcoRI digestion of pMP7424 | 10 |
| pMP7444 | pRL1063a containing pip and flanking regions; Kmr | This study |
| pMP7447 | PtacphzR; pBBR1MCS-5 containing the phzR gene of PCL1391 under control of the Ptac promoter, inserted between the XhoI and EcoRI sites | 10 |
| pMP7451 | pMP5285 containing an internal 350-bp PCR fragment of pip | This study |
| pMP7455 | Ptacpip; pBBR1MCS-5 containing the pip gene of PCL1391 under control of the Ptac promoter | This study |
| pMP7465 | PtacpsrA; pBBR1MCS-5 containing the psrA gene of PCL1391 under control of the Ptac promoter | 10 |
| pMP7487 | Ppippip; pBBR1MCS-5 containing the pip gene of PCL1381 under control of its own promoter | This study |
See Materials and Methods for an explanation of the notation.
Isolation and sequence analysis of chromosomal regions flanking the transposon in the pip mutant PCL1114.
A transposon library was obtained by transformation of strain PCL1391 with the plasmid pRL1063a (6). The Tn5 transposon of pRL1063a contains an origin of replicationthat functions in E. coli (36). Chromosomal DNA was isolated from PCL1114, digested with EcoRI, religated, and transferred into E. coli by transformation. One clone was picked among the colonies obtained after kanamycin resistance selection. The plasmid containing the regions flanking the transposon was named pMP7444 and sequenced using primers oMP458 (5′-TACTAGATTCAATGCTATCAATGAG-3′) and oMP459 (5′-AGGAGGTCACATGGAATATCAGAT-3′). Similarity and domain searches were performed using BLAST (http://www.ncbi.nih.gov/BLAST). A search for bacterial promoters and terminators was done using Softberry (http://www.softberry.com). Alignments of amino acid sequences were obtained using the ClustalW software (http://www.ch.embnet.org/software/ClustalW.html).
Recombinant DNA techniques.
General DNA techniques were performed as described previously (31). PCRs were carried out with Super Taq enzyme (Enzyme Technologies Ltd., United Kingdom). Only for the construction of pip under control of the Ptac or Ppip promoter were PCRs performed using Phusion from Finnzymes (Bioké, Leiden, The Netherlands). Primers were synthesized by Isogen Life Science (Maarssen, The Netherlands). Restriction enzymes were purchased from New England BioLabs, Inc. (Westburg, Leusden, The Netherlands) and T4 DNA ligase was from Promega (Leiden, The Netherlands).
Construction of plasmids and PCL1391 mutant strains.
In order to construct a suicide plasmid for disruption of pip by single homologous recombination, an internal pip fragment of 350 bp was obtained by PCR on PCL1391 chromosomal DNA with the primers oMP814 (5′-ATATATGAATTCCCGGCGCTCGGGTGGATGCC-3′) and oMP815 (5′-ATATATGAATTCTCTCGCCCAGGGCATGGAGG-3′). The PCR fragment was cloned in the EcoRI site of the vector pMP5285. The obtained suicide vector was named pMP7451 and introduced into PCL1391 by triparental mating using the helper plasmid pRK2013. The resulting mutant was named PCL2008. PCL2008 is impaired in PCN and C6-HSL production, like PCL1114 (data not shown), confirming that the phenotype of PCL1114 is not due to a secondary mutation in the genome.
In order to constitutively express pip, a plasmid was constructed harboring pip under control of the constitutive Ptac promoter. Two primers were designed according to the pip sequence obtained from pMP7444: oMP816 (5′-ATATATGAATTCTTGACAATTAATCATCGGCTCGTATAATGTGTGGAATTGTGAGCGGATAACAATTTTCACACAGGAAACAGCTAAATGACAATGACCACAGAACTCTCCGTAGTGCCC-3′), which contained the Ptacpromoter, and oMP817 (5′-ATATATGAATTCAGGATGCGGTTGAAACCCTGTGCCGCG-3′).These primers were used for PCR on chromosomal DNA of PCL1391. The obtained fragment was cloned in the EcoRI site of pBBR1MCS-5. The resulting vector was named pMP7455 and was introduced into PCL1114 by triparental mating to obtain PCL2012. The cloning vector pBBR1MCS-5 was introduced into PCL1114 in order to obtain the control strain PCL2011.
For overexpression of phzR, PCL1114 was transformed with pMP7447 (10) to obtain PCL2013. For overexpression of rpoS, PCL1114 was transformed with pMP7420 (10) to obtain PCL2036. For overexpression of psrA, PCL1114 was transformed with pMP7465 (10) to obtain PCL2046. In order to study the effect of overexpression of pip on phzR expression, PCL1104 (6) was transformed with pMP7455, which resulted in PCL2035.
The pip gene was also constitutively expressed in wild-type, psrA, and rpoS mutant backgrounds. Therefore, PCL1391, PCL1111 (7), and PCL1954 (10) were transformed with the plasmid pMP7455 by triparental mating, and the resulting strains were named PCL2019, PCL2038, and PCL2040, respectively.
Primers oMP1045 (5′-ATATATGAATTCGAGGTCAGCCGGGCCAAGGAG-3′) and oMP817 were used for PCR on chromosomal DNA of PCL1391 with Phusion enzyme (Finnzymes) to obtain pip with 424 nucleotides of the sequence upstream of its start codon. The 1.1-kb product was cloned in the EcoRI site of pBBR1MCS-5. The orientation of the insert was verified by PCR, and a clone was selected in which pip and the β-galactosidase gene of pBBR1MCS-5 have opposite directions of transcription. This plasmid was named pMP7487 and verified by sequencing. Strains PCL1391, PCL1114, PCL1104, and PCL1111 (pip, phzR, and psrA mutants, respectively) were transformed with pMP7487 to obtain strains PCL2089, PCL2085, PCL2086, and PCL2087, respectively.
Extraction and analysis for phenazine and N-AHL.
