Abstract
Although numerous bacteria possess genes annotated iol in their genomes, there have been very few studies on the possibly associated myo-inositol metabolism and its significance for the cell. We found that Corynebacterium glutamicum utilizes myo-inositol as a carbon and energy source, enabling proliferation with a high maximum rate of 0.35 h−1. Whole-genome DNA microarray analysis revealed that 31 genes respond to myo-inositol utilization, with 21 of them being localized in two clusters of >14 kb. A set of genomic mutations and functional studies yielded the result that some genes in the two clusters are redundant, and only cluster I is necessary for catabolizing the polyol. There are three genes which encode carriers belonging to the major facilitator superfamily and which exhibit a >12-fold increased mRNA level on myo-inositol. As revealed by mutant characterizations, one carrier is not involved in myo-inositol uptake whereas the other two are active and can completely replace each other with apparent Kms for myo-inositol as a substrate of 0.20 mM and 0.45 mM, respectively. Interestingly, upon utilization of myo-inositol, the l-lysine yield is 0.10 mol/mol, as opposed to 0.30 mol/mol, with glucose as the substrate. This is probably not only due to myo-inositol metabolism alone since a mixture of 187 mM glucose and 17 mM myo-inositol, where the polyol only contributes 8% of the total carbon, reduced the l-lysine yield by 29%. Moreover, genome comparisons with other bacteria highlight the core genes required for growth on myo-inositol, whose metabolism is still weakly defined.
Inositol is a building block of plants and is thus probably one of the sources of traces of myo-inositol, or its phosphorylated derivative myo-inositol hexakisphosphate, in soil (26). Accordingly, there are indications that a number of microorganisms are able to utilize myo-inositol. For instance, the soil-inhabiting Rhizobiaceae family members Sinorhizobium fredii (16) and Rhizobium leguminosarum (9) have the ability to catabolize or even grow on myo-inositol and this feature may increase their fitness for better nodulating the host plant (10). Also, Klebsiella (Aerobacter) aerogenes is able to utilize myo-inositol (19) and early biochemical work with this organism established how the polyol could be metabolized (2) (Fig. 1).
FIG. 1.
Assumed pathway for myo-inositol degradation. The pathway is in part speculative and largely based on enzymological studies of K. aerogenes (2, 19), studies of B. subtilis (27-30), and genome comparisons. The genes and the enzymes encoded are given; those with a presumed assignment are in parentheses.
As can be seen from their genome sequences, a large number of bacteria have genes which are annotated as iol genes. These are often clustered, for example, the iolDEB genes in R. leguminosarum, which are required for growth on inositol (9), or in Clostridium perfringens, where a cluster of 13 genes is induced by myo-inositol with the participation of the regulator IolR (17). In Bacillus subtilis, there is an iol divergon comprising iolABCDEFGHIJ and iolRS whose repression by glucose is in part CcpA dependent (29, 30). Relatively few studies have been done to demonstrate the participation of the iol genes in inositol metabolism, and there are a very limited number of biochemical studies on their enzyme function. A myo-inositol dehydrogenase has been identified that initiates myo-inositol metabolism. The enzyme is encoded by iolG of B. subtilis (10) or idhA in Sinorhizobium meliloti (11), respectively, as well as S. fredii (16). In S. meliloti, its inactivation was found to disable myo-inositol utilization (11). The second gene function identified is that of iolE, which encodes a 2-keto-myo-inositol dehydratase (31). Import of the polyol is known to be catalyzed by transporters belonging to the major facilitator superfamily. In B. subtilis, two such myo-inositol uptake carriers are present (28), with iolF located within the iolABCDEFGHIJ operon representing the minor transporter for uptake. The second transporter, iolT, is located elsewhere in the chromosome, and its inactivation causes reduced growth on a number of carbon sources, with the most pronounced effect on myo-inositol (28).
We are interested in Corynebacterium glutamicum, an apathogenic bacterium of industrial interest used for the large-scale production of amino acids, in particular, l-glutamate and l-lysine (8). Together with Mycobacterium tuberculosis, for instance, this bacterium belongs to the suborder Corynebacterineae, characterized among others by possessing myo-inositol as a cellular building block, as is the case in eukaryotes (20, 22). The inositol is required for the synthesis of phosphatidyl-myo-inositol, which is an abundant phospholipid in the cytoplasmic membrane and which in turn is also a precursor of more-complex cellular glycolipids in Corynebacterineae such as lipomannans and lipoarabinomannans (5). Furthermore, myo-inositol is also a building block for mycothiol (5, 7, 22), which is a low-molecular-mass thiol specific to Corynebacterineae and necessary for protection against the damaging effects of reactive oxygen species, similar to glutathione in eukaryotes and gram-negative bacteria. In C. glutamicum, we detected a number of genes annotated as iol and wished to know whether their presence enables C. glutamicum to grow on this polyol with an additional focus on the transport of myo-inositol and the physiological consequences of its utilization.
MATERIALS AND METHODS
Bacteria, plasmids, oligonucleotides, and culture conditions.
All of the strains, plasmids, and oligonucleotides used in this study are described in Table 1. The minimal medium used for C. glutamicum was CGXII (8), which contained the carbon source glucose or myo-inositol autoclaved separately. C. glutamicum was grown as 50-ml cultures in 500-ml baffled Erlenmeyer flasks on a rotary shaker at 120 rpm. Growth of the bacteria was monitored by measuring the optical density at 600 nm (OD600).
TABLE 1.
Strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotidea | Relevant characteristic(s) or sequenceb | Source, reference, or purpose |
|---|---|---|
| C. glutamicum strains | ||
| ATCC 13032 | WT | Culture collection |
| ATCC 21527 | Lysine producer obtained by undirected mutagenesis | Culture collection |
| MH20-22B | Lysine producer obtained by undirected mutagenesis | 24 |
| DM1730 | Lysine producer; pycP458S homV59A lysCT311I zwfA243T | 13 |
| WTΔiolD | In-frame deletion of iolD | This work |
| WT::piolG′ | Vector integrated into iolG | This work |
| WTΔoxiII | In-frame deletion of oxiC-oxiE | This work |
| WTΔiolII | In-frame deletion of adhA-oxiE | This work |
| WTΔoxiII::piolG′ | In-frame deletion of oxiC-oxiE and vector integrated into iolG | This work |
| WTΔiolT1 | In-frame deletion of iolT1 | This work |
| WTΔiolT2 | In-frame deletion of iolT2 | This work |
| WTΔiolT1ΔiolT2 | In-frame deletion of iolT1 and iolT2 | This work |
| Plasmids | ||
| pK19mobsacB | Kmr; mobilizable (oriT); oriV | 23 |
| pT18mob2 | Tetr; mobilizable (oriT); oriV | 23 |
| pK19mobsacBΔiolD | Vector enabling deletion of 1,861 bp of iolD | This work |
| pT18mob2::piolG′ | Vector used to integrate 408 bp into iolG | This work |
| pK19mobsacBΔoxiII | Vector enabling deletion of 4,072 bp of cluster II | This work |
| pK19mobsacBΔiolII | Vector enabling deletion of 8,799 bp of cluster II | This work |
| pK19mobsacBΔiolT1 | Vector enabling deletion of 1,422 bp of iolT1 | This work |
| pK19mobsacBΔiolT2 | Vector enabling deletion of 1,472 bp of iolT2 | This work |
| piolG′ | Vector pK19mobsacB with internal fragment of iolG enabling its disruption | This work |
| Oligonucleotides | ||
| iolD_No | 5′-CGCGGATCCAAGTAATCACCCCAGGTGAAAACTGGAG-3′ | Primer for 1,861-bp iolD deletion (BamHI) |
| iolD_Co | 5′-CGCGGATCCAACGAGGTGCTCAGCACCCAGC-3′ | Primer for1,861-bp iolD deletion (BamHI) |
| iolD_Ni | 5′-CCCATCCACTAAACTTAAACATCTCTTCGTTTCAGCCATGAAATTTTA-3′ | Primer for 1,861-bp iolD deletion |
| iolD_Ci | 5′-TGTTTAAGTTTAGTGGATGGGAAAAACCAAGCCCTCCAGCGTCC-3′ | Primer for 1,861-bp iolD deletion |
| iolG_U1B | 5′-CGC GGA TCC GCG CGG CGA AGC TGG CGA ACT GC-3′ | Primer for iolG disruption (BamHI) |
| iolG_L1B | 5′-CGCGGATCCGCGCGGTAGCGAAACGGGTGGTGA-3′ | Primer for iolG disruption (BamHI) |
| oxiII_No | 5′-TCCCCCCGGGGGATCGCCGCTGTAGGAGCAC-3′ | Primer for 4,072-bp deletion (SmaI) |
| oxiII_Co | 5′-TCCCCCCGGGGGTTAGGCAGGATGAGGTTGAGAa-3′ | Primer for 4,072-bp deletion (SmaI) |
| oxiII_Ni | 5′-CCCATCCACTAAACTTAAACAAATTTTTTGATCACTCATGGGAATTCT-3′ | Primer for 4,072-bp deletion |
| oxiII_Ci | 5′-TGTTTAAGTTTAGTGGATGGGCCAGTTGAGGTGCGTGCGCTG-3′ | Primer for 4,072-bp deletion |
| iol II_No | 5′-CGCGGATCCGATACGAGCATTCGGAACGGGA-3′ | Primer for 8,799-bp deletion (BamHI) |
| iolII_Co | 5′-GCGGGATCCCAGTCCGAGCTTTGAGATGTTC-3′ | Primer for 8,799-bp deletion (BamHI) |
| iolII_Ni | 5′-CCCATCCACTAAACTTAAACAGATCCGGCAGTTCTTAGCGCA-3′ | Primer for 8,799-bp deletion |
| iolII_Ci | 5′-TGTTTAAGTTTAGTGGATGGGCCAGTTGAGGTGCGTGCGCT-3′ | Primer for 8,799-bp deletion |
| iolT1_No | 5′-CGCGGATCCCTGAGTCGTCGTATTATTGCGTATTTT-3′ | Primer for 1,422-bp deletion (BamHI) |
| iolT1_Co | 5′-CGCGGATCCACATTAGGATCTTTAAGCAGTGAATGA-3′ | Primer for 1,422-bp deletion (BamHI) |
| iolT1_Ni | 5′-CCCATCCACTAAACTTAAACAAAAGGAAAGGTGCACTAAAAACCCAG-3′ | Primer for 1,422-bp deletion |
| iolT1_Ci | 5′-TGTTTAAGTTTAGTGGATGGGTTTCAGGGCTGTCGGCCTGAATGA-3′ | Primer for 1,422-bp deletion |
| iolT2_No | 5′-CCGGAATTCTGCTTTGGCCAAACCTATGGTGGA-3′ | Primer for 1,472-bp deletion (BamHI) |
| iollT2_Co | 5′-CCGGAATTCACGGCTAAACAGGTTGTCTTGGGTA-3′ | Primer for 1,472-bp deletion (BamHI) |
| iolTII_Ni | 5′-CCCATCCACTAAACTTAAACAATCTTCAAGAAGGCTTAAACCCCCT-3′ | Primer for 1,472-bp deletion |
| iolTII_Ci | 5′-TGTTTAAGTTTAGTGGATGGGGGCCGATGTACTTGATGTGGCCTT-3′ | Primer for 1,472-bp deletion |
No, N-terminal outer primer; Co, C-terminal outer primer; Ni, N-terminal inner primer; Ci, C-terminal inner primer.
Restriction sites in the oligonucleotides are underlined.
Recombinant DNA work.
Standard protocols were applied for the generation of fragments via PCR, ligation, and restriction (21), with each plasmid made verified by sequencing. The chromosomal mutations of C. glutamicum were made by introducing the nonreplicative plasmids via electroporation. For in-frame deletions, the clones with an integrated vector were subsequently selected for absence of the vector due to the lethal sacB function of pK19mobsacB (23).
Preparation of total RNA and DNA microarray analyses.
