Abstract
The phytopathogenic bacterium Pseudomonas syringae pv. glycinea infects soybean plants and causes bacterial blight. In addition to P. syringae, the human pathogen Pseudomonas aeruginosa and the soil bacterium Azotobacter vinelandii produce the exopolysaccharide alginate, a copolymer of d-mannuronic and l-guluronic acids. Alginate production in P. syringae has been associated with increased fitness and virulence in planta. Alginate biosynthesis is tightly controlled by proteins encoded by the algT-muc regulatory gene cluster in P. aeruginosa and A. vinelandii. These genes encode the alternative sigma factor AlgT (σ22), its anti-sigma factors MucA and MucB, MucC, a protein with a controversial function that is absent in P. syringae, and MucD, a periplasmic serine protease and homolog of HtrA in Escherichia coli. We compared an alginate-deficient algT mutant of P. syringae pv. glycinea with an alginate-producing derivative in which algT is intact. The alginate-producing derivative grew significantly slower in vitro growth but showed increased epiphytic fitness and better symptom development in planta. Evaluation of expression levels for algT, mucA, mucB, mucD, and algD, which encodes an alginate biosynthesis gene, showed that mucD transcription is not dependent on AlgT in P. syringae in vitro. Promoter mapping using primer extension experiments confirmed this finding. Results of reverse transcription-PCR demonstrated that algT, mucA, and mucB are cotranscribed as an operon in P. syringae. Northern blot analysis revealed that mucD was expressed as a 1.75-kb monocistronic mRNA in P. syringae.
Pseudomonas syringae undergoes diverse host-specific interactions with plants and is subdivided into >50 different pathovars based on host specificity (29). P. syringae pv. glycinea PG4180 infects soybean plants (Glycine max) and induces bacterial blight disease. The infection process involves epiphytic colonization, entry into the plant apoplast, multiplication within host tissue, and manifestation of disease symptoms (1, 29). Different cellular determinants, e.g., exopolysaccharides (EPS) or toxins, help P. syringae to cope with its diverse natural niches, host responses, and environmental conditions (4).
Exopolysaccharides are carbohydrate polymers that are secreted by a wide variety of bacteria and form a loosely associated extracellular slime or remain closely attached to cells as capsules (64). EPS have multiple functions, including water absorption, accumulation of minerals and nutrients, and protection from hydrophobic and toxic macromolecules (14). P. syringae produces at least two EPS: (i) levan, a β-(2,6) polyfructan with extensive branching through β-(1,4) linkages; and (ii) alginate, a copolymer of O-acetylated β-(1,4)-linked d-mannuronic acid and its C-5 epimer, l-guluronic acid (25, 28, 38, 49). Previous studies on EPS produced by P. syringae indicated that alginate was the major exopolysaccharide produced in planta (17). Alginate production has been associated with increased epiphytic fitness, resistance to desiccation and toxic molecules, the induction of water-soaked lesions, and the colonization and/or dissemination of P. syringae in planta (17, 58, 69). A direct correlation between virulence of P. syringae and the quantity of alginate produced in planta has been demonstrated (24, 49). Bacterial alginate is produced by Azotobacter vinelandii and several species of Pseudomonas, where it is widespread in the rRNA homology group I (18, 44).
Alginate biosynthesis has been studied extensively in Pseudomonas aeruginosa, where it functions as a major virulence factor in strains infecting the lungs of cystic fibrosis patients (55). Biosynthesis of alginate in P. syringae is similar to that established for P. aeruginosa (18, 19, 54, 69). Genes required for alginate biosynthesis in P. aeruginosa and P. syringae are organized in a chromosomal operon (algD-algA), with the algC biosynthetic gene located at a different position on the chromosome (11, 53, 54, 70). The first gene of the operon, algD, encodes GDP-mannose dehydrogenase, whose enzymatic activity is the rate-limiting step in the alginate biosynthetic pathway (63).
An important feature of alginate production by P. aeruginosa is that alginate biosynthetic genes are normally silent but are activated at the stage of chronic infection in the cystic fibrotic lung, which results in a mucoid phenotype (40). Although wild-type P. aeruginosa strains have the genetic capacity to synthesize alginate, they normally produce only very small amounts of this polymer (2, 37, 56). Similarly, most phytopathogenic strains of P. syringae, including PG4180, are normally nonmucoid (35). In P. aeruginosa the alginate biosynthetic operon is tightly controlled by several two-component regulatory systems and the alternative sigma factor, AlgT (44, 62). AlgT functions as a global stress response sigma factor that induces a number of other genetic traits in P. aeruginosa (21, 22, 60). AlgT (σ22), which is functionally homologous to RpoE (σE) from Escherichia coli, is encoded by the first gene in a cluster comprising the main switch controlling alginate biosynthesis. AlgT activates its own transcription and that of algD in P. aeruginosa, P. syringae, and A. vinelandii (27, 34, 43, 66). The algT (rpoE) gene cluster is conserved in several other bacteria, including A. vinelandii, Photobacterium, and E. coli (10, 13, 43, 47). Interestingly, the algT-mucABCD gene cluster of P. aeruginosa and A. vinelandii harbors five genes, whereas P. syringae lacks mucC. In Photobacterium and E. coli, there are no mucD homologs directly associated with the rpoE gene cluster (23, 33, 43, 44). Instead, htrA, the E. coli mucD homolog, maps at a different position in the genome, and transcription of the monocistronic htrA mRNA depends on RpoE (51). MucA, MucB, and MucD were shown to act as negative regulators of AlgT in P. aeruginosa and A. vinelandii (6, 43, 45, 48, 57, 61). MucD belongs to the HtrA family of periplasmic serine proteases, which are effectors of the extreme stress response and degrade improperly folded proteins in the periplasm (6, 51).
In the present study, we investigated the role of algT in P. syringae pv. glycinea in alginate production and its effect on growth in vitro and in planta. Transcriptional analysis revealed that AlgT activates mucA, mucB, algD, and its own transcription, whereas mucD transcription is not dependent on AlgT. Using primer extension experiments, we mapped a separate promoter for mucD, thus providing evidence that mucD in P. syringae pv. glycinea is not cotranscribed with the algT-muc operon but instead is transcribed as a monocistronic mRNA.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli and Pseudomonas aeruginosa strains were maintained at 37°C on Luria-Bertani medium (59).Azotobacter vinelandii was maintained at 28°C on Burk's medium (50). P. syringae was maintained at 28°C on mannitol-glutamate (MG) medium (32). For liquid cultures of P. syringae incubated at 18 or 28°C, bacteria were grown in Hoitink-Sinden minimal (HSC) medium (52). Bacterial growth was monitored by measuring the optical density at 600 nm (OD600). Antibiotics were added at the following concentrations (in micrograms/milliliter): ampicillin, 50; kanamycin, 25; and tetracycline, 25.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Escherichia coli | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 59 |
| HB101 | supE44 hsdS20(rB mB) recA13 ara-14proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | 5 |
| Pseudomonas syringae pv. glycinea | ||
| PG4180 | Wild type, produces small amounts of alginate | R. Mitchell |
| PG4180.muc | Emerged spontaneously from PG4180, produces vast amounts of alginate | This study |
| Azotobacter vinelandii | ||
| wtC15 | Produces alginate | Tonje Strommen, NTNU,b Trondheim, Norway |
| Pseudomonas aeruginosa | ||
| Pa8821 | Alg+ (unstably mucoid) | 12 |
| Pa8822 | Alg− (nonmucoid revertant of Pa8821) | 12 |
| Pa8830 | Alg+ (stably mucoid) | 12 |
| Plasmids | ||
| pBluescript SK(+) | Apr | Stratagene |
| pBlueSK:AXSalgTop | Apr; contains an 8,860-bp SalI fragment carrying the algT-mucABD gene cluster region from PG4180 cloned in pBluescript SK(+) | This study |
| pRK2013 | Kmr; helper plasmid | 20 |
| pBBR1MCS-3 | Tcr; ColE1 origin | 36 |
| pBBR3-AXSalgTop | Tcr; contains a 5,096-bp XbaI-HindIII PCR fragment (consisting of 988-bp upstream algT promoter region, 582-bp algT reading frame, 591-bp mucA reading frame, 960-bp mucB reading frame, and 1,440-bp mucD reading frame) from PG4180.muc cloned in pBBR1MCS-3 | This study |
| pBBR3-AXSalgT | Tcr; contains a 1,837-bp XbaI-PmlI PCR fragment (consisting of 988-bp upstream algT promoter region, 582-bp algT reading frame, and 270-bp downstream algT region) from PG4180.muc cloned in pBBR1MCS-3 | This study |
Ap, ampicillin; Km, kanamycin; Tc, tetracycline.
