Abstract
Our current understanding of segregation of prokaryotic plasmids has been derived mainly from the study of the gram-negative bacterial plasmids. We previously reported a replicon of the cryptic plasmid from a gram-positive bacterium, Leifsonia xyli subsp. cynodontis. The replicon contains a putative plasmid partition cassette including a Walker-type ATPase followed by open reading frame 4 without sequence homologue. Here we reported that the orf4 gene was essential for maintaining the plasmid stability in L. xyli subsp. cynodontis. Furthermore, the purified orf4 protein specifically and cooperatively bound to direct repeat sequences located upstream of the parA gene in vitro, indicating that orf4 is a parB gene and that the direct repeat DNA sequences constitute a partition site, parS. The location of parS and the features of ParA and ParB proteins suggest that this plasmid partition cassette belongs to type Ib, representing the first type Ib cassette identified from a gram-positive bacterial plasmid.
Bacterial plasmids maintain their presence within a growing bacterial population depending on different stabilization systems. While high-copy-number plasmids rely on random distribution among daughter cells, the stabilization of low-copy-number plasmids can be achieved through the mechanisms of plasmid multimer resolution, active partitioning, and postsegregational killing (14, 16, 30, 32).
The typical active partitioning cassettes of low-copy-number plasmids are composed of two genes that are organized as an operon and a cis-acting DNA sequence, termed the plasmid partition site or centromere-like region (2). The upstream gene to be transcribed gives rise to an ATPase belonging to a superfamily of phylogenetically related proteins that are involved in plasmid and chromosome segregation, which is called the ParA superfamily. The downstream gene encodes a DNA-binding factor called ParB, which interacts directly with the plasmid partition sites that are normally composed of direct or inverted iterated sequences (36). ParA and DNA-binding proteins assemble on the partition site to form a nucleoprotein complex that is required for the directional movement of paired plasmids away from the cell median (18, 31).
The active partitioning systems can be broadly classified into two types depending on the type of ATPase encoded by the upstream gene. Type I ParA contains the Walker-type ATPase motif, while the type II ATPase belongs to the actin/Hsp70 superfamily (14, 23). The type I partition systems are further divided into type Ia and type Ib subgroups based on the features of the ParB protein, the localization of the partition site, and the mechanism of the transcriptional regulation. In comparison with type Ia systems (3, 7, 19, 28, 33), the type Ib ATPases lack the N-terminal DNA-binding domain that appears to be required for autoregulation of the transcription of the active partition operon. The downstream parB genes of type Ia and type Ib partitioning systems are phylogenetically unrelated, and normally type Ia ParB proteins are larger than proteins from subgroup Ib (14).
The role of the parB gene in partitioning type Ia plasmids has been well characterized (6, 12, 19, 27, 33), while few studies have focused on type Ib partitioning. Several type Ib ParB-like proteins have recently been identified in plasmids TP228 (1, 4, 5, 15, 17), pVS1 (20), pTAR (21), pB171 (10), pVT745 (13), and pRA2 (24). However, these plasmids are exclusively from gram-negative bacteria (11).
Several active partitioning systems from gram-positive bacterial plasmids have been identified previously (9, 22, 26, 35), but very few studies have been carried out to characterize the mechanism of the partitioning. A recent study showed that δ and ω genes located in the streptococcal plasmid pSM19035 constitute a novel partitioning system, although these two genes are transcribed separately (9). In this system, the δ gene encodes a Walker-type ATPase and the ω-encoded protein is a DNA-binding protein that functions as a global regulator of plasmid functions, including replication, copy number control, and postsegregational killing (8).
Leifsonia xyli subsp. cynodontis, originally named Clavibacter xyli subsp. cynodontis subsp. nov., is a gram-positive bacterium isolated from the xylem of Bermuda grass (Cynodontis dactylon L. Per.) (25). A cryptic plasmid about 51 kb in size, namely, pCXC100, was harbored by some L. xyli subsp. cynodontis isolates but not by others (29). The 5-kb replicon of pCXC100, in which a parA homologue and a downstream open reading frame (orf4) that has no homologues with any ParB sequence are present, has been identified previously (26). In this study, we aimed to study the function of orf4 in plasmid maintenance. We found that orf4 is essential for the plasmid stability in L. xyli subsp. cynodontis. Furthermore, the orf4 protein was expressed in Escherichia coli and in vitro purified. This purified protein specifically and cooperatively binds to the direct repeats located upstream of the parA gene, called parS. Therefore, the protein encoded by orf4 is a functional homologue of the ParB protein. The features of parS gene products, as well as those of ParA and ParB proteins, further suggest that the active partition cassette of pCXC100 belongs to type Ib.