Cultures were grown in CGXII minimal medium containing 40 g liter−1 myo-inositol or glucose. In the exponential growth phase at an OD600 of 4 to 6, 25 ml of each culture was used for the preparation of total RNA as previously described (27). Isolated RNA samples were analyzed for quantity and quality by UV spectrophotometry and denaturing formaldehyde agarose gel electrophoresis (21), respectively, and stored at −70°C until use. The generation of whole-genome DNA microarrays, synthesis of fluorescently labeled cDNA from total RNA, microarray hybridization, washing, and data analysis were performed as described previously (27). Genes that exhibited significantly changed mRNA levels (P < 0.05 by Student's t test) by at least a factor of 2.8 were determined in independent growth experiments with subsequent hybridizations (Table 2).
TABLE 2.
Genes of C. glutamicum whose average mRNA ratio was altered ≥2.8-fold (P ≤ 0.05) in myo-inositol-grown cells compared to that in glucose-grown cells in at least three independent cultivations
| NCBIa designation | Open reading frame | Annotation | Gene | mRNA ratiob
|
|
|---|---|---|---|---|---|
| Inositol vs Glucose | BHI vs CGXII | ||||
| NCgl0029 | Cgl0030 | ABC transporter/periplasmic d-ribose-binding protein | rbsB | 0.19 | 0.38 |
| NCgl0155 | Cgl0158 | myo-Inositol catabolism, carbohydrate kinase | iolC | 4.94 | 3.07 |
| NCgl0157 | Cgl0160 | myo-Inositol catabolism, aldehyde dehydrogenase | iolA | 16.65 | 7.16 |
| NCgl0158 | Cgl0161 | myo-Inositol catabolism | iolB | 16.55 | 15.04 |
| NCgl0159 | Cgl0162 | myo-Inositol catabolism, thiamine pyrophosphate-requiring enzyme | iolD | 12.01 | 4.66 |
| NCgl0160 | Cgl0163 | 2-Keto-myo-inositol dehydratase | iolE | 18.64 | 13.93 |
| NCgl0161 | Cgl0164 | myo-Inositol dehydrogenase, oxidoreductase | iolG | 16.55 | 7.38 |
| NCgl0162 | Cgl0165 | myo-Inositol catabolism, isomerases/epimerase | iolH | 8.32 | 3.69 |
| NCgl0163 | Cgl0166 | Efflux carrier, major facilitator superfamily; MFS1 | 17.33 | 2.33 | |
| NCgl0164 | Cgl0167 | myo-Inositol dehydrogenase, oxidoreductase | oxi1 | 2.85 | 0.88 |
| NCgl0167 | Cgl0170 | Transcriptional regulator, LacI type; LacI1 | 7.93 | 4.07 | |
| NCgl0168 | Cgl0171 | Putative oxidoreductase dehydrogenases | oxiB | 9.37 | 5.14 |
| NCgl0178 | Cgl0181 | myo-Inositol transporter | iolT1 | 12.86 | 7.17 |
| NCgl0697 | Cgl0727 | Trehalose/maltose-binding protein; malE | 0.25 | 2.53 | |
| NCgl0916 | Cgl0954 | γ-Glutamyltransferase; ggt | 0.34 | 1.34 | |
| NCgl0933 | Cgl0972 | Porin | porB | 0.25 | 2.52 |
| NCgl1368 | Cgl1423 | Putative acetyltransferase | 0.21 | 3.36 | |
| NCgl1917 | Cgl1992 | ABC transporter/oligopeptide permease; oppC | 0.29 | 2.63 | |
| NCgl2477 | Cgl2566 | Succinyl-CoA synthetase (beta chain) | sucC | 0.23 | 1.53 |
| NCgl2865 | Cgl2967 | Secreted multicopper oxidase; cumA | 4.40 | 1.29 | |
| NCgl2894 | Cgl2996 | myo-Inositol-1-phosphate synthase | ips | 0.09 | 0.17 |
| NCgl2904 | Cgl3007 | Malic enzyme | mez | 0.33 | 3.27 |
| NCgl2951 | Cgl3056 | Hydroxyquinol 1,2-dioxygenase; catA | 9.48 | 2.12 | |
| NCgl2952 | Cgl3057 | Iron-containing alcohol dehydrogenase, oxidoreductase; adh1 | 7.56 | 7.27 | |
| NCgl2953 | Cgl3058 | myo-Inositol transporter | iolT2 | 13.94 | 6.19 |
| NCgl2955 | Cgl3060 | myo-Inositol dehydrogenase | oxiC | 18.83 | 9.85 |
| NCgl2956 | Cgl3061 | myo-Inositol catabolism, sugar phosphate isomerase/epimerase | 21.02 | 8.67 | |
| NCgl2957 | Cgl3062 | myo-Inositol dehydrogenase | oxiD | 26.83 | 9.53 |
| NCgl2958 | Cgl3063 | myo-Inositol dehydrogenase | oxiE | 12.54 | 3.04 |
| NCgl2959 | Cgl3064 | Secreted phosphoesterase | 4.34 | 0.6 | |
| NCgl2961 | Cgl3066 | Proline/ectoine carrier | proP | 0.30 | 0.83 |
Numbers for the corresponding open reading frames of the C. glutamicum genome NC_003450 are given. NCBI, National Center for Biotechnology Information.
The first column gives the mRNA ratio for genes of C. glutamicum ATCC 13032 grown on myo-inositol to that of cells grown on glucose. The far right column gives the ratios for BHI- versus CGXII-glucose grown cells, and data are limited to those genes where myo-inositol utilization already revealed altered mRNA levels.
Uptake measurements.
For determination of myo-inositol uptake, cultures grown overnight on CGXII with myo-inositol were diluted in fresh medium and grown to an OD600 of approximately 4 to 6. Cells were washed twice with cold CGXII medium without myo-inositol, the OD600 was adjusted to 5 (corresponding to 1.2 mg [dry weight] ml−1), and the cells were stored on ice. A 0.9-ml volume of this cell suspension was equilibrated for 4 min at 25°C by stirring in a water bath. Uptake was initiated by the addition of 100 μl of an inositol mixture containing 5 to 500 nmol myo-inositol and 5 μl myo-[1,2-3H(N)]-inositol (1 mCi/ml; Biotrend Chemicals, Cologne, Germany). Aliquots of 100 μl were taken after 10, 25, 40, and 55 s, and individually processed by drawing through a prewetted glass fiber filter (Millipore catalog no. APFF02500) placed on a vacuum manifold. Cells were immediately washed with 2 × 2.5 ml ice-cold 1 M LiCl. Radioactivity was quantified by liquid scintillation counting on a Tri-Carb 1600CA in Instant Scin-Gel Plus (Packard catalog no. 6013398). The uptake was usually linear over time, but for concentrations below 0.25 mM myo-inositol, only uptake up to 40 s was considered. The kinetic data were analyzed via nonlinear regression according to the Michaelis-Menten equation.