NTNU, Norwegian University of Science and Technology.
Molecular genetic techniques.
Plasmid isolation, restriction enzyme digests, agarose gel electrophoresis, Southern blots, electroporation, PCR, and other routine molecular methods were performed using standard protocols (59). Isolation of plasmid DNA from P. syringae was accomplished by the method of Kado and Liu (31). Genomic DNA from P. syringae was isolated as described by Ausubel et al. (3).
Cloning and sequencing of the algT-mucABD gene cluster of PG4180.
A clone containing the algT-mucABD gene cluster was identified from a genomic library of PG4180 (28) by Southern hybridization with a 1.1-kb BamHI fragment containing the mucD gene. An 8.9-kb SalI fragment, containing the algT-muc gene cluster, was subcloned into pBluescript SK(+) (Stratagene, Heidelberg, Germany), yielding plasmid pBlueSK:AXSalgTop, and was sequenced commercially (MWG Biotech, Ebersberg, Germany). Comparative sequence analysis of PG4180 and PG4180.muc was done using primers alg_seq1, alg_seq2, alg_seq3, and alg_seq4 (Table 2).
TABLE 2.
Oligonucleotide primers used in this study
| Oligonucleotide primer | Nucleotide sequencea (5′ to 3′) |
|---|---|
| alg-Operon_fwd | TGCTCTAGAGGCGCGCTGTTCAAGCAGTTCGAT |
| alg-Operon_rev | CCCAAGCTTCGATGATGGAGAAATTGCGG |
| alg_seq1 | TGACGCTCGCAATGTCTTC |
| alg_seq2 | AACACGCCGTTGGCCATA |
| alg_seq3 | CAGGTGAAGTTCAAGGCCG |
| alg_seq4 | TTCAGTACCTGCGCCCAAG |
| algT_fwd | CGGATCCCTTAACGAGGAGTGTTCATG |
| algT_revT7 | TAATACGACTCACTATAGGGAGGACGGTACCAACAGGACACTG |
| mucA_fwd | TCCGCGGTACTGGATAACGAAGC |
| mucA_revT7 | TAATACGACTCACTATAGGGAGGACGCCGTTGGCCATAGGG |
| mucB_fwd | TTCTGCGTGTTGACGGGCTT |
| mucB_revT7 | TAATACGACTCACTATAGGGAGGACATCAGATGGGTAACGG |
| mucB2_fwd | TGATGTATGGCGACGGTCTGGC |
| mucB2_revT7 | TAATACGACTCACTATAGGGAGGGTCACCATCATGTCGCCC |
| mucD_fwd | CGAATTTCTCGAGCGCAGCATGC |
| mucD_revT7 | TAATACGACTCACTATAGGGAGGGGAGCGGGTAAATATCTGCG |
| mucD_fwd_deg | TCGCCGGCGGTGGTCAATATCAGTAC |
| mucD_revT7_deg | TAATACGACTCACTATAGGGAGGCGAACCGATGGCCAGGACCCATTC |
| algD_fwd | GCGTATCAGCATATTTGGTTTGGG |
| algD_revT7 | TAATACGACTCACTATAGGGAGGCGTTGCAGGTGTACTTGATCA |
| mucD-Ps-3_pe | GAGCACAGCGGCAATCAGTG |
Restriction sites incorporated in primers are underlined; TCTAGA, XbaI; AAGCTT, HindIII. T7 RNA polymerase promoter sequences incorporated in primers are italics
Construction of pBBR3-AXSalgTop and pBBR3-AXSalgT.
A 5.1-kb fragment containing the algT-mucABD gene cluster was amplified by PCR from genomic DNA of PG4180.muc using primers alg-Operon_fwd and alg-Operon_rev (Table 2). The product was digested with XbaI-HindIII and ligated into the broad-host-range vector pBBR1MCS-3 (36), resulting in pBBR3-AXSalgTop. The 5.1-kb PCR product was also digested with XbaI-PmlI, yielding a 1,837-bp fragment, which contained the 988-bp algT promoter region, the 579-bp algT coding region, and 270 nucleotides (nt) downstream of algT (including a truncated mucA). This fragment was cloned into pBBR1MCS-3, yielding plasmid pBBR3-AXSalgT. Plasmids were mobilized into P. syringae strains by triparental matings (20).
Isolation and quantification of alginate.
Bacteria were grown on MG agar at 28°C for 96 h. Cells were washed from the plates and resuspended in 0.9% NaCl. Alginate isolation and quantification were performed as described by May and Chakrabarty (46), and alginic acid from seaweed (Macrocystis pyrifera; Sigma Chemical Co., St. Louis, Mo.) was used as a standard. The experiment was repeated twice, and mean values were expressed as the quantity of alginate produced per milligram of protein.
RNA isolation.
Bacteria were cultured in HSC medium at 28°C to an OD600 of 1.0 (early to mid-exponential growth phase) and harvested by mixing 15 ml of culture with an equal volume of chilled killing buffer (20 mM Tris-HCl [pH 7.5], 20 mM NaN3). This mixture was then centrifuged at 4°C for 15 min at 3,220 × g. Total RNA was isolated from the cell pellets by acid phenol-chloroform extraction as described by Majumdar et al. (41). The RNA concentration was determined by measuring the absorbance at 260 nm (59).
RNA spot blot and Northern blot analyses.
For RNA spot blot analysis, aliquots of total RNA (200 ng per spot) were transferred to nylon membranes (Pall, Dreieich, Germany) using the Minifold I Spot-Blot System (Schleicher & Schuell BioScience, Dassel, Germany). For Northern blot analysis, aliquots of total RNA (1.5 μg per lane) and an RNA size standard (0.24- to 9.5-kb RNA ladder; 3 μg per lane; Invitrogen, Karlsruhe, Germany) were separated by denaturing glyoxal RNA agarose gel electrophoresis as described by Burnett (9) and transferred to nylon membrane as described by Ingelbrecht et al. (30). Successful transfer of the RNA was verified by reversible staining of the membrane with methylene blue (26). Transcript sizes were estimated by comparison with an RNA size standard and with 16S rRNA and 23S rRNA bands.
