Abstract
The bldB gene of Streptomyces coelicolor encodes the best-characterized member of a family of small proteins that have low isoelectric points but that lack any previously characterized sequence motifs. BldB is dimeric and is required for the efficient production of antibiotics and spore-forming cells, called aerial hyphae, by growing colonies. The mechanism of action of BldB and its relatives is unknown. Here, we have explored amino acids in BldB that either are highly conserved or have been implicated in function genetically. We show that five amino acids are important for its function at physiological expression levels. Mutations in three of these amino acids gave rise to proteins that were either monomeric or unstable in vivo, while two others are not. We find that overexpression of bldB in S. coelicolor blocks sporulation prior to sporulation-specific septation but permits the formation of aerial hyphae. Vegetative septation was apparently normal in both the bldB null mutant and the bldB overexpression strain. To our surprise, overexpression of the dimerization-competent but functionally defective alleles caused a dramatic acceleration of sporulation. Our results suggest that BldB makes at least one important contact with another subcellular constituent and that a loss or alteration of this interaction impairs the phenotypic properties of the organism.
The bacterium Streptomyces coelicolor is the best-characterized model organism for the spore-forming Streptomyces genus. These organisms are filamentous bacteria that are characterized by a robust secondary metabolism that has been exploited as a source of antibiotics, chemotherapeutic agents, antifungal drugs, immune suppressants, and other medicinally and agriculturally important molecules (3). The life cycle of this organism is especially complex for a bacterium. When spores germinate, they give rise to filamentous cells referred to as “substrate hyphae.” These substrate hyphae grow by elongating and branching and form septal cross walls at infrequent intervals such that each cell is an elongated compartment housing multiple chromosomes. After ∼48 h (under laboratory conditions), a second filamentous cell type appears on the colony surface and grows up into the air, forming a white layer referred to as an “aerial mycelium.” Individual aerial hyphae adopt a coiled shape as they mature. Antibiotic production commences at around the same time as the formation of the aerial mycelium. In S. coelicolor, secondary metabolism is readily visible, as two of the antibiotics produced by this organism are pigmented: the polyketide actinorhodin is blue, and the tripyrrole undecylprodigiosin is red. Spore formation occurs exclusively in the aerial hyphae, and this too be can be readily visualized as a gray polyketide pigment is deposited in the maturing spore wall (12).
Mutations that interrupt this life cycle can be divided into several categories. The bld mutants block the formation of the aerial hyphae, giving rise to colonies that lack the fuzzy surface layer (13). Some bld mutations also block the production of antibiotics such that colonies lack the red and blue pigmentation that is characteristic of wild-type S. coelicolor. Mutations in the whi genes permit the propagation of the aerial mycelium but prevent the normal maturation of spores such that colonies fail to acquire their usual gray pigmentation and instead maintain a white aerial mycelium (5). Finally, genes that, when mutated, impair secondary metabolism but permit the formation of the aerial mycelium and the production of spores have been identified previously (see, e.g., reference 1).
The bldB gene encodes a small acidic protein (pI ∼4.2) that has a monomeric molecular mass of 10.9 kDa and that is normally dimeric (7, 9, 21). Null mutations in bldB confer a profound defect in both the formation of aerial hyphae and the production of antibiotics (7, 17). Indeed, while the phenotypic defects of many bld mutations can be at least partially reversed by cultivation on media that have poor carbon sources, the defects conferred by bldB mutations cannot (22). Furthermore, there is evidence that strains bearing mutations in bldB are defective in normal carbon metabolism, such that genes that are normally repressed by the presence of glucose are expressed (22).