RESULTS
Growth on myo-inositol.
In microarray studies with C. glutamicum with the transcriptional regulator lysG deleted (4), we occasionally noted altered mRNA levels of genes putatively related to myo-inositol utilization. We therefore assayed C. glutamicum for the ability to utilize this polyol. When C. glutamicum was inoculated into the salt medium CGXII containing 40 g liter−1 myo-inositol as the sole carbon and energy source, after a short lag, a high growth rate of 0.35 h−1 was obtained (Fig. 2A). This was close to that on glucose (0.41 h−1). The short lag phase observed on myo-inositol is due to the use of glucose-grown cells for inoculation, since it was absent in cells pregrown in inositol (data not shown). On a mixture of both sugars, each at 2 g liter−1, the growth rate was 0.41 h−1 with no observable diauxie. This indicates an efficient molecular and enzymological machinery of C. glutamicum for using myo-inositol as efficiently as more common carbon sources in laboratory use like glucose, ribose, acetate, or lactate, for instance.
FIG. 2.
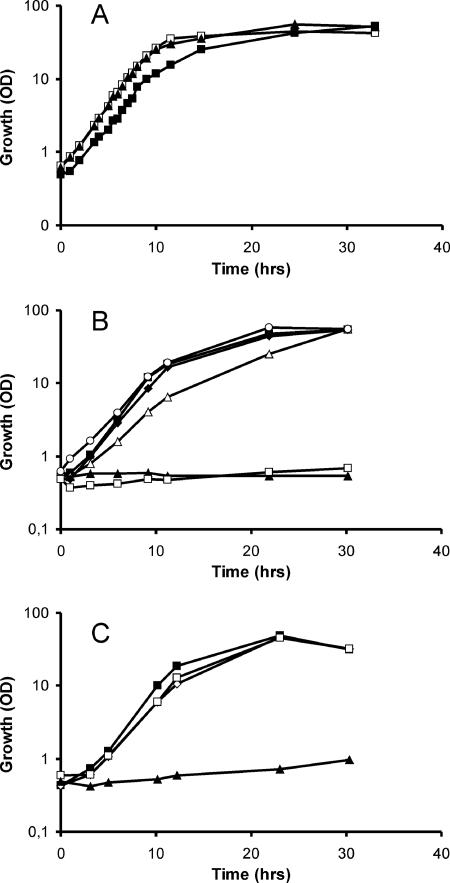
Growth of C. glutamicum WT and mutants generated in this study on myo-inositol and glucose. (A) Growth of the WT on 4% glucose (□), on 4% myo-inositol (▪), and on a mixture (2% plus 2%) of the two sugars (▴). (B) Growth of the ΔiolD mutant on myo-inositol (□) and glucose (○). Growth of the ΔoxiII mutant (⧫), the piolG′ mutant (▵), and the ΔoxiII::piolG′ double mutant (▴) on myo-inositol is compared to that of the WT on myo-inositol (▪). (C) Growth of the transporter deletion mutants ΔiolT1 (□), ΔiolT2 (⋄), and ΔiolT1ΔiolT2 (▴) compared to that of the WT on myo-inositol (▪).
Transcriptome analysis.
In order to determine the effect of myo-inositol utilization on global gene expression, whole-genome DNA microarrays of C. glutamicum were used (27). The determination of the mRNA population of cells grown with myo-inositol compared to cells grown with glucose resulted in a relatively small number of 31 genes exhibiting an at least 2.8-fold change in their transcript level. Among those reduced are mez, encoding malic enzyme, and sucC, encoding the β-chain of succinyl-coenzyme A (CoA) synthetase (Table 2). When the threshold was set to 2 (data not shown), also ptsS, encoding the sucrose-specific IIABC component of the phosphotransferase system, and ptsM, which encodes the glucose-specific IIABC component, exhibited a significantly reduced level, a fact indicating significant carbon source-dependent regulation of the genes of the central metabolism.
The most strongly reduced mRNA level during growth on myo-inositol was determined for the myo-inositol-1-phosphate synthase gene, ips (20). This gene is required in Corynebacterineae for glucose 6-phosphate conversion to myo-inositol, which is a constituent of mycothiol and phosphatidyl-myo-inositol. A regulation of ips is not yet known, but its repression is consistent with the fact that an external supply of myo-inositol makes its cellular synthesis dispensable.
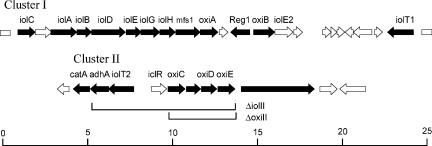
Twenty-one genes showed an up to 18-fold increase in mRNA level, indicative of a possible function within myo-inositol catabolism (Table 2). All but one of these genes are located in two large clusters. The exception is NCgl2865, predicted to encode a secreted protein containing three copper oxidase-like domains. The genome organization of the two clusters is given in Fig. 3, where genes which showed a ≥2.8-fold increase in mRNA level are marked in black. The core of cluster I spans about 16 kb and comprises 13 genes (Fig. 3). It partly resembles the iol cluster of B. subtilis (29), and the iol gene annotations were used according to that introduced for this organism, although definite functions are unknown in almost all cases. The mRNA level of NCgl0156, located between iolC and iolA, was also slightly increased but less than 2.8-fold. An orthologue of this gene is not present in B. subtilis. It is most likely that the 10 genes of C. glutamicum from iolC to oxiA are cotranscribed as an operon. In contrast, the LacI-type regulator encoded by NCgl0167 (Reg1 in Fig. 3) and divergently transcribed oxiB, as well as remotely located iolT1, all three with an mRNA level of ≥7, might be separately transcribed.
FIG. 3.