Phenazine extraction was carried out on supernatants of 10-ml liquid MVB1 cell cultures at regular time points during growth and/or after overnight growth as described previously (34). For N-AHL extraction, supernatants from 50-ml MVB1 cultures were harvested at an optical density at 620 nm (OD620) of 3.0 and mixed with 0.7 volume of dichloromethane and shaken for 45 min, after which the organic phase was collected. The extract was dried using a rotary evaporator. The dried residue was redissolved in 25 μl of acetonitrile and spotted on RP-C18 thin-layer chromatography (TLC) plates (Merck, Darmstadt, Germany). The TLC plates were developed in methanol-water (60:40, vol:vol). For detection of N-AHLs, the TLC was overlaid with 0.8% agar LC containing a 10-fold dilution of overnight culture of the Chromobacterium violaceum indicator strain CV026 (24) and kanamycin (50 μg/ml). After incubation for 48 h at 28°C, chromatograms were analyzed for the appearance of violet spots, indicating the presence of N-AHLs.
Expression analysis of bioluminescent Tn5luxAB reporter strains.
Expression of pip was monitored in various derivatives making use of the luxAB reporter genes of the Tn5 derivative in PCL1114. Expression was determined by quantification of bioluminescence during growth. Cells from overnight MVB1 cultures were washed with fresh medium and diluted to an OD620 of 0.1 in 10 ml of fresh MVB1 medium. During growth, the OD620 was measured at regular intervals, and 100-μl samples were taken in duplicate to quantify luminescence. A volume of 100 μl of N-decyl-aldehyde substrate solution (0.2% N-decyl-aldehyde [Sigma, St. Louis, MO] in a 2% bovine serum albumin solution) was added, and after 5 min of incubation at room temperature bioluminescence was determined with a MicroBeta 1450 TriLux luminescence counter (Wallac, Turku, Finland) and normalized to the luminescence per OD620 unit.
Western blot analysis.
Ten milliliters of MVB1 medium was inoculated with an overnight culture washed with fresh medium at an OD620 of 0.1. Cells were harvested at an OD620 of 1.0 or 2.2 in volumes corrected for equal cell amounts. Cell pellets were resuspended in 200 μl of cracking buffer (50 mM Tris-HCl, pH 6.8, 1% sodium dodecyl sulfate, 2 mM EDTA, 10% glycerol, 0.01% bromophenol blue, 1% β-mercaptoethanol) and boiled for 3 min. The samples were subsequently loaded on a 10% sodium dodecyl sulfate-polyacrylamide gel, and proteins were separated and blotted following a standard Western blotting procedure (1). A dried aliquot of RpoS antibody was kindly provided by K. Tanaka (Tokyo, Japan). This sample was resuspended in 100 μl of phosphate-buffered saline, diluted 1,000-fold, and allowed to react with the blot. The blots were subsequently incubated with peroxidase-labeled goat anti-rabbit antiserum (Amersham Biosciences, Roosendaal, The Netherlands). Finally, blots were incubated in a solution of 250 μM sodium luminol (Sigma) in 0.1 M Tris-HCl, (pH 8.6), and 0.01% H2O2 mixed with 60 μl of enhancer solution (67 μM p-hydroxy coumaric acid [Sigma] in dimethyl sulfoxide). Protein bands were detected using a Super R-X photographic film (Fujifilm, Düsseldorf, Germany).
Shift assays.
A PCR product of the upstream region of pip was obtained using the primers oMP1116 (5′-CCAAGTTGTAGGAGTTTCGTAAC-3′) and oMP1117 (5′-TGTGGTCATTGTCATTCTGGG-3′) with pMP7444 as template and the Phusion DNA polymerase (Finnzymes). After purification on QIAquick columns (QIAGEN, Westburg, Leusden, The Netherlands), the PCR product was labeled with [γ-32P]ATP using polynucleotide kinase (Fermentas, St. Leon-Rot, Germany) and purified over MicroSpin S-200 HR columns (GE Healthcare, Roosendaal, The Netherlands).
Cellular extracts of several P. chlororaphis derivatives were produced using the following method. Fifty milliliters of fresh MVB1 medium was inoculated with washed cells from overnight cultures at an initial OD620 of 0.1. Cells were harvested by spinning down cultures at an OD620 of 1.0 for 15 min at 3,000 rpm at 4°C. The pellets were resuspended in 2 ml of B-PER bacterial protein extraction reagent (Pierce, Perbio Science, Etten-Leur, The Netherlands) and gently shaken at room temperature for 15 min. The samples were centrifuged at 25,000 rpm for 30 min at 4°C in a Centrikon T-2070 ultracentrifuge (Kontron Instruments, Beun-De Ronde, Abcoude, The Netherlands). Supernatants (S30 fractions) were collected and frozen at −80°C in 10% glycerol for later use in binding reactions.
Reactions were performed in a 10-μl final volume, containing 50 mM Tris-HCl (pH 7.6), 60 mM NH4Cl, 7 mM MgCl2, 0.9 ng of 32P-labeled PCR product (1 nM), and purified Pip-His6 protein and/or S30 extracts as indicated. A 1,000-fold excess of genomic DNA was present in the samples to avoid nonspecific DNA-protein interactions. After a 20-min incubation at room temperature, samples were supplemented with 10% glycerol and loaded on an 8% polyacrylamide gel electrophoresis gel in 20 mM Tris-borate (pH 7.6) and run in the same buffer. Radioactivity was visualized by phosphor imaging (Bio-Rad, Veenendaal, The Netherlands).
Computational prediction of Pip target genes.