RNA hybridization probes were generated by in vitro transcription of PCR products. Gene-specific primers (Table 2) were used to amplify PCR products from genomic DNA of P. syringae, P. aeruginosa, or A. vinelandii. The reverse primers carried a T7 promoter sequence at their 5′ ends. Digoxigenin-labeled RNA probes were synthesized using the Strip-EZ RNA Probe Synthesis and Removal kit (Ambion Europe, Cambridgeshire, United Kingdom) and digoxigenin-11-UTP (Roche Diagnostics, Mannheim, Germany), yielding hybridization probes of the following sizes internal to the structural genes: for algT, 536 nucleotides (nt) (algT_fwd, algT_revT7); mucA, 431 nt (mucA_fwd, mucA_revT7); mucB, 501 nt (mucB_fwd, mucB_revT7); mucD, 511 nt (mucD_fwd, mucD_revT7); algD, 641 nt (algD_fwd, algD_revT7); mucDA.vinelandii, 426 nt (mucD_fwd_deg, mucD_revT7_deg); mucDP.aeruginosa, 325 nt (mucD_fwd, mucD_revT7_deg). Hybridization, washing steps, antibody reactions, and signal detection and quantification using an FLA-3000 phosphoimager (Raytest, Straubenhardt, Germany) were done according to standard procedures (59) and the manufacturer's recommendations.
RT-PCR fragment analysis.
Total RNA was treated with RNase-free DNase I (Ambion Europe, Cambridgeshire, United Kingdom). A phenol-chloroform/isoamyl alcohol (25:24:1) and successive chloroform/isoamyl alcohol (24:1) extraction step was used to remove DNase I. The RNA was recovered from the aqueous phase by adding ammonium acetate (final concentration of 0.5 M) and 2.5 volumes of ethanol. RNA was then precipitated by incubating the solution at −80°C for 1 h and centrifuging for 20 min at 16,100 × g and 4°C. The pellet was washed twice by adding 1 ml 75% ethanol and centrifuging for 10 min at 16,100 × g and 4°C. RNA was resuspended in RNase-free water, and the concentration was photometrically determined (59). Synthesis of cDNA was performed using SuperScript II Reverse Transcriptase (Invitrogen, Karlsruhe, Germany) with 5 μg of DNA-free total RNA, 250 ng random hexamer primers (Roche, Mannheim, Germany), and SUPERASE-In (Ambion Europe, Cambridgeshire, United Kingdom). The cDNA was used for PCRs, amplifying regions within the algT-mucABD gene cluster by using the following primer combinations: (1) algT_fwd, algT_revT7; (2) algT_fwd, mucA_revT7; (3) algT_fwd, mucB_revT7; (4) algT_fwd, mucB2_revT7; (5) algT_fwd, mucD_revT7_deg; (6) mucA_fwd, mucD_revT7_deg; (7) mucB_fwd, mucD_revT7_deg; (8) mucB2_fwd, mucD_revT7_deg; and (9) mucD_fwd, mucD_revT7_deg. Genomic DNA isolated from PG4180.muc served as a positive control. A reverse transcription (RT) reaction, where reverse transcriptase was omitted, served as negative control.
Determination of transcriptional start site.
For primer extension analysis, 32P-labeled primer mucD-Ps-3_pe (4 pmol) was annealed with 10 μg of total RNA, and reverse transcription was performed with M-MLV Reverse Transcriptase (Invitrogen). Nucleotide sequencing of 5 μg plasmid pBBR3-AXSalgTop with primer mucD-Ps-3_pe was done with the Sequenase version 2.0 DNA sequencing kit (USB) according to the manufacturer's recommendation. The extension product and sequencing reaction were resolved on a 6% polyacrylamide sequencing gel.
Determination of bacterial growth in planta.
In planta growth of PG4180 and PG4180.muc was evaluated on soybeans (Glycine max cv. Choska). Soybean seedlings were maintained in a growth chamber at 24 to 25°C, 30 to 40% relative humidity (RH), with a 12-h photoperiod. Three- to 4-week-old plants were incubated at ≥92% RH for 48 h before inoculation. PG4180 and PG4180.muc were incubated for 48 h at 28°C on MG agar. Cells were suspended in distilled water, adjusted to an OD600 of 0.05 (approximately 5.0 × 107 CFU/ml), and applied to leaves with an airbrush (∼8 lb/in2) until the leaf surfaces were uniformly wet. Inoculated plants were grown in the greenhouse (20 to 25°C), and growth of bacterial strains was monitored by removing random leaf samples at 0 to 10 days postinoculation (three replicates per time point). Leaves were weighed separately and macerated in 5 ml sterile distilled water. Bacterial counts (CFU/gram fresh weight) were determined by plating dilutions of leaf homogenate onto MG agar and counting colonies after a 96-h incubation period.
Nucleotide sequence accession number.
The nucleotide sequence of the algT-mucABD gene cluster of P. syringae pv. glycinea PG4180 was deposited in GenBank under accession no. DQ991248.
RESULTS
Alginate production of P. syringae pv. glycinea PG4180 and PG4180.muc.
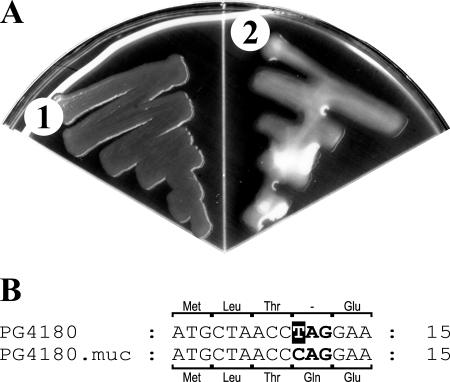
Strain P. syringae pv. glycinea PG4180 did not produce visible amounts of alginate when cultured on MG agar plates (Fig. 1A). In the process of screening for mucoid derivatives of PG4180, we identified PG4180.muc (Fig. 1A), which emerged spontaneously from a PG4180 culture. Quantification of alginate levels of both strains grown at 28°C for 96 h showed that PG4180.muc was able to produce larger amounts of alginate (106 μg alginate/mg protein) than nonmucoid PG4180 (2 μg alginate/mg protein, which was a negligible amount at the detection limit of the assay).
FIG. 1.
(A) Phenotype of P. syringae pv. glycinea strains PG4180 (1) and PG4180.muc (2) grown on MG agar plates at 28°C for 6 days. (B) Nucleotide sequence alignment and the corresponding amino acids encoded by the algT genes of PG4180 and PG4180.muc for the 5′ end of algT. The fourth codon (boldface letters) codes for glutamine (Gln) in PG4180.muc, whereas the respective codon in PG4180 leads to a nonsense (amber) mutation.
Nucleotide sequence analysis of the algT-mucABD gene cluster of PG4180 and PG4180.muc.
To find out whether the phenotypic difference between PG4180 and PG4180.muc was due to changes in the algT-mucABD regulatory gene cluster, the nucleotide sequence of the insert of pBlueSK:AXSalgTop was determined (accession number DQ991248). The insert contained the coding regions of algT, mucA, mucB, mucD, and the upstream region of algT, which included the nadB gene. Sequence alignments with known algT-muc gene clusters showed the same general arrangement. In contrast to P. aeruginosa (7) and A. vinelandii (43), mucC was lacking in PG4180, PG4180.muc, and theclosely related P. syringae strains FF5 (33) and DC3000 (8). Instead of mucC, PG4180 harbors an intergenic region of 283 bp between mucB and mucD, whereas in P. aeruginosa and A. vinelandii the intergenic region between mucC and mucD is just 40 bp and 11 bp, respectively. The PG4180 algT homologue is 582 bp (193 amino acids) and is closely related to algT from FF5, DC3000, and P. aeruginosa (99.5%, 99.0%, and 89.6% amino acid identity, respectively). Interestingly, the algT gene of PG4180 contained a single-nucleotide change at position 10, which resulted in a nonsense (amber) mutation in the fourth codon and thus the absence of a functional AlgT gene product (Fig. 1B). Sequencing of the algT, mucA, mucB, and mucD genes from PG4180.muc showed that the sequence was identical to that of PG4180, except that algT of PG4180.muc lacked the nonsense mutation. Therefore, this variant is likely to synthesize a functional AlgT gene product (Fig. 1B).
