Abstract
Quorum sensing is a community behavior that bacteria utilize to coordinate a variety of population density-dependent biological functions. In Agrobacterium tumefaciens, quorum sensing regulates the replication and conjugative transfer of the tumor-inducing (Ti) plasmid from pathogenic strains to nonpathogenic derivatives. Most of the quorum-sensing regulatory proteins are encoded within the Ti plasmid. Among these, TraR is a LuxR-type transcription factor playing a key role as the quorum-sensing signal receptor, and TraM is an antiactivator that antagonizes TraR through the formation of a stable oligomeric complex. Recently, a second TraM homologue called TraM2, not encoded on the Ti plasmid of A. tumefaciens A6, was identified, in addition to a copy on the Ti plasmid. In this report, we have characterized TraM2 and its interaction with TraR and solved its crystal structure to 2.1 Å. Like TraM, TraM2 folds into a helical bundle and exists as homodimer. TraM2 forms a stable complex (Kd = 8.6 nM) with TraR in a 1:1 binding ratio, a weaker affinity than that of TraM for TraR. Structural analysis and biochemical studies suggest that protein stability may account for the difference between TraM2 and TraM in their binding affinities to TraR and provide a structural basis for L54 in promoting structural stability of TraM.
Many bacterial species employ intercellular communication to couple their population density with the expression of specific genes via the production, release, and perception of diffusible signal molecules termed autoinducers (15). This phenomenon, designated quorum sensing, allows bacteria to coordinate their own behaviors to adapt to rapidly changing environmental conditions, compete against other microorganisms for the same nutrients, and escape the immune response of host organisms during infection (6). The first described example of this system is the LuxI/LuxR signaling circuit identified in the marine bacterium Vibrio fischeri. To date, similar proteins have been documented in more than 70 species of proteobacteria, making quorum sensing a paradigm of bacterial communication systems (1, 4, 7, 18).
The gram-negative bacterium Agrobacterium tumefaciens is one of the most extensively studied model systems for microbe-host interactions. A. tumefaciens infects a broad spectrum of more than 60 different families of plants by responding to a combination of phytochemicals secreted by target tissues (14). This infection ultimately causes the ubiquitous plant disease crown gall, which results in substantial losses of agricultural production worldwide and represents a remarkable example of interkingdom gene transfer. During the infection, a DNA fragment (T-DNA), copied from the tumor-inducing (Ti) plasmid of the pathogen, is transferred into the plant host cell and integrated into the chromosomal DNA (13, 27, 28). In addition to T-DNA, which contains more than 10 genes that ultimately are delivered to infected plants, many of the genes implicated in regulation and facilitation of T-DNA transfer are also located on the Ti plasmid (27). Many environmental agrobacterial isolates do not harbor the Ti plasmid and are avirulent. Conjugative transfer of the Ti plasmid from the pathogenic strains to avirulent, plasmid-free strains could play a key role in maintaining and expanding populations of infectious A. tumefaciens. The copy number and conjugative transfer of the Ti plasmid are regulated by an acylhomoserine lactone (AHL)-type quorum-sensing system (19, 25). The quorum-sensing transcription factor TraR is a LuxR homologue, an AHL-responsive transcription factor that binds to an 18-bp inverted symmetric DNA sequence called the tra box when associated with its cognate ligand, the AHL signal N-3-oxooctanoyl-l-homoserine lactone (8, 10, 19). The TraR protein is the only LuxR-type protein to have been structurally defined. In the crystal structure, TraR binds to the tra box as a homodimer, and each monomer consists of two functional domains, with AHL being deeply buried inside the N-terminal domain of each monomer and the C-terminal domains interacting with the tra box DNA via a helix-turn-helix motif. A flexible linker of ∼10 residues tethers the two domains (23, 26).
The activity of TraR is directly antagonized by two antiactivators: TrlR and TraM. TrlR is a truncated version of TraR, comprising 1 to 181 residues of the 234-amino-acid TraR, and exerts its antiactivation function by forming an inactive TraR-TrlR heterodimer (2, 17). While TrlR exists only in octopine-type strains, TraM is a conserved quorum-sensing modulator in both octopine- and nopaline-type strains of A. tumefaciens. In A. tumefaciens R10 (an octopine strain) and C58 (a nopaline strain), expression of the traM gene, located on the Ti plasmid, is activated by TraR (9, 11). TraM exists as a homodimer, and the crystal structure reveals that the dimer folds into a coiled-coil structure (3, 22). It has been shown that TraM plays a crucial role in controlling the quorum-sensing threshold for the initiation of replication and conjugative transfer of the Ti plasmid (20). The antiactivator modulates quorum sensing by binding directly to TraR and forms a stable antiactivation TraR-TraM complex with a stoichiometry of 1:1 (3, 22). The critical role of TraM is further enforced by our recent finding, which shows that a spontaneous point mutation in TraM (L54P) could abolish its antiactivator function and results in overproduction of the AHL quorum-sensing signal and constitutive transfer of the Ti plasmid (24).