Search for conserved motifs in the upstream region of pip orthologues of P. aeruginosa PAO1, P. chlororaphis PCL1391, P. fluorescens Pf-5, P. fluorescens PfO1, Pseudomonas putida KT2440, Pseudomonas syringae B728a, and P. syringae pv. tomato str. DC3000, was performed using the MEME program (2) (available at http://meme.sdsc.edu/meme/meme.html). A 47-nucleotide (nt) sequence (with the consensus CGCCATCGCGGCTTCCTTCGCTGGGCGGCGCGCCCCATAATCGCCCG) was proposed as the best-conserved motif among orthologous pip upstream regions. In order to identify similar conserved patterns and therefore potential Pip target genes in genomes of pseudomonads, we generated a position weight matrix from the set of the seven conserved sequences (see Table S1 in the supplemental material) deduced from the MEME program using the Target Explorer web tool (33) (available at http://trantor.bioc.columbia.edu/Target_Explorer/). The maximum score that a 47-nt sequence could obtain with the “Pip” scoring matrix is 39.44 bits, and the minimum score is −95.04 bits. We used the generated matrix to scan the partial genome of P. chlororaphis PCL1391, and scoring of potential binding sites is based on the program PATSER (14). The cutoff score was fixed to 10 bits, 10.23 bits below the minimum score for the training set of sequences (20.23 bits for Pip of P. putida KT2440) in order to allow identification of sequences with several mismatches versus the consensus described above. No sequences with significant scores (similar to those obtained by the 47-nt motifs of upstream pip genes) were recovered. Identical results were obtained from P. aeruginosa PAO1 and P. fluorescens Pf-5 genome scans. As an alternative, the same in silico approach was applied using the Predict Regulon Server (37) (available at http://210.212.212.6/cgi-bin/regsites/predictregulonv1.pl). This program, although similar to Target Explorer, is less restrictive, but predictions contain many more false-positive hits than the former one. This program predicted in P. aeruginosa PAO1 eight sequences that contain patterns similar to the one observed upstream of Pip (see Table S2 in the supplemental material).
Nucleotide sequence accession number.
The pip sequence determined in this study was given accession number DQ311664.
RESULTS
For clarity in the presentation of results, strains will be described as, for example, the phzR Ptac pip strain, where the first gene indicates a genomic modification and the second is the gene cloned in the vector pBBR1MCS-5 and added in trans. In general, genomic mutations are due to transposon insertion of the luxAB gene, via the plasmid pRL1063a (Table 1). Some mutations were made by single homologous recombination (SHR) and are indicated as, for example, rpoSSHR (Table 1).
General characteristics of pip. (i) Isolation of mutant unable to produce PCN (strain PCL1114).
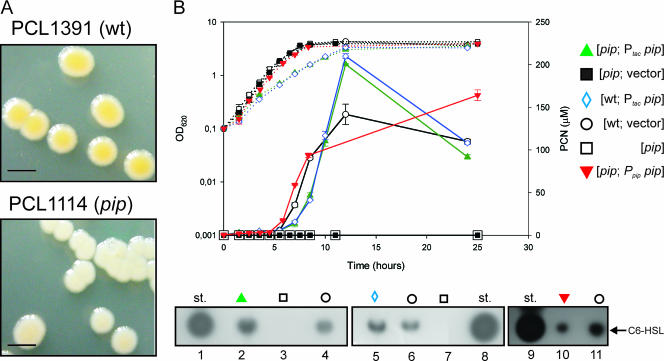
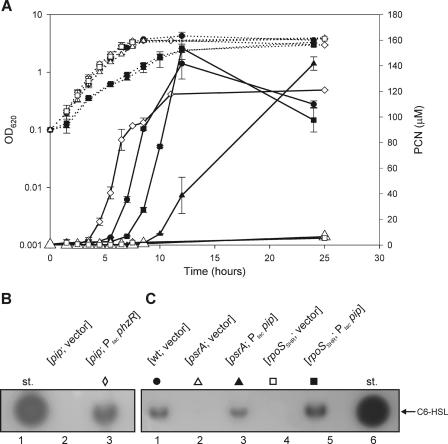
A transposon library containing 18,000 mutants of P. chlororaphis PCL1391, established using pRL1063a (6), was screened for mutants exhibiting reduced PCN production. After growth on LC agar, mutant colonies producing PCN appear yellow. Among 20 white transposon colonies, one mutant (Fig. 1A) was tested for PCN production after growth in liquid, complex LC medium. Quantitative high-pressure liquid chromatography (HPLC) analysis shows that this mutant, named PCL1114, is severely affected in PCN production (<1% of wild-type [wt] strain PCL1391 production). When mutant PCL1114 was grown in King's B medium, another complex medium, PCN production was reduced to 2.5% compared to wt. PCN production by PCL1114 was not detected (<1% of wt) during growth in the poorer synthetic MVB1 medium (Fig. 1B), which was used as the standard growth medium in subsequent experiments. For convenience, the results of the experiment in Fig. 1 are summarized in Table 2. In addition, analysis of N-AHL production showed that C6-HSL could not be detected in the supernatant of PCL1114 (Fig. 1C, lane 7). The PCL1114 mutant is therefore unable to synthesize both PCN and its associated N-AHL signaling molecule, suggesting a mutation within a gene involved in the signaling cascade.
FIG. 1.
Analysis of PCN and N-AHL production by P. chlororaphis PCL1391 and PCL1114 derivatives. (A) Phenotypic aspect of PCL1391 and PCL1114 colonies. Bar, 5 mm. (B) Extractions were made from at least three independent cultures in 10 ml of MVB1 medium in a time course, and the PCN production level was determined by HPLC. The error bars indicate the standard deviations. On each graph, the OD620 (left axis; dotted lines) and the PCN concentration (right axis; solid lines) are plotted. The symbol for the pip strain (□) was magnified for better visualization. Below the graph is the result of C8 reverse-phase TLC analysis of N-AHL production by various PCL1391 derivatives at an OD620 of 3.0. st, standard of 2.5 nmol of synthetic C6-HSL. For details, see Materials and Methods.
TABLE 2.
Overview of the results for PCN and N-AHL production by various derivative strains
| Metabolite | Production with the indicated mutation in:a
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
psrA
|
rpoS
|
pip
|
|||||||
| Empty vector | Ptacpip strain | Empty vector | Ptacpip strain | Empty vector | PtacpsrA strain | PtacrpoS strain | Ptacpip strain | PtacphzR strain | |
| PCN | − | + | − | + | − | − | − | + | + |
| N-AHL | − | + | − | + | − | − | − | + | + |
Cloned genes were added in trans. See Materials and Methods for an explanation of the notation.
(ii) Pip is essential to PCN synthesis.
