Abstract
The barA and uvrY genes of Escherichia coli encode a two-component sensor kinase and a response regulator, respectively. Although this system plays a major role in the regulation of central carbon metabolism, motility, and biofilm formation by controlling the expression of the CsrB and CsrC noncoding RNAs, the environmental conditions and the physiological signal(s) to which it responds remain obscure. In this study, we explored the effect of external pH on the activity of BarA/UvrY. Our results indicate that a pH lower than 5.5 provides an environment that does not allow activation of the BarA/UvrY signaling pathway.
The BarA (bacterial adaptive response) sensor kinase of Escherichia coli is a member of the subclass of tripartite sensor kinases that contain a primary transmitter domain, a central receiver domain, and a C-terminal phosphotransfer domain (12). BarA is the cognate kinase of UvrY, a response regulator belonging to the FixJ family (14). The designation uvrY is derived from the close linkage on a bicistronic mRNA with the uvrC gene, which encodes a subunit of the UvrABC DNA repair complex. However, mutations in uvrY have no effect on the UV-light-induced DNA repair response (11).
The E. coli BarA/UvrY two-component system (TCS) and its homologues in other gram-negative bacteria, such as BarA/SirA of Salmonella, ExpS/ExpA of Erwinia, VarS/VarA of Vibrio, and GacS/GacA of Pseudomonas species, have been shown to positively control expression of noncoding RNAs, including CsrB and CsrC in E. coli (9, 16, 23). These small regulatory RNAs together with the 6.8-kDa CsrA protein constitute the Csr (carbon storage regulation) system of E. coli, which has a major impact on regulation of carbon metabolism pathways (16, 23). Consequently, deletion of the barA or uvrY gene in E. coli has drastic effects on the ability of the bacteria to successfully grow in competition with the wild-type strain depending on the carbon source in the medium, suggesting that BarA and UvrY are crucial for efficient adaptation to different metabolic pathways (13). Mutations in the Csr system in E. coli and Salmonella enterica also cause other phenotypic effects, including changes in motility, adhesion, and biofilm formation (1, 7, 18, 19, 22). Furthermore, the BarA/UvrY TCS and its homologues have a clear link to genes involved in bacterial virulence (1-3, 6, 15, 20).
The environmental conditions and the physiological signal(s) to which BarA and its homologue sensor kinases respond have not been identified. It has been suggested that BarA may respond to the sensing of the host organism by the bacteria during an infection (24), as many of the target genes are involved in pathogenesis. Experimental evidence, however, shows that this system is active in the absence of cell attachment and in the absence of any host organism (13, 18).
External pH can be shifted substantially by bacterial metabolism. For instance, growth on sugars, especially if oxygen becomes limiting, produces organic acids that are excreted and lead to a low pH, whereas growth on amino acids generates alkaline amines, producing the opposite effect. This fact together with the observation that BarA/UvrY is needed for efficient switching between glycolytic and gluconeogenic carbon sources (13) encouraged us to explore the effect of external pH on the activity of this TCS.
Effect of external pH on the activity of BarA/UvrY.
The activity of the BarA/UvrY TCS was tested using the widest pH range (pH 5.0 to 9.0) in which cultures maintained reasonable generation times by monitoring the expression of a chromosomal Φ(csrB-lacZ) transcriptional fusion.