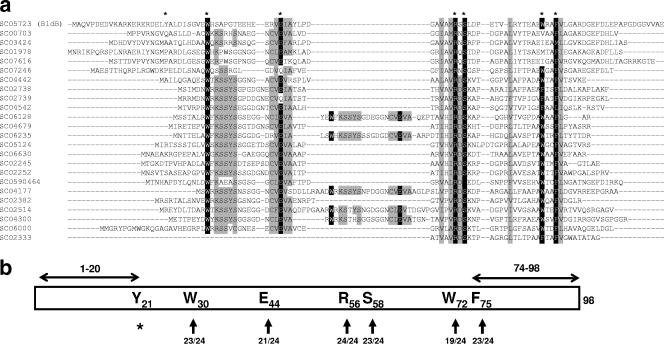
BldB lacks obvious sequence motifs of known function. It is, however, related to the products of at least 24 (Fig. 1a) other genes in the S. coelicolor genome. Included among these are the product of an open reading frame in the abaA locus (SCO0703), a protein that has been implicated in the production of several antibiotics, and the product of an open reading frame (SCO4542) lying immediately downstream of whiJ, which is important for spore formation by aerial hyphae. We have conducted an exhaustive search of DNA sequence databases and discovered that the bldB-like family of genes is restricted to filamentous actinomycetes. We have explored the role of six highly conserved amino acids in BldB and one amino acid that was previously implicated in function. Of these seven amino acids, we found that two are unimportant for function in vivo and that the other five are critical. Of these five amino acids, we show that two give rise to stable proteins that nevertheless fail to function at normal physiological levels. We find that the overexpression of bldB blocks sporulation in aerial hyphae but that the overexpression of the two dimerization-competent but defective alleles causes accelerated sporulation. These data suggest that BldB must have at least one interaction with another cellular constituent that is important for its biological function.
FIG. 1.
Highly conserved residues in members of the BldB family. (a) Sequence alignment of BldB homologues in Streptomyces coelicolor. Homologues were identified using PSI-BLAST (NCBI) and are identified by the SCO numbers (2). Conserved residues are emphasized by gray (moderately conserved) or black (nearly universally conserved) shading. BldB residues that were mutated in this work are indicated by asterisks. An internal repeat within five of the protein sequences is evident. (b) Cartoon of BldB emphasizing residues 21, 30, 44, 56, 58, 72, and 75, which were mutated in this work, and the N- and C-terminal truncations that can be made without compromising dimer formation (7). The degree of conservation of these residues in the 24 BldB homologues of S. coelicolor is indicated.
MATERIALS AND METHODS
Sequence alignment.
The full-length BldB amino acid sequence was used as a query in a PSI-BLAST search at the NCBI website. Repeated iterations yielded a list of 89 homologues from which the S. coelicolor proteins were extracted. Alignments were performed using ClustalX (24) followed by manual adjustment. To attempt to rule out the existence of bldB-like genes in the Mycobacteria and Corynebacteria, we conducted more targeted BLAST searches of their genomes, none of which revealed any statistically relevant homologues.
Site-directed mutagenesis.
Mutagenesis was carried out on plasmid pRA1 (the plasmids and strains used in this work are listed in Tables 1 and 2, respectively) at the amino acid residues indicated by boxes in Fig. 1 by using the oligonucleotides listed in Table 3 and Quickchange mutagenesis (Stratagene). Each codon of interest was altered to code for an alanine residue. The same oligonucleotides were used to introduce the desired changes into the two-hybrid plasmids pT18NHB and pT25NHB (7). All mutations were confirmed by DNA sequencing.
TABLE 1.
Plasmids used in this study
| Plasmid | Descriptiona | Phenotypeb | Source or reference |
|---|---|---|---|
| S. coelicolor | |||
| pIJ486 | ter neo tsr ori pIJ101 rep pIJ101 MCS | Thior | 14 |
| pIJ486BB | ter neo tsr ori pIJ101 rep pIJ101 MCS bldB lacZα aac(3)IV ori pUC18 | Aprr/Thior | This work |
| pRA1 | lacZα aac(3)IV ori pUC18 oriT (RK2) int ϕC31 φC31 attP MCS bldB + 304-bp promoter region | Aprr | 7 |
| E. coli | |||
| pT18NHB | bla ori colE1 f1 origin T18 MCS 0.32-kb KpnI amplicon from pBB801 containing bldB inserted into pT18 | Ampr | 6 |
| pT25NHB | cat ori p15A T25 MCS 0.32-kb KpnI amplicon from pBB801 containing bldB inserted into pT25 | Chlr | 7 |
MCS, multiple cloning site.
Antibiotic resistance markers are apramycin (Aprr), ampicillin (Ampr), thiostrepton (Thior), and chloramphenicol (Chlr).
TABLE 2.