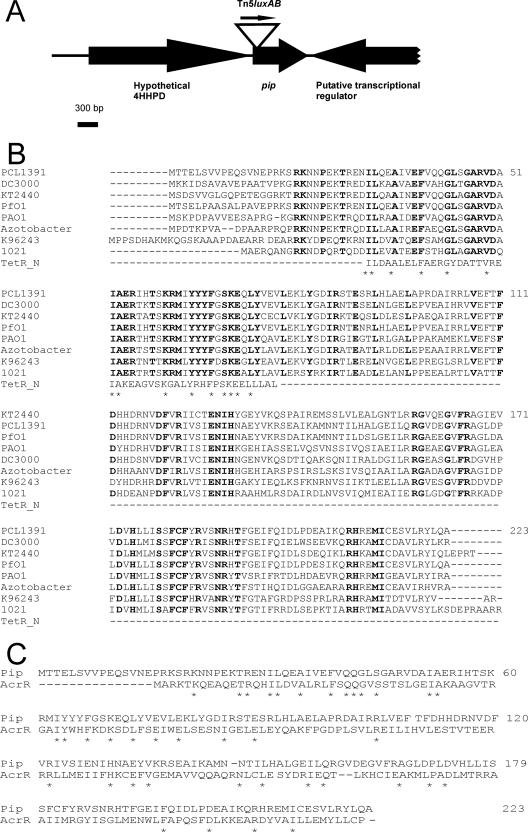
Plasmid rescue from chromosomal DNA of PCL1114 showed that the Tn5luxAB transposon is inserted in a small open reading frame (ORF) of 669 bp in between positions 71 and 72 (Fig. 2A). The gene corresponding to this ORF was named pip. The protein that shows highest overall similarity with Pip is a putative transcriptional regulator of the TetR family in P. fluorescens (ZP_00262623). Orthologues of Pip were found in other Pseudomonas species (94% homology in P. fluorescens Pf-5, 83% homology in P. putida KT2440, and 79% homology in P. aeruginosa PAO1), as well as homologues in a large variety of other gram-negative species, such as Azotobacter vinelandii, Burkholderia spp., Sinorhizobium meliloti, Agrobacterium tumefasciens, and Ralstonia spp. To our knowledge, no function has been published for any of these homologues. A domain homology search on the Pip sequence showed that Pip is homologous to members of the TetR/AcrR family, which are transcriptional regulators (11). In E. coli, TetR regulates a pump involved in tetracycline resistance (3), and AcrR regulates a pump involved in multidrug resistance (26). A multiple alignment between Pip orthologues and the TetR N-terminal domain (Pfam 00440) shows that many amino acids are conserved within the region that contains a helix-turn-helix motif (Fig. 2B). Although no homology with the TetR C-terminal effector-binding domain was found, full-length alignments are possible with AcrR-like proteins, and Pip has an overall similarity of 42% with AcrR (accession number AAC73566) (Fig. 2C). A putative promoter sequence was found upstream of the pip ORF (putative −10 box [GCCCATAAT] and −35 box [TTTCCT]). No rho-independent terminator could be detected downstream of pip, although a putative gene is located there in the opposite direction of transcription (see below).
FIG. 2.
In silico analysis of the pip gene. (A) Genomic organization of the chromosomal region of P. chlororaphis PCL1391 surrounding pip. Each ORF is represented by an arrow which indicates the direction of transcription. The putative transcriptional regulator was not completely sequenced. The position of the transposon insertion is shown as an arrowhead at the beginning of pip. 4-HHPD, 4-hydroxyphenylpyruvate dioxygenase. (B) Alignment of Pip homologues from various bacterial species with the TetR N-terminal domain of E. coli. Homologues of Pip from P. chlororaphis PCL1391 were found in P. syringae pv. tomato str. DC3000 (NP_792164), P. putida KT2440 (NP_745664), P. fluorescens PfO-1 (ZP_00262623), P. aeruginosa PAO1 (AAG03632), A. vinelandii (ZP_00091468), Burkholderia pseudomallei K96243 (YP_111478), and S. meliloti 1021 (NP_436576). The amino acids that are conserved in all the Pip homologues are indicated in bold. The amino acids that are conserved in all the Pip homologues and in the TetR N terminus are indicated by asterisks. The numbers at right indicate the amino acid numbering of Pip in strain PCL1391. (C) Alignment of Pip and AcrR from E. coli (AAC73566). Conserved amino acids are indicated by asterisks. The numbers at right indicate the amino acid numbering of Pip.
The chromosomal organization around pip was determined by sequencing and analyzed by BLAST search. An ORF of 1,908 nucleotides is present upstream of pip (Fig. 2A), which has the same transcription orientation as pip and encodes a protein showing 92% homology to a putative 4-hydroxyphenylpyruvate dioxygenase of P. fluorescens PfO1 (accession number ZP_00262624). Computer analysis shows the presence of a putative rho-independent transcription terminator for the 4-hydroxyphenylpyruvate dioxygenase gene, seven nucleotides downstream of its stop codon (GTAACGGCGGCGGCAAAGGGCCGCCGTCCTGC), followed by the putative promoter sequence upstream of the pip ORF. Downstream of pip, an ORF is present (Fig. 2A), of which the predicted protein product shows 58% homology with a putative transcriptional regulator of C. violaceum ATCC 12472 (accession number AAQ59192).
In order to test if the inhibition of PCN production was indeed due to the defect in pip, we tested whether PCL1114 could be complemented by expression of pip. The pip Ptac pip strain is PCL1114 harboring pip under control of the tac promoter in trans. This strain produced 1.4-fold more PCN (Fig. 1B) than wt (empty vector) strain as analyzed after 12 h of growth. The pip Ptac pip strain also produces high amounts of C6-HSL (Fig. 1C, lane 2). The pip gene was also expressed under its own promoter in trans in the pip Ppip pip mutant strain and showed restored production of PCN and N-AHL (Fig. 1B and C). These results clearly show that the impaired production of PCN and N-AHL in PCL1114 is only caused by disruption of pip.
Role of Pip in the regulation of PCN synthesis. (i) Autoregulation of pip expression.
