Abstract
In cyanobacteria and many chemolithotrophic bacteria, the CO2-fixing enzyme ribulose 1,5-bisphosphate carboxylase/oxygenase (RubisCO) is sequestered into polyhedral protein bodies called carboxysomes. The carboxysome is believed to function as a microcompartment that enhances the catalytic efficacy of RubisCO by providing the enzyme with its substrate, CO2, through the action of the shell protein CsoSCA, which is a novel carbonic anhydrase. In the work reported here, the biochemical properties of purified, recombinant CsoSCA were studied, and the catalytic characteristics of the carbonic anhydrase for the CO2 hydration and bicarbonate dehydration reactions were compared with those of intact and ruptured carboxysomes. The low apparent catalytic rates measured for CsoSCA in intact carboxysomes suggest that the protein shell acts as a barrier for the CO2 that has been produced by CsoSCA through directional dehydration of cytoplasmic bicarbonate. This CO2 trap provides the sequestered RubisCO with ample substrate for efficient fixation and constitutes a means by which microcompartmentalization enhances the catalytic efficiency of this enzyme.
A wide variety of bacterial species package some metabolically important enzymes into polyhedral microcompartments that are bounded by shells comprised of three to seven highly conserved proteins (reviewed in reference 20). Microcompartmentalization is thought to enhance the catalytic efficacy of the enzyme(s) sequestered inside by providing metabolic channeling through enzyme colocalization and by reducing the effects of product/substrate diffusion (47). The structures may also protect the enzyme(s) from inhibitors (13) or prevent escape of products that are toxic to other portions of cellular metabolism (19). The best-studied example of these microbial microcompartments is the carboxysome, found in many chemolithotrophic bacteria and in all cyanobacteria thus far studied (Fig. 1). Carboxysomes are organelles that derive their name from their content, the CO2-fixing enzyme of the Calvin-Benson-Bassham cycle, ribulose 1,5-bisphosphate carboxylase/oxygenase (RubisCO) (40). Genetic and metabolic studies have revealed that carboxysomes are required for growth at normal atmospheric CO2 levels (reviewed in reference 13). In some species, the number of carboxysomes per cell and the proportion of cellular RubisCO packaged into the microcompartments are regulated in a manner dependent on the level of inorganic carbon available to the cells (9, 15, 29, 32, 52). However, in some cyanobacteria, such as Synechocystis 6803, the number of carboxysomes seems to remain constant regardless of available inorganic carbon (Ci) levels (48).
FIG. 1.
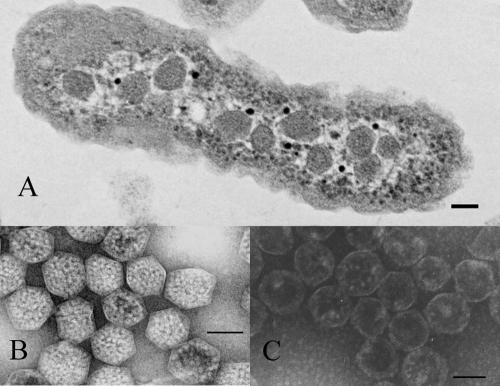
Carboxysomes of H. neapolitanus. (A) Transmission electron micrograph of an H. neapolitanus cell containing carboxysomes. (B) Purified, negatively stained intact carboxysomes. (C) Negatively stained carboxysomes after rupture by freeze-thaw treatment. In all panels, the bar represents 100 nm.
The carboxysome shell polypeptides of Halothiobacillus neapolitanus are encoded by a cluster of genes that are arranged in an apparent operon (cso), which includes cbbL and cbbS, the genes for the large subunit and the small subunit, respectively, of RubisCO (12-14). A similar organization of carboxysome genes has been observed in all carboxysome-bearing chemolithotrophs thus far examined and in α-cyanobacteria (5, 14), exemplified by the genus Prochlorococcus, which dominates the oligotrophic oceans (47). In contrast, the carboxysome genes of β-cyanobacteria are arranged in a less-defined cluster termed ccmKLMN, which is typically located upstream from the genes encoding the large and small subunits of RubisCO (6).
It has been proposed that carboxysomes play a role in an inducible CO2-concentrating mechanism (CCM) in some cyanobacteria (reviewed in references 4, 6, and 24). In the β-cyanobacteria Synechocystis and Synechococcus, as many as five energy-dependent Ci accumulation modes have been described, which result in cytoplasmic concentrations of HCO3− that are 1,000 times higher than the concentrations of Ci on the outside of the cell (reviewed in references 4, 17, and 24). Since the substrate for RubisCO is CO2 and the uncatalyzed rate of HCO3− dehydration to CO2 is much lower than needed to sustain the observed photosynthetic rates, it is generally accepted that a carbonic anhydrase (CA) is colocalized with RubisCO inside the carboxysome. Bicarbonate is thought to be able to diffuse into the carboxysome, where its conversion to CO2 is catalyzed by the resident CA. Based on calculations from two quantitative models, it has been proposed that some feature of the carboxysome structure limits the outward diffusion of CO2, although no direct experimental results supporting a molecular mechanism that could give rise to this purported characteristic have been put forth so far (11, 24, 37, 38).