Strains used in this study
| Strain | Descriptiona | Phenotype | Source or reference |
|---|---|---|---|
| S. coelicolor | |||
| M145 | Prototroph SCP1− SCP2− | Lab collection | |
| N985 | bldB::aphI SCP1− SCP2− | Bld | 7 |
| N17 | bldK::Specr SCP1− SCP2− | Bld | 19 |
| N376 | ramR::AprrSCP1− SCP2− | Bld | 20 |
| J2401 | whiA::Hygr SCP1− SCP2− | Whi | 8 |
| J2402 | whiB::Hygr SCP1− SCP2− | Whi | 8 |
| J2400 | whiG::HygrSCP1− SCP2− | Whi | 8 |
| J2210 | whiH::Hygr SCP1− SCP2− | Whi | 8 |
| J2407 | sigF::TsrrSCP1− SCP2− | Whi | 8 |
| GSB1 | ssgB::Aprr SCP1− SCP2− | Whi | 11 |
| E. coli | |||
| Er2-1 | F′ lacIQleuB6 thi-1 fhuA31 lacY1 tsx-78 galK2 galT22 supE44 hisG4 rpsL136 (Strr) xyl-5 mtl-1 dam13::Tn9 (Camr) dcm-6 mcrB1 mcrA hsdR2 (rK− mK+) | 16 | |
| XL-1 Blue | recA1 endA1 gyrA96 thi-1 glnV44 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
Antibiotic resistance markers are apramycin (Aprr), ampicillin (Ampr), hygromycin (Hygr), kanamycin (Kanr), and spectinomycin (Specr).
TABLE 3.
Oligonucleotides used for mutagenesis in this study
| Oligonucleotide | Sequence (5′-3′) | End use |
|---|---|---|
| Y21P1 | CGAGCTGGCCGCGCTCGAC | Y21A mutagenesis |
| Y21P2 | GTCGAGCGCGGCCAGCTCG | Y21A mutagenesis |
| W30P1 | CTCGACATCTCGGGTGTGGAGGCGCACAGCGCG | W30A mutagenesis |
| W30P2 | CGCGCTGTGCGCCTCCACACCCGAGATGTCGAG | W30A mutagenesis |
| E44P1 | GGAACACGAGGAGCGGGTGGCGATCGCCTATC | E44A mutagenesis |
| E44P2 | GATAGGCGATCGCCACCCGCTCCTCGTGTTCC | E44A mutagenesis |
| R56P1 | CCGACGGAGCCGTGGCCATGGCGTCGTCGCTG | R56A mutagenesis |
| R56P2 | CAGCGACGACGCCATGGCCACGGCTCCGTCGG | R56A mutagenesis |
| S58P1 | GAGCCGTGGCCATGCGGTCGGCGCTGGATCC | S58A mutagenesis |
| S58P2 | GGATCCAGCGCCGACCGCATGGCCACGGCTC | S58A mutagenesis |
| W72P1 | CTGCGGTACACCGAGGCGGAGGCGCGGGCTTTC | W72A mutagenesis |
| W72P2 | GAAAGCCCGCGCCTCCGCCTCGGTGTACCGCAG | W72A mutagenesis |
| F75P1 | CCGAGGCGGAGTGGCGGGCTGCCGTCCTGGGTG | F75A mutagenesis |
| F75P2 | CACCCAGGACGGCAGCCCGCCACTCCGCCTCGG | F75A mutagenesis |
Cloning of the BldB overexpression vector pIJ486BB.
The high-copy-number Streptomyces vector pIJ486 was isolated from Streptomyces lividans and fused to a fragment of pRA1 that included an Escherichia coli origin of replication, the bldB gene and promoter, and an apramycin resistance gene to generate pIJ486BB. Plasmid pRA1 was digested with BamHI and NheI, liberating a DNA fragment that included the bldB gene, the apramycin resistance gene, and the pUC18 origin of replication. pIJ486 was cut with BamHI and HindIII to generate a fragment bearing the pIJ486 origin of replication, and the two fragments were ligated and transformed into E. coli. The same cloning procedure was repeated for all of the point mutants in pRA1, generating overexpression plasmids for wild-type bldB and for all of the mutant forms of the gene.
Electron microscopy.