In the process of characterizing a TraM spontaneous mutant in the octopine-type A. tumefaciens strains A6 and Ach5, we discovered a second TraM (designated TraM2), encoded in the genome but not on the Ti plasmid (24). TraM2 shares over 60% sequence identity with TraM encoded by the Ti plasmid. Genetic analysis showed that TraM2 is also implicated in modulation of quorum sensing. In this report, we characterized the biochemistry of TraM2, including its crystal structure and the interaction with TraR. The data demonstrate that like TraM, TraM2 is a potent inhibitor of TraR, and the findings also provide an explanation for a modest decrease in the affinity of TraM2 for TraR.
MATERIALS AND METHODS
Plasmid construction, protein expression, and purification.
The 309-bp complete traM2 coding sequence was amplified from the genomic DNA of Agrobacterium tumefaciens strain A6 by PCR with two synthetic oligonucleotides: 5′-GGAATTCCATATGGATTTGAAAGATTCAG (NdeI site is underlined) and 5′-CGGGATCCTTATCAGTTGACCGAAACTTTCGGG (BamHI site is underlined). The resulting DNA fragment was digested with NdeI and BamHI and ligated with NdeI-BamHI-digested pET15b cloning vector (Novagen). The N-terminal six-His-tagged TraM protein was expressed in Escherichia coli BL21λ DE3 Codon Plus (Novagen). Cells were grown in terrific broth medium supplemented with 100 μg/ml ampicillin at 37°C to an optical density at 600 nm of 0.8 and induced with 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 5 h. Cells were lysed at 4°C in 50 mM sodium phosphate, pH 8.0, 300 mM NaCl, and 5 mM imidazole, using a continuous flow microfluidizer (Microfluidics). Clear cell lysate was subjected to a Ni-nitrilotriacetic acid (QIAGEN) column, fractionated further by Fast Q ion-exchange chromatography (Amersham Biosciences), followed by a Superdex 75 16/60 gel filtration column (Amersham Biosciences).
TraR and TraM were purified as described previously (3). To prepare complexes, TraR was mixed with an excess amount of TraM2 or TraM in TENDG buffer (50 mM Tris-Cl, pH 8.0, 0.5 mM EDTA, 200 mM NaCl, 1 mM dithiothreitol, and 5% glycerol) and incubated at 4°C for at least 12 h. The elution was performed in a preequilibrated Superdex 200 16/60 column (Amersham Biosciences) at 4°C at a flow rate of 0.5 ml/min.
Cross-linking assay.
A volume of 25 μl of the purified TraM2 at 4 mg/ml was dialyzed overnight against conjugation buffer of 50 mM sodium phosphate, pH 8.0, and then incubated with either 10 μl of 25 mM disuccinimidyl suberate (DSS; Pierce) dissolved in dimethyl sulfoxide or 10 μl of conjugation buffer for 30 min at room temperature. The reaction was quenched by adding 5 μl of 1 M Tris-Cl, pH 8.0, and incubated for 30 min. A volume of 10 μl of the reaction mixture was then subjected to a 15% Tris-Tricine gel stained with Coomassie brilliant blue.
Dynamic light scattering.
Protein samples were dialyzed overnight to 50 mM sodium phosphate, pH 8.0, and concentrated to 10 mg/ml using a Centriprep YM-10 (Millipore). Dynamic light scattering measurements were carried out at 25°C in a Zetasizer Nano S (Malvern) instrument, and data were analyzed using Zetasizer Nano S software. The molecular weight was estimated using the specific volume calculated from the specific volume of individual amino acids and the hydrophobic radius, assuming the particles are spherical.
ITC.
The protein samples were dialyzed overnight to 50 mM HEPES, pH 7.5, 200 mM NaCl, and 0.5 mM EDTA and equilibrated to room temperature overnight. Isothermal titration calorimetry (ITC) analysis was performed in a VP-ITC system (MicroCal) at 25°C by 30 injections of 10 μl of TraR (172 μM) into 1.4 ml of TraM2 (14.7 μM) or TraM (14.7 μM), with 4-min intervals. In parallel, TraM2 (115 μM) or TraM (115 μM) was injected into 1.4 ml of 10 μM TraR. Thermodynamic parameters of the binding process were derived using Origin ITC software (Origin Lab) by fitting the corrected binding isotherm to a single-site binding model.
CD.
Circular dichroism (CD) measurements were carried out in a J-715 spectropolarimeter (JASCO), which is equipped with a PTC-343 Peltier-type cell holder for temperature control. Both TraM and TraM2 protein samples of 7.5 μM were prepared in 50 mM sodium phosphate buffer, pH 8.0. CD spectra from 200 nm to 280 nm were recorded in a 0.5-cm-path-length cell between 4°C and 95°C. Thermally induced unfolding was performed at a scan rate of 1°C/min, and an average of five spectra was used to build each curve profile.
Protein crystallization, structure determination, and refinement.