Genome maps of the C. glutamicum regions functioning in myo-inositol catabolism. Cluster I shows the genome organization of the nucleotide sequence from 167,768 to 193,453, and cluster II shows the sequence from 3,257,372 to 3,272,564 of the C. glutamicum genome NC_003450. Genes exhibiting a ≥2.8-fold increase in the mRNA ratio during growth on myo-inositol compared to glucose are in black. Below cluster II, the two genomic deletions ΔoxiII and ΔiolII, respectively, are indicated. The values on the scale bar are in kilobases.
Cluster II spans about 14.6 kb and consists of eight genes which also exhibited a ≥2.8-fold increase in mRNA level. Only the level of the putative regulator gene iclR, located in front of the cluster of genes encoding three oxidoreductases, was not increased during growth on myo-inositol.
We also wanted to know whether mRNA levels of cells grown on complex medium consisting of brain heart extract (BHI) also influence iol gene expression and compared the mRNA levels of cells grown on BHI with those of cells grown on CGXII salt medium containing glucose (Table 2). Indeed, a number of iol genes responded and ips was downregulated, suggesting that inositol utilization is part of cell mass generation during growth on the complex medium BHI. This is in agreement with the detection of two Iol proteins of C. glutamicum grown on complex medium (15).
Characterization of selected iol mutants.
The large number of genes apparently related to myo-inositol catabolism in C. glutamicum is intriguing. Unfortunately, functional studies of enzymes and genes of inositol metabolism are scarce (Fig. 1). However, cleavage of the putative intermediate 2-deoxy-5-keto-d-gluconic acid is considered a key step in myo-inositol metabolism, as shown in the early studies of K. aerogenes by Anderson and Magasanik (2). Since cleavage of α-ketols in carbohydrate metabolism is typically performed by thiamine-pyrophosphate-dependent enzymes and the iolD gene product of C. glutamicum possesses a corresponding binding site (not shown), a vector was constructed to delete this gene from the wild type (WT) of C. glutamicum (see Materials and Methods). The resulting mutant, WTΔiolD, was unable to grow on myo-inositol, whereas growth on glucose was hardly affected (Fig. 2B). Thus, iolD within cluster I is essential for myo-inositol utilization by C. glutamicum.
As many as six genes for oxidoreductases have increased expression levels upon myo-inositol utilization. PFAM analysis (3) identified three oxidoreductases (iolG, oxiA, oxiB) within cluster I and an additional three in cluster II (oxiC, oxiD, oxiE). Since a iolG orthologue is present within the B. subtilis iol operon (39% identities), we inactivated the orthologue to generate strain WT::piolG′. As shown in Fig. 2B, its growth on inositol was reduced compared to that of the control but was still possible. Therefore, we considered that one of the other oxidoreductases might partially substitute for the function of iolG. IolG exhibits identities of 27% to OxiE, 22% to OxiD, and still 19% to OxiC over the entire lengths of the proteins, whereas identities to OxiA and OxiB were significantly less. The oxidoreductase encoded by idhA of S. meliloti (11) is, apart from iolE of B. subtilis (31), the only functionally identified gene of bacterial inositol metabolism, and OxiE, OxiD, and OxiC in cluster II exhibit high identities to IdhA but low identities to OxiA and OxiB. We therefore considered that the three oxidoreductases in cluster II might have some overlapping activity with the function of iolG and constructed plasmid pK19mobsacBΔoxiII to delete the 4.072-kb region of cluster II encompassing oxiC to oxiE (Fig. 3). The growth of generated strain WTΔoxiII is shown in Fig. 2B. Growth on neither inositol nor glucose (not shown) was influenced by the deletion. We subsequently inactivated iolG in the strain with the four genes of cluster II deleted to generate WTΔoxiII::piolG′. This strain was no longer able to grow on myo-inositol. This result indicates that the genes of cluster II play a subordinate role in myo-inositol utilization and that apparently a number of overlapping oxidoreductase activities exist in C. glutamicum which can be used to enable growth on this carbon source. Due to the growth characteristics of WTΔoxiII::piolG′, we hypothesized that further genes of cluster II might be redundant or unnecessary for myo-inositol utilization. To this end, plasmid pK19mobsacBΔiolII was constructed and the 8.799-kb chromosomal region extending from adhA to oxiE (Fig. 3) was deleted to generate WTΔiolII. Growth on myo-inositol was not reduced by this deletion either (not shown), which indicates that cluster II is dispensable for myo-inositol metabolism and that the corresponding genes might play an as-yet-undiscovered role or indicate ongoing evolution although the data show that they are at least in part functional.
Characterization of transporter mutants.
There are three carrier genes which exhibit an at least 12-fold increase in the mRNA level upon myo-inositol utilization: mfs in cluster I, remote gene iolT1, and iolT2 in cluster II (Fig. 3). All transporters belong to the major facilitator superfamily and exhibit identities to sugar transporters or, in the case of mfs, also identities to annotated efflux carriers. The presence of three transporters resembles the situation in B. subtilis, which possesses two transporters for inositol uptake (28). To move toward an analysis of the function of the transporters in C. glutamicum, we inactivated each of these three genes individually in strain ATCC 21527, an l-lysine producer, but growth was not hampered on CGXII plates containing as a carbon source sorbitol, glucose, ribose, fructose, arabitol, gluconate, or saccharose (each at 40 g liter−l) compared to that of controls (not shown). Following this observation, we deleted the transporter genes iolT1 and iolT2 in C. glutamicum strain ATCC 13032 (WT) individually and together. The growth of the resulting strains is shown in Fig. 2C. Whereas the growth of WTΔiolT1 and WTΔiolT2 on myo-inositol or glucose was not influenced, the growth of WTΔiolT1ΔiolT2 on myo-inositol was disabled but its growth on glucose was not. Consequently, both the iolT1- and iolT2-encoded carriers appear to catalyze inositol uptake, whereas mfs1, although located within the putative iolA-oxiA operon (Fig. 3), does not.
Kinetic characterization of IolT1 and IolT2.