N985, a bldB null mutant (7) containing pIJ486, and wild-type strain M145 containing pIJ486, pIJ486BB, and the pIJ486BBF75A bldB(F75A) overexpression vector were cultivated on R2YE solid medium (14) for 48 h and prepared for electron microscopy.
Cells were fixed by the addition of a layer of 2% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) for 30 min at room temperature. The samples were then rinsed in 0.2 M sodium cacodylate buffer (pH 7.4). Once rinsed, they were postfixed in 1% osmium tetraoxide in 0.1 M sodium cacodylate buffer (pH 7.4) for 30 min at room temperature. Upon completion of the fixation procedure, the samples were isolated and dehydrated in an ethanol series of 50, 70, 95, and 100% ethanol.
For transmission electron microscopy, samples that were first dehydrated with ethanol were further dehydrated with propylene oxide, infiltrated with Spurr's resin, and polymerized in 100% Spurr's resin at 60°C overnight. The samples were then cut into thin (70-nm) sections and stained with uranyl acetate and lead citrate. After staining, the samples were imaged using a JEOL 1200EX Toyoko transmission electron microscope.
For scanning electron microscopy, ethanol-dehydrated samples were critical point dried and gold coated to 20 to 40 nm. Once coated with gold, samples were viewed on the JEOL 840 Toyoko scanning electron microscope.
RESULTS AND DISCUSSION
We have conducted an extensive search for genes encoding proteins homologous to BldB using repeated iterations of PSI-BLAST. In total, there were 89 BldB-like proteins encoded in four bacterial genera. These included 24 clearly BldB-like gene products in the S. coelicolor genome (Fig. 1a), 25 in Streptomyces avermitilis, 5 in other streptomycetes whose genomes are only partly sequenced (2, 10), and 3 in other actinomycetes. There were 11 BldB-like proteins in the moderately thermophilic organism Thermobifida fusca, 7 in the human pathogen Nocardia farcinica, and 16 in the N2-fixing endosymbiont Frankia. The unifying feature of this remarkably diverse group of organisms is that they are all filamentous (23). Three of them are also spore-forming bacteria, and while N. farcinica is not, its vegetative hypha cells undergo a morphological transition (fragmentation) that similarly results in dispersal and smaller units (23). Intriguingly, searches of the genomes of two other actinomycetes, Mycobacteria and Corynebacteria, did not reveal any genes encoding BldB-like proteins. In contrast to the other four, these actinomycetes are rod-shaped bacteria and do not produce spores, suggesting that the bldB family of genes may be restricted to filamentous actinomycetes. The significance of this is unknown but will surely be relevant to future investigation.
Alignment of the BldB-like protein sequences (see Fig. 1a for S. coelicolor and Fig. S1 in the supplemental material for the entire BldB-like family) revealed four blocks of relatively conserved sequence. In the S. coelicolor group, which includes representatives of all major features, 17 sequences exhibit the highly conserved WXK/RSSYS and CVEV/IA sequences (motifs I and II) near their amino termini. Five of these sequences have a second repeat of these motifs further into the protein (motifs I′ and II′ in Fig. 1a). BldB has a good match for motif II but not motif I, although it does have a W at residue 30, which is universally conserved in the family. The conserved sequences V/IA/HV/IRDSK (motif III) and AW/FXXFV/L (motif IV) were found progressively further towards the C termini, and BldB exhibits reasonable matches to both. We examined BldB in the context of these conserved blocks of sequence and identified six residues that were of particular interest (Fig. 1b). In addition to being highly conserved, these residues had hydrophilic side chains (E44, R56, and S58) or aromatic side chains (W30, W72, and F75), some of which might be surface exposed. We were less interested in conserved L, I, and V residues, assuming that these residues are more likely to be buried folding determinants. We targeted W30, E44, R56, S58, W72, and F75 for alanine substitution in the context of a previously constructed bldB complementation plasmid, pRA1 (7). We also constructed an alanine substitution at Y21, a residue previously demonstrated to be important for bldB function, as it is a frequent site of defective alleles of the gene (21). pRA1 is a low-copy-number vector that integrates into the S. coelicolor genome via a bacteriophage integrase mechanism (4). Between one and three copies of this vector integrate at a time (6).