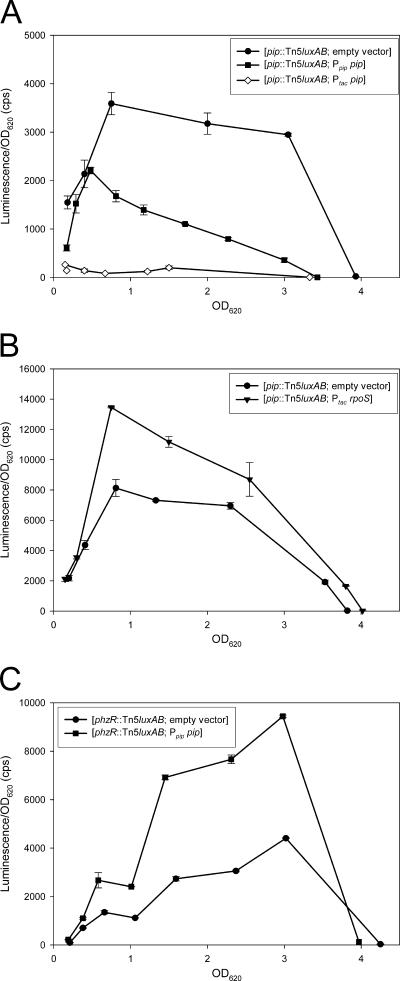
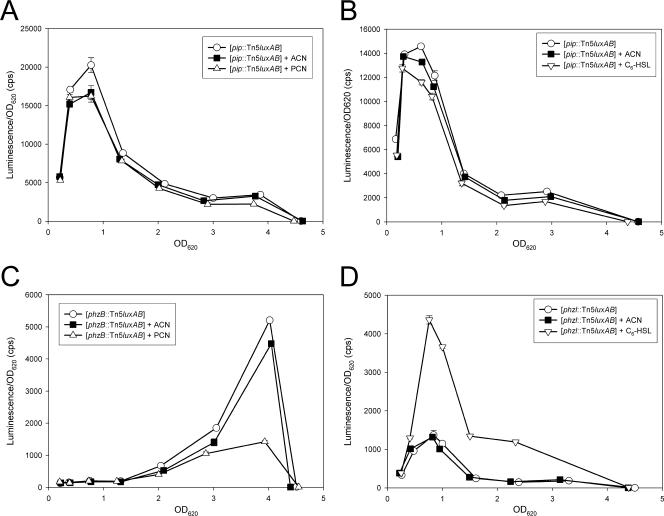
Since both TetR and AcrR repress their own expression (15, 21), we tested whether Pip shows a similar autoregulatory mechanism. Analysis of the orientation of the Tn5luxAB in PCL1114 showed that the luxAB genes and pip have the same direction of transcription, which allows measurements of pip transcription by quantifying the luxAB activity. The expression of pip was measured in three pip derivatives containing Ptac pip, Ppip pip, or the empty cloning vector (Fig. 3A). The pip Ppip pip strain showed an intermediate lux activity (2,210 ± 67 cps) compared to the pip (empty vector) strain (3,590 ± 231 cps) and to the pip Ptac pip strain (262 ± 1 cps). These results suggest that Pip, like TetR and AcrR, represses its own transcription.
FIG. 3.
Expression analyses of P. chlororaphis PCL1391 pip, psrA, and phzR derivative strains. Each panel corresponds to a particular chromosomal background, and the genes expressed in the different backgrounds are indicated in the legend. Cell cultures were grown in 10 ml of MVB1 medium, and samples were taken at regular time intervals. Activity of the luxAB reporter was determined by quantifying bioluminescence. Measurements were performed in duplicate, and averages are plotted. The bars represent the standard deviation. The strains used in these experiments are as indicated on the graphs.
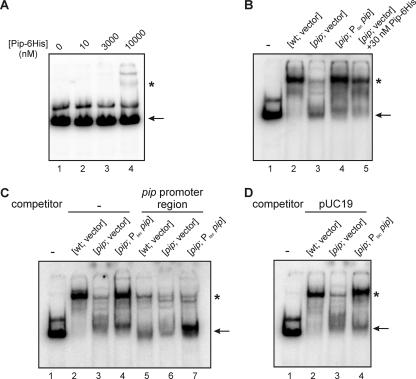
Since Pip shows homologies with DNA-binding transcriptional regulators and autoregulates its own expression, we searched for a Pip-specific cis-acting element within its upstream region. An in silico search for conserved motifs in several upstream regions of pip orthologues retrieved a 47-nt sequence (see Materials and Methods) located 36 bp upstream of the pip start codon as the best-conserved motif among orthologous pip upstream regions. The putative −35 and −10 boxes suggested above are included in this sequence. A 120-bp PCR product of the Pip upstream region containing the conserved sequence was obtained and used for band-shift analysis. Purified Pip-His6 did not seem to be able to bind and shift the labeled DNA (Fig. 4A). However, S30 fractions (see Materials and Methods) of wt and of the pip Ptac pip strain were able to bind the 32P-labeled PCR product as shown by band retardation, whereas the S30 fraction of the pip mutant was not (Fig. 4B, lanes 2, 4, and 3, respectively). Interestingly, the addition of purified Pip-His6 to the S30 fraction of the pip mutant resulted in the shifting of the DNA (Fig. 4B, lane 5). Competition with the unlabeled PCR product inhibited the shift, whereas competition with unlabeled pUC19 plasmid did not (Fig. 4C and D, respectively). These results show that Pip is able to specifically recognize and bind a DNA sequence within the 120 bp upstream of the start codon. However, an additional factor present in S30 fractions obtained from P. chlororaphis cultures is apparently necessary for DNA-binding activity of Pip.
FIG. 4.
Binding of Pip to its own promoter region. One nanogram of a 32P-labeled DNA fragment of 120 bp, corresponding to the pip promoter region, was used as a probe for band shift assays with either purified Pip protein (A) with cell extracts from different strains (B, C, and D) or a combination of both (B). Competition assays were performed with 50 ng of the pip promoter region (C) and with 50 ng of pUC19 plasmid (D). Samples were separated by 8% native polyacrylamide gel electrophoresis at 120 V for 20 min, and bands were visualized by phosphor imaging. The arrows indicate the positions of the free probes and the asterisks indicate the positions of the complex.
(ii) Position of Pip in the regulatory network of PCN synthesis.
Several genes, including psrA, rpoS, and phzI/phzR, are known to play a role in the regulation of PCN synthesis (6, 10), and therefore experiments were conducted in order to understand how pip fits into the PCN biosynthesis signaling cascade.