The existence of a CCM in chemoautotrophic bacteria is supported by the fact that H. neapolitanus accumulates Ci as HCO3− in its cytoplasm, although at levels below those observed in cyanobacteria with a functional CCM (22). An energy-dependent CCM is also present in the deep sea hydrothermal vent sulfur bacterium Thiomicrospira crunogena and probably in other autotrophs (16). The role—if any—of carboxysomes in the CCM of chemoautotrophs has not been established to date. An alternative hypothesis suggests that the carboxysomal shell plays a role as a selective permeability barrier favoring the influx of HCO3− into the carboxysome and limiting the diffusion of O2, the competitive inhibitor of CO2 fixation by RubisCO. The recent report (25) describing the crystal structure of the major carboxysomal shell protein from Synechocystis suggests a role for the carboxysomal shell in the regulation of metabolite passage into and out of the microcompartments. This role is consistent with the alternative hypothesis but to date cannot explain how this control can be achieved through structures and interactions of the five putative α-carboxysomal shell proteins.
We have identified one of the carboxysomal shell proteins, CsoS3 (referred to here as CsoSCA for carboxysomal shell CA), as a CA that appears to be limited to organisms containing α-carboxysomes (47). The novel primary structure of CsoSCA led us to propose a new class of CA that we termed ɛ in keeping with the nomenclature for the four other known classes of CA (α, β, γ, and δ) (50). Previous structural studies using immunogold electron microscopy suggested that CsoS3 is a carboxysomal shell protein (8), and CA assays of purified carboxysome shells confirmed a tight association of the enzyme with the shell (47). It is not clear at this point if CsoSCA is embedded in the shell and perhaps plays a role in facilitating the diffusion of HCO3− into the carboxysome or if it is tightly bound to the inside surface of the structure, converting bicarbonate within the interior of the particle to CO2 for use by the carboxysomal RubisCO. Recent solution of the crystal structure of CsoSCA has revealed a remarkable similarity to the secondary and tertiary structure of β-CAs and a near-identical three-dimensional arrangement of active-site residues, suggesting that the enzyme is best described as a subclass of β-CAs (39). In this study, we have utilized stopped-flow spectrophotometry to assess the activity of CsoSCA in carboxysomes and as purified recombinant protein.
MATERIALS AND METHODS
Metals analysis.
Semiquantitative and quantitative metals analyses were performed by inductively coupled plasma atomic emission spectroscopy at the Center for Trace Analysis, Department of Marine Science, The University of Southern Mississippi.
Carboxysome purification.
A chemostat culture of H. neapolitanus was grown as previously described (15) with a dilution rate of 0.08 h−1 and at a pH of 6.4. Cells (5 to 8 g wet weight) were harvested by centrifugation and resuspended into 20 ml of TEMB buffer (10 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 20 mM NaHCO3, 1 mM EDTA) before disruption by sonication as previously reported. The resulting cell break was mixed with 1 volume of BPER II (Pierce Biotechnology, Inc., Rockford, IL), incubated with shaking at room temperature for 30 min, and clarified by centrifugation at 12,000 × g for 10 min. The supernatant was then centrifuged at 48,000 × g for 30 min to sediment the carboxysomes, which were resuspended and purified by sucrose density gradient centrifugation as previously described (47).
Carboxysome shell disruption.
Homogeneous preparations of carboxysomes (50 to 100 μl) at a protein concentration of 6 to 8 mg ml−1 in TEMB were collected as a pellet by centrifugation at 14,000 × g for 30 min. The resulting supernatant was carefully removed and the remaining semidry pellet frozen at −20°C for 10 to 15 min. The pellet was then rapidly resuspended in 50 to 100 μl of the desired buffer (typically TEMB). Transmission electron microscopy of the preparation revealed that more than 95% of the carboxysome shells were disrupted, allowing most of the RubisCO to be released from the carboxysome interior into the surrounding buffer. The shells retained their polyhedral shape, with some fraction of RubisCO always evident lining the inside.
Expression and purification of CsoSCA.
Recombinant histidine-tagged CsoSCA (rCsoSCA) was expressed from plasmid pProExCsoSCA in Escherichia coli and purified by affinity chromatography on a Ni column as previously described (39).
Measurement of CA activity.
Steady-state CA activity was measured by stopped-flow spectrophotometry using the changing pH indicator method of Khalifah (26). Assays were performed at 25°C with an Applied Photophysics SX.18MV stopped-flow spectrophotometer/fluorometer. The buffer/indicator pairs and wavelengths used were MES (morpholineethanesulfonic acid)/chlorophenol red (A574) at pH 5.5 to 6.8; MOPS (morpholinepropanesulfonic acid)/p-nitrophenol (A400) at pH 6.8 to 7.5; HEPES/phenol red (A557) at pH 7.5 to 8.0; and TAPS [N-tris(hydroxymethyl)methyl-3-aminopropanesulfonic acid]/m-cresol purple (A578) at pH 8.0 to 9.0. For measurements of CO2 hydration reactions, the appropriate amount of water saturated with CO2 (32.9 mM) was diluted with N2-saturated water to yield, when mixed in the stopped-flow cell, final dissolved CO2 concentrations from 5 to 25 mM. For measurements of bicarbonate dehydration reactions, KHCO3 solutions in final assay concentrations in the range of 10 to 100 mM were used. In both instances, sufficient sodium sulfate was added to maintain an ionic strength of 150 mM. Initial reaction rates were obtained from progress curves, corrected by subtracting the uncatalyzed rate, and fitted to the Michaelis-Menten equation using KaleidaGraph (Synergy Software).