The crystallization of TraM2 was carried out in hanging drops by vapor diffusion. Crystals of TraM2 were obtained by mixing 25 mg/ml TraM2 in buffer containing 50 mM Tris-Cl, pH 8.0, 200 mM NaCl, 0.5 mM EDTA, and 1 mM dithiothreitol with 25% (wt/vol) ethylene glycol at a protein/well solution ratio of 1:1 (vol/vol). Crystals were flash cooled in 50% (wt/vol) ethylene glycol at 100 K. Data for TraM2 crystals were collected using synchrotron radiation at Beamline 5.0.2 of Advance Light Source (Berkeley, CA). Diffraction data were integrated and scaled using the HKL2000 software suite. The crystals belong to P43 with the following cell dimensions: a = 79.52 Å; b = 79.52 Å; c = 90.23 Å; α = β = γ = 90°. The structure was solved by molecular replacement using TraM from A. tumefaciens strain R10 as the search model (Protein Data Bank identification [ID] code 1RFY) (3). Interactive model building was carried out in O (12) between rounds of simulated annealing and positional and B-factor refinement performed in a crystallography and NMR system using the maximum-likelihood target on amplitudes. The final model at 2.1 Å includes residues 12 to 100 for three monomers and 12 to 101 for one monomer and 307 molecules of H2O. The structures shown in the figures were generated using the program PyMOL (5).
Coordinates.
Atomic coordinates and structure factors have been deposited in the Protein Data Bank (accession code 2HJD).
RESULTS
Biochemical properties of TraM2.
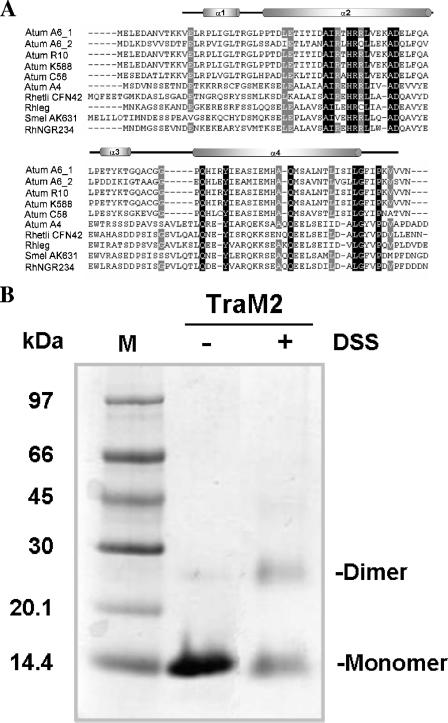
Sequence alignment shows that TraM2 shares about 64% sequence identity with TraM at the amino acid sequence level (Fig. 1A). Except for two conservative mutations (L29I and R42Q), TraM2 contains 20 out of 22 residues conserved among all other sequenced TraM orthologues of the Rhizobiaceae family and all of the residues crucial for the dimerization except for a few conservative mutations (L14V, I32L, and I70L) (Fig. 1A) (3). The calculated molecular mass of TraM2 is 13.3 kDa (including a six-His tag), but the molecular mass for the native protein was estimated to be 33 kDa by gel filtration chromatography and 31.3 kDa by dynamic light scattering (data not shown), suggesting a dimeric conformation. In addition, a dimeric molecular species of TraM2 was captured in the presence of a cross-linker, DSS (Fig. 1B). Thus, as with TraM from A. tumefaciens R10, the A6 TraM2 exists as a homodimer in solution (3).
FIG. 1.
Sequence alignment and structural analysis of TraM2. (A) Sequence alignment of TraM proteins using GeneDoc (16). Amino acid sequences are from the following bacteria: (i) Atum A6_1, A. tumefaciens A6; (ii) Atum A6_2, A. tumefaciens A6, traM2; (iii) Atum R10, A. tumefaciens R10; (iv) Atum K588, A. tumefaciens K588; (v) Atum C58, A. tumefaciens C58; (vi) Atum A4, A. tumefaciens A4; (vii) Rhetli CFN42, Rhizobium etli CFN42; (viii) Rhleg, Rhizobium leguminosarum; (ix) Smel AK631, Sinorhizobium meliloti AK631; and (x) RhNGR234, Rhizobium sp. NGR234. Invariant residues are highlighted in black, and highly conserved resides are shaded in gray. Secondary structure elements of TraM2 are indicated above. (B) Cross-linking of TraM2 with DSS. TraM2 at a final concentration of 2.86 mg/ml was incubated in the presence (+) or absence (−) of 25 mM DSS for 30 min at room temperature and quenched with 1 M Tris-Cl, pH 8.0, and then subjected to 15% Tris-Tricine gel electrophoresis. The protein bands were visualized by staining with Coomassie brilliant blue. The bands corresponding to monomer and dimer molecules are indicated. M, molecular weight marker.
Oligomeric state of the TraM2-TraR complex.