In MVB1 medium the psrA and rpoS genes positively regulate PCN and N-AHL production (10). To test whether Pip could regulate PCN and N-AHL production downstream of psrA and/or rpoS, pip was overexpressed in strains PCL1111 (psrA mutant) and PCL1954 (rpoS mutant). Both strains showed restored production of PCN (Fig. 5A) and C6-HSL (Fig. 5C; compare lanes 3 and 2 and 5 and 4). In addition, constitutive expression of rpoS in the mutant pip Ptac rpoS strain resulted in a 25% increase of lux activity compared to the pip (empty vector) strain (Fig. 3B), showing that RpoS can influence pip expression. In contrast, constitutive production of PsrA and RpoS in the pip mutant PCL1114 (pip Ptac psrA strain and pip Ptac rpoS strain, respectively) was not able to restore PCN production (results not shown). Western blot analysis confirmed that similar amounts of RpoS were isolated in the extracts from the wt and pip mutant (see Fig. S1, lanes 1 and 2, in the supplemental material). These cross-complementation assays position Pip downstream of PsrA and RpoS in the regulatory pathway leading to PCN production. For convenience, results of the experiment shown in Fig. 5 are summarized in Table 2.
FIG. 5.
Analysis of PCN and N-AHL production in P. chlororaphis PCL1391 derivative strains. (A) Extractions were carried out from at least three independent cultures in 10 ml of MVB1 medium in a time course, and the PCN concentration was determined by HPLC analysis. On each graph, the absorbance is plotted along the left axis (dotted lines), and the PCN concentration is plotted along the right axis (solid lines). The error bars indicate the standard deviations. Symbols correspond to those of panels B and C. The symbol for the psrA strain (empty vector) (▵) was magnified for better visualization. (B and C) C18 reverse-phase TLC analysis of N-AHL produced by the pip (empty vector) strain (lane 2) and other derivatives. st, 2.5 nmol of synthetic C6-HSL. Extractions were performed on supernatants of cultures that reached an OD620 value of 3.0. For the detection of N-AHL, see Materials and Methods.
To test the relationship between Pip and quorum sensing, a pip mutant derivative was constructed that constitutively expresses phzR. The resulting pip Ptac phzR strain showed restored production of both PCN and C6-HSL (Fig. 5A and B). Transformation with a plasmid containing pip under its own promoter showed a positive effect on phzR::Tn5luxAB expression (Fig. 3C). These results confirm that Pip regulates PCN synthesis via the PhzI/PhzR quorum-sensing system, downstream of psrA and rpoS. Pip is therefore the fifth transcription factor shown to be involved in the regulation of PCN biosynthesis in strain PCL1391, along with GacA, PsrA, RpoS, and PhzR.
(iii) Pip, an efflux-pump regulator?
Based on the homology between Pip and AcrR/TetR, we considered the hypothesis that Pip might directly regulate the expression of a gene encoding an efflux pump, analogously to AcrR and TetR. Results described above could suggest that this pump would secrete PCN or even more likely N-AHL. To test this hypothesis, the effect of PCN and N-AHL on pip transcription was measured, since it was shown that the expression of acrR and tetR is under the regulation of the molecules secreted by their target pumps. Results show that neither PCN nor N-AHL regulates the activity of Ppip (Fig. 6A and B). The same results were obtained with an intact pip gene in trans by using the pip::Tn5luxAB Ppip pip strain (data not shown). Importantly, the concentrations of PCN and N-AHL we used were sufficient to inhibit transcription of phzB (Fig. 6C) and stimulate that of phzI (Fig. 6D). These results indicate that the transcription of pip is not influenced by N-AHL or PCN.
FIG. 6.
Influence of C6-HSL and PCN on Ppip activity. Cell cultures were grown in 10 ml of MVB1 medium supplemented with acetonitrile (ACN), 2 μM PCN, or 5 μM C6-HSL. Samples were taken at regular time intervals. Activity of the luxAB reporter was determined by quantifying bioluminescence. Measurements were performed in duplicate, and averages are plotted. The bars represent the standard deviations, and some are too small to be seen. The following strains were used in these experiments: the pip::Tn5luxAB strain (A and B), the phzB::Tn5luxAB strain (C), and the phzI::Tn5luxAB strain (D).
DISCUSSION
In this study we identified a new gene (pip) required for the production of the antifungal metabolite PCN in P. chlororaphis PCL1391. A pip mutant does not produce any detectable amounts of PCN, while increasing the copy number of pip results in an increased transcription of the PCN activator phzR (Fig. 3C), which is now confirmed by preliminary microarray data analyses (data not shown). The identity of Pip as a transcriptional regulator was suggested in silico (AcrR/TetR family) and demonstrated in vitro (Fig. 4) and in vivo (luxAB expression), which enables us to insert a new control point into the signaling pathway leading to PCN production.
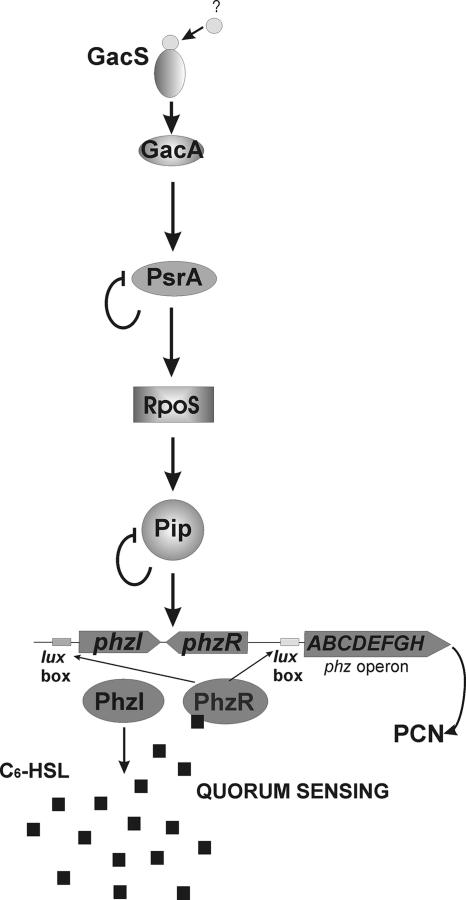
Several experiments were conducted to assess where Pip was positioned in the regulatory cascade of PCN synthesis. The relative position of Pip was deduced from the ability of Pip to restore in trans PCN production in mutants of other genes known to be involved in the control of PCN production, i.e., psrA (activator of rpoS transcription) and rpoS (stationary-phase sigma factor). Opposite in trans complementation assays were also performed for testing the ability of PsrA, RpoS, and PhzR to restore PCN production in the pip mutant. Production of PCN and N-AHL was fully restored in psrA and rpoS mutants constitutively expressing pip, while the pip mutation could only be suppressed by constitutive production of Pip or PhzR (Table 2). These results justify the position of Pip downstream of PsrA and RpoS and upstream of PhzR in our model (Fig. 7).