MATERIALS AND METHODS
Bacteria and plasmids.
All E. coli strains were grown in LB medium at 37°C. L. xyli subsp. cynodontis was grown on solid medium (DM agar) at 28°C as previously described (29). Antibiotics for selection of resistant E. coli cells were added to final concentrations of 60 μg/ml of ampicillin, 25 μg/ml of chloramphenicol, and 10 μg/ml of tetracycline. Tetracycline at 2 μg/ml was used for selections in L. xyli subsp. cynodontis. E. coli strain DH5α (Gibco BRL) was used as a host for creating subclones of the pCXC100 replicon. L. xyli subsp. cynodontis strain no. 3, lacking the 51-kb native plasmid, was transformed by plasmid containing derivatives of the pCXC100 replicon by use of an electroporation method (29).
YB411 was constructed for overexpression of the ParB protein. The parB gene was amplified by PCR using primers dparB1 (5′-CTG GAA TTC ATG GCT GAT CGC ACG GTT GC; is the EcoRI restriction site; underlined) and dparB2 (5′-GCC AAG CTT AGC TTC CCA GTG GGC GCC; the HindIII restriction site is underlined) and cloned into expression vector pMAL_c2x (New England Biolabs) to produce the maltose-binding protein (MBP)-ParB fusion protein.
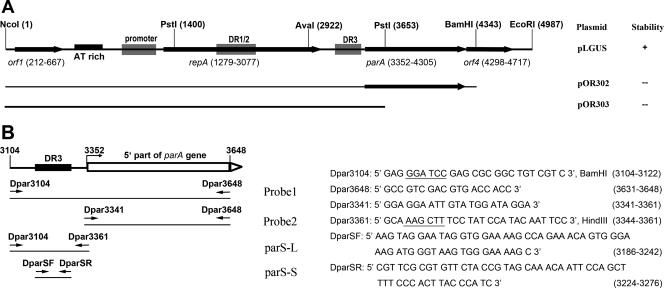
pLGUS is a pBR325-based plasmid containing the full pCXC100 replicon (Fig. 1A), which served as the positive control in the plasmid stability assay. This plasmid was constructed as described elsewhere (T.-Y. Li et al., submitted for publication). The plasmids pOR302 and pOR303 were constructed by cloning an NcoI-BamHI (positions 1 to 4343) fragment and an NcoI-PstI (positions 1 to 3653) fragment, respectively, into pBR325 (Fig. 1A). All regular DNA manipulations were carried out by following methods described by Sambrook et al. (34).
FIG. 1.
(A) Physical map of the pCXC100 replicon and the composition of the pCXC100 replicon in the pBR325-based plasmids used in this study. Plasmid pLGUS contains the complete replicon, while pOR302 contains a deleted replicon lacking the majority of orf4 and pOR303 lacks both the main body of parA and the whole orf4. Other data shown in this study indicate that orf4 encodes a ParB analogue that has specific DNA-binding activity. Whether each plasmid stably propagated in L. xyli subsp. cynodontis is listed on the right: +, the plasmid was stable when propagating in the bacterium; −, the plasmid was not stable. The details of the plasmid stability are shown in Table 1. (B) Probes used in this study to assess the DNA-binding activity encoded by orf4. The positions and sequences of primers used for PCR amplification of the corresponding DNA fragments are indicated. Restriction sites for Dpar3104 and Dpar3361, given after the sequences, are underlined. DR, direct repeat.
Protein expression and purification.