Complementation studies with algT.
To determine if lack of alginate production in PG4180 is due to the nonsense mutation in algT, we cloned the intact algT gene, including its 988-bp upstream region from PG4180.muc, into vector pBBR1MCS-3, yielding pBBR3-AXSalgT. Plasmids pBBR1MCS-3 and pBBR3-AXSalgT were introduced into PG4180 and PG4180.muc. Transconjugants with plasmid pBBR3-AXSalgT clearly showed an alginate-overproducing phenotype when cultured on MG agar plates, whereas bacteria carrying vector pBBR1MCS-3 showed the same phenotype as their plasmid-free parental strains. Quantitative analysis of alginate levels confirmed that PG4180(pBBR3-AXSalgT) produced alginate (80.7 μg alginate/mg protein) in contrast to the control PG4180(pBBR1MCS-3) (3.7 μg alginate/mg protein). PG4180.muc(pBBR-AXSalgT) produced about 3.7-fold more alginate (353.6 μg alginate/mg protein) than PG4180.muc(pBBR1MCS-3) (94.4 μg alginate/mg protein). These results indicated that algT of PG4180.muc confers alginate synthesis.
Influence of functional algT on transcript levels of target genes.
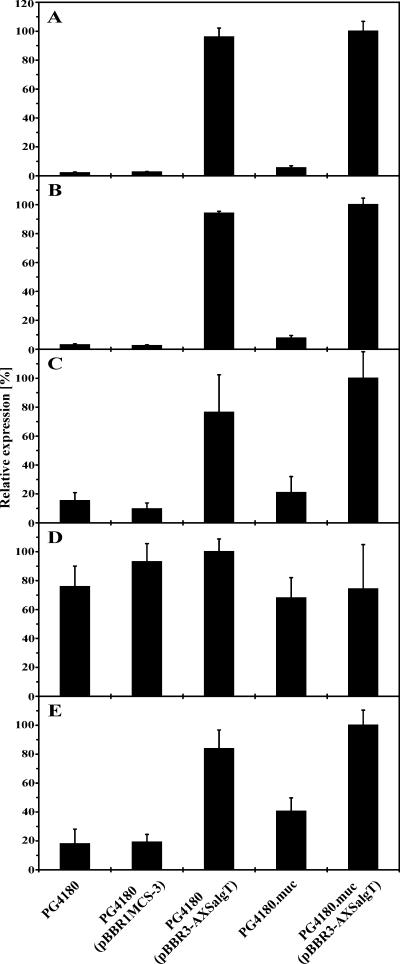
In P. aeruginosa, transcriptional activation of the biosynthetic algD operon and the algT-mucABD gene cluster depends on expression of AlgT (15, 21, 22, 27, 66). Whether AlgT of P. syringae pv. glycinea activates transcription of the algT-mucABD gene cluster and the algD operon was investigated by RNA spot blot analysis. Total RNA was isolated from PG4180, PG4180.muc, PG4180(pBBR3-AXSalgT), PG4180.muc(pBBR3-AXSalgT), and PG4180(pBBR1MCS-3) and analyzed with antisense RNA probes specific for algT, mucA, mucB, mucD, or algD, respectively (Fig. 2). Compared to its algT-deficient parent, the algT+ strain PG4180.muc showed a transcriptional induction of 2.7-fold for algT, 2.6-fold for mucA, 1.4-fold for mucB, and 2.3-fold for algD. Interestingly, no induction for mucD was observed. Transconjugants PG4180(pBBR3-AXSalgT) and PG4180.muc(pBBR3-AXSalgT) showed a transcriptional induction of 37-fold for algT, 35-fold for mucA, 8-fold for mucB, and 4.4-fold for algD. As observed above, no significant induction for mucD was observed. These results suggested that the exceptionally high induction of algT and mucA in transconjugants carrying plasmid pBBR3-AXSalgT might be due to additional mRNA copies containing algT. Our results indicated that AlgT in P. syringae induces transcription of the algT-mucAB gene cluster and the algD operon, whereas transcription of the mucD gene does not depend on AlgT.
FIG. 2.
Relative transcript abundance of the genes algT (A), mucA (B), mucB (C), mucD (D), and algD (E) in dependence of different algT genetic backgrounds. PG4180, PG4180(pBBR1MCS-3), PG4180.muc, PG4180(pBBR3-AXSalgT), and PG4180.muc(pBBR3-AXSalgT) were cultured in HSC medium at 28°C to an OD600 of 1.0. Total RNA was isolated and subjected to quantitative RNA spot blot analysis with algT-, mucA-, mucB-, mucD-, and algD-specific antisense RNA probes. The data represent the mean relative expression ± standard deviations (n = 4). Data were normalized to the highest expression value, which was set to 100%.
Transcriptional organization of the algT-mucABD gene cluster.
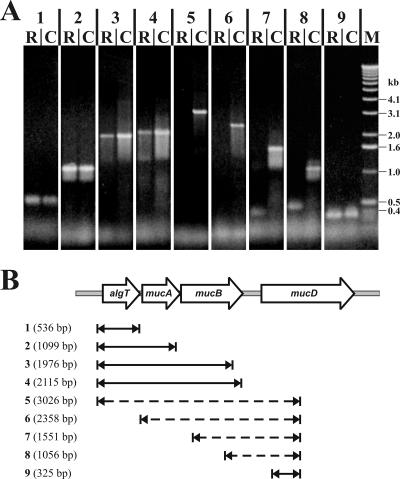
Differences in the transcriptional levels of algT, mucA, and mucB as opposed to that of mucD in PG4180.muc indicated that the transcriptional organization might differ from the previously proposed operon structure (22). To investigate this, an RT-PCR-based approach was used to amplify different regions within the algT-mucABD gene cluster from PG4180.muc total RNA (Fig. 3B). DNA-free total RNA was transcribed into cDNA by use of reverse transcriptase. The cDNA was then used as template in PCR amplification reactions employing different combinations of primers (Fig. 3A). PCR products were obtained for those primer pairs which were localized either within the algT-mucA-mucB or the mucD gene regions. No products were amplified spanning the region between the mucB and mucD genes. These results suggested that algT, mucA, and mucB are located on a polycistronic mRNA, whereas mucD is transcribed into an independent mRNA.
FIG. 3.
RT-PCR analysis of the algT-mucABD gene cluster in P. syringae pv. glycinea. (A) Agarose gel with PCR products obtained from PG4180.muc cDNA (R) and PG4180.muc genomic DNA (C) as a positive control. For size estimation, a DNA size standard (M) was included. Numbers 1 to 9 represent the PCR products obtained by different primer combinations as described in panel B. (B) Schematic overview of length and location of the PCR products (1 to 9) within the algT-mucABD gene cluster. A solid arrow indicates that a PCR product was obtained from PG4180.muc cDNA, whereas a dashed arrow indicates that no PCR product could be obtained.