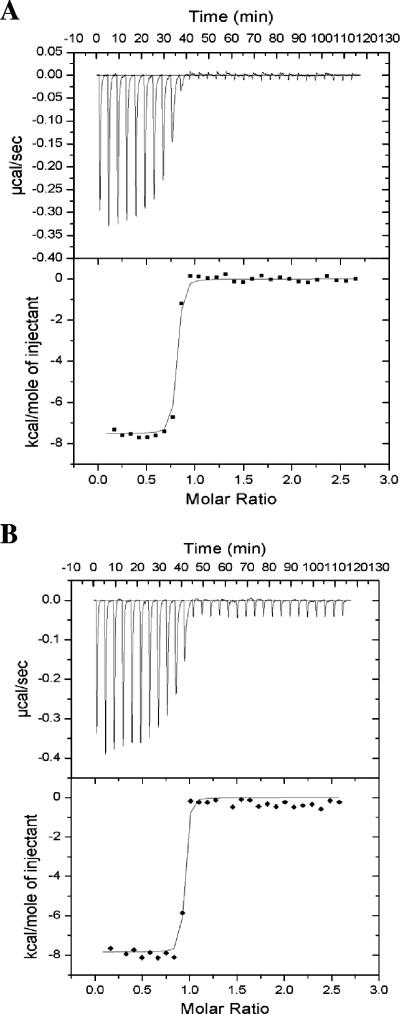
Our previous gel shift assay showed that TraM2 inhibits the activity of TraR, albeit at a concentration eightfold higher (16 μM) than that of TraM (2 μM) (24). Although both TraM2 and TraM have high affinities for TraR, ITC measurements showed that the dissociation constant (Kd) of the TraM2-TraR complex is 8.6 nM while that of the TraM-TraR complex is of 3.7 nM (Fig. 2A); the latter finding is consistent with that obtained from surface plasmon resonance (Kd = 1 to 4 nM) (21). Thus, the higher concentration of TraM2 required in the gel shift assay may be accounted for by its lower affinity for TraR. ITC analysis also indicated that the formation of both complexes is enthalpy driven (−7.5 kcal/mol for the TraM2-TraR complex and −7.8 kcal/mol for the TraM-TraR complex).
FIG. 2.
ITC analysis of TraR interaction with (A) TraM2 and (B) TraM. ITC experiments were performed at 25°C by 30 injections of 10 μl of TraR (172 μM) into 1.4 ml of TraM2 (1.47 μM) or TraM (1.47 μM), with 4-min intervals. The protein samples were in buffer containing 50 mM HEPES, pH 7.5, 200 mM NaCl, and 0.5 mM EDTA. Top panels, each peak represents a single injection, and a negative injection peak indicates that the reaction is exothermic; bottom panels, the area enclosed within each peak is integrated and is represented by ▪ or ♦. The data were then fit into a single-site binding model (shown in line), and thermodynamic parameters (ΔH, Kd, and binding stoichiometry n) were derived.
The TraM2-TraR complex migrates as a 160-kDa molecular entity in a gel filtration column, which is consistent with the results from our analytical ultracentrifugation using the sedimentation equilibrium method (data not shown) and from dynamic light scattering measurements (22). The binding stoichiometry of the TraM2-TraR complex approximates 1 (0.87) based on ITC analysis (Fig. 2A), which is similar to that of 0.91 for the TraM-TraR complex (Fig. 2B). These findings are consistent with the model in which the TraM2-TraR complex consists of two dimeric TraM2 and two dimeric TraR molecules. Taken together, these data indicate that the TraM2-TraR complex resembles the TraM-TraR complex and is a hetero-octomer.
Crystal structure of TraM2.
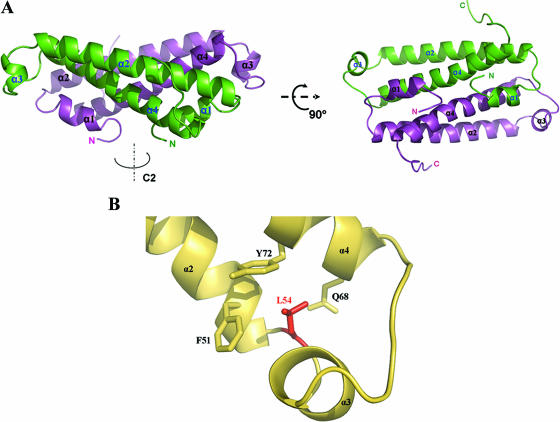
The crystal structure of TraM2 was solved by molecular replacement using TraM as the search model (Protein Data Bank ID 1RFY) (3) and refined to 2.1 Å. Table 1 summarizes the crystallographic data and refinement statistics. There are four TraM2 molecules per asymmetric unit, packed into two separate dimers. The model consists of residues 12 to 100 (or 101), with the first 11 and the last 1 or 2 residues unresolved. Like TraM, TraM2 folds into a coiled-coil structure (Fig. 3A). Each TraM2 consists of four helices, two long α-helices (α2, residues 25 to 54 for one monomer and 27 to 52 for the other; and α4, residues 66 to 94 and 67 to 94, respectively), and two short α-helices (α1, residues 18 to 21 and 16 to 22; and α3, residues 57 to 60 and 55 to 61). The two long helices form an antiparallel coiled-coil structure. To form the homodimer, the α2-α4 coiled coil from each monomer further intertwines into a four-helix bundle related by a twofold rotational symmetry.
TABLE 1.