The ability of rCsoSCA to hydrolyze p-nitrophenylacetate was tested at 25°C following the method of Armstrong et al. (3). The enzyme was incubated with 1.5 mM p-nitrophenylacetate in 100 mM potassium phosphate buffer, pH 7.2, and the change in A348 was followed over time.
RESULTS
The carboxysomal CA CsoSCA is tightly associated with and therefore believed to be an integral component of the carboxysome shell (8, 47). Upon disruption of carboxysomes by freezing and thawing, the CA activity copurifies with the shell fraction through differential and density gradient centrifugation steps (47). To further characterize the carboxysomal enzyme, attempts were made to purify CsoSCA from the carboxysomes under conditions that would preserve its enzymatic activity. Ion-exchange and immunoaffinity chromatography of disrupted carboxysomes did not yield preparations of active CsoSCA protein (data not shown), presumably because of the protein's tight interaction with other carboxysome shell components. When these interactions were disrupted by treatment of shell-enriched carboxysome fractions with strong denaturants such as urea or guanidinium hydrochloride, CsoSCA could be purified by denaturing size-exclusion chromatography, but it was not possible to recover enzymatic activity after removal of the denaturants by dialysis. Although the CA activity of CsoSCA is easily measurable in purified carboxysomes or in isolated shells, the protein represents only a small portion of the total shell protein (approximately 8%) (15, 20, 41). To be able to characterize a homogeneous preparation of the carboxysomal carbonic anhydrase, it was therefore necessary to produce the recombinant enzyme in E. coli and purify the recombinant protein by affinity chromatography. The basic catalytic parameters determined for rCsoSCA were compared with those measured with purified carboxysomes and purified shells and were corrected for the total protein amounts.
Expression and purification of recombinant CsoSCA.
The carboxysomal CA CsoSCA from H. neapolitanus was produced in E. coli by use of several different expression systems (47). For this study, the csoS3 expression construct in the pProEx vector was used exclusively, because the resulting recombinant protein rCsoSCA, which carries an N-terminal hexahistidine tag to facilitate purification, was soluble and typically yielded 10 mg of homogeneously pure protein from 1 liter of E. coli culture (Fig. 2). The purified protein had an enzymatic activity similar to that of recombinant enzyme produced in the IMPACT system, which was devoid of any extraneous amino acids but could be obtained only in considerably lower yields (47). One of the clones obtained was subsequently found to contain a mutation that resulted in the replacement of a phenylalanine residue at position 92 with a histidine. This change created a second Zn2+-binding site, which likely represents a vestigial active site that does not contribute to the enzymatic activity of the protein (39). Care was taken to ensure that all results reported here were obtained from a clone representing wild-type CsoSCA.
FIG. 2.
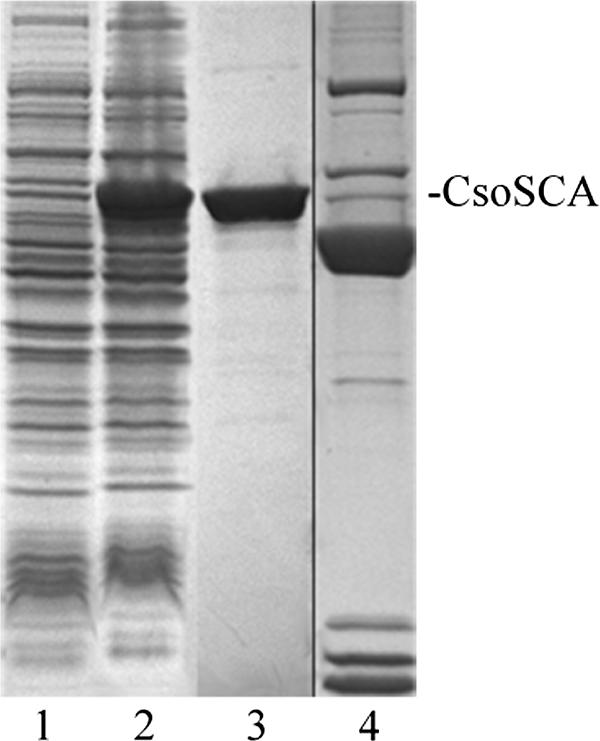
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel of heterologously expressed CsoSCA. Lanes: 1, 20 μg of cell extract from uninduced E. coli cells containing pProExCsoSCA; 2, 20 μg of cell extract of induced cells; 3, 2 μg of rCsoSCA protein eluted from a Ni2+ affinity column; 4, purified carboxysomes (from a separate gel).
Metals analysis.