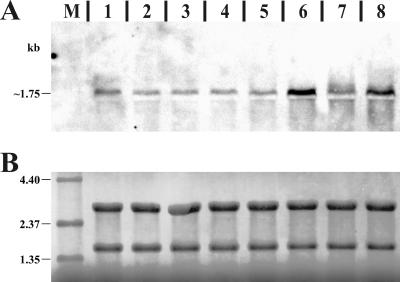
To gain further evidence for a separate mucD transcript, Northern blot analysis was carried out. In addition to the P. syringae pv. glycinea strains, we included one Azotobacter vinelandii and three Pseudomonas aeruginosa strains, which all possess an algT-mucABCD gene cluster. For increased and specific signals, equal mixtures of mucD antisense RNA probes, specific for mucD from P. syringae, A. vinelandii, and P. aeruginosa, were used for Northern blot hybridization. All strains showed a clear band of about 1.75 kb (Fig. 4). Comparison of the sizes of the predicted mucD open reading frames of A. vinelandii (1,422 bp), P. aeruginosa (1,422 bp), and P. syringae pv. glycinea (1,437 bp) with the observed mucD transcript sizes (∼1,750 bp) indicated that mucD is likely to be transcribed as a monocistronic mRNA in all investigated strains. Interestingly, PG4180 showed a slightly higher mucD transcript level than PG4180.muc. Moreover, in both PG4180 and PG4180.muc, there was more mucD transcript observed when total RNA was obtained from cells grown at 28°C compared to cells grown at 18°C.
FIG. 4.
(A) Northern blot membrane probed with a mucD-specific antisense RNA. The lanes contain 1.5 μg total RNA isolated from cultures grown to an OD600 of 1.0 from A. vinelandii wtC15 at 28°C (lane 1), P. aeruginosa Pa8821 at 37°C (lane 2), P. aeruginosa Pa8822 at 37°C (lane 3), P. aeruginosa Pa8830 at 37°C (lane 4), P. syringae PG4180 at 18°C (lane 5) or 28°C (lane 6), and P. syringae PG4180.muc at 18°C (lane 7) or 28°C (lane 8). The obtained signal, slightly above the 16S rRNA band, is marked with the estimated size. (B) Methylene blue-stained membrane prior to Northern blot hybridization for transcript size estimation, control of total RNA quantity, and successful Northern transfer. The same region of the membrane as that shown in panel A is shown. Distinct bands in lanes 1 to 8 represent those of the 23S (upper) and 16S rRNA (lower) or of the RNA size standard (3.0 μg) in lane M.
Transcriptional start site of mucD in P. syringae.
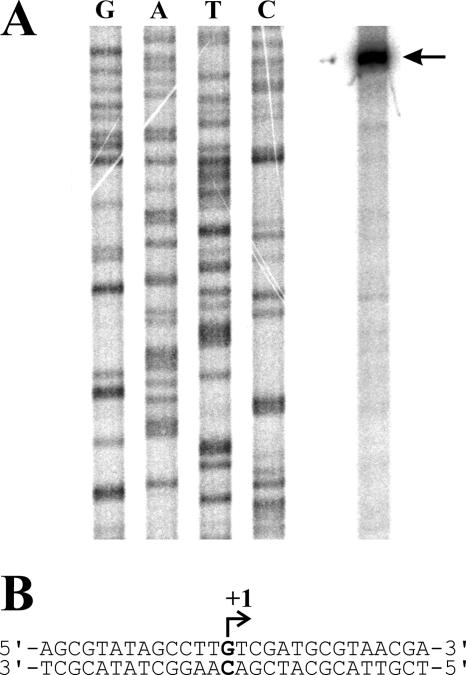
Results of RT-PCR and Northern blot analyses prompted us to identify the transcriptional start site of mucD in PG4180. For this, primer extension experiments using total RNA from PG4180 and primer mucD-Ps-3_pe were carried out, resulting in a clear signal at nucleotide position −320 upstream of the translational start codon of mucD (Fig. 5). Interestingly, the potential transcriptional start site of mucD in PG4180 is located 37 nucleotides upstream of the mucB stop codon. It is further noteworthy that the 283-bp intergenic region between mucB and mucD is unique to various P. syringae strains but is not found in P. aeruginosa or A. vinelandii. Results were verified with two additional primers identifying the same nucleotide as the transcriptional start site of mucD in PG4180 (data not shown).
FIG. 5.
Determination of the transcriptional start site of mucD in PG4180. (A) Separation of the results of nucleotide sequencing and primer extension reaction (black arrow) on a 6% polyacrylamide gel. (B) Nucleotide sequence surrounding the transcriptional start site (+1) of mucD.
In vitro and in planta growth of PG4180 and PG4180.muc.
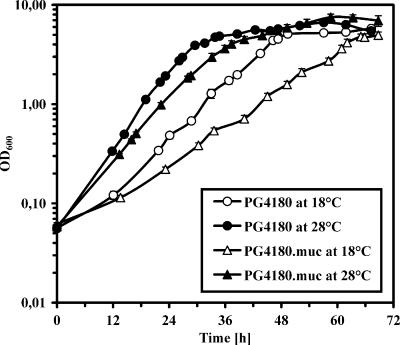
Alginate synthesis in P. syringae is up-regulated by AlgT. Whether or not this influences in vitro or in planta growth was studied using strains PG4180 and PG4180.muc. For in vitro growth studies, bacteria were cultured in HSC medium at 18 and 28°C; 28°C represents the optimal growth temperature, while 18°C is a temperature where P. syringae pv. glycinea is most virulent. The OD600 was monitored continuously until cultures entered the late stationary phase (Fig. 6). PG4180.muc was impaired in its in vitro growth at 18 and 28°C compared to PG4180. This was most apparent in the extended doubling times (td) for PG4180.muc (td at 18°C, 8.9 h; td at 28°C, 5.4 h) compared to PG4180 (td at 18°C, 6.4 h; td at 28°C, 4.4 h).
FIG. 6.
In vitro growth of P. syringae pv. glycinea PG4180 and PG4180.muc in HSC medium at 18°C and 28°C as monitored by OD600. Data represent the mean values of three independent cultures ± standard deviations.
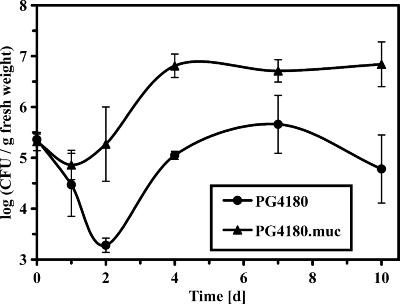
In planta growth was evaluated by monitoring bacterial populations (Fig. 7) of PG4180 and PG4180.muc on soybean plants for 10 days. Within 10 days postinoculation, PG4180.muc induced clear symptoms in the form of water soaking followed by necrosis, which were significantly more pronounced than those induced by PG4180 (data not shown). Population dynamics showed that both strains started with about the same number of CFU (105.4 CFU/g fresh weight) (Fig. 7). Afterwards, this number declined for PG4180 at day 2 by more than two orders of magnitude (103.3 CFU/g fresh weight). The difference in population size between PG4180 and PG4180.muc was most significant at day 2 postinoculation with almost two orders of magnitude and remained like this with little alteration until the end of the sampling period. The results suggested that PG4180.muc had a clear advantage in its ability to survive and maintain its population size on soybean leaves, in contrast to PG4180. The actual in planta multiplication rates during exponential growth (day 2 to 4) showed no significant difference. In summary, our results indicated that the presence of a functional algT gene is disadvantageous for the ability to grow in vitro but strongly promotes in planta survival of P. syringae pv. glycinea.
FIG. 7.
In planta growth of P. syringae pv. glycinea PG4180 and PG4180.muc on soybean leaves. Bacterial strains (107 CFU/ml) were spray inoculated (∼8 lb/in2) onto soybean leaves until surfaces were uniformly wet. Data represent the mean values of three independent leaf samples ± standard deviations.