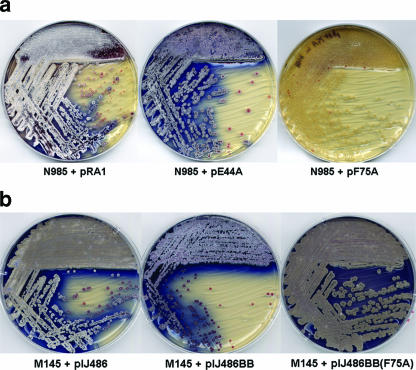
We introduced each mutation into pRA1 and then inserted the resulting plasmids, designated pY21A through pW75A, into a bldB null mutant strain, N985, and its developmentally competent parent, M145, to assess the ability of each allele to drive aerial mycelium formation relative to the wild-type gene. At low copy numbers, none of these alleles had any effect on growth or development in M145 (data not shown); however, there were a number of important effects when the complementing alleles were introduced into the bldB null mutant (Fig. 2). As expected, pRA1 restored aerial mycelium formation to N985 such that development was normal (Fig. 2a, left plate). The mutations in amino acids E44 (Fig. 2a, middle plate) and S58 (data not shown) had little or no effect on gene function: both supported the formation of an aerial mycelium at levels similar to those of the wild-type gene, suggesting that these residues are relatively unimportant for function in vivo. In contrast, mutations that changed R56, Y21, W30, W72, and F75 (Fig. 2a, right plate) did not restore antibiotic-associated pigmentation or the formation of aerial hyphae to the mutant, suggesting that these residues are important for BldB function.
FIG. 2.
Effect of bldB mutations and overexpression on S. coelicolor. (a) Complementation of the bldB null mutant (N985) with the wild-type, the E44A, and the F75A bldB alleles at low copy number (one to three copies/chromosome). The wild-type and E44A alleles have clearly restored the formation of aerial hyphae and the production of pigmented antibiotics. In contrast, the presence of the F75A allele had no effect on the mutant, which remained bald and unpigmented. (b) Effect of the same alleles at high copy number. Cells were grown for 48 h on R2YE medium.
We have previously used two-hybrid analysis to demonstrate that BldB (7) and one of its homologues, the product of the gene SCO0703 in the abaA locus (M. Eccleston and J. R. Nodwell, unpublished observations), form homodimers. To determine whether these mutations had any effect on dimer formation, we introduced each sequence change into the bait and target plasmids that we constructed previously and assessed the ability of each mutant to dimerize in E. coli. Consistent with the fact that both alleles functioned normally in morphogenesis, BldB(E44A) and BldB(S58A) were both able to dimerize. In contrast, three of the defective alleles, W30, R56, and W72 exhibited defects in dimer formation, suggesting that these residues may form part of the dimer interface. We did not confirm, however, that these three alleles gave rise to stable proteins in vivo, so it is also possible that they simply encode proteins that fail to fold correctly. More importantly, two of the defective alleles, Y21A and F75A, encoded proteins that were able to homodimerize in vivo. The two-hybrid system that we used for this analysis is not quantitative; however, the behavior of these strains was indistinguishable from the behavior of those strains encoding the wild-type alleles of BldB. These data suggest that Y21 and F75 are important for BldB function but dispensable for dimer formation. The properties of all bldB alleles, including those described below, are summarized in Table 4.
TABLE 4.
Effects of bldB mutations on function in vivoa
| Allele | Dimerization | Single copy | Overexpression |
|---|---|---|---|
| Wild type | + | + | Whi |
| Y21A | + | − | Rsp |
| W30A | − | − | — |
| E44A | + | + | NT |
| R56A | − | − | — |
| S58A | + | + | NT |
| W72A | − | − | — |
| F75A | + | − | Rsp |
The capacity or inability of each allele to dimerize or complement in single copy is indicated by a “+” or “−,” respectively. The phenotype conferred by each overexpressed allele is indicated by “—” (no effect), NT (not tested), Whi (white), or Rsp (rapid sporulation).