MBP and MBP-ParB fusion protein were expressed and purified according to an established protocol (New England Biolabs), with some modifications. The 1-liter culture containing the YB411 plasmid was grown to an optical density at 600 nm of 0.6 to 0.8 and then was induced for 3 h at 37°C by adding IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 0.4 mM. Cells were harvested by centrifugation and resuspended in 100 ml buffer A (20 mM Tris, pH 8.0, 100 mM NaCl). The collected cells were then sonicated and centrifuged. To remove contamination, the supernatant containing soluble MBP or MBP-ParB protein was loaded onto an SP-Sepharose Fast Flow column (Amersham Pharmacia). The flow fraction was collected and then loaded onto 6-ml amylose columns (New England Biolabs) equilibrated with the same buffer. The unbound proteins were washed away with 20 volumes of the same buffer, and the column-retained products were released by adding 10 mM maltose to the washing buffer. The recovered protein was dialyzed against 1 liter of buffer containing 10 mM Tris-HCl (pH 7.6) and 100 mM NaCl and concentrated by Amicon ultrafiltration (Millipore). A bicinchoninic acid assay kit (Pierce) was used to determine protein concentration.
Plasmid stability assay.
Plasmid stability was examined as previously described (26). Plasmid-containing L. xyli subsp. cynodontis cells were spread and grown on solid DM agar in the absence of antibiotics for 7 days at 28°C, which corresponded to a minimum of 20 generations of growth. Then, cells from the culture lawn were collected, diluted, and spread on fresh solid DM agar to grow for another 7 days free of antibiotics to cumulate the minimum of 40 generations of growth. The subculture was repeated one more time to reach a minimum of 60 generations of growth free of antibiotics. Before the start of growth and after each of the 7-day growth cycles, the culture was diluted and plated on DM agar in the presence or absence of tetracycline to calculate the plasmid retention rate. About 100 colonies were picked up from the antibiotic-free plate to further confirm the plasmid loss (26).
Electrophoretic mobility shift assay.
All of the DNA fragments indicated in Fig. 1B were PCR amplified and radiolabeled at the 5′ end with [γ-32P]ATP (5,000 Ci/mmol; Furui Biotech, Beijing, China) by T4 polynucleotide kinase (Promega). The DNA probe (about 15 nM) was incubated with the indicated amounts of the purified proteins in a final volume of 10 μl containing 15 mM Tris-HCl (pH 7.6), 3 mM MgCl2, 0.3 mM dithiothreitol, 50 mM KCl, 1 mM ATP, 0.25 μg poly(dI · dC), and 2% glycerol for 30 min at 30°C, followed by addition of an equal volume of the loading dye containing 50% glycerol and then electrophoresis at 4°C on 5% native polyacrylamide gels containing 5% glycerol in 0.5× Tris-borate-EDTA buffer (0.045 M Tris-borate and 1 mM EDTA at pH 8.0) at ∼20 V/cm for about 3 h. Gels were exposed to phosphorimager screens and analyzed by a Typhoo 9200 variable scanner (Amersham Pharmacia).
DNase I footprinting assay.
DNA fragment parS-L was amplified by using primers Dpar3104 and Dpar3361 (Fig. 1B) and then cleaved by endonucleases BamHI or HindIII (Takara), and the sticky 5′ end was labeled with [α-32P]dATP (Furui Biotech, Beijing, China) or [α-32P]dCTP (Amersham Pharmacia Biotech, United Kingdom) by Klenow fragment (Takara). The DNA probe was incubated with the indicated amounts of the purified proteins in a final volume of 20 μl at 30°C for 30 min in buffer containing 15 mM Tris-Hcl (pH 7.6), 3 mM MgCl2, 0.3 mM dithisthreitol, 50 nM KCl, 1 mM ATP, and 2% glycerol. After incubation, DNase I (Takara) and 20 μl of the reaction buffer (10 mM MgCl2 and 5 mM CaCl2) were added. Incubation was continued for 3 min, and an equal volume of stop buffer (1% sodium dodecyl sulfate, 200 mM NaCl, and 20 mM EDTA) was added. The reaction mixture was extracted with phenol-chloroform and then ethanol precipitated and resuspended in 8 μl H2O. The samples were run on a denatured, 8% polyacrylamide sequence gel.
RESULTS AND DISCUSSION
orf4 is essential for plasmid stability in L. xyli subsp. cynodontis.