DISCUSSION
Most isolates of P. syringae, including P. syringae pv. glycinea PG4180, are nonmucoid (35). In P. aeruginosa, alginate genes are normally silent but can be activated by mutational changes in the algT-mucABCD gene cluster (43, 44). A respective model of a transmembrane apparatus consisting of the sigma factor, AlgT, its anti-sigma factor, MucA, and the anti-anti-sigma factor, MucB, had been proposed (44) and is widely accepted. In the present study, we analyzed how the non-alginate-producing strain PG4180 converted into its alginate-producing derivative, PG4180.muc. Sequence analysis of the algT-mucABD gene cluster of PG4180 showed the same gene organization as that of the closely related P. syringae pv. syringae strain, FF5 (33). Interestingly, P. syringae strains lack mucC, in contrast to P. aeruginosa and A. vinelandii (8, 33). In P. aeruginosa, mucC was required for survival in the presence of high salt and elevated temperatures, and in A. vinelandii it functions as a negative regulator for alginate biosynthesis (7, 48).
Absence of alginate production in PG4180 was due to a nonsense mutation in algT, which spontaneously reverted into an intact open reading frame resulting in a functional AlgT protein in the alginate-producing derivative, PG4180.muc. Complementation with algT cloned from PG4180.muc turned PG4180 into an alginate-producing strain, suggesting that alginate synthesis in PG4180.muc is solely due to a functional algT gene. The alternative sigma factor, AlgT, controls alginate biosynthesis in P. aeruginosa, A. vinelandii, and P. syringae pv. syringae (27, 34, 40, 43, 44, 66). While alginate production in PG4180 was turned off by a mutation in algT, in P. aeruginosa conversion to mucoid phenotype is often due to mutations in the anti-sigma factor mucA or mucB (44). As expected, additional plasmid-borne copies of algT converted PG4180.muc into an alginate overproducer, possibly due to an altered ratio of sigma factor versus anti-sigma factor. This result is in line with the broadly accepted model for induction and autoregulation of AlgT (57).
Based on different algT backgrounds, we studied transcription of selected genes known to depend on AlgT (16, 22). As expected, P. syringae transcription levels showed an autoregulation of algT, mucA, and mucB in bacteria producing AlgT, which furthermore resulted in activation of the alginate biosynthetic operon as measured by algD transcript levels. Our data are thus consistent with expression data obtained for P. aeruginosa and A. vinelandii (6, 51). Transcript levels for mucD were slightly higher in the algT mutant background, which contradicts the model according to which mucD transcription depends on AlgT and mucD resides on the same transcript as algT and mucABC.
The transcriptional organization of mucD within the algT-muc gene cluster was studied in more detail in A. vinelandii, P. aeruginosa, and P. syringae. Our primer extension experiment suggested that mucD is not part of the algT-mucAB operon in P. syringae but has its own transcriptional start site, which in fact is located 320 bp upstream of the mucD translational start site and within the 3′ end of the mucB coding sequence. The intergenic region between mucB and mucD is unique to P. syringae, suggesting that the situation might be different in P. aeruginosa and A. vinelandii. According to the mucD transcript sizes predicted in this study via Northern blot analyses and RT-PCR experiments, putative mucD promoters in A. vinelandii and P. aeruginosa might be located within the annotated mucC gene. For A. vinelandii, it had previously been proposed that algT-mucABCD is an operon, but no direct evidence was given (43). Recently, Firoved and Deretic (22) reported microarray expression data for AlgT-dependent genes in P. aeruginosa, where a significant gradient in expression levels for the algT-mucABCD gene cluster was observed. Highest expression levels for algT and lowest levels for mucD were interpreted as mRNA degradation starting from the 3′ end of the polycistronic transcript (22, 43). When those authors compared mucD transcription of the wild type and an algT mutant, mucD showed only a low 2.2-fold induction, while algT was induced 49.2-fold when AlgT was present. Our data furthermore confirmed results made recently by Wood and Ohman (65), who also reported an expression of mucD independent from that of algT-mucABC in P. aeruginosa. All these former results are therefore supportive to our findings, which allow us to speculate on the presence of two independent transcripts for algT-mucAB and mucD in P. syringae.
Elevated levels of mucD transcription in P. syringae at 28°C as opposed to 18°C indicated a potential role for MucD as a temperature-induced periplasmic protease (39). This is consistent with our preliminary observation that a P. syringae PG4180 mucD mutant is hindered in its in vitro growth at elevated temperatures (A. Schenk and M. Ullrich, unpublished data). Our findings support the hypothesis that mucD might act on AlgT or its regulators in an indirect way by removing periplasmic signals that activate the stress response system (6, 42, 67, 68). In consequence, the previously found impact of mucD mutations on the AlgT-dependent regulation of alginate synthesis (6, 43) might be rather indirect and might not be associated with the colocalization of mucD and algT-muc in the genome of alginate-producing bacteria. This speculation is supported by the fact that the mucD homolog htrA is located far distant from the algT homolog, rpoE, in the genome of E. coli (47).
Alginate production had previously been associated with several fitness traits in the plant-microbe interaction (17, 58, 69). Herein, we evaluated the impact of a functional algT gene on in vitro and in planta growth. The AlgT+ strain, PG4180.muc, was impaired in growth in liquid medium, which was manifested by longer doubling times. In contrast, the algT-deficient strain, PG4180, showed better growth in vitro. Interestingly, results obtained in planta showed a clear advantage to survive and multiply for AlgT-producing bacteria. This is consistent with an earlier study where an alginate-deficient mutant of P. syringae pv. syringae was impaired in epiphytic fitness (69). Whether the advantage of PG4180.muc is only due to its ability to produce alginate or to other regulatory effects of AlgT needs to be addressed in more detail in future studies. AlgT promotes in planta survival of P. syringae, correlating with better bacterial growth, and therefore may be an indispensable regulator of natural fitness of P. syringae. According to long-lasting experience with PG4180, this strain was nonmucoid ever since it had been isolated from soybean plants in 1975 (R. E. Mitchell, unpublished data). We speculate that alginate production in P. syringae pv. glycinea might not be beneficial for all stages during the plant-microbe interaction. Synthesis of an exopolysaccharide is energy expensive and might hinder bacteria during particular periods, i.e., the onset of infection or rapid multiplication. During such periods, a single point mutation in algT might have rendered PG4180 ecologically fit and thus isolatable. It could be hypothesized that mixed populations of alginate-positive and alginate-negative cells might be of advantage for the complex infection process. Whether or not a typical genotype conversion takes place in P. syringae pv. glycinea and whether it is limited to mutations in algT remains to be elucidated and will be addressed in future studies.
Acknowledgments
We thank H. Weingart, A. Smirnova, A. Wensing, Y. Braun, L. Steil, M. Winterhalter, and U. Schwaneberg for stimulating discussions and critical comments.
This work was supported by grants from the Max-Planck Society and the Deutsche Forschungsgemeinschaft (M.U.) and grant AI 43311 from the National Institutes of Health (C.L.B.).
Footnotes
Published ahead of print on 29 September 2006.