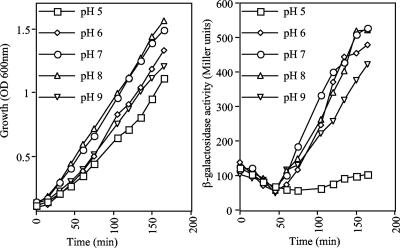
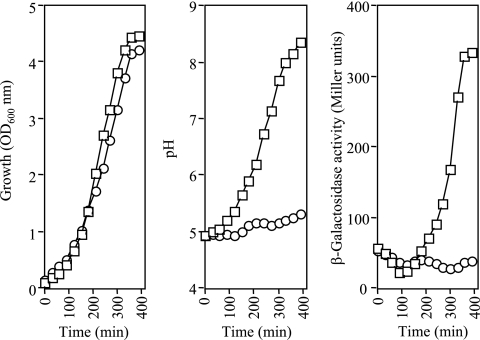
The csrB-lacZ-carrying strain KSB837 (4) was grown in Luria-Bertani (LB) broth, the pH of which was adjusted and buffered to pH 5.0, 6.0, 7.0, 8.0, and 9.0, and the growth and expression of the reporter were monitored (Fig. 1). As expected, the growth rates of cultures at pH 7 and 8 were somewhat higher than the growth rates of cultures at pH 6 and 9, while the growth rate of the culture at pH 5 was slightly lower than the growth rates of cultures at pH 6 and 9. On the other hand, the expression of Φ(csrB-lacZ) was similar in all cultures in the initial phase (first 50 min) of growth, exhibiting a small decrease, indicating that the BarA/UvrY system was not active. However, at an optical density at 600 nm of approximately 0.5, reporter expression resumed and increased constantly in cultures grown at pH 6, 7, 8, and 9, indicating that there was activation of the BarA/UvrY system. In contrast, reporter expression did not resume at pH 5, even though the culture reached an optical density greater than 1.0 (Fig. 1), suggesting that the BarA/UvrY system was not operative. The expression of Φ(csrB-lacZ) in barA and uvrY mutant strains BAKSB837 and UYKSB837 (18) under the pH conditions described above was also tested. No induction of the reporter was observed under any condition (data not shown).
FIG. 1.
Effect of external pH on the activity of BarA/UvrY. Strain KSB837, carrying the Φ(csrB-lacZ) transcriptional fusion, was grown in LB medium whose pH had been adjusted and buffered to 5, 6, 7, 8, and 9. The following buffers were used at a concentration of 0.1 M: for pH 5, homopiperazine-N,N′-bis-2-(ethanesulfonic acid) (HOMOPIPES); for pH 6, 2-(N-morpholino)ethanesulfonic acid (MES); for pH 7, 3-(N-morpholino)propanesulfonic acid (MOPS); for pH 8, 3-[N-tris(hydroxymethyl)methylamino]propanesulfonic acid (TAPS); and for pH 9, 3-[(1,1-dimethyl-2-hydroxyethyl)amino]-2-hydroxypropanesulfonic acid (AMPSO). (Left panel) Optical density at 600 nm (OD 600nm) as a function of time. (Right panel) β-Galactosidase activity expressed from the csrB-lacZ transcriptional fusion. The experiment was repeated three times in its entirety, and essentially identical results were obtained.
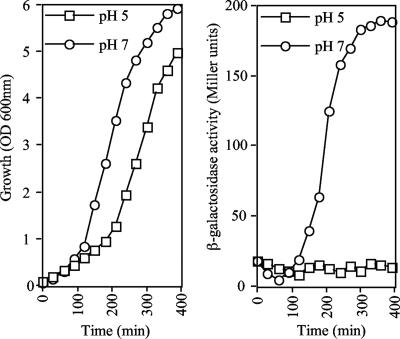
We next argued that if the observed pH regulation were BarA/UvrY dependent, the expression of CsrC, another BarA/UvrY-controlled noncoding RNA, should behave like the expression of CsrB. To test this hypothesis, the Φ(csrC-lacZ)-carrying strain GS1114 (23) was grown at pH 5 and 7, and the expression of the reporter was monitored at 30-min intervals (Fig. 2). No induction of the csrC-lacZ expression was observed in the culture grown at pH 5, whereas robust induction was observed in the culture grown at pH 7. Therefore, the most appealing interpretation is that pH regulation of CsrB and CsrC expression is BarA/UvrY dependent and that the BarA/UvrY TCS is inactive at low pH values.
FIG. 2.
Effect of external pH on csrC-lacZ expression. Strain GS1114, carrying the Φ(csrC-lacZ) transcriptional fusion, was grown in LB medium whose pH had been adjusted and buffered to 5 and 7. (Left panel) Optical density at 600 nm (OD 600nm) as a function of time. (Right panel) β-Galactosidase activity expressed from the csrC-lacZ transcriptional fusion. The experiment was repeated three times in its entirety, and essentially identical results were obtained.