Our previously published results indicate that the par locus of pCXC100, containing the putative parA gene and orf4, is essential for the plasmid stability in L. xyli subsp. cynodontis (26). No sequence homologue of orf4 has been found from the protein database, and it was unclear whether orf4 plays a role in maintaining the plasmid stability (26). In this study, we constructed two deletion mutants, one with both parA and orf4 being deleted (pOR303) and the other with orf4 being deleted (pOR302) (Fig. 1A). In comparison with the plasmid containing the wild-type par locus, both deletion mutants resulted in a dramatically decreased stability of the corresponding plasmids when propagated in L. xyli subsp. cynodontis, and pOR303 was less stable than pOR302 (Fig. 1A; Table 1). These results suggest that orf4 is essential for stabilizing plasmid in L. xyli subsp. cynodontis, while parA plays an additive role.
TABLE 1.
Stability of plasmids in L. xyli subsp. cynodontisa
| Plasmid (genotype) | Avg retention rate (%) at no. of generations
|
|||
|---|---|---|---|---|
| 0 | 20 | 40 | 60 | |
| pLGUS (parA orf4) | 100 | 100 | 100 | 100 |
| pOR302 (parA Δorf4) | 100 | 96.9 | 22.8 | 6.4 |
| pOR303 (ΔparA Δorf4) | 91.5 | 78.7 | 13.8 | 5.2 |
The plasmid stability was analyzed as described in Materials and Methods. Two parallel experiments were performed for each plasmid, and the percentages of recovered colonies that contain plasmid are shown.
The parA gene of the pCXC100 par locus encodes a typical Walker-type ATPase, indicating that pCXC100 may employ an active partition mechanism to stabilize itself in host cells, consistent with the fact that this cryptic plasmid is a low-copy-number plasmid (26, 29). The parB gene is typically located downstream of parA in the active partitioning operon. Although no sequence homologue of orf4 was found within the known parB gene, the physical location and function in maintaining the plasmid stability suggest that orf4 could encode a ParB function.
orf4 specifies a novel ParB protein with sequence-specific DNA-binding activity.
The DNA-binding function of orf4 was assessed in vitro by expressing orf4 from the pMAL_C2X expression vector, in which orf4 was fused downstream of MBP. Fusion with MBP both increased the protein solubility and provided a convenient means of purifying the orf4 protein (data not shown).
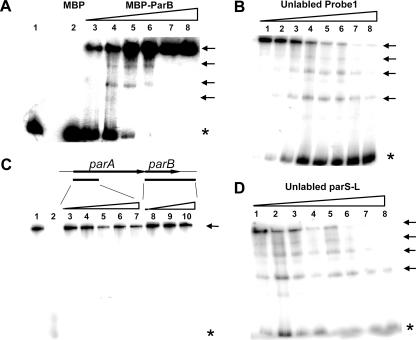
The ParB protein in an active partitioning cassette is expected to interact directly with the plasmid partition sites that are normally composed of direct or inverted iterated sequences (36). A previous study has shown that an AT-rich region located upstream of the parA gene of the pCXC100 replicon contains direct repeat sequences (26), providing a candidate site for orf4 protein binding if it encodes ParB function. Probe 1 (bp 3104 to 3668), covering the AT-rich region and the 5′ part of the parA gene, was used to assess the binding activity of orf4 (Fig. 1B). Figure 2A shows that only the fusion protein and not MBP alone bound to the radiolabeled DNA fragment and resulted in DNA species with lower gel mobility, reflecting the formation of DNA-protein complexes. A competition experiment showed that the labeled DNA-protein complex could be competed by the same unlabeled DNA fragment but not by DNA fragments amplified from the orf4 gene (Fig. 2B and C), indicating that the binding is sequence specific. These results suggest that orf4 of the pCXC100 par locus encodes a novel ParB analogue. This finding agrees with the notion that type Ib ParB lacks a sequence homologue but shares a conserved DNA-binding function and that the diversity of the ParB proteins adds a layer of specificity to the macromolecular interactions inside the nucleoprotein complex driving plasmid segregation (11).
FIG. 2.