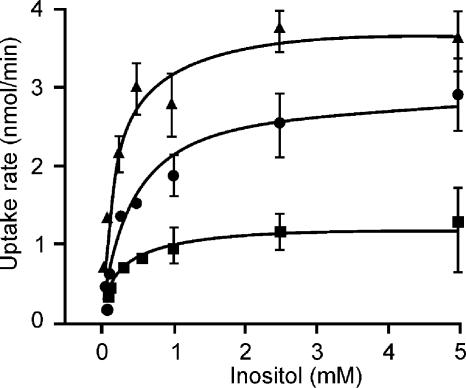
Cells were grown on CGXII with myo-inositol up to an OD600 of about 4 to 6, washed twice with CGXII without a carbon source, and stored on ice to quantify myo-[1,2-3H]inositol uptake in a rapid filtration assay. Saturation curves were obtained from the initial linear uptake rate (at least three data points over 40 s) at each substrate concentration. For the WT, nonlinear regression analysis yielded an apparent Michaelis constant (Km) of 0.20 ± 0.04 mM myo-inositol concentration and a Vmax value of 3.79 ± 0.19 nmol min−1 (mg cells)−1 (Fig. 4). The mutants with individually deleted IolT1 or IolT2 were analyzed in an identical manner. Assuming that in WTΔiolT2 only IolT1 is active, as is indicated by the inability of the double mutant to utilize myo-inositol, IolT1 is characterized by a Km of 0.22 ± 0.04 mM myo-inositol and a Vmax value of 1.22 ± 0.05 nmol min−1 (mg cells)−1 and IolT2 is characterized by a Km of 0.45 ± 0.09 mM myo-inositol and a Vmax value of 2.90 ± 0.18 nmol min−1 (mg cells)−1. Thus, both carriers have comparable kinetic constants, which is probably not surprising since both share a high degree of identity of 55%, although two insertions of up to 15 aminoacyl residues are present in IolT1.
FIG. 4.
Substrate saturation kinetics of myo-inositol uptake with C. glutamicum. Uptake of myo-[1,2-3H]inositol by C. glutamicum cells was monitored between myo-inositol concentrations of 0.05 and 5 mM for 55 s in CGXII by the rapid-filtration technique. From the initial uptake rate at each substrate concentration, the Michaelis-Menten plots were derived by nonlinear regression analysis. Uptake by the WT (▴), the ΔiolT1 mutant (▪), and the ΔiolT2 mutant (•) is shown.
Effect of myo-inositol utilization on l-lysine accumulation.
C. glutamicum is used to satisfy the worldwide demand for l-lysine of more than 600,000 tons per year (8), and it is well known that with the use of fructose or sucrose, l-lysine accumulation is reduced compared to that achieved with glucose (13, 18). Although myo-inositol catabolism is only partially known (2, 19, 31), it does not involve complete glycolysis (Fig. 1) and therefore represents an entirely different flux and energetic situation compared to glucose or fructose, for instance. We grew l-lysine-producing strain C. glutamicum DM1730 (13) on salt medium CGXII with glucose and myo-inositol as a carbon source, as well as on mixtures of these compounds. After 48 h, the substrates were consumed and the l-lysine concentration was 22.7 mM with myo-inositol (Table 3). However, with glucose it was 63.6 mM, which might indicate a reduced supply of the l-lysine building blocks oxaloacetate and pyruvate for l-lysine synthesis. Also with the classically derived l-lysine producer MH20-22B (24), the yields were drastically reduced on myo-inositol compared to those achieved with glucose (not shown). Interestingly, even the smallest concentration of 16.6 mM myo-inositol added to 186.8 mM glucose reduced the l-lysine yield by 29% (yield of 0.22 instead 0.31), although myo-inositol contributed only 8.2% of the total carbon, illustrating a possible regulatory influence of the polyol.
TABLE 3.
l-Lysine formation with C. glutamicum strain DM1730 as a function of the substrates glucose and myo-inositol and mixtures of these substrates
| Timea | Glucose concn (mM) | Inositol concn (mM) | Growth (OD) | l-Lysineb concn (mM) | Yieldc (mol/mol) |
|---|---|---|---|---|---|
| I | 202.0 | 0 | 14 | 37.8 | 0.19 |
| I | 186.8 | 16.6 | 24 | 40.5 | 0.20 |
| I | 176.7 | 27.7 | 23 | 38.1 | 0.19 |
| I | 0 | 222.0 | 17 | 23.0 | 0.10 |
| II | 202.0 | 0 | 21 | 63.6 | 0.31 |
| II | 186.8 | 16.6 | 23 | 44.1 | 0.22 |
| II | 176.7 | 27.7 | 24 | 38.3 | 0.19 |
| II | 0 | 222.0 | 19 | 22.7 | 0.10 |
Time I gives growth, l-lysine concentration, and yield after 24 h, whereas time II gives the values after 48 h, when all of the sugar was consumed.
All data are mean values of at least three independent cultivations with errors of <5% for l-lysine concentrations and yields.
Molar yields are given as moles of l-lysine per mole of carbon source consumed.
DISCUSSION
It was surprising to find that myo-inositol enables such excellent growth of C. glutamicum, since similar growth has hardly been reported elsewhere, with the exception of K. (Aerobacter) aerogenes (19). Indeed, the mRNA populations quantified indicate well-balanced growth conditions similar to that on glucose, since only a very limited number of genes are differentially expressed and genes of ribosomal proteins, which often respond to starvation conditions, are absent (14). The most strongly downregulated gene is ips, and it is logical to assume that this is a direct consequence of the presence of myo-inositol. Although the mechanism of regulation is still to be discovered, regulation of ips opens up the possibility of controlling lipomannan and lipoarabinomannan synthesis as an interesting target for reducing the viability of Corynebacterineae such as M. tuberculosis (5) by the synthesis of these myo-inositol-containing glycolipids.
The number of genes exhibiting increased expression upon myo-inositol catabolism was puzzling at the beginning of our work, in particular since knowledge of myo-inositol utilization in general is rather limited. Thus, according to the array analysis, in principle, as many as six oxidoreductases could be required for myo-inositol utilization, as well as three transporters and two isomerase-epimerases (Table 2). However, the deletion of all of cluster II and growth of the corresponding mutant WTΔiolII are strong evidence that cluster II encodes redundant functions of myo-inositol utilization as specifically demonstrated for uptake and oxidation steps within the catabolism of the polyol. Moreover, the genomic region of cluster II encompassing the adjacent catA-adh region exhibits amazingly high identities at the nucleotide level of up to 74% to NCgl1112 and NCgl1113, located elsewhere in the chromosome, showing that gene duplication within C. glutamicum might also be involved in the formation of cluster II. Also the high identity of 30% between oxiD and oxiE at the protein level and even at the nucleotide level (not shown) illustrates that this genomic region does not belong to regions encoding conserved cellular core functions like cell wall synthesis, for instance (1, 12). Instead, this region appears to be rather the result of a more recent event of genome alteration. This is in full accord with the absence of iol genes in C. efficiens, C. diphtheriae, and C. jeikeium indicating a specific and fortuitous acquisition of these genes by C. glutamicum.