FIG. 7.
Schematic model showing the role of Pip in the genetic cascade regulating PCN synthesis in P. chlororaphis PCL1391 and in the presence of several stress factors. The regulatory cascade of PCN starts with the sensing of an as yet unidentified environmental signal by GacS and subsequent activation of GacA. The TetR homologue PsrA regulates rpoS, probably by binding to its promoter. The alternative sigma factor RpoS positively regulates pip, the product of which stimulates expression of the quorum-sensing system phzI/phzR. Both pip and psrA exhibit negative autoregulation. PhzI is responsible for the production of C6-HSL, which is supposed, in turn, to bind to PhzR. The PhzR-C6-HSL complex binds to lux boxes in the promoter sequences of phzI and the phz operon. Subsequently, phzI is upregulated, and expression of the phz operon is switched on, which finally results in the synthesis of PCN.
This new model raises the questions of why there are so many checkpoints for PCN biosynthesis and, in the focus of this particular study, what the effector molecule of Pip is and how the information further is delivered to initiate PCN production. The identification of a direct Pip target gene should provide crucial information to answer these questions. A first hypothesis is that Pip could regulate an efflux pump, as the TetR and AcrR regulators do. To test if Pip, like TetR, regulates an antibiotic efflux-pump, the resistance of pip to several antibiotics was compared to that of the wt, but no difference could be shown (data not shown). In analogy to AcrR/TetR, the molecules secreted by the Pip-regulated pump would modulate, in turn, the activity of Pip. It was shown that a factor present in the cell is necessary for Pip to bind DNA (Fig. 4A and B). However, it cannot be either N-AHL or PCN, since both are absent in a pip mutant background, the cellular extract of which seems to contain the predicted Pip-interacting factor (Fig. 4B, lane 5). In addition, we showed that the expression of pip is not under the influence of PCN or N-AHL metabolites (Fig. 6). Taken together, these results do not support the notion that pip directly regulates a pump for N-AHL or PCN; rather, they suggest either that an additional metabolite participates in this regulation or that Pip does not directly control genes encoding an efflux pump.
As a first attempt to identify putative Pip target genes, we used the conserved sequence upstream of pip, shown to interact with Pip, to generate a position weight matrix and scan Pseudomonas genomes for similar DNA patterns. Computational prediction programs predicted in P. aeruginosa PAO1 eight sequences that contain patterns similar to the one observed upstream of pip (see Table S3 in the supplemental material). The best scoring hit is located 83 nt upstream of the epd gene coding for the d-erythrose 4-phosphate dehydrogenase that connects the pentose-phosphate pathway to the vitamin B6 metabolism. The other seven potential target genes all encode hypothetical proteins with unknown functions. It is also possible that the motif recognized by Pip is not well conserved, as is the case for other known transcriptional regulators, for example LasR in P. aeruginosa (32). A large number of additional experiments will be required to precisely identify the primary target of Pip. Particularly, identification of possible partner(s) of Pip, as indicated by the shift experiments (Fig. 4), is crucial to a better understanding of how Pip interacts with target DNA. Future work could also include a broader analysis of genes regulated by pip, using the microarray developed for strain PCL1391 (10). We are currently investigating preliminary data that suggest that Pip connects the PCN biosynthetic pathway to the stress response in P. chlororaphis PCL1391.
Acknowledgments
We thank Daan van den Broek and Thomas Chin-A-Woeng for screening the transposon bank and isolating PCL1114 and K. Tanaka (Institute of Molecular and Cellular Biosciences, University of Tokyo, Japan) for providing the anti-RpoS rabbit serum.
This project was financially supported by the European Union FW6 Research and Development project QRLT-2002-00914 (“Pseudomics”) and by the BioScience Initiative from Leiden University. S. Barends was supported by a VENI grant from the Netherlands Organization for Scientific Research (NWO).
Footnotes
Published ahead of print on 22 September 2006.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1997. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers, p. 28-36. In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, Calif. [PubMed]
- 3.Beck, C. F., R. Mutzel, J. Barbe, and W. Muller. 1982. A multifunctional gene (tetR) controls Tn10-encoded tetracycline resistance. J. Bacteriol. 150:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chin-A-Woeng, T. F. C., G. V. Bloemberg, I. H. Mulders, L. C. Dekkers, and B. J. J. Lugtenberg. 2000. Root colonization by phenazine-1-carboxamide-producing bacterium Pseudomonas chlororaphis PCL1391 is essential for biocontrol of tomato foot and root rot. Mol. Plant-Microbe Interact. 13:1340-1345. [DOI] [PubMed] [Google Scholar]
- 5.Chin-A-Woeng, T. F. C., G. V. Bloemberg, A. J. van der Bij, K. M. G. M. van der Drift, J. Schripsema, B. Kroon, R. J. Scheffer, C. Keel, P. A. H. M. Bakker, H. V. Tichy, F. J. de Bruijn, J. E. Thomas-Oates, and B. J. J. Lugtenberg. 1998. Biocontrol by phenazine-1-carboxamide-producing Pseudomonas chlororaphis PCL1391 of tomato root rot caused by Fusarium oxysporum f. sp. radicis-lycopersici. Mol. Plant-Microbe Interact. 11:1069-1077. [Google Scholar]
- 6.Chin-A-Woeng, T. F. C., D. van den Broek, G. de Voer, K. M. van der Drift, S. Tuinman, J. E. Thomas-Oates, B. J. J. Lugtenberg, and G. V. Bloemberg. 2001. Phenazine-1-carboxamide production in the biocontrol strain Pseudomonas chlororaphis PCL1391 is regulated by multiple factors secreted into the growth medium. Mol. Plant-Microbe Interact. 14:969-979. [DOI] [PubMed] [Google Scholar]