Electrophoretic mobility shift assay of the specific DNA-binding activity of the ParB protein in vitro. All DNA probes used here were PCR amplified and radiolabeled at the 5′ end with [γ-32P]ATP. Probes are defined in Fig. 1B. (A) Dose-dependent binding of the ParB protein with probe 1. Increasing amounts of MBP-ParB fusion protein (from lane 3 to lane 8, 100, 200, 400, 800, 1,600, and 1,900 ng, respectively) were incubated with a fixed amount of radiolabeled probe 1 DNA fragment. Control samples in lanes 1 and 2 contained 1,000 ng of BSA protein and MBP, respectively. (B) Assay of the competition ability of the unlabeled DNA probe 1 with the labeled probe 1. The ratios of the radiolabeled to unlabeled DNA for the reactions were (from lanes 1 to 8, respectively) 1:0, 1:1, 1:2.5, 1:5, 1:7.5, 1:10, 1:15, and 1:27. (C) Assay of the ability of the unlabeled probe 2 (5′ part of parA) and parB (containing the full parB gene and 280 bp of the downstream sequence) to compete with the labeled probe 1. The ratios of labeled probe 1 to unlabeled probe 2 (from lanes 3 to 7, respectively) were 1:1, 1:5, 1:10, 1:19, and 1:37, and those to the unlabeled parB gene (from lanes 8 to 10, respectively) were 1:1, 1:5, and 1:19. Lane 1 contained no unlabeled DNA. The reaction shown in lane 2 was conducted in the same manner as that shown in lane 8 of panel B. (D) Assay of the competition ability of the unlabeled DNA parS-L with the labeled probe 1. The ratios of the radiolabeled to unlabeled DNA were the same as those given for panel B. The amount of MBP-ParB fusion protein for each reaction shown in panels B to D was 200 ng. Asterisks indicate unbound DNA, and arrows indicate different forms of protein-DNA complexes.
Four shifted DNA species were evident on native gels (Fig. 2A, B, and D), and the most slowly mobilized species corresponding to the largest DNA-protein complex was dominant. This observation raises the possibility that there are at least four ParB binding sites on the DNA sequence and that the fusion protein binds to the DNA in a highly cooperative manner.
To further examine if the ParB binding site parS is located solely in the AT-rich region where the direct repeats are present, unlabeled parS-L, containing an AT-rich region, and probe 2, containing the 5′ part of parA, were separately added to the competition reaction mixtures. It is shown that the unlabeled probe 2 did not compete with probe 1 for the binding of ParB at all, while parS-L efficiently competed the binding (Fig. 2C and D), supporting the theory that parS is located in the AT-rich region upstream of the parA gene.
The partition sites are usually located upstream of the parA gene or downstream of the parB gene in an active partition cassette (14). However, some cassettes, such as that in plasmid pB171, contain two partition sites, with one being located upstream of the parA gene and the other downstream of the parB gene (10, 18). The presence of parS upstream of the parA gene in the pCXC100 partitioning cassette has been demonstrated clearly thus far in this report. Meanwhile, the result shown in Fig. 2C argues against the presence of an additional parS gene downstream of parB, because the inclusion of the 280-bp sequence downstream of the parB gene in the parB unlabeled probe showed no activity in competing with the upstream parS gene for ParB binding (Fig. 2C, lanes 8 to 10).
parS is the direct repeat sequence located upstream of the parA gene.
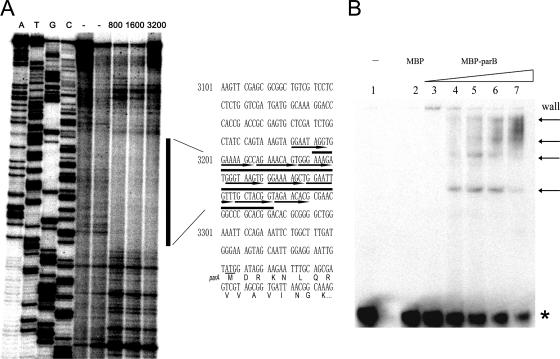
It was shown above that multiple ParB binding sites are present upstream of the parA gene. A DNase I footprinting assay was carried out to further examine the ParB binding sequence. The labeled parS-L fragment (Fig. 1B) was incubated with ParB protein and then cleaved by DNase I. It is clearly shown that the direct repeat sequence in the AT-rich region was protected from DNase I nuclease cleavage by MBP-ParB fusion protein but not by MBP or bovine serum albumin (BSA) protein (Fig. 3A; also data not shown), indicating that ParB protein bound to the direct repeats. We noticed that not all of the nine predicted direct repeats were protected from DNase I cleavage by ParB binding (Fig. 3A).