Cluster I encodes relevant functions for myo-inositol catabolism in C. glutamicum, as evident from the consequences of iolD inactivation and the iolG mutation in the ΔoxiII background. As evident from the early biochemical work of Magasanik and coworkers (2, 19), myo-inositol catabolism might involve oxidative steps, epimerization, phosphorylation of a linear diketo-deoxy-inositol, cleavage, and a further oxidative step to yield acetyl-CoA and dihydroxyacetone-P (Fig. 1). Interestingly, orthologues of six genes of cluster I are present and largely syntenic to the organization of C. glutamicum in B. subtilis (29), B. halodurans, Clostridium perfringens (17), and Yersinia pseudotuberculosis. Therefore, these genes are likely to encode the key functions to catabolize myo-inositol, as discussed in detail by Magasanik. This relates to iolG and iolE, whose functions have been identified (10, 31); to iolC, whose structural characteristics according to PFAM analysis (3) indicate that it encodes a 5-dehydro-2-deoxygluconokinase; iolD, which encodes a thiamine pyrophosphate-dependent enzyme typically cleaving sugar phosphates; and iolA, which encodes an aldehyde dehydrogenase. The iolB-encoded protein is also conserved but does not have a PFAM entry, consequently representing a protein with no structural counterpart in the current databases. The initial oxidative steps to form a cleavable diketo intermediate from myo-inositol might differ among the bacteria. At least the genome comparisons did not allow us to specify a core requirement for oxidoreductases.
Uptake of myo-inositol to sustain maximal growth is possible by either IolT1 or IolT2, and the genes for both are expressed to similar degrees (Table 2). They also have comparable kinetic properties, which is probably not surprising due to their high sequence identity of 55%. The IolT proteins are 12-membrane spanners belonging to the major facilitator superfamily. Also, B. subtilis has two inositol uptake carriers that are similar in structure, but these share fewer sequence identities than do the C. glutamicum proteins. In marked contrast to C. glutamicum, in B. subtilis neither transporter can be substituted for the other since they represent a minor and a major myo-inositol transporter (28). It is surprising that the third transporter of C. glutamicum analyzed in the present study is part of cluster I but nevertheless is not involved in myo-inositol uptake. Instead, it shares identities with efflux pumps, indicating together with its absence in the iol locus of other bacteria, its fortuitous presence in cluster I.
The l-lysine formation data showed a reduced yield with myo-inositol as a carbon source compared to glucose. It is known that the amino acid yield strongly depends on the type of substrate. The highest l-lysine yields with C. glutamicum are obtained with glucose, and the lowest are obtained with fructose (13, 18). Thus, myo-inositol ranks among the substrates giving rather low yields. This could be due to the fact that pyruvate and oxaloacetate, required as building blocks for l-lysine, probably do not result from myo-inositol catabolism (2, 19). Surprising is the fact that there is a significant yield reduction when myo-inositol is present at relatively low supplementary concentrations in addition to glucose. Although myo-inositol contributes about 8% of the total carbon in a mixture with glucose, the resulting yield is reduced by as much as 29% (Table 3). The effect of cells grown in different complex media and used as inoculum on the final yield is well established (25), and the present study, as well as a proteome study (15), shows that iol genes are expressed on complex media usually used to derive the inoculum. However, in our experiments the preculture medium for deriving the inoculum was identical to the culture medium where the yields were determined. We envisage two possibilities. Although we did no DNA microarray analysis for cells grown on a glucose-inositol mixture, myo-inositol undoubtedly affects expression of genes of central metabolism, like components of the phosphotransferase system (see above), as well as mez, malic enzyme, or sucC, encoding a succinyl-CoA synthetase subunit (Table 2). Moreover, enzyme activities could be influenced by different metabolite concentrations. The other possibility is that due to the presence of myo-inositol the cell wall and its lipomannan composition are influenced so as to reduce l-lysine export. It is known that in C. glutamicum the cellular lipid composition influences amino acid efflux properties (6).
Footnotes
Published ahead of print on 22 September 2006.
REFERENCES
- 1.Alderwick, L. J., M. Seidel, H. Sahm, G. S. Besra, and L. Eggeling. 2006. Identification of a novel arabinofuranosyl transferase (AftA) involved in cell wall arabinan biosynthesis in Mycobacterium tuberculosis. J. Biol. Chem. 281:15653-15661. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, W. A., and B. Magasanik. 1971. The pathway of myo-inositol degradation in Aerobacter aerogenes. J. Biol. Chem. 246:5662-5675. [PubMed] [Google Scholar]
- 3.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellmann, A., M. Vrljic, M. Patek, H. Sahm, R. Krämer, and L. Eggeling. 2001. Expression control and specificity of the basic amino acid exporter LysE of Corynebacterium glutamicum. Microbiology 147:1765-1774. [DOI] [PubMed] [Google Scholar]
- 5.Brennan, P. J., and H. Nikaido. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 64:29-63. [DOI] [PubMed] [Google Scholar]
- 6.Eggeling, L., and H. Sahm. 2001. The cell wall barrier of Corynebacterium glutamicum and amino acid efflux. J. Biosci. Bioeng. 92:201-213. [Google Scholar]
- 7.Eggeling, L., and H. Sahm. 1985. The formaldehyde dehydrogenase of Rhodococcus erythropolis, a trimeric enzyme requiring a cofactor and active with alcohols. Eur. J. Biochem. 150:129-134. [DOI] [PubMed] [Google Scholar]
- 8.Eggeling, L., and M. Bott. 2005. Handbook of Corynebacterium glutamicum. CRC Press, Inc., Boca Raton, Fla.