REFERENCES
- 1.Alfano, J. R., and A. Collmer. 1996. Bacterial pathogens in plants: life up against the wall. Plant Cell 8:1683-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anastassiou, E. D., A. C. Mintzas, C. Kounavis, and G. Dimitracopoulos. 1987. Alginate production by clinical nonmucoid Pseudomonas aeruginosa strains. J. Clin. Microbiol. 25:656-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 4.Beattie, G. A., and S. E. Lindow. 1994. Epiphytic fitness of phytopathogenic bacteria: physiological adaptations for growth and survival. Curr. Top. Microbiol. Immunol. 192:1-27. [DOI] [PubMed] [Google Scholar]
- 5.Bender, C. L., H. Liyanage, D. Palmer, M. Ullrich, S. Young, and R. Mitchell. 1993. Characterization of the genes controlling the biosynthesis of the polyketide phytotoxin coronatine including conjugation between coronafacic and coronamic acid. Gene 133:31-38. [DOI] [PubMed] [Google Scholar]
- 6.Boucher, J. C., J. Martinezsalazar, M. J. Schurr, M. H. Mudd, H. Yu, and V. Deretic. 1996. Two distinct loci affecting conversion to mucoidy in Pseudomonas aeruginosa in cystic fibrosis encode homologs of the serine protease HtrA. J. Bacteriol. 178:511-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boucher, J. C., M. J. Schurr, H. Yu, D. W. Rowen, and V. Deretic. 1997. Pseudomonas aeruginosa in cystic fibrosis - role of mucC in the regulation of alginate production and stress sensitivity. Microbiology 143:3473-3480. [DOI] [PubMed] [Google Scholar]
- 8.Buell, C. R., V. Joardar, M. Lindeberg, J. Selengut, I. T. Paulsen, M. L. Gwinn, R. J. Dodson, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, S. Daugherty, L. Brinkac, M. J. Beanan, D. H. Haft, W. C. Nelson, T. Davidsen, N. Zafar, L. Zhou, J. Liu, Q. Yuan, H. Khouri, N. Fedorova, B. Tran, D. Russell, K. Berry, T. Utterback, S. E. Van Aken, T. V. Feldblyum, M. D'Ascenzo, W. L. Deng, A. R. Ramos, J. R. Alfano, S. Cartinhour, A. K. Chatterjee, T. P. Delaney, S. G. Lazarowitz, G. B. Martin, D. J. Schneider, X. Tang, C. L. Bender, O. White, C. M. Fraser, and A. Collmer. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 100:10181-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burnett, W. V. 1997. Northern blotting of RNA denatured in glyoxal without buffer recirculation. BioTechniques 22:668-671. [DOI] [PubMed] [Google Scholar]
- 10.Chi, E., and D. H. Bartlett. 1995. An rpoE-like locus controls outer membrane protein synthesis and growth at cold temperatures and high pressures in the deep-sea bacterium Photobacterium sp. strain SS9. Mol. Microbiol. 17:713-726. [DOI] [PubMed] [Google Scholar]
- 11.Chitnis, C. E., and D. E. Ohman. 1993. Genetic analysis of the alginate biosynthetic gene cluster of Pseudomonas aeruginosa shows evidence of an operonic structure. Mol. Microbiol. 8:583-593. [DOI] [PubMed] [Google Scholar]
- 12.Darzins, A., and A. M. Chakrabarty. 1984. Cloning of genes controlling alginate biosynthesis from a mucoid cystic fibrosis isolate of Pseudomonas aeruginosa. J. Bacteriol. 159:9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delaspenas, A., L. Connolly, and C. A. Gross. 1997. The sigma(E)-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of sigma(E). Mol. Microbiol. 24:373-385. [DOI] [PubMed] [Google Scholar]
- 14.Denny, T. P. 1995. Involvement of bacterial polysaccharides in plant pathogenesis. Annu. Rev. Phytopathol. 33:173-197. [DOI] [PubMed] [Google Scholar]
- 15.DeVries, C. A., and D. E. Ohman. 1994. Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternate sigma factor, and shows evidence for autoregulation. J. Bacteriol. 176:6677-6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fakhr, M. K., A. Penaloza-Vazquez, A. M. Chakrabarty, and C. L. Bender. 1999. Regulation of alginate biosynthesis in Pseudomonas syringae pv. syringae. J. Bacteriol. 181:3478-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fett, W. F., and M. F. Dunn. 1989. Exopolysaccharides produced by phytopathogenic Pseudomonas syringae pathovars in infected leaves of susceptible hosts. Plant Physiol. 89:5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fett, W. F., C. Wijey, and E. R. Lifson. 1992. Occurrence of alginate gene sequences among members of the pseudomonad rRNA homology groups I-IV. FEMS Microbiol. Lett. 78:151-157. [DOI] [PubMed] [Google Scholar]
- 19.Fialho, A. M., N. A. Zielinski, W. F. Fett, A. M. Chakrabarty, and A. Berry. 1990. Distribution of alginate gene sequences in the Pseudomonas rRNA homology group I-Azomonas-Azotobacter lineage of superfamily B procaryotes. Appl. Environ. Microbiol. 56:436-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Firoved, A. M., J. C. Boucher, and V. Deretic. 2002. Global genomic analysis of AlgU (σE)-dependent promoters (sigmulon) in Pseudomonas aeruginosa and implications for inflammatory processes in cystic fibrosis. J. Bacteriol. 184:1057-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Firoved, A. M., and V. Deretic. 2003. Microarray analysis of global gene expression in mucoid Pseudomonas aeruginosa. J. Bacteriol. 185:1071-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Govan, J. R., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gross, M., and K. Rudolph. 1987. Studies on the extracellular polysaccharides (EPS) produced in vitro by Pseudomonas syringae pv. phaseolicola. II. Characterization of levan, alginate, and LPS. J. Phytopathol. 119:206-215. [Google Scholar]
- 25.Gross, M., and K. Rudolph. 1987. Studies on the extracellular polysaccharides (EPS) produced in vitro by Pseudomonas syringae pv. phaseolicola. II. Indications for a polysaccharide resembling alginic acid in seven P. syringae pathovars, and LPS. J. Phytopathol. 118:276-287. [Google Scholar]
- 26.Herrin, D. L., and G. W. Schmidt. 1988. Rapid, reversible staining of northern blots prior to hybridization. BioTechniques 6:196-200. [PubMed] [Google Scholar]
- 27.Hershberger, C. D., R. W. Ye, M. R. Parsek, Z. D. Xie, and A. M. Chakrabarty. 1995. The algT (algU) gene of Pseudomonas aeruginosa, a key regulator involved in alginate biosynthesis, encodes an alternative sigma factor (σE). Proc. Natl. Acad. Sci. USA 92:7941-7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hettwer, U., F. R. Jaeckel, J. Boch, M. Meyer, K. Rudolph, and M. S. Ullrich. 1998. Cloning, nucleotide sequence, and expression in Escherichia coli of levansucrase genes from the plant pathogens Pseudomonas syringae pv. glycinea and P. syringae pv. phaseolicola. Appl. Environ. Microbiol. 64:3180-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirano, S. S., and C. D. Upper. 2000. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae-a pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 64:624-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ingerecht, I. L., C. I. Mandelbaum, and T. E. Mirkov. 1998. Highly sensitive northern hybridization using a rapid protocol for downward alkaline blotting of RNA. BioTechniques 25:420-426. [DOI] [PubMed] [Google Scholar]
- 31.Kado, C. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keane, P. J., A. Kerr, and P. B. New. 1970. Crown gall of stone fruit. II. Identification and nomenclature of Agrobacterium isolates. Aust. J. Biol. Sci. 23:585-595. [Google Scholar]
- 33.Keith, L. M., and C. L. Bender. 2001. Genetic divergence in the algT-muc operon controlling alginate biosynthesis and response to environmental stress in Pseudomonas syringae. DNA Seq. 12:125-129. [DOI] [PubMed] [Google Scholar]
- 34.Keith, L. M. W., and C. L. Bender. 1999. AlgT (sigma22) controls alginate production and tolerance to environmental stress in Pseudomonas syringae. J. Bacteriol. 181:7176-7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kidambi, S. P., G. W. Sundin, D. A. Palmer, A. M. Chakrabarty, and C. L. Bender. 1995. Copper as a signal for alginate synthesis in Pseudomonas syringae pv. syringae. Appl. Environ. Microbiol. 61:2172-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 37.Lam, J., R. Chan, K. Lam, and J. W. Costerton. 1980. Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect. Immun. 28:546-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, H., and M. S. Ullrich. 2001. Characterization and mutational analysis of three allelic lsc genes encoding levansucrase in Pseudomonas syringae. J. Bacteriol. 183:3282-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lipinska, B., S. Sharma, and C. Georgopoulos. 1988. Sequence analysis and regulation of the htrA gene of Escherichia coli: a sigma 32-independent mechanism of heat-inducible transcription. Nucleic Acids Res. 16:10053-10067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Majumdar, D., Y. J. Avissar, and J. H. Wyche. 1991. Simultaneous and rapid isolation of bacterial and eukaryotic DNA and RNA: a new approach for isolating DNA. BioTechniques 11:94-101. [PubMed] [Google Scholar]
- 42.Martin, D. W., M. J. Schurr, H. Yu, and V. Deretic. 1994. Analysis of promoters controlled by the putative sigma factor AlgU regulating conversion to mucoidy in Pseudomonas aeruginosa: relationship to sigma E and stress response. J. Bacteriol. 176:6688-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinezsalazar, J. M., S. Moreno, R. Najera, J. C. Boucher, G. Espin, G. Soberonchavez, and V. Deretic. 1996. Characterization of the genes coding for the putative sigma factor AlgU and its regulators MucA, MucB, MucC, and MucD in Azotobacter vinelandii and evaluation of their roles in alginate biosynthesis. J. Bacteriol. 178:1800-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mathee, K., A. Kharazami, and N. Hoiby. 2002. Role of exopolysaccharide in biofilm matrix formation: the alginate paradigm, p. 23-55. In M. R. J. C. (ed.), Molecular ecology of biofilms. Horizon Scientific Press, Norfolk, England.