The lack of BarA/UvrY activity at pH 5 could be due to either the absence of a BarA-stimulating signal or the formation of a BarA-inhibiting signal. An alternative explanation is that expression of barA and/or uvrY is repressed at pH 5. A third possibility is that low pH affects the activities of all TCSs and not specifically the activity of BarA/UvrY.
Low pH does not affect the activities of all TCSs.
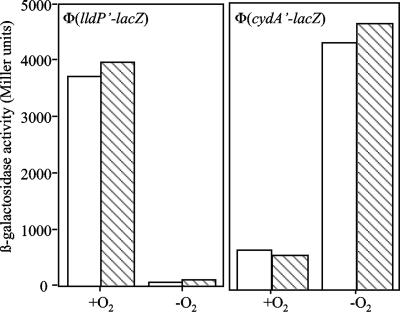
To address the question of whether a low pH influences the activities of TCSs in general, we tested the effect of low pH on the activity of the ArcB/ArcA system. Under anaerobic conditions, ArcB autophosphorylates and transphosphorylates ArcA. Phosphorylated ArcA, in turn, positively or negatively modulates the expression of many operons. For instance, during anaerobiosis ArcA-P positively regulates the cydAB operon and negatively regulates the lldPRD operon (10).
Strains ECL5002 and ECL5003 (8), carrying chromosomal Φ(cydA′-lacZ) and Φ(lldP′-lacZ) reporter fusions, respectively, were grown aerobically or anaerobically in LB broth whose pH had been adjusted and buffered to pH 5 or 7. In the mid-exponential growth phase, the expression of the reporters was assayed, and it was found that under anaerobic conditions, the Arc system was able to activate cydA′-lacZ expression and to repress lldP′-lacZ expression equally well at pH 5 and pH 7 (Fig. 3). As expected, no regulation of cydA′-lacZ and lldP′-lacZ expression was observed in arcB mutant strains ECL5004 and ECL5012 (8) under the two pH conditions tested (data not shown). Thus, low pH does not provide an environment that influenced the activity of the Arc system, suggesting that the observed pH effect on the activity of the BarA/UvrY system is rather specific and not general for all TCSs.
FIG. 3.
Effect of external pH on the activity of ArcB/ArcA. The Φ(cydA′-lacZ)-carrying strain ECL5002 (right panel) and the Φ(lldP′-lacZ)-carrying strain ECL5003 (left panel) were grown aerobically or anaerobically in buffered LB medium at pH 5 (open bars) or pH 7 (striped bars). In the mid-exponential growth phase, β-galactosidase activity was assayed and the results were expressed in Miller units. The data are averages from four experiments (the variations were less than 5% of the means).
Low pH does not affect the expression of barA or uvrY.