Typically, rCsoSCA was purified from E. coli by use of buffers without any added metal ions, but no special provisions were made to eliminate existing trace metals. Addition of Zn2+, Co2+, or Fe2+ to a final concentration of 1 mM did not significantly alter the CA activity (Table 1). However, extended dialysis of the purified recombinant enzyme against buffer that had been treated with Chelex-100 to remove traces of metal ions resulted in the loss of nearly 75% of its activity compared to what was seen for samples that were dialyzed against buffers containing 1 mM ZnCl2. The addition of 1 mM ZnCl2 to the metal-depleted samples restored the CA activity to nearly the level observed for nondialyzed samples, while neither CoCl2 nor MgCl2 was able to reverse the inactivation of the enzyme (Table 1).
TABLE 1.
Metal dependence of recombinantly expressed carboxysomal carbonic anhydrase rCsoSCA from H. neapolitanus
| Metal added (1 mM) | Relative activity (%)a |
|---|---|
| None | 100 |
| Zn2+ | 92.8 |
| Co2+ | 101.8 |
| Fe2+ | 98.1 |
| Mg2+ | 98.4 |
| Chelex-treated buffer | 28.4 |
| Chelex-treated buffer + Zn2+ | 84.6 |
| Chelex-treated buffer + Co2+ | 25.9 |
| Chelex-treated buffer + Fe2+ | 33.4 |
| Chelex-treated buffer + Mg2+ | 26.7 |
CO2 hydration by rCsoSCA (0.5 μM) was measured by stopped-flow spectrophotometry at pH 8 (buffer/indicator pair, TAPS/m-cresol purple). Values are averages of three determinations with maximum variations of less than 12%.
Qualitative analysis of the purified enzyme by inductively coupled plasma atomic emission spectroscopy clearly revealed the presence of Zn2+ in the purified enzyme but not in buffer blanks. In addition, low levels of both Co2+ and Cr2+ were observed in the enzyme preparation. Quantitative inductively coupled plasma atomic emission spectroscopy analysis yielded Zn2+ contents of between 0.8 and 1.1 mol per mole of the rCsoSCA 57-kDa monomer, with an average value of 0.9 mol of Zn2+ per mol of monomer. This is consistent with the existence of a single Zn2+-binding site per protein monomer, as predicted from the protein's crystal structure (39).
Inhibitor effects.
The effects of some common CA inhibitors on rCsoSCA activity were assessed by stopped-flow spectrometry at 25°C to confirm results from earlier electrometric and mass spectrometric assays (Table 2). The recombinant carboxysomal enzyme was inhibited by the dithiol dithiothreitol (DTT) (50% inhibitory concentration [IC50] = 2.6 × 10−4 M) and, to a lesser extent, by the monothiol β-mercaptoethanol (IC50 = 3.4 × 10−3 M), suggesting two different mechanisms of inhibition for these two reducing agents. As previously reported, ethoxyzolamide inhibits rCsoSCA but to an extent that is significantly lower than that observed for other bacterial enzymes (35, 44, 47), while the effect of cyanide on rCsoSCA was similar to that on the chloroplast CA Cah6 from Chlamydomonas (30). Recombinant CsoSCA was remarkably resistant to azide, with an IC50 value of 4.1 mM, which is in the range of those reported for the β-CA and the γ-CA of the archaea Methanobacterium thermoautotrophicum and Methanosarcina thermophila, respectively (1, 43). Sodium chloride, Na2SO4, and KCl at concentrations up to 100 mM did not affect the activity of rCsoSCA. Unlike many α-CAs from animal sources, rCsoSCA exhibited no detectable esterase activity when assayed spectrophotometrically with p-nitrophenylacetate as a substrate.
TABLE 2.
Susceptibility of the H. neapolitanus carboxysomal carbonic anhydrase rCsoSCA to inhibitors
| Inhibitor | IC50 (M)a |
|---|---|
| Dithiothreitol | 2.4 × 10−4 |
| 2-Mercaptoethanol | 3.4 × 10−3 |
| Ethoxyzolamide | 1.9 × 10−3 |
| KCl (<100 mM) | No effect |
| Na2SO4 (<100 mM) | No effect |
| Sodium azide | 4.1 × 10−3 |
| Sodium cyanide | 2 × 10−6 |
IC50 is the molar inhibitor concentration that reduced the CO2 hydration activity of rCsoSCA by 50% at pH 8. Initial rates of CO2 hydration activity were determined by stopped-flow spectrophotometry in the presence of the indicated inhibitors. The IC50 values were calculated from plots of relative activity versus inhibitor concentrations. Individual data points were averaged from three to five independent determinations that did not vary more than 10%.
Enzymatic activity of rCsoSCA.
The CsoSCA protein from H. neapolitanus and its homologues in other α-carboxysome-producing autotrophs represent a novel CA class (ɛ) based on their unique primary structure. Their secondary and tertiary structures, however, strongly suggest that they constitute a variant of the β-class of these enzymes. The CA activity of the carboxysomal enzyme was demonstrated electrometrically with the classical Wilber-Andersen assay and with the mass spectrometry-based 18O exchange assay (47). However, under the conditions employed neither of these methods yields data that are suitable for calculations of rate parameters. The kinetic constants for rCsoSCA were determined with the colorimetric stopped-flow method of Khalifah (26), which allows one to follow the progress of the hydration of CO2 and, under some conditions, the dehydration of bicarbonate. At pH 8.0, a kcat of 8.9 × 104 s−1 was determined for the hydration of CO2 by rCsoSCA, with an apparent Km for CO2 of 3.2 mM (Table 3). The kcat for the dehydration of HCO3− was measured to be 4.6 × 104 s−1 at pH 7.0, with an apparent Km of 9.3 mM for bicarbonate.