FIG. 3.
(A) DNase I footprinting assay of the exact ParB binding sites. The DNA fragment parS-L, labeled at one of the two 5′ ends, was incubated with the indicated amounts of the purified proteins, and the reactions were followed by DNase I cleavage and gel electrophoresis. The negative-control lane (−) on the left contained 2,000 ng BSA, and that on the right contained the same amount of MBP. The amounts of MBP-ParB in the far-right lanes ranged from 800 to 3,200 ng. The vertical black line shows the DNA region protected by the ParB fusion protein, and the protected sequence is shown (overlined) on the right. Black arrows overline the direct repeats (26). DNA ladders generated as described previously (34) are shown on the left lanes. (B) Electrophoretic mobility shift assay of the MBP-ParB fusion protein binding with direct repeat sequences in the AT-rich region in vitro. Lanes 1 and 2 contained 1,000 ng of BSA and MBP, respectively. Lanes 3 to 7 contained 100, 200, 400, 800, and 1,600 ng of the ParB fusion protein, respectively. The asterisk indicates unbound DNA, and arrows indicate different forms of protein-DNA complexes.
To confirm the results, the direct repeat DNA fragment parS-S (∼90 bp) was synthesized (Fig. 1B) (see Materials and Methods) and the ParB binding property of this fragment was studied. Figure 3B shows that ParB effectively and cooperatively bound to this 90-bp fragment, as it did to the large DNA fragment probe 1 (Fig. 2A), and that four similarly shifted bands were evident. We thus concluded that the direct repeat sequences located upstream of the parA gene, namely, parS, constitute the ParB binding sites (Fig. 4).
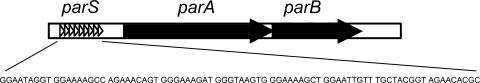
FIG. 4.
Overall structure of the active partition cassette (type Ib) of the plasmid pCXC100. Open triangles indicate nine direct repeats located in the partition site (parS), and the corresponding sequence is shown below.
The partition cassette of plasmid pCXC100 belongs to type Ib.
Clearly, the replicon of pCXC100 has a complete active partition cassette containing a parS site upstream of the parA gene, and the parB gene is downstream of the parA gene, with an 8-bp overlap (Fig. 1A and 4). It has been demonstrated that this cassette is responsible for stabilizing the plasmid in its native host L. xyli subsp. cynodontis, a gram-positive bacterium (26; this study). The feature that ParA is a typical Walker-type ATPase shows that the pCXC100 active partition system belongs to a type I cassette. Furthermore, the ParA protein of the pCXC100 partition cassette has no HTH motif (26), and its ParB protein is short (139 amino acids). Moreover, parS of the pCXC100 par locus is located upstream of the parA gene. This evidence further categorizes the partition cassette of pCXC100 into type Ib. Binding of the ParB protein with the parS site is expected to function in both partitioning of the pCXC100 plasmid and autoregulation of the operon expression. Prior to this study, four out of the five reported type Ib parS genes have been determined to be direct repeat sequences (11). This study adds a new type Ib parS composed of direct repeats (Fig. 4).
In recent years, study of the plasmid partitioning mechanisms has been extended beyond type Ia partition cassettes. For example, the recently studied plasmids from gram-negative bacteria, including plasmid TP228 (1, 4, 5, 15, 17), pVS1 (20), pTAR (21), pB171 (10), pVT745 (13), and pRA2 (24), all contain type Ib partition cassettes (14). However, the partitioning mechanisms utilized by this new type of cassette are unclear. This study reports that the native plasmid pCXC100 from a gram-positive bacterium harbors a type Ib partition cassette. Further characterization of the mechanisms of this partitioning system should provide key insights into the understanding of type Ib plasmid partitioning. Study of the molecular basis for active partitioning of plasmid in L. xyli subsp. cynodontis should also expand our understanding of chromosome segregation in gram-positive bacteria.
Acknowledgments
We thank L. Cheng and H. Lin for useful discussions.
This work is supported by the National Natural Science Foundation of China through grants 30270749 and 30470942 (to T.-Y. Li) and by Wuhan University through grant 0000028 (to Y. Zhang).