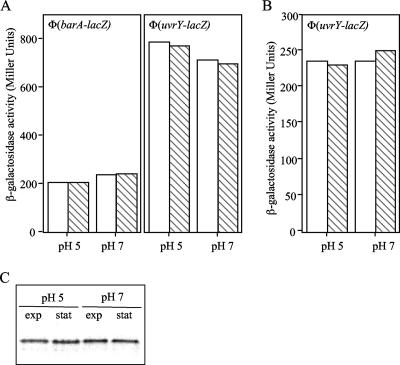
We next asked whether a low external pH affects the expression of the barA and uvrY genes. To determine this, we constructed strains IFC601 and IFC602, which carried a Φ(barA′-lacZ) transcriptional fusion and a Φ(uvrY′-lacZ) transcriptional fusion, respectively. The promoter regions of barA and uvrY were amplified by PCR using total genomic DNA as the template along with primers 5′-GGAATTCCCGACCACGGCAGC-3′ and 5′-CGGGATCCCATGGAGTTCCGTTATGGGAC-3′ and primers 5′-GGAATTCGAAAGTCTGATGG-3′ and 5′-CGGGATCCTCAAAGGAATATCTCC-3′, respectively. The PCR product was digested with EcoRI-BamHI and cloned between the EcoRI-BamHI-digested lacZ operon fusion vector pRS415 (17), generating plasmids pMX601 and pMX602. The barA′-lacZ and uvrY′-lacZ fusions were then transferred to the λ transducing phage λRS45 (17). High-titer lysates were used to lysogenize a wild-type strain, and single lysogens were selected, yielding strains IFC601 and IFC602. The constructed strains were grown in buffered LB broth at pH 5 or pH 7, and their β-galactosidase activities were determined in the mid-exponential and stationary growth phases (Fig. 4A). We found that neither the pH nor the growth stage had a significant effect on the transcription of the two reporters. The efficiency of translation of the two genes was examined by using strain KSY009, which carries a chromosomal Φ(uvrY′-lacZ) translational fusion (18), and by Western blot analysis using BarA-specific antibodies (21) and crude extracts of cells grown under the conditions described above. No differences in translation of the uvrY reporter (Fig. 4B) or the BarA protein levels were observed (Fig. 4C). Thus, low pH does not affect the expression of barA or uvrY but rather provides a nonstimulating environment for BarA/UvrY that most probably affects the activity of the BarA sensor kinase.
FIG. 4.
Effect of external pH on the expression of barA and uvrY. (A and B) Strains IFC601 and IFC602 carrying the Φ(barA′-lacZ) and Φ(uvrY′-lacZ) transcriptional fusions, respectively (A), and strain KSY009 carrying the Φ(uvrY′-lacZ) translational fusion (B) were cultured in buffered LB medium containing 0.1 M HOMOPIPES (pH 5) or 0.1 M MOPS (pH 7), and the β-galactosidase activity was assayed in the mid-exponential growth phase (open bars) or stationary growth phase (striped bars). The averages of four experiments are shown (the variations were less than 5% of the means). (C) Western blot analysis. Equal numbers of bacteria, grown as described above, were harvested and solubilized by incubation at 95°C for 5 min in 2× sodium dodecyl sulfate sample buffer. Samples were subjected to electrophoresis in a sodium dodecyl sulfate-10% polyacrylamide gel, and the resolved proteins were electrotransferred to a Hybond-ECL filter (Amersham). Immunoblot analyses were subsequently performed, using BarA polyclonal antibodies as previously described (21). exp, exponential growth phase; stat, stationary growth phase.
pH lower than 5.5 does not allow activation of the BarA/UvrY TCS.
Finally, we attempted to identify with more precision the pH that allows activation of the BarA/UvrY signaling cascade. To do this, the Φ(csrB-lacZ)-carrying strain KSB837 was grown in unbuffered or buffered [with 0.1 M homopiperazine-N,N′-bis-2-(ethanesulfonic acid) (HOMOPIPES)] LB medium (initial pH 5), and the expression of the reporter and the external pH were monitored at 30-min intervals (Fig. 5). As expected, the pH of the buffered culture remained reasonably constant, reaching a maximum of 5.4 at the end of growth. Also, in agreement with the results shown in Fig. 1, no activation of csrB-lacZ expression was observed, despite the fact that the optical density at 600 nm of the culture reached a value higher than 4. In contrast, the pH of the culture grown in unbuffered LB medium progressively increased to more than pH 8. Interestingly, the slight decline in csrB-lacZ expression during the initial growth was reversed at pH 5.6 ± 0.1, and reporter expression steadily increased until the culture reached the stationary phase. Therefore, we conclude that pH 5.5 provides an environmental threshold for BarA activation.
FIG. 5.
Growth of E. coli KSB837 and effects of the culture pH. Strain KSB837, carrying the Φ(csrB-lacZ) transcriptional fusion, was grown in unbuffered LB medium having an initial pH of 5 (○) or LB medium buffered with 0.1 M HOMOPIPES at pH 5 (□). (Left panel) Optical density at 600 nm (OD600 nm) as a function of time. (Middle panel) Culture pH as a function of time. (Right panel) β-Galactosidase activity of csrB-lacZ expressed in Miller units. The experiment was repeated three times in its entirety, and essentially identical results were obtained.