TABLE 3.
Kinetic constants of the H. neapolitanus carboxysomal carbonic anhydrase CsoSCA
| Fraction | Kinetic constant value for:
|
|||
|---|---|---|---|---|
| Hydration reactiona
|
Dehydration reactionb
|
|||
| kcat (s−1) | Km (mM) | kcat (s−1) | Km (mM) | |
| rCsoSCA | (8.9 ± 0.5) × 104 | 3.2 (± 0.4) | (4.6 ± 1.2) × 104 | 9.3 (± 2.1) |
| Broken carboxysomes | (6.5 ± 0.9) × 104 | 3.6 (± 0.7) | (3.1 ± 0.8) × 104 | 10.1 (± 3.2) |
| Intact carboxysomes | (1.8 ± 0.2) × 104 | 12.2 (± 2.3) | ND | ND |
The hydration reaction was performed at pH 8.0.
The dehydration reaction was performed at pH 7.0. ND, not determined.
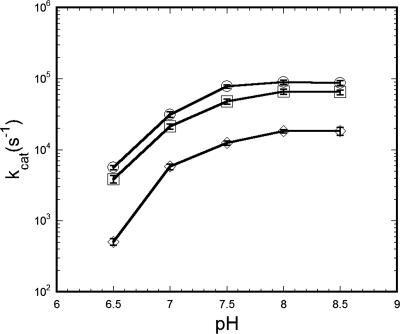
The rate of CO2 hydration by rCsoSCA was dependent on pH, and kcat increased over a pH range from 6.5 to 8.5. The marked inflection between pH 7.0 and 6.5 implicated a histidine residue participating in the catalytic chemistry, which is consistent with the previously proposed mechanism for this enzyme (39) (Fig. 3). The dehydration rate was also pH dependent but showed a decrease in kcat with increasing pH over the pH range of 6.0 to 7.5. At pH values outside of this range, measurement of the dehydration progress curves was unreliable.
FIG. 3.
pH dependence of CsoSCA kcat for CO2 hydration. Turnover numbers were calculated by fitting initial rates (8 to 15 independent determinations for each data point) to the Michaelis-Menten equation as described in Materials and Methods and were plotted as log(kcat) versus pH (○, rCsoSCA; □, broken carboxysomes; ⋄, intact carboxysomes). The buffer/indicator pairs and wavelengths used were MES/chlorophenol red (A574) at pH 5.5 to 6.8; MOPS/p-nitrophenol (A400) at pH 6.8 to 7.5; HEPES/phenol red (A557) at pH 7.5 to 8.0; and TAPS/m-cresol purple (A578) at pH 8.0 to 9.0.
The efficiency (kcat/Km) of CsoSCA for the hydration of CO2 at pH 8.0 was calculated to be 2.8 × 107 M−1 s−1 (Table 3), while a kcat/Km of 4.9 × 106 M−1 s−1 was determined for the dehydration of bicarbonate at pH 7.0. The cytoplasmic pH of H. neapolitanus and related thiobacilli has been estimated to be between 7 and 8 (16, 22), a pH range for which the efficiency of HCO3− dehydration is predicted to be approximately three- to fivefold lower than that of CO2 hydration. To date, no estimate of the pH of the carboxysome interior, which might be different from that of the cytoplasm, has been reported. However, recent studies of Penrod and Roth (33) examining the effects of CO2 and acetaldehyde exchange on wild-type Salmonella enterica and on various S. enterica eut mutants suggest that the microcompartments in that organism may provide a low-pH subcellular compartment that prevents loss of volatile metabolites.
In situ activity of native CsoSCA suggests shell limits substrate access.