In parallel with this work, we assessed the effects of placing bldB, under the control of its own promoter, onto a high-copy-number vector resulting in the overexpression in S. coelicolor (Fig. 2b). The high-copy-number vector lacking bldB had no effect on the growth or development of M145 (Fig. 2b, left plate). To our surprise, we found that when the wild-type allele was present at high copy numbers in wild-type S. coelicolor, colonies exhibited a phenotype reminiscent of that of the whi mutants, suggesting a defect in sporulation (Fig. 2b, middle plate). The aerial mycelium formed under these conditions was somewhat sparse compared to that of phenotypically wild-type strains, but it was reproducible. Antibiotic-associated pigmentation appeared normal. We observed a similar defect when we introduced the bldB overexpression plasmid into bldB null mutant strain N985 and into another morphologically wild-type strain, M600 (data not shown). While development did not proceed any further in any of the three strains with continued incubation, M145 containing the control plasmid sporulated to completion within 4 days.
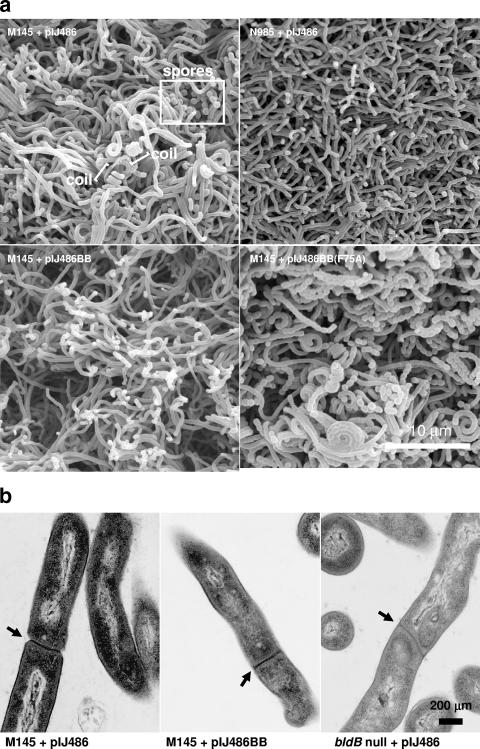
To determine the point in development where a high copy number of bldB induced arrest, we conducted scanning electron microscopy (Fig. 3). The surface of M145 colonies bearing a control plasmid and grown for 48 h had clearly initiated the formation of aerial hyphae, as demonstrated by the presence of coiled filaments and some mature spores. In contrast, M145 overexpressing bldB was blocked prior to sporulation septation: all hyphae were smooth and lacked the regularly spaced perforations of septating hyphae and there were no coiled filaments or mature spores. We also noticed a tendency for these aerial hyphae to be clumped in some places. Cells on the surface of bldB null mutant colonies did not exhibit any of the properties of aerial hyphae. We also conducted transmission electron microscopy on these strains to determine whether they were able to carry out vegetative septation (Fig. 3b). Vegetative septa were detectable in both the wild-type strain containing a high copy number of bldB and in the bldB null mutant. The developmental block caused by the overexpression of wild-type bldB therefore appears to be later than the erection of the aerial mycelium but earlier than the sporulation septation step.
FIG. 3.
Electron microscopy of phenotypes conferred by bldB overexpression. (a) Scanning electron microscopy of the surfaces of colonies of wild-type strain M145 containing a high-copy-number control plasmid (pIJ486), the bldB overexpression plasmid (pIJ486BB), and the bldB(F75A) plasmid [pIJ486BB(F75A)]. As an additional control, the bldB null mutant N985 containing the high-copy-number control plasmid (pIJ486) is also shown. Examples of coiled aerial filaments and mature spores are indicated. (b) Transmission electron microscopy of M145 containing a high-copy-number control plasmid, M145 containing the bldB overexpression plasmid, and the bldB null mutant N985 containing the high-copy-number control plasmid. Septa are indicated by arrows.
To determine the effects of bldB mutations on this dominant effect, we constructed identical overexpression vectors for each of the five defective alleles listed in Table 4 and introduced these vectors into M145. The bldB(W30A), bldB(R56A), and bldB(W72A) alleles had no effect on morphogenesis, consistent with the fact that their products were otherwise devoid of activity. The fact that the increased copy number of these alleles had no effect on development demonstrated that the phenotypic effects of overexpression of the wild-type gene were due to the BldB protein and not due to the titration of transcription factors by bldB promoter DNA.