- 9.Fry, J., M. Wood, and P. S. Poole. 2001. Investigation of myo-inositol catabolism in Rhizobium leguminosarum bv. viciae and its effect on nodulation competitiveness. Mol. Plant-Microbe Interact. 14:1016-1025. [DOI] [PubMed] [Google Scholar]
- 10.Fujita, Y., K. Shindo, Y. Miwa, and K. Yoshida. 1991. Bacillus subtilis inositol dehydrogenase-encoding gene (idh): sequence and expression in Escherichia coli. Gene 108:121-125. [DOI] [PubMed] [Google Scholar]
- 11.Galbraith, M. P., S. F. Feng, J. Borneman, E. W. Triplett, F. J. de Bruijn, and S. Rossbach. 1998. A functional myo-inositol catabolism pathway is essential for rhizopine utilization by Sinorhizobium meliloti. Microbiology 144:2915-2924. [DOI] [PubMed] [Google Scholar]
- 12.Gande, R., K. J. Gibson, A. K. Brown, K. Krumbach, L. G. Dover, H. Sahm, S. Shioyama, T. Oikawa, G. S. Besra, and L. Eggeling. 2004. Acyl-CoA carboxylases (accD2 and accD3), together with a unique polyketide synthase (Cg-pks), are key to mycolic acid biosynthesis in Corynebacterineae such as Corynebacterium glutamicum and Mycobacterium tuberculosis. J. Biol. Chem. 279:44847-44857. [DOI] [PubMed] [Google Scholar]
- 13.Georgi, T., D. Rittmann, and V. F. Wendisch. 2005. Lysine and glutamate production by Corynebacterium glutamicum on glucose, fructose and sucrose: roles of malic enzyme and fructose-1,6-bisphosphatase. Metab. Eng. 7:291-301. [DOI] [PubMed] [Google Scholar]
- 14.Gourse, R. L., T. Gaal, M. S. Bartlett, J. A. Appleman, and W. Ross. 1996. rRNA transcription and growth rate-dependent regulation of ribosome synthesis in Escherichia coli. Annu. Rev. Microbiol. 50:645-677. [DOI] [PubMed] [Google Scholar]
- 15.Hansmeier, N., T. C. Chao, A. Pühler, A. Tauch, and J. Kalinowski. 2006. The cytosolic, cell surface and extracellular proteomes of the biotechnologically important soil bacterium Corynebacterium efficiens YS-314 in comparison to those of Corynebacterium glutamicum ATCC 13032. Proteomics 6:233-250. [DOI] [PubMed] [Google Scholar]
- 16.Jiang, G., A. H. Krishnan, Y.-W. Kim, T. J. Wacek, and H. B. Krishnan. 2001. A functional myo-inositol dehydrogenase gene is required for efficient nitrogen fixation and competitiveness of Sinorhizobium fredii USDA191 to nodulate soybean (Glycine max [L.] Merr.). J. Bacteriol. 183:2595-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawsar, H. I., K. Ohtani, K. Okumura, H. Hayashi, and T. Shimizu. 2004. Organization and transcriptional regulation of myo-inositol operon in Clostridium perfringens. FEMS Microbiol. Lett. 235:289-295. [DOI] [PubMed] [Google Scholar]
- 18.Kiefer, P., E. Heinzle, O. Zelder, and C. Wittmann. 2004. Comparative metabolic flux analysis of lysine-producing Corynebacterium glutamicum cultured on glucose or fructose. Appl. Environ Microbiol. 70:229-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magasanik, B. 1953. Enzymatic adaptation in the metabolism of cyclitols in Aerobacter aerogenes. J. Biol. Chem. 205:1007-1018. [PubMed] [Google Scholar]
- 20.Norman, R. A., M. S. McAlister, J. Murray-Rust, F. Movahedzadeh, N. G. Stoker, and N. Q. McDonald. 2002. Crystal structure of inositol 1-phosphate synthase from Mycobacterium tuberculosis, a key enzyme in phosphatidylinositol synthesis. Structure 10:393-402. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Sareen, D., G. L. Newton, R. C. Fahey, and N. A. Buchmeier. 2003. Mycothiol is essential for growth of Mycobacterium tuberculosis Erdman. J. Bacteriol. 185:6736-6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 24.Schrumpf, B., L. Eggeling, and H. Sahm. 1992. Isolation and prominent characteristics of an l-lysine hyperproducing strain of Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 37:566-571. [Google Scholar]
- 25.Sonntag, K., L. Eggeling, A. A. De Graaf, and H. Sahm. 1993. Flux partitioning in the split pathway of lysine synthesis in Corynebacterium glutamicum. Quantification by 13C- and 1H-NMR spectroscopy. Eur. J. Biochem. 213:1325-1331. [DOI] [PubMed] [Google Scholar]
- 26.Turner, B. L., and A. L. Richardson. 2004. Identification of scyllo-inositol phosphates in soil by solution phosphorus-31 nuclear magnetic resonance spectroscopy. Soil Sci. Soc. Am. J. 68:802-808. [Google Scholar]
- 27.Wendisch, V. F. 2003. Genome-wide expression analysis in Corynebacterium glutamicum using DNA microarrays. J. Biotechnol. 104:273-285. [DOI] [PubMed] [Google Scholar]
- 28.Yoshida, K., Y. Yamamoto, K. Omae, M. Yamamoto, and Y. Fujita. 2002. Identification of two myo-inositol transporter genes of Bacillus subtilis. J. Bacteriol. 184:983-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshida, K.-I., D. Aoyama, I. Ishio, T. Shibayama, and Y. Fujita. 1997. Organization and transcription of the myo-inositol operon, iol, of Bacillus subtilis. J. Bacteriol. 179:4591-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida, K., K. Kobayashi, Y. Miwa, C.-M. Kang, M. Matsunaga, H. Yamaguchi, S. Tojo, M. Yamamoto, R. Nishi, N. Ogasawara, T. Nakayama, and Y. Fujita. 2001. Combined transcriptome and proteome analysis as a powerful approach to study genes under glucose repression in Bacillus subtilis. Nucleic Acids Res. 29:683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshida, K., M. Yamaguchi, H. Ikeda, K. Omae, K. Tsurusaki, and Y. Fujita. 2004. The fifth gene of the iol operon of Bacillus subtilis, iolE, encodes 2-keto-myo-inositol dehydratase. Microbiology 150:571-580. [DOI] [PubMed] [Google Scholar]