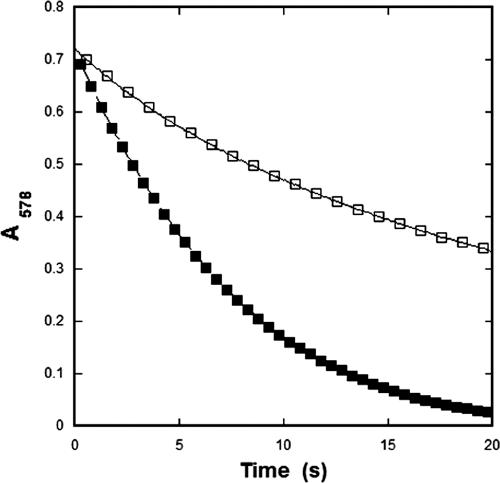
To compare the enzymatic characteristics of the carboxysomal CA within the confines of the carboxysome shell to those of the recombinant enzyme, steady-state kinetic parameters for CsoSCA were calculated from assays performed with intact and broken carboxysomes. The numerical values for the kinetic constants of the enzyme determined in situ are based on the amount of total carboxysome protein, which can be accurately measured and related to the well-established percentage of total particle protein represented by CsoSCA (15, 20). As expected, the carboxysome was capable of hydrating CO2 and dehydrating HCO3−. Using broken carboxysomes that had been disrupted by freezing and thawing (47), the rates of CO2 hydration and bicarbonate dehydration were comparable to, albeit slightly lower than, those measured for purified rCsoSCA (Table 3). Surprisingly, it was observed that the rate of CO2 hydration was significantly lower in assays with intact carboxysomes than in those employing purified rCsoSCA or carboxysomes ruptured by a freeze-thaw cycle (Fig. 1 and 4; Table 3). The calculated kcat for CO2 hydration at pH 8.0 by the CA in intact carboxysomes (1.8 × 104 s−1) was at least threefold lower than that determined for the enzyme in ruptured carboxysomes (6.45 × 104 s−1) and almost fivefold lower than that calculated for purified rCsoSCA (8.9 × 104 s−1). Since freezing and thawing of rCsoSCA under the same conditions that were used to disrupt the carboxysome did not affect the activity of the isolated enzyme, the lower turnover number determined for the enzyme in intact carboxysomes is very likely related in some manner to the structural integrity of the carboxysomal shell. The general trend of HCO3− dehydration was also much lower in assays with intact carboxysomes than in those with broken carboxysomes or with recombinant enzyme. However, the poor signal obtained for such low levels of activity made it impossible to reliably fit the dehydration kinetics to Michaelis-Menten equations and prevented comparisons to results obtained with disrupted carboxysomes and rCsoSCA (Table 3). Since the rate of the CA-catalyzed reaction is influenced by the pH of its environment, as indicated by the dependence of kcat on pH (Fig. 3), the kcat for the hydration reaction was measured over the pH range from 6.5 to 8.5 for intact and broken carboxysomes (Fig. 3). An internal carboxysome pH that is significantly lower than that of the surrounding buffer could explain the difference in CA activity observed between disrupted and intact carboxysomes; however, the pH profiles of CA activity plotted in Fig. 3 for all three forms of CsoSCA assayed did not display significant differences in shape.
FIG. 4.
Reaction progress of CO2 hydration with intact and ruptured carboxysomes. Samples of carboxysomes were identical, except that the ruptured sample had been subjected to one round of freeze-thaw treatment before the assay was performed as described in Materials and Methods. The amount of carboxysome protein used in these assays corresponded to 0.25 μM CsoSCA (□, intact carboxysomes; ▪, broken carboxysomes).
DISCUSSION
We have previously shown that the carboxysomal shell protein encoded by csoS3 from a number of α-carboxysome-containing prokaryotes is a carbonic anhydrase (47). The protein is a minor component of the carboxysome, comprising approximately 2.3% of total carboxysomal protein by weight. This value corresponds to 80 copies of the monomer per carboxysome, compared to the estimated 270 molecules of RubisCO holoenzyme per particle (15, 20, 41). The lack of significant primary sequence homology of CsoS3 with representatives of the other CA classes (α, β, γ, and δ) led us to propose a new class (ɛ) for the carboxysomal enzyme (47). However, subsequent elucidation of the protein's crystal structure established unequivocally that CsoS3 and its homologues are unique variants of β-CAs (39). To signify the connection of these CAs with the carboxysomal shell, the term CsoS3 has been replaced by CsoSCA. The association of CsoSCA with the carboxysomal shell is so tight that it is impossible to remove the protein from shell-enriched fractions of broken carboxysomes without the use of denaturants. Since the treatment needed to completely disassemble the shell irreversibly inactivates the CA activity, CsoSCA from H. neapolitanus was expressed heterologously in E. coli, purified, and characterized for this study. Consistent with its crystal structure (39), CsoSCA was found to bind one Zn2+ per monomer. Removal of the metal by dialysis or by treatment with chelators resulted in a loss of enzymatic activity that could not be restored by other divalent metal ions. Interestingly, CsoSCA was potently inhibited by the dithiol reducing agent DTT (reference 47 and this study) and to a lesser extent by the monothiol β-mercaptoethanol. The inhibition by DTT is similar to that reported for the cyanobacterial β-CA CcaA, which shares some sequence homology with higher plant chloroplast carbonic anhydrases and may be the carboxysome-associated enzyme in Synechocystis PCC 6803 and other β-cyanobacteria (49). The mechanism of inhibition by DTT is generally assumed to be the destabilization of tertiary or quaternary protein structure through the disruption of disulfide bonds (23, 28). However, the low concentration of DTT, compared to that of β-mercaptoethanol, required to bring about a 50% inhibition of CsoSCA suggests that DTT might interact directly with an active-site residue, such as one of the Zn2+-coordinating cysteines (39), that is not reactive with β-mercaptoethanol. The sulfanilamide ethoxyzolamide inhibited rCsoSCA as it does all known carbonic anhydrases, presumably through binding the active-site Zn2+. The ethoxyzolamide concentration necessary to effect a 50% reduction of enzymatic activity, however, is orders of magnitude higher than that expected for most bacterial carbonic anhydrases (1, 2, 35, 44, 51), suggesting that the Zn2+ in CsoSCA may be blocked from interaction with the inhibitor.