In marked contrast, overexpression of the bldB(Y21A) and bldB(F75A) alleles caused a dramatic acceleration of morphogenesis such that within 48 h of growth, the colonies were completely covered in a mature, deep gray aerial mycelium (Fig. 2a, right plate). When viewed under the scanning electron microscope (Fig. 3a), the surfaces of these strains were almost completely covered with mature spores, and development was clearly accelerated relative to the control-plasmid-containing strain.
These dominant effects of bldB overexpression on morphogenesis by wild-type colonies suggested that while excess levels of the wild-type protein inhibited sporulation, similar levels of dimerization-competent, morphogenesis-defective proteins caused it to proceed in an accelerated manner, reminiscent of the “rapid aerial mycelium” or Ram phenotype conferred by overexpression of the ram genes (15, 18). We introduced the overexpression plasmids for the Y21A and F75A mutants into strains bearing null mutations in the genes bldK, ramR, whiA, whiB, whiG, whiH, sigF, and ssgB, all of which interrupt development at various stages. Neither overexpression plasmid caused a bypass of the effects of any of the mutations. This suggests that the accelerated spore formation brought about by the overexpression of bldB(Y21A) or bldB(F75A) is dependent on the normal genetic machinery involved in morphogenesis.
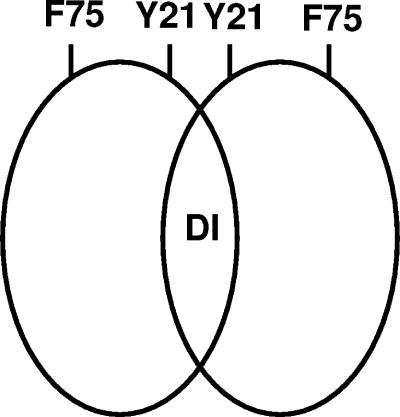
The fact that the Y21A and F75A mutations confer identical phenotypes under a variety of expression conditions suggests to us that these residues fulfill related roles for the protein. An obvious explanation is that they are located close to each other in the three-dimensional structure of the protein (Fig. 4) and that they constitute part of an interaction surface for the binding of another molecule, most likely another protein. Altering either residue might therefore compromise or alter this interaction, bringing about dramatic changes in the sporulation properties of the organism. F75 is located in the most C terminal of the conserved blocks of sequence in the BldB family (Fig. 1a). Indeed, this residue is found in almost all family members (see Fig. S1 in the supplemental material), suggesting perhaps that many or most of these proteins make similar use of this motif. Identifying this BldB partner protein promises to provide major insights into sporulation in S. coelicolor and into the mechanism of action of the BldB family of proteins.
FIG. 4.
BldB is a dimeric protein with at least one interaction surface. The shared phenotypes conferred by mutations in Y21 and F75 suggest that these residues fulfill similar functions. We suggest that this is an interaction and that these residues are part of the same surface. BldB must also have a dimerization interface (DI), and it is possible that one or more of the highly conserved residues W30, R56, or W72 make up part of this surface.
Acknowledgments
This work was supported by an Ontario Graduate Scholarship to Marcus Eccleston and by operating grant 217540-99 from the Natural Science and Engineering Research Council of Canada.