Footnotes
Published ahead of print on 22 September 2006.
REFERENCES
- 1.Barilla, D., M. F. Rosenberg, U. Nobbmann, and F. Hayes. 2005. Bacterial DNA segregation dynamics mediated by the polymerizing protein ParF. EMBO J. 24:1453-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bignell, C., and C. M. Thomas. 2001. The bacterial ParA-ParB partitioning proteins. J. Biotechnol. 91:1-34. [DOI] [PubMed] [Google Scholar]
- 3.Bouet, J. Y., and B. E. Funnell. 1999. P1 ParA interacts with the P1 partition complex at parS and an ATP-ADP switch controls ParA activities. EMBO J. 18:1415-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carmelo, E., D. Barilla, A. P. Golovanov, L. Y. Lian, A. Derome, and F. Hayes. 2005. The unstructured N-terminal tail of ParG modulates assembly of a quaternary nucleoprotein complex in transcription repression. J. Biol. Chem. 280:28683-28691. [DOI] [PubMed] [Google Scholar]
- 5.Dagmara, J., M. Sebastien, Z.-C. Jolanta, and F. C. Keith. 2006. Developmental control of a parAB promoter leads to formation of sporulation-associated ParB complexes in Streptomyces coelicolor. J. Bacteriol. 188:1710-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis, M. A., and S. J. Austin. 1988. Recognition of the P1 plasmid centromere analog involves binding of the ParB protein and is modified by a specific host factor. EMBO J. 7:1881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis, M. A., K. A. Martin, and S. J. Austin. 1992. Biochemical activities of the ParA partition protein of the P1 plasmid. Mol. Microbiol. 6:1141-1147. [DOI] [PubMed] [Google Scholar]
- 8.de la Hoz, A. B., S. Ayora, I. Sitkiewicz, S. Fernandez, R. Pankiewicz, J. C. Alonso, and P. Ceglowski. 2000. Plasmid copy-number control and better-than-random segregation genes of pSM19035 share a common regulator. Proc. Natl. Acad. Sci. USA 97:728-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dmowski, M., I. Sitkiewicz, and P. Ceglowski. 2006. Characterization of a novel partition system encoded by the δ and ω genes from the streptococcal plasmid pSM19035. J. Bacteriol. 188:4362-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebersbach, G., and K. Gerdes. 2001. The double par locus of virulence factor pB171: DNA segregation is correlated with oscillation of ParA. Proc. Natl. Acad. Sci. USA 98:15078-15083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fothergill, T., D. Barilla, and F. Hayes. 2005. Protein diversity confers specificity in plasmid segregation. J. Bacteriol. 187:2651-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Funnell, B. E. 1988. Mini-P1 plasmid partitioning: excess ParB protein destabilizes plasmids containing the centromere parS. J. Bacteriol. 170:954-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galli, D. M., J. Chen, K. F. Novak, and D. J. Leblanc. 2001. Nucleotide sequence and analysis of conjugative plasmid pVT745. J. Bacteriol. 183:1585-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerdes, K., J. Moller-Jensen, and R. B. Jensen. 2000. Plasmid and chromosome partition: surprises from phylogeny. Mol. Microbiol. 37:455-466. [DOI] [PubMed] [Google Scholar]
- 15.Golovanov, A. P., D. Barilla, M. Golovanova, F. Hayes, and L. Y. Lian. 2003. ParG, a protein required for active partition of bacterial plasmids, has a dimeric ribbon-helix-helix structure. Mol. Microbiol. 50:1141-1153. [DOI] [PubMed] [Google Scholar]
- 16.Hayes, F. 2000. The partition system of multidrug resistance plasmid TP228 includes a novel protein that epitomizes an evolutionarily distinct subgroup of the ParA superfamily. Mol. Microbiol. 37:528-541. [DOI] [PubMed] [Google Scholar]
- 17.Hayes, F. 2003. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301:1496-1499. [DOI] [PubMed] [Google Scholar]