Crystallographic data collection and refinement statisticsa
| Parameter (unit) | Result for TraM2 |
|---|---|
| X-ray diffraction data | |
| Unit cell dimensions (Å) | |
| a | 79.52 |
| b | 79.52 |
| c | 90.23 |
| Space group | P43 |
| Data collection | |
| Resolution (Å) | 40-2.1 |
| No. of reflections | |
| Total | 420,815 |
| Unique | 32,614 |
| Redundancy | 5.5 (5.5) |
| Completeness (%) | 99.7 (99.4) |
| Rsym (%) | 2.9 (20.3) |
| I/σ | 55.0 (7.5) |
| Refinement statistics | |
| No. of reflections: | 31,523 |
| Working set | 28,998 |
| Test set | 2,525 |
| Rwork (%) | 22.9 |
| Rfree (%) | 26.7 |
| Final model | |
| No. of nonhydrogen atoms | 2,655 |
| No. of water atoms | 307 |
| Avg area (Å2) of B factors in: | |
| Protein | 45.5 |
| Water | 51.7 |
| RMS deviation | |
| Bond length (Å) | 0.005 |
| Bond angle (°) | 1.0 |
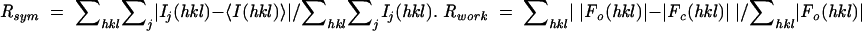 (crystallographic R factor).
(crystallographic R factor). 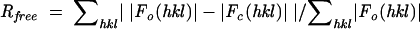 (calculated from ∼8% of the total reflections, which belong to a test set of randomly selected data). The numbers in parentheses refer to the last shell (2.18-2.10 Å) in the structural refinement.
(calculated from ∼8% of the total reflections, which belong to a test set of randomly selected data). The numbers in parentheses refer to the last shell (2.18-2.10 Å) in the structural refinement.
FIG.3.
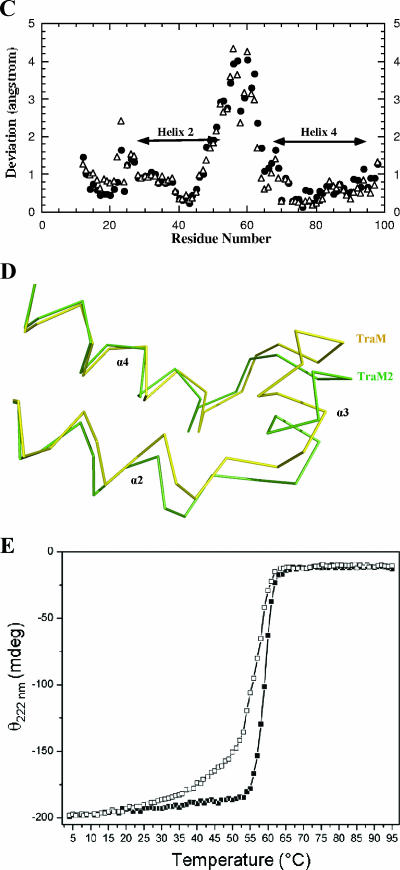
Structural analysis of TraM2. (A) Dimeric structure of TraM2. Each monomer consists of two long helices (α2 and α4) coiled together, and this intramolecular two-helix coil is further bundled with its counterpart in the other monomer. (B) Interaction network around L54 of TraM (Protein Data Bank ID 1RFY) (3). L54 is sandwiched between F51/Y72 and Q68. When aligned with α2, α4 has an additional turn, which is brought on to continue the hydrophobic stacking between α2 and α4 by the side chain of L54 interacting with that of Q68. (C) Structural variations between TraM2 and TraM. The Cα atoms of the intermolecular coiled structures of TraM2 and TraM (formed by α2 and α4 of each monomer) are superimposed by least-squares fitting, and the corresponding Cα atoms of the residues are calculated. ▵, one monomer; •, the second monomer. (D) Structural comparison between TraM2 (green) and TraM (yellow) in the region, including α3, with maximal structural deviation. (E) Thermal unfolding analysis of TraM and TraM2 monitored by CD. CD spectra were recorded from 200 nm to 280 nm between 4°C and 95°C. Shown here are the changes of ellipticity (θ) at 222 nm of TraM (▪) and TraM2 (□) as a function of temperature. mdeg, millidegree.
α2 is a typical amphipathetic helix, and its hydrophilic side consists largely of charged or polar residues (D28, E30, T31, N35, R38, R41, Q42, E45, D48, E49, and Q52) and is solvent exposed. The hydrophobic face of the helix is buried via the formation of the α2-α4 coiled-coil structure. Arrays of hydrophobic residues from both helices align with their hydrophobic side chains stacked upon each other across the ∼40-Å coiled structure. These residues include I29, L32, A36, I37, H40, L43, L44, A47, L50, and F51 from α2 and Q68, H69, A75, M76, M79, Q82, M83, V86, L89, V90, and L93 from α4. Also included in this interface are two charged residues, T33 (α2) and Y72 (α4); however, their hydroxyl groups are projecting into solvent. The polar residues in α4 (Q68 and Q82) bury their side chain polarity through hydrogen bonding to the backbone N or O atoms in the α2 helix (H40 and A36 or V53, respectively), strengthening the intrahelical interaction. The sequestered molecular surface within the α2-α4 coiled-coil structure is 1,431.8 Å2 or 32.8% of the total surface of these two helices.
The long hydrophobic stacking tower within the α2-α4 helical interface is fostered by two clusters of hydrophobic interaction located at either end of the coiled-coil structure. One such cluster consists of L54, I58, and H69 located between α2 and α4 helices, including α3. Of particular note, L54 is sandwiched between Y72/F51 and Q68 (Fig. 3B), suggesting that L54 may contribute its side chain to secure the stacking structure at the end of the coiled-coil structure. The other cluster includes the C terminus of the protein, with residues F95, I96, P97, and V99 interacting with L92, L93, and I29.