Concluding remarks.
The data presented in this report demonstrate that pH values lower than 5.5 provide an environment that does not allow activation of the BarA/UvrY signaling pathway. The observed pH effect appears to be specific for the BarA/UvrY TCS and does not seem to be exerted through modulation of the expression of barA or uvrY. Therefore, we propose that low-pH growth conditions affect BarA directly by not permitting activation of its kinase activity. Such an effect could be the result of the absence of a stimulating signal or the presence of an inhibitory signal. Although the chemical nature of such a signal remains to be discovered, it is interesting that a solvent-extractable extracellular signal, capable of inducing moderate activation of GacS/GacA-regulated targets, was isolated from stationary-phase cultures of Pseudomonas fluorescens (5, 25). This finding suggests that GacS, and most likely its orthologous sensor kinases, could respond to an extracellular molecule that is produced or accumulates in stationary phase. In view of the data presented here, the stability of such a molecule should be compromised and/or its synthesis should be blocked at a low external pH. However, an alternative explanation that cannot be excluded at this stage is that BarA senses directly the pH difference across the inner cell membrane. More detailed studies directed toward understanding the nature of the stimulus for BarA need to be performed. Our finding that the BarA/UvrY system is kept silent under low-pH growth conditions provides an important physiological tool for further experimentation in this direction.
Acknowledgments
We thank Claudia Rodriguez for technical assistance and Diego González-Halphen and Bertha Michel for critically reading the manuscript.
This work was supported by grant 37342-N from the Consejo Nacional de Ciencia y Tecnología (CONACyT), by grant IN221106/17 from DGAPA-PAPIIT, UNAM, by grant MEST-CT-2004-8475 from Marie Curie, and by grant GM59969 from NIH.
Footnotes
Published ahead of print on 15 September 2006.
REFERENCES
- 1.Altier, C., M. Suyemoto, A. I. Ruiz, K. D. Burnham, and R. Maurer. 2000. Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol. Microbiol. 35:635-646. [DOI] [PubMed] [Google Scholar]
- 2.Chan, K., C. C. Kim, and S. Falkow. 2005. Microarray-based detection of Salmonella enterica serovar Typhimurium transposon mutants that cannot survive in macrophages and mice. Infect. Immun. 73:5438-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eriksson, A. R., R. A. Andersson, M. Pirhonen, and E. T. Palva. 1998. Two-component regulators involved in the global control of virulence in Erwinia carotovora subsp. carotovora. Mol. Plant-Microbe Interact. 11:743-752. [DOI] [PubMed] [Google Scholar]
- 4.Gudapaty, S., K. Suzuki, X. Wang, P. Babitzke, and T. Romeo. 2001. Regulatory interactions of Csr components: the RNA binding protein CsrA activates csrB transcription in Escherichia coli. J. Bacteriol. 183:6017-6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heeb, S., C. Blumer, and D. Haas. 2002. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J. Bacteriol. 184:1046-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heeb, S., and D. Haas. 2001. Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. Mol. Plant-Microbe Interact. 14:1351-1363. [DOI] [PubMed] [Google Scholar]
- 7.Jackson, D. W., K. Suzuki, L. Oakford, J. W. Simecka, M. E. Hart, and T. Romeo. 2002. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 184:290-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwon, O., D. Georgellis, A. S. Lynch, D. Boyd, and E. C. Lin. 2000. The ArcB sensor kinase of Escherichia coli: genetic exploration of the transmembrane region. J. Bacteriol. 182:2960-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lenz, D. H., M. B. Miller, J. Zhu, R. V. Kulkarni, and B. L. Bassler. 2005. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol. Microbiol. 58:1186-1202. [DOI] [PubMed] [Google Scholar]