The molecular mass of CsoSCA calculated from its primary structure is 57.3 kDa, which agrees well with denaturing polyacrylamide gel electrophoresis-based molecular weight estimates for the carboxysome-associated enzyme from H. neapolitanus and for rCsoSCA without the hexahistidine tag. Other CAs from autotrophic microorganisms are usually considerably smaller and fall into the range of 30 kDa. The notable exception is the β-CA from Porphyridium purpureum, which has a molecular weight of approximately 60,000 and is comprised of two very similar (70% sequence similarity) domains that probably arose through a gene duplication event (31, 51). CsoSCA also has two domains that are likely to have arisen from gene duplication but have diverged considerably more than those of the P. purpureum enzyme, such that only one of the domains contains a functional Zn2+-binding site. This is consistent with the single Zn2+ ion per monomer that was predicted from the protein's crystal structure (39) and was detected in this study.
The catalytic ability of rCsoSCA, as indicated by its kcat and efficiency of CO2 hydration, is well within the range of values reported for other β-CAs (1.7 × 104 to 2.3 × 105 s−1) (21, 27, 44, 45). If the H. neapolitanus carboxysome functions as the terminal component of a CCM, as is hypothesized for the carboxysomes of β-cyanobacteria (6, 36), the presumed role of CsoSCA would be the conversion of abundant cytoplasmic HCO3− to CO2 within the carboxysome. RubisCO inside the microcompartments would fix the CO2 onto ribulose 1,5-bisphosphate (RubP) and produce 3-phosphoglycerate (3-PG), which would be returned to the cytoplasm for completion of the Calvin-Benson-Bassham cycle. If this is the role of the carboxysomal CA, one might expect that CsoSCA would favor the dehydration of HCO3− to CO2 in vitro (42). Although our results indicate that this is not the case, considering the low turnover rate of RubisCO (26 s−1) (46) and the ratio of 80 CsoSCA molecules to 270 RubisCO holoenzyme molecules per carboxysome (15, 20, 41), the calculated kcat values of HCO3− dehydration for rCsoSCA and for native enzyme in disrupted carboxysomes suggest that the CA is more than capable of providing the necessary CO2 for efficient fixation by RubisCO.
Most puzzling are the differences in CA activity observed with intact and broken carboxysomes. The rate of bicarbonate dehydration in broken carboxysomes was comparable to that observed with rCsoSCA, but in intact carboxysomes the rate was very low and not consistently reproducible. The ability to measure the dehydration reaction by the changing indicator method is limited by the low signal-to-noise ratio at the pH values relevant to these studies. In our hands, the signal obtained with intact carboxysomes was so low that it was impossible to calculate rate parameters from the data collected. However, the rate of CO2 hydration in intact carboxysomes could be reproducibly measured, and it was found to be approximately threefold lower than that measured with broken carboxysomes. We considered the possibility that the internal pH of intact carboxysomes is lower than that of the buffer used in the assay, thereby reducing the activity of the intact particles compared to that observed with free rCsoSCA or ruptured carboxysomes. However, when kcat values for all three samples were compared over a range of pH values there was no significant difference in the shapes of the activity curves, and the activity observed with the intact particles was consistently lower than those for the other two forms of the enzyme. This suggests that there was no difference in the internal and external pHs of isolated intact carboxysomes. Since the only obvious difference in the two samples is the structural integrity of the shell, the most straightforward explanation for these observations is that the protein shell limits the availability of HCO3− and CO2 to the shell-bound CsoSCA. This explanation does not seem consistent with the CCM proposed for β-carboxysomes, for which a location of the CA inside the microcompartment is posited (37, 38), and an alternative scenario must be considered. Perhaps the carboxysome shell offers diffusional resistance to gases, and the orientation of CsoSCA in the shell allows the enzyme to bind HCO3− on the cytoplasmic side, convert the bicarbonate to CO2, and release the product of catalysis to the inside of the carboxysome. In the CA assays employed in this study, as HCO3− on the outside of the particle is dehydrated and passed to the inside, CO2 would accumulate rapidly, and mass action would slow further dehydration of HCO3− and release of CO2 into the particle's interior. Since RubisCO is not active in these assays, the slow diffusion of CO2 out of the carboxysome would then be the rate-limiting step for the conversion of HCO3− to CO2 and the concurrent release of CO2 into the carboxysome. This would account for the significantly lower apparent catalytic rate observed with intact carboxysomes than that observed with ruptured carboxysomes, where CO2 and HCO3− equilibrate rapidly the inside and the outside of the microcompartment. The measured reduction in the hydration rate could be explained in the same manner if it is assumed that CO2 must first diffuse into the carboxysome before it reaches the side of the shell-bound CsoSCA that is capable of binding and converting it to HCO3−.