Footnotes
Published ahead of print on 8 September 2006.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Adamidis, T., P. Riggle, and W. Champness. 1990. Mutations in a new Streptomyces coelicolor locus which globally block antibiotic biosynthesis but not sporulation. J. Bacteriol. 172:2962-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 3.Berdy, J. 2005. Bioactive microbial metabolites. J. Antibiot. (Tokyo) 58:1-26. [DOI] [PubMed] [Google Scholar]
- 4.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 5.Chater, K. F. 2001. Regulation of sporulation in Streptomyces coelicolor A3(2): a checkpoint multiplex? Curr. Opin. Microbiol. 4:667-673. [DOI] [PubMed] [Google Scholar]
- 6.Combes, P., R. Till, S. Bee, and M. C. Smith. 2002. The Streptomyces genome contains multiple pseudo-attB sites for the φC31-encoded site-specific recombination system. J. Bacteriol. 184:5746-5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eccleston, M., R. A. Ali, R. Seyler, J. Westpheling, and J. Nodwell. 2002. Structural and genetic analysis of the BldB protein of Streptomyces coelicolor. J. Bacteriol. 184:4270-4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flärdh, K., K. C. Findlay, and K. F. Chater. 1999. Association of early sporulation genes with suggested developmental decision points in Streptomyces coelicolor A3(2). Microbiology 145:2229-2243. [DOI] [PubMed] [Google Scholar]
- 9.Harasym, M., L. H. Zhang, K. Chater, and J. Piret. 1990. The Streptomyces coelicolor A3(2) bldB region contains at least two genes involved in morphological development. J. Gen. Microbiol. 136:1543-1550. [DOI] [PubMed] [Google Scholar]
- 10.Ikeda, H., J. Ishikawa, A. Hanamoto, M. Shinose, H. Kikuchi, T. Shiba, Y. Sakaki, M. Hattori, and S. Omura. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21:526-531. [DOI] [PubMed] [Google Scholar]
- 11.Keijser, B. J., E. E. Noens, B. Kraal, H. K. Koerten, and G. P. van Wezel. 2003. The Streptomyces coelicolor ssgB gene is required for early stages of sporulation. FEMS Microbiol. Lett. 225:59-67. [DOI] [PubMed] [Google Scholar]
- 12.Kelemen, G. H., P. Brian, K. Flardh, L. Chamberlin, K. F. Chater, and M. J. Buttner. 1998. Developmental regulation of transcription of whiE, a locus specifying the polyketide spore pigment in Streptomyces coelicolor A3(2). J. Bacteriol. 180:2515-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelemen, G. H., and M. J. Buttner. 1998. Initiation of aerial mycelium formation in Streptomyces. Curr. Opin. Microbiol. 1:656-662. [DOI] [PubMed] [Google Scholar]
- 14.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Centre, Norwich, England.
- 15.Ma, H., and K. Kendall. 1994. Cloning and analysis of a gene cluster from Streptomyces coelicolor that causes accelerated aerial mycelium formation in Streptomyces lividans. J. Bacteriol. 176:3800-3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCormick, J. R., E. P. Su, A. Driks, and R. Losick. 1994. Growth and viability of Streptomyces coelicolor mutant for the cell division gene ftsZ. Mol. Microbiol. 14:243-254. [DOI] [PubMed] [Google Scholar]
- 17.Merrick, M. J. 1976. A morphological and genetic mapping study of bald colony mutants of Streptomyces coelicolor. J. Gen. Microbiol. 96:299-315. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen, K. T., J. M. Willey, L. D. Nguyen, L. T. Nguyen, P. H. Viollier, and C. J. Thompson. 2002. A central regulator of morphological differentiation in the multicellular bacterium Streptomyces coelicolor. Mol. Microbiol. 46:1223-1238. [DOI] [PubMed] [Google Scholar]
- 19.Nodwell, J. R., K. McGovern, and R. Losick. 1996. An oligopeptide permease responsible for the import of an extracellular signal governing aerial mycelium formation in Streptomyces coelicolor. Mol. Microbiol. 22:881-893. [DOI] [PubMed] [Google Scholar]
- 20.O'Connor, T. J., P. Kanellis, and J. R. Nodwell. 2002. The ramC gene is required for morphogenesis in Streptomyces coelicolor and expressed in a cell type-specific manner under the direct control of RamR. Mol. Microbiol. 45:45-57. [DOI] [PubMed] [Google Scholar]
- 21.Pope, M. K., B. Green, and J. Westpheling. 1998. The bldB gene encodes a small protein required for morphogenesis, antibiotic production, and catabolite control in Streptomyces coelicolor. J. Bacteriol. 180:1556-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pope, M. K., B. D. Green, and J. Westpheling. 1996. The bld mutants of Streptomyces coelicolor are defective in the regulation of carbon utilization, morphogenesis and cell-cell signalling. Mol. Microbiol. 19:747-756. [DOI] [PubMed] [Google Scholar]
- 23.Society for Actinomycetes Japan. 1997. Atlas of actinomycetes. Asakura Publishing Co., Ltd., Tokyo, Japan.
- 24.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]