- 10.Malpica, R., G. R. Sandoval, C. Rodriguez, B. Franco, and D. Georgellis. 2006. Signaling by the arc two-component system provides a link between the redox state of the quinone pool and gene expression. Antioxid. Redox Signal. 8:781-795. [DOI] [PubMed] [Google Scholar]
- 11.Moolenaar, G. F., C. A. van Sluis, C. Backendorf, and P. van de Putte. 1987. Regulation of the Escherichia coli excision repair gene uvrC. Overlap between the uvrC structural gene and the region coding for a 24 kD protein. Nucleic Acids Res. 15:4273-4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagasawa, S., S. Tokishita, H. Aiba, and T. Mizuno. 1992. A novel sensor-regulator protein that belongs to the homologous family of signal-transduction proteins involved in adaptive responses in Escherichia coli. Mol. Microbiol. 6:799-807. [DOI] [PubMed] [Google Scholar]
- 13.Pernestig, A. K., D. Georgellis, T. Romeo, K. Suzuki, H. Tomenius, S. Normark, and O. Melefors. 2003. The Escherichia coli BarA-UvrY two-component system is needed for efficient switching between glycolytic and gluconeogenic carbon sources. J. Bacteriol. 185:843-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pernestig, A. K., O. Melefors, and D. Georgellis. 2001. Identification of UvrY as the cognate response regulator for the BarA sensor kinase in Escherichia coli. J. Biol. Chem. 276:225-231. [DOI] [PubMed] [Google Scholar]
- 15.Rahme, L. G., F. M. Ausubel, H. Cao, E. Drenkard, B. C. Goumnerov, G. W. Lau, S. Mahajan-Miklos, J. Plotnikova, M. W. Tan, J. Tsongalis, C. L. Walendziewicz, and R. G. Tompkins. 2000. Plants and animals share functionally common bacterial virulence factors. Proc. Natl. Acad. Sci. USA 97:8815-8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romeo, T. 1998. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol. Microbiol. 29:1321-1330. [DOI] [PubMed] [Google Scholar]
- 17.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki, K., X. Wang, T. Weilbacher, A. K. Pernestig, O. Melefors, D. Georgellis, P. Babitzke, and T. Romeo. 2002. Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J. Bacteriol. 184:5130-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teplitski, M., R. I. Goodier, and B. M. Ahmer. 2003. Pathways leading from BarA/SirA to motility and virulence gene expression in Salmonella. J. Bacteriol. 185:7257-7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomenius, H., A. K. Pernestig, K. Jonas, D. Georgellis, R. Mollby, S. Normark, and O. Melefors. 2006. The Escherichia coli BarA-UvrY two-component system is a virulence determinant in the urinary tract. BMC Microbiol. 6:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomenius, H., A. K. Pernestig, C. F. Mendez-Catala, D. Georgellis, S. Normark, and O. Melefors. 2005. Genetic and functional characterization of the Escherichia coli BarA-UvrY two-component system: point mutations in the HAMP linker of the BarA sensor give a dominant-negative phenotype. J. Bacteriol. 187:7317-7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei, B. L., A. M. Brun-Zinkernagel, J. W. Simecka, B. M. Pruss, P. Babitzke, and T. Romeo. 2001. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol. Microbiol. 40:245-256. [DOI] [PubMed] [Google Scholar]
- 23.Weilbacher, T., K. Suzuki, A. K. Dubey, X. Wang, S. Gudapaty, I. Morozov, C. S. Baker, D. Georgellis, P. Babitzke, and T. Romeo. 2003. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol. Microbiol. 48:657-670. [DOI] [PubMed] [Google Scholar]
- 24.Zhang, J. P., and S. Normark. 1996. Induction of gene expression in Escherichia coli after pilus-mediated adherence. Science 273:1234-1236. [DOI] [PubMed] [Google Scholar]
- 25.Zuber, S., F. Carruthers, C. Keel, A. Mattart, C. Blumer, G. Pessi, C. Gigot-Bonnefoy, U. Schnider-Keel, S. Heeb, C. Reimmann, and D. Haas. 2003. GacS sensor domains pertinent to the regulation of exoproduct formation and to the biocontrol potential of Pseudomonas fluorescens CHA0. Mol. Plant-Microbe Interact. 16:634-644. [DOI] [PubMed] [Google Scholar]