Helix α4, however, is largely hydrophobic throughout, and one of the hydrophobic sides is concealed within the intramolecular interface with α2 as described above. The other side of the helix is buried primarily by forming the intermolecular contacts with α1 and secondarily with α2 of the other monomer. More specifically, residues I70, I73, I77, and A81 from α4 interact with residues V14, L17, and L24 of α1 and L32 of α2 of the other protomer in a leucine zipper-like manner. The preservation of the hydrophobic character in the dimeric interface is critical in maintaining the structural stability, as demonstrated by our previous mutational analysis of TraM (3). The total buried molecular surface as a result of dimer formation is 1,999.8 Å2 or ∼20.6% of the total molecular surface (9,710.1 Å2).
Structural stability of TraM2 and TraM.
By least-squares fitting of the Cα atoms of the intermolecular coiled-coil structures (formed by α2 and α4 of each monomer) of the two structures, TraM2 can be superimposed with TraM with a root mean square (RMS) deviation of 1.5 Å. The most variation lies in the region between α2 and α4 (including α3), with the RMS deviation being 2.1 Å (Fig. 3C). A closer look at this variable region (Fig. 3D) reveals that compared to that of TraM, the C terminus of TraM2 α2 is stretched slightly outward whereas α3 is shifted ∼2 Å away from α2. As a result, the interaction between TraM2 helices may be less stable due to compromised structural packing. To test this hypothesis, we carried out thermal unfolding experiments with TraM and TraM2 monitored by CD. As shown in Fig. 3E, the midpoint of unfolding transition for TraM2 is 56°C, which is lower than the 59°C midpoint for TraM.
Impact of the L54P mutation.
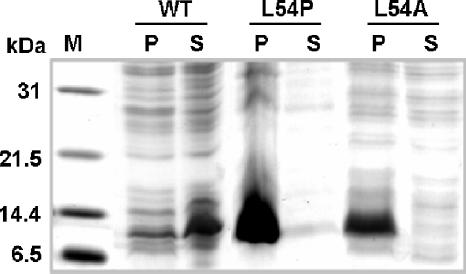
The above-described crystal structure analysis suggests that residue L54 may be important for stacking the side chains of the hydrophobic residues in the α2-α4 coiled coil. This may explain why the L54P or L54A mutation in TraM causes a drastic decrease in its antiactivator activity (21, 24). To test this possibility, we cloned the genes encoding the L54P and L54A variants of TraM and expressed them individually in E. coli. None of the TraM mutant proteins were soluble when overexpressed (Fig. 4), suggesting that structural instability is most likely responsible for the loss of function in the mutated proteins. The structural importance of a long side chain at position 54 will be discussed in the following section.
FIG. 4.
Substitution of L54 affects the solubility of TraM. Wild-type traM (WT) and its variants encoding L54P or L54A substitutions were expressed in the E. coli BL21λ DE3 strain under the control of the T7 promoter in expression vector pET15b with IPTG induction. Lane 1, molecular weight marker; lanes 2 and 3, cell debris and clear lysate of wild-type TraM; lanes 4 and 5, cell debris and clear lysate of the L54P mutation; lanes 6 and 7, cell debris and clear cell lysate of the L54A mutation. The dark bands (∼11 kDa) are overexpressed TraM or variants. P, pellet; S, supernatant.
DISCUSSION
In Agrobacterium tumefaciens, several regulatory proteins play critical roles in modulating the replication and conjugative transfer of the Ti plasmid. The first one is the transcriptional regulator TraR, which, by interacting with its quorum-signaling ligand, N-3-oxooctanoyl-l-homoserine lactone, activates the expression of the tra regulon implicated in Ti plasmid replication and conjugative transfer. The second is the antiactivator TraM, which prevents TraR from binding to the promoter of the tra regulon. The genes encoding TraR and TraM are located in the Ti plasmid. Recently, a TraM-like antiactivator (termed TraM2) was discovered in the genome of certain closely related octopine-type strains, although the general biological significance of this observation remains to be addressed. In this study, we demonstrate that like TraM, TraM2 folds into a coiled-coil structure, exists as a homodimer in solution, and forms a stable complex with TraR to inhibit TraR from binding to DNA. Structural analysis, supported by biochemical results, of TraM and TraM2 suggests that structural stability may account for the subtle difference in their affinities for TraR. We also show that structural instability caused by the L54P mutation in TraM accounts for the constitutive quorum-sensing phenomenon in the nopaline strain K588 (24), and we propose a structural basis for the crucial role of L54 in the maintenance of the structural stability of TraM.
Structurally, TraM2 resembles TraM: the dimeric TraM2 folds into a four-helix coiled-coil structure and can be superimposed with TraM. For both molecules, the dimer interfaces are extensive and comparable in size and shape (buried molecular surface for TraM is 2,086 Å2 and for TraM2 is 2,000 Å2). The interface within both molecules is hydrophobic in nature. In particular, nine hydrophobic residues (L14, L17, L20, L24, I32, I70, I73, I77, and A81) located within the dimer interface of TraM are shown to be critical in maintaining the solubility and structure of TraM by mutagenesis study (3); six of them are conserved in TraM2, and the other three (L17V, I32L, and I70L) are conservatively replaced. Based on structural similarities in the dimer interfaces of these two proteins, it is most likely that a heterodimer of TraM-TraM2 can be readily formed and that its stability is similar to those of homodimeric TraM and TraM2.