- 7.Chin-A-Woeng, T. F. C., D. van den Broek, B. J. J. Lugtenberg, and G. V. Bloemberg. 2005. The Pseudomonas chlororaphis PCL1391 sigma regulator psrA represses the production of the antifungal metabolite phenazine-1-carboxamide. Mol. Plant-Microbe Interact. 18:244-253. [DOI] [PubMed] [Google Scholar]
- 8.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuqua, C., M. R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35:439-468. [DOI] [PubMed] [Google Scholar]
- 10.Girard, G., E. T. van Rij, B. J. J. Lugtenberg, and G. V. Bloemberg. 2006. Roles of psrA and rpoS in phenazine-1-carboxamide synthesis by Pseudomonas chlororaphis PCL1391. Microbiology 152:43-58. [DOI] [PubMed] [Google Scholar]
- 11.Grkovic, S., M. H. Brown, and R. A. Skurray. 2002. Regulation of bacterial drug export systems. Microbiol. Mol. Biol. Rev. 66:671-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 13.Heeb, S., C. Blumer, and D. Haas. 2002. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J. Bacteriol. 184:1046-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hertz, G. Z., and G. D. Stormo. 1999. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics 15:563-577. [DOI] [PubMed] [Google Scholar]
- 15.Hillen, W., and C. Berens. 1994. Mechanisms underlying expression of Tn10-encoded tetracycline resistance. Annu. Rev. Microbiol. 48:345-369. [DOI] [PubMed] [Google Scholar]
- 16.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simpl e media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 17.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 18.Kuiper, I., G. V. Bloemberg, S. Noreen, J. E. Thomas-Oates, and B. J. J. Lugtenberg. 2001. Increased uptake of putrescine in the rhizosphere inhibits competitive root colonization by Pseudomonas fluorescens strain WCS365. Mol. Plant-Microbe Interact. 14:1096-1104. [DOI] [PubMed] [Google Scholar]
- 19.Lau, G. W., D. J. Hassett, H. Ran, and F. Kong. 2004. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol. Med. 10:599-606. [DOI] [PubMed] [Google Scholar]
- 20.Laville, J., C. Voisard, C. Keel, M. Maurhofer, G. Defago, and D. Haas. 1992. Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco. Proc. Natl. Acad. Sci. USA 89:1562-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma, D., M. Alberti, C. Lynch, H. Nikaido, and J. E. Hearst. 1996. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol. Microbiol. 19:101-112. [DOI] [PubMed] [Google Scholar]
- 22.Marsh, J. L., M. Erfle, and E. J. Wykes. 1984. The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene 32:481-485. [DOI] [PubMed] [Google Scholar]
- 23.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 24.Milton, D. L., A. Hardman, M. Camara, S. R. Chhabra, B. W. Bycroft, G. S. Stewart, and P. Williams. 1997. Quorum sensing in Vibrio anguillarum: characterization of the vanI/vanR locus and identification of the autoinducer N-(3-oxodecanoyl)-l-homoserine lactone. J. Bacteriol. 179:3004-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ochsner, U. A., and J. Reiser. 1995. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 92:6424-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olliver, A., M. Valle, E. Chaslus-Dancla, and A. Cloeckaert. 2004. Role of an acrR mutation in multidrug resistance of in vitro selected fluoroquinolone-resistant mutants of Salmonella enterica serovar Typhimurium. FEMS Microbiol. Lett. 238:267-272. [DOI] [PubMed] [Google Scholar]
- 27.Pearson, J. P., E. C. Pesci, and B. H. Iglewski. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179:5756-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pessi, G., and D. Haas. 2000. Transcriptional control of the hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the quorum-sensing regulators LasR and RhlR in Pseudomonas aeruginosa. J. Bacteriol. 182:6940-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reimmann, C., M. Beyeler, A. Latifi, H. Winteler, M. Foglino, A. Lazdunski, and D. Haas. 1997. The global activator GacA of Pseudomonas aeruginosa PAO1 positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol. Microbiol. 24:309-319. [DOI] [PubMed] [Google Scholar]
- 30.Sacherer, P., G. Defago, and D. Haas. 1994. Extracellular protease and phospholipase C are controlled by the global regulatory gene gacA in the biocontrol strain Pseudomonas fluorescens CHA0. FEMS Microbiol. Lett. 116:155-160. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Schuster, M., M. L. Urbanowski, and E. P. Greenberg. 2004. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc. Natl. Acad. Sci. USA 101:15833-15839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sosinsky, A., C. P. Bonin, R. S. Mann, and B. Honig. 2003. Target Explorer: an automated tool for the identification of new target genes for a specified set of transcription factors. Nucleic Acids Res. 31:3589-3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Rij, E. T., M. Wesselink, Chin-A-Woeng, T. F. C., G. V. Bloemberg, and B. J. J. Lugtenberg. 2004. Influence of environmental conditions on the production of phenazine-1-carboxamide by Pseudomonas chlororaphis PCL1391. Mol. Plant-Microbe Interact. 17:557-566. [DOI] [PubMed] [Google Scholar]
- 35.Winson, M. K., M. Camara, A. Latifi, M. Foglino, S. R. Chhabra, M. Daykin, M. Bally, V. Chapon, G. P. Salmond, and B. W. Bycroft. 1995. Multiple N-acyl-l-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 92:9427-9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolk, C. P., Y. Cai, and J. M. Panoff. 1991. Use of a transposon with luciferase as a reporter to identify environmentally responsive genes in a cyanobacterium. Proc. Natl. Acad. Sci. USA 88:5355-5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yellaboina, S., J. Seshadri, M. S. Kumar, and A. Ranjan. 2004. PredictRegulon: a web server for the prediction of the regulatory protein binding sites and operons in prokaryote genomes. Nucleic Acids Res. 32:W318-W320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zuber, S., F. Carruthers, C. Keel, A. Mattart, C. Blumer, G. Pessi, C. Gigot-Bonnefoy, U. Schnider-Keel, S. Heeb, C. Reimmann, and D. Haas. 2003. GacS sensor domains pertinent to the regulation of exoproduct formation and to the biocontrol potential of Pseudomonas fluorescens CHA0. Mol. Plant-Microbe Interact. 16:634-644. [DOI] [PubMed] [Google Scholar]