- 45.Mathee, K., C. J. McPherson, and D. E. Ohman. 1997. Posttranslational control of the algT (algU)-encoded sigma22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN). J. Bacteriol. 179:3711-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.May, T. B., and A. M. Chakrabarty. 1994. Isolation and assay of Pseudomonas aeruginosa alginate. Methods Enzymol. 235:295-304. [DOI] [PubMed] [Google Scholar]
- 47.Missiakas, D., M. P. Mayer, M. Lemaire, C. Georgopoulos, and S. Raina. 1997. Modulation of the Escherichia coli sigma(E) (RpoE) heat-shock transcription-factor activity by the RseA, RseB and RseC proteins. Mol. Microbiol. 24:355-371. [DOI] [PubMed] [Google Scholar]
- 48.Nunez, C., R. Leon, J. Guzman, G. Espin, and G. Soberon-Chavez. 2000. Role of Azotobacter vinelandii mucA and mucC gene products in alginate production. J. Bacteriol. 182:6550-6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Osman, S. F., W. F. Fett, and M. L. Fishman. 1986. Exopolysaccharides of the phytopathogen Pseudomonas syringae pv. glycinea. J. Bacteriol. 166:66-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Page, W. J., and H. L. Sadoff. 1976. Physiological factors affecting transformation of Azotobacter vinelandii. J. Bacteriol. 125:1080-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pallen, M. J., and B. W. Wren. 1997. The HtrA family of serine proteases. Mol. Microbiol. 26:209-221. [DOI] [PubMed] [Google Scholar]
- 52.Palmer, D. A., and C. L. Bender. 1993. Effects of environmental and nutritional factors on production of the polyketide phytotoxin coronatine by Pseudomonas syringae pv. glycinea. Appl. Environ. Microbiol. 59:1619-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Penaloza-Vazquez, A., M. K. Fakhr, A. M. Bailey, and C. L. Bender. 2004. AlgR functions in algC expression and virulence in Pseudomonas syringae pv. syringae. Microbiology 150:2727-2737. [DOI] [PubMed] [Google Scholar]
- 54.Penaloza-Vazquez, A., S. P. Kidambi, A. M. Chakrabarty, and C. L. Bender. 1997. Characterization of the alginate biosynthetic gene cluster in Pseudomonas syringae pv. syringae. J. Bacteriol. 179:4464-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pier, G. B. 1998. Pseudomonas aeruginosa: a key problem in cystic fibrosis. ASM News 64:339-347. [Google Scholar]
- 56.Pier, G. B., D. Desjardins, T. Aguilar, M. Barnard, and D. P. Speert. 1986. Polysaccharide surface antigens expressed by nonmucoid isolates of Pseudomonas aeruginosa from cystic fibrosis patients. J. Clin. Microbiol. 24:189-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rowen, D. W., and V. Deretic. 2000. Membrane-to-cytosol redistribution of ECF sigma factor AlgU and conversion to mucoidy in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Mol. Microbiol. 36:314-327. [DOI] [PubMed] [Google Scholar]
- 58.Rudolph, K. W. E., M. Gross, and F. Ebrahim-Nesbat. 1994. The role of extracellular polysaccharides as virulence factors for phytopathogenic pseudomonads and xanthomonads. In C. I. Kado and J. H. Cross (ed.), Molecular mechanisms for bacterial virulence. Kluwer, Dordrecht, The Netherlands.
- 59.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 60.Schurr, M. J., and V. Deretic. 1997. Microbial pathogenesis in cystic fibrosis - co-ordinate regulation of heat-shock response and conversion to mucoidy in Pseudomonas aeruginosa. Mol. Microbiol. 24:411-420. [DOI] [PubMed] [Google Scholar]
- 61.Schurr, M. J., H. Yu, J. M. Martinez-Salazar, J. C. Boucher, and V. Deretic. 1996. Control of AlgU, a member of the sigma E-like family of stress sigma factors, by the negative regulators MucA and MucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J. Bacteriol. 178:4997-5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schurr, M. J., H. Yu, J. M. Martinez-Salazar, N. S. Hibler, and V. Deretic. 1995. Biochemical characterization and posttranslational modification of AlgU, a regulator of stress response in Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 216:874-880. [DOI] [PubMed] [Google Scholar]
- 63.Tatnell, P. J., N. J. Russell, and P. Gacesa. 1994. GDP-mannose dehydrogenase is the key regulatory enzyme in alginate biosynthesis in Pseudomonas aeruginosa: evidence from metabolite studies. Microbiology 140:1745-1754. [DOI] [PubMed] [Google Scholar]
- 64.Whitfield, C. 1988. Bacterial extracellular polysaccharides. Can. J. Microbiol. 34:415-420. [DOI] [PubMed] [Google Scholar]
- 65.Wood, L. F., and D. E. Ohman. 2006. Independent regulation of MucD, an HtrA-like protease in Pseudomonas aeruginosa, and the role of its proteolytic motif in alginate gene regulation. J. Bacteriol. 188:3134-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wozniak, D. J., and D. E. Ohman. 1994. Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J. Bacteriol. 176:6007-6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu, H., J. C. Boucher, N. S. Hibler, and V. Deretic. 1996. Virulence properties of Pseudomonas aeruginosa lacking the extreme-stress sigma factor AlgU (σE). Infect. Immun. 64:2774-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu, H., M. J. Schurr, and V. Deretic. 1995. Functional equivalence of Escherichia coli sigma E and Pseudomonas aeruginosa AlgU: E. coli rpoE restores mucoidy and reduces sensitivity to reactive oxygen intermediates in algU mutants of P. aeruginosa. J. Bacteriol. 177:3259-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu, J., A. Penaloza-Vazquez, A. M. Chakrabarty, and C. L. Bender. 1999. Involvement of the exopolysaccharide alginate in the virulence and epiphytic fitness of Pseudomonas syringae pv. syringae. Mol. Microbiol. 33:712-720. [DOI] [PubMed] [Google Scholar]
- 70.Zielinski, N. A., A. M. Chakrabarty, and A. Berry. 1991. Characterization and regulation of the Pseudomonas aeruginosa algC gene encoding phosphomannomutase. J. Biol. Chem. 266:9754-9763. [PubMed] [Google Scholar]