Both sequence and structural analyses suggest that TraM2, like TraM, functions as an antiactivator to TraR. The TraM2-TraR complex shares the same stoichiometry (1:1) as the TraM-TraR complex, and the two complexes have the same hydrodynamic volume (elution volume in gel filtration chromatography). In comparison with TraM (Kd of 3.7 nM), TraM2 binds to TraR with a slightly weaker affinity (Kd of 8.6 nM). This decrease in binding affinity is consistent with the observation in the gel shift assay that a higher concentration of TraM2 is required to outcompete formation of the TraR-DNA complex (24). Residues in the C terminus, such as L93, G94, and P97, have been implicated for functional importance (21), and they are conserved between TraM2 and TraM. However, the overall structures of both C termini do not deviate significantly, and the last few residues (101 to 102 in TraM2 and 99 to 102 in TraM) are disordered in either structure and cannot be compared.
Alternatively, our structural analysis suggests that overall protein stability may account for the subtle difference in the TraR binding affinities of TraM2 and TraM. The region between α2 and α4, including α3 of TraM2, is less optimally packed than that in TraM, which may compromise the overall structural stability of TraM2. As predicted, our thermal unfolding studies indicate that TraM2 is less stable than TraM, as it unfolds at a lower temperature and has a lower midpoint temperature. We thus propose that this weakened structural stability of TraM2, due to a less compact structural packing, is responsible for the modest decrease in the affinity of TraM2 for TraR, compared to that of TraM for TraR.
L54 interacts extensively with residues located at the end of the α2/α4 coiled-coil structure by being sandwiched between Y72/F51 and Q68. Interestingly, all three residues are absolutely conserved in the sequences of TraM and TraM2 known so far. Gln has a high preference (second only to Glu) found at the beginning of a helix; Q68 together with E67 is well positioned for initiation of α4. Phe, as well as Gln, is favorable at the end of a helix; thus, F51 and Q52 are primed for termination of α2. As shown in Fig. 3B, the N terminus of α4 and the C terminus of α2 are not aligned; the additional turn of α4 is fastened by the extensive contact of Q68 with the side chain of L54. Consistent with this observation, mutations to residues with either a short or absent side chain, such as Ala or Pro, respectively, destabilize the structure and lead to the loss of function. Glu is also found at this position (Fig. 1A); the charge carried by Glu can be neutralized by potential hydrogen bonds to the backbones of neighboring residues.
The discovery of a second antiactivator for TraR-dependent quorum sensing that is not encoded on the Ti plasmid is intriguing. The available evidence indicates that TraM2 may provide yet another layer of control to prevent the quorum-sensing transcription factor TraR from premature activation of tra and rep regulons, encoding genes responsible for Ti plasmid conjugative transfer and increased copy number. In the derivatives of the octopine-type A. tumefaciens A6 and Ach5 strains engineered to harbor mutations or deletions in traM2, TraR is inhibited by the Ti plasmid-encoded TraM as it is in the strains that do not harbor a traM2 gene in their genome (e.g., other octopine-type strains or nopaline-type strains), whereas in the mutants of strains A6 and Ach5 carrying loss-of-function mutations (e.g., L54P) or deletions in the Ti plasmid-encoded TraM, the antiactivation of TraR is carried out effectively by TraM2 (24). Disruption of both TraM and TraM2 is required in A6 and Ach5 to abolish the tight control of TraR and generates the constitutive quorum-sensing phenotype observed for single TraM disruptions in other strains of A. tumefaciens (23). The presence of TraM2 is reminiscent of that of TrlR, an additional TraR inhibitor which exists only in certain octopine-type strains (2, 17). The identification of TraM2 as a strain-specific quorum-sensing antiactivator seems to highlight another dimension of variation of the conserved quorum-sensing paradigm in A. tumefaciens, and the significance of this additional layer of regulation remains to be addressed in further studies.
Acknowledgments
We would like to thank Jay Nix for assistance in the data collection at Beamline 5.0.2, ALS, Berkeley, CA.
This work is supported by grants MCB-0416447 from the NSF and GM065260-01 from the NIH to L.C.
Footnotes
Published ahead of print on 22 September 2006.
REFERENCES
- 1.Bassler, B. L. 1999. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2:582-587. [DOI] [PubMed] [Google Scholar]
- 2.Chai, Y., J. Zhu, and S. C. Winans. 2001. TrlR, a defective TraR-like protein of Agrobacterium tumefaciens, blocks TraR function in vitro by forming inactive TrlR:TraR dimers. Mol. Microbiol. 40:414-421. [DOI] [PubMed] [Google Scholar]
- 3.Chen, G., J. W. Malenkos, M. R. Cha, C. Fuqua, and L. Chen. 2004. Quorum-sensing antiactivator TraM forms a dimer that dissociates to inhibit TraR. Mol. Microbiol. 52:1641-1651. [DOI] [PubMed] [Google Scholar]
- 4.de Kievit, T. R., and B. H. Iglewski. 2000. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68:4839-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeLano, W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, Calif.