- 18.Hayes, F., and D. Barilla. 2006. The bacterial segrosome: a dynamic nucleoprotein machine for DNA trafficking and segregation. Nat. Rev. Microbiol. 4:133-143. [DOI] [PubMed] [Google Scholar]
- 19.Hayes, F., L. Radnedge, M. A. Davis, and S. J. Austin. 1994. The homologous operons for P1 and P7 plasmid partition are autoregulated from dissimilar operator sites. Mol. Microbiol. 11:249-260. [DOI] [PubMed] [Google Scholar]
- 20.Heeb, S., Y. Itoh, T. Nishijyo, U. Schnider, C. Keel, J. Wade, U. Walsh, F. O'Gara, and D. Haas. 2000. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. Mol. Plant-Microbe Interact. 13:232-237. [DOI] [PubMed] [Google Scholar]
- 21.Kalnin, K., S. Stegalkina, and M. Yarmolinsky. 2000. pTAR-encoded proteins in plasmid partitioning. J. Bacteriol. 182:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kearney, K., G. F. Fitzgerald, and J. F. Seegers. 2000. Identification and characterization of an active plasmid partition mechanism for the novel Lactococcus lactis plasmid pCI2000. J. Bacteriol. 182:30-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koonin, E. V. 1993. A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res. 21:2541-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwong, S. M., C. C. Yeo, and C. L. Poh. 2001. Molecular analysis of the pRA2 partitioning region: ParB autoregulates parAB transcription and forms a nucleoprotein complex with the plasmid partition site, parS. Mol. Microbiol. 40:621-633. [DOI] [PubMed] [Google Scholar]
- 25.Lee, I.-M., I. M. Bartoszyk, D. E. Gundersen-Rindal, and R. E. Davis. 1997. Phylogeny and classification of bacteria in the genera Clavibacter and Rathayibacter on the basis of 16S rRNA gene sequence analyses. Appl. Environ. Microbiol. 63:2631-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, T. Y., P. Yin, Y. Zhou, Y. Zhang, Y. P. Zhang, and T. A. Chen. 2004. Characterization of the replicon of a 51-kb native plasmid from the gram-positive bacterium Leifsonia xyli subsp. cynodontis. FEMS Microbiol. Lett. 236:33-39. [DOI] [PubMed] [Google Scholar]
- 27.Li, Y., A. Dabrazhynetskaya, B. Youngren, and S. Austin. 2004. The role of Par proteins in the active segregation of the P1 plasmid. Mol. Microbiol. 53:93-102. [DOI] [PubMed] [Google Scholar]
- 28.Libante, V., L. Thion, and D. Lane. 2001. Role of the ATP-binding site of SopA protein in partition of the F plasmid. J. Mol. Biol. 314:387-399. [DOI] [PubMed] [Google Scholar]
- 29.Metzler, M. C., Y.-P. Zhang, and T.-A. Chen. 1992. Transformation of the gram-positive bacterium Clavibacter xyli subsp. cynodontis by electroporation with plasmids from the IncP incompatibility group. J. Bacteriol. 174:4500-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moller-Jensen, J., R. B. Jensen, and K. Gerdes. 2000. Plasmid and chromosome segregation in prokaryotes. Trends Microbiol. 8:313-320. [DOI] [PubMed] [Google Scholar]
- 31.Niki, H., and S. Hiraga. 1997. Subcellular distribution of actively partitioning F plasmid during the cell division cycle in E. coli. Cell 90:951-957. [DOI] [PubMed] [Google Scholar]
- 32.Nordstrom, K., and S. J. Austin. 1989. Mechanisms that contribute to the stable segregation of plasmids. Annu. Rev. Genet. 23:37-69. [DOI] [PubMed] [Google Scholar]
- 33.Radnedge, L., B. Youngren, M. A. Davis, and S. A. Austin. 1998. Probing the structure of complex macromolecular interactions by homolog specificity scanning: the P1 and P7 plasmid partition systems. EMBO J. 17:6076-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Simpson, A. E., R. A. Skurray, and N. Firth. 2003. A single gene on the staphylococcal multiresistance plasmid pSK1 encodes a novel partitioning system. J. Bacteriol. 185:2143-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Surtees, J. A., and B. E. Funnell. 2003. Plasmid and chromosome traffic control: how ParA and ParB drive partition. Curr. Top. Dev. Biol. 56:145-180. [DOI] [PubMed] [Google Scholar]