- 6.Fuqua, C., and E. P. Greenberg. 1998. Self perception in bacteria: quorum sensing with acylated homoserine lactones. Curr. Opin. Microbiol. 1:183-189. [DOI] [PubMed] [Google Scholar]
- 7.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50:727-751. [DOI] [PubMed] [Google Scholar]
- 8.Fuqua, W. C., S. C. Winans, and E. P. Greenberg. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hwang, I., D. M. Cook, and S. K. Farrand. 1995. A new regulatory element modulates homoserine lactone-mediated autoinduction of Ti plasmid conjugal transfer. J. Bacteriol. 177:449-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hwang, I., P. L. Li, L. Zhang, K. R. Piper, D. M. Cook, M. E. Tate, and S. K. Farrand. 1994. TraI, a LuxI homologue, is responsible for production of conjugation factor, the Ti plasmid N-acylhomoserine lactone autoinducer. Proc. Natl. Acad. Sci. USA 91:4639-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang, I., A. J. Smyth, Z. Q. Luo, and S. K. Farrand. 1999. Modulating quorum sensing by antiactivation: TraM interacts with TraR to inhibit activation of Ti plasmid conjugal transfer genes. Mol. Microbiol. 34:282-294. [DOI] [PubMed] [Google Scholar]
- 12.Jones, T. A., J. Y. Zou, S. W. Cowan, and Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. Sect. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 13.Kado, C. I. 1998. Agrobacterium-mediated horizontal gene transfer. Genet. Eng. 20:1-24. [DOI] [PubMed] [Google Scholar]
- 14.Kalogeraki, V. S., and S. C. Winans. 1998. Wound-released chemical signals may elicit multiple responses from an Agrobacterium tumefaciens strain containing an octopine-type Ti plasmid. J. Bacteriol. 180:5660-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 16.Nicholas, K. B., H. B. Nicholas, Jr., and D. W. Deerfield II. 1997. GeneDoc: analysis and visualization of genetic variation. EMBNEW News 4:14. [Google Scholar]
- 17.Oger, P., K. S. Kim, R. L. Sackett, K. R. Piper, and S. K. Farrand. 1998. Octopine-type Ti plasmids code for a mannopine-inducible dominant-negative allele of traR, the quorum-sensing activator that regulates Ti plasmid conjugal transfer. Mol. Microbiol. 27:277-288. [DOI] [PubMed] [Google Scholar]
- 18.Parsek, M. R., and E. P. Greenberg. 2000. Acyl-homoserine lactone quorum sensing in gram-negative bacteria: a signaling mechanism involved in associations with higher organisms. Proc. Natl. Acad. Sci. USA 97:8789-8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piper, K. R., S. Beck von Bodman, and S. K. Farrand. 1993. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature 362:448-450. [DOI] [PubMed] [Google Scholar]
- 20.Piper, K. R., and S. K. Farrand. 2000. Quorum sensing but not autoinduction of Ti plasmid conjugal transfer requires control by the opine regulon and the antiactivator TraM. J. Bacteriol. 182:1080-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swiderska, A., A. K. Berndtson, M. R. Cha, L. Li, G. M. Beaudoin III, J. Zhu, and C. Fuqua. 2001. Inhibition of the Agrobacterium tumefaciens TraR quorum-sensing regulator. Interactions with the TraM anti-activator. J. Biol. Chem. 276:49449-49458. [DOI] [PubMed] [Google Scholar]
- 22.Vannini, A., C. Volpari, and S. Di Marco. 2004. Crystal structure of the quorum-sensing protein TraM and its interaction with the transcriptional regulator TraR. J. Biol. Chem. 279:24291-24296. [DOI] [PubMed] [Google Scholar]
- 23.Vannini, A., C. Volpari, C. Gargioli, E. Muraglia, R. Cortese, R. De Francesco, P. Neddermann, and S. Di Marco. 2002. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 21:4393-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang, C., H.-B. Zhang, G. Chen, L. Chen, and L.-H. Zhang. 2006. Dual control of quorum sensing by two TraM-type antiactivators in Agrobacterium tumefaciens octopine strain A6. J. Bacteriol. 188:2435-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang, L., P. J. Murphy, A. Kerr, and M. E. Tate. 1993. Agrobacterium conjugation and gene regulation by N-acyl-l-homoserine lactones. Nature 362:446-448. [DOI] [PubMed] [Google Scholar]
- 26.Zhang, R. G., T. Pappas, J. L. Brace, P. C. Miller, T. Oulmassov, J. M. Molyneaux, J. C. Anderson, J. K. Bashkin, S. C. Winans, and A. Joachimiak. 2002. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature 417:971-974. [DOI] [PubMed] [Google Scholar]
- 27.Zhu, J., P. M. Oger, B. Schrammeijer, P. J. Hooykaas, S. K. Farrand, and S. C. Winans. 2000. The bases of crown gall tumorigenesis. J. Bacteriol. 182:3885-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zupan, J., T. R. Muth, O. Draper, and P. Zambryski. 2000. The transfer of DNA from Agrobacterium tumefaciens into plants: a feast of fundamental insights. Plant J. 23:11-28. [DOI] [PubMed] [Google Scholar]