In vivo under physiological conditions, the fixation of CO2 onto RubP and the subsequent conversion to 3-PG within the carboxysome would lower the CO2 concentration, allowing more cytoplasmic HCO3− to be converted to CO2 by CsoSCA and be “transported” into the microcompartment. As long as bicarbonate remained plentiful in the cytoplasm, this hypothetical scenario would effectively constitute an “on-demand” CO2 supply system for RubisCO within the carboxysome. This model of carboxysome function requires that there be a means of rapid “transport” of RubP into the carboxysome and of 3-PG out of the microcompartment. While it may seem difficult to envision a protein barrier to CO2 that permits facile passage of negatively charged organic molecules such as RubP and 3-PG, it must be noted that another carboxysomal shell protein, CsoS2, might be a candidate for interaction with the negatively charged phosphates of RubP and 3-PG. This protein is the only shell component with a basic isoelectric point and a positive net charge at the presumed cytosolic pH of 7 to 8 (16, 22). Little is known about the structure of CsoS2 other than that it migrates as two polypeptides during denaturing gel electrophoresis. The one of higher apparent molecular weight is either a homodimer or a glycosylation variant of the lower-molecular-weight band (7).
While it is generally accepted that the carboxysome functions in cyanobacteria and autotrophic bacteria to localize RubisCO and CA in a common structure that maximizes the availability of CO2 for fixation by RubisCO, the mechanics of how this is accomplished are not completely understood. Previous quantitative models of CCM systems from cyanobacteria suggest that some component or substructure of the carboxysome is responsible for preventing CO2 efflux from the particle before the kinetically challenged RubisCO can catalyze its fixation. The carboxysome shell and the geometric arrangement of CA and RubisCO within the microcompartment have been hypothesized to function in this capacity in a manner analogous to that of the bundle sheath of C4 plants (34). Given the fact that in H. neapolitanus the carboxysomal CA is tightly associated with the shell (47) and that the intact shell appears to limit the accessibility of exogenously supplied substrate to carboxysomal CA, it would seem likely that the shell is the microcompartment's primary diffusion barrier.
It is interesting to note in this context the widespread occurrence of homologues of carboxysome shell protein genes in a large number of heterotrophic bacteria (10, 20), suggesting that these microcompartments might function as general metabolic organizers throughout the microbial world. In the one other case in which limited biochemical analysis of isolated microcompartments has been performed (18, 19), the shell of the polyhedral organelles formed in Salmonella enterica was also proposed to act as a diffusion barrier to propionaldehyde, which is thought to remain inside the microcompartment in close proximity to the enzyme that utilizes the compound as a substrate. The full range of biological roles played by polyhedral microcompartments is far from being understood, but it is clear that they have important and diverse functions in many aspects of microbial metabolism.
Acknowledgments
This work was supported by grants MCB-0444568 and DMR-0213883 from the National Science Foundation.
Footnotes
Published ahead of print on 29 September 2006.
REFERENCES
- 1.Alber, B. E., C. M. Colangelo, J. Dong, C. M. V. Stalhandske, T. T. Baird, C. Tu, A. Fierke, D. N. Silverman, R. A. Scott, and J. G. Ferry. 1999. Kinetic and spectroscopic characterization of the gamma carbonic anhydrase from the methanoarchaeon Methanosarcina thermophila. Biochemistry 38:13119-13128. [DOI] [PubMed] [Google Scholar]
- 2.Alber, B. E., and J. G. Ferry. 1996. Characterization of heterologously produced carbonic anhydrase from Methanosarcina thermophila. J. Bacteriol. 178:3270-3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong, J. M., D. V. Myers, J. A. Verpoorte, and J. T. Edsall. 1966. Purification and properties of human erythrocyte carbonic anhydrases. J. Biol. Chem. 241:5137-5149. [PubMed] [Google Scholar]
- 4.Badger, M. R. 2003. The roles of carbonic anhydrases in photosynthetic CO2 concentrating mechanisms. Photosynth. Res. 77:83-94. [DOI] [PubMed] [Google Scholar]
- 5.Badger, M. R., D. Hanson, and G. D. Price. 2002. Evolution and diversity of CO2 concentrating mechanisms in cyanobacteria. Funct. Plant Biol. 29:161-173. [DOI] [PubMed] [Google Scholar]
- 6.Badger, M. R., and G. D. Price. 2003. CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J. Exp. Bot. 54:609-622. [DOI] [PubMed] [Google Scholar]
- 7.Baker, S. H., S. C. Lorbach, M. Rodriguez-Buey, D. S. Williams, H. C. Aldrich, and J. M. Shively. 1999. The correlation of the gene csoS2 of the carboxysome operon with two polypeptides of the carboxysome in Thiobacillus neapolitanus. Arch. Microbiol. 172:233-239. [DOI] [PubMed] [Google Scholar]
- 8.Baker, S. H., D. S. Williams, H. C. Aldrich, A. C. Gambrell, and J. M. Shively. 2000. Identification and localization of the carboxysome peptide CsoS3 and its corresponding gene in Thiobacillus neapolitanus. Arch. Microbiol. 173:278-283. [DOI] [PubMed] [Google Scholar]
- 9.Beudeker, R. F., G. C. Cannon, J. G. Kuenen, and J. M. Shively. 1980. Relations between d-ribulose-1,5-bisphosphate carboxylase, carboxysomes, and CO2 fixing capacity in the obligate chemolithotroph Thiobacillus neapolitanus grown under different limitations in the chemostat. Arch. Microbiol. 124:185-189. [Google Scholar]
- 10.Bobik, T. A. 2006. Polyhedral organelles compartmenting bacterial metabolic processes. Appl. Microbiol. Biotechnol. 70:517-525. [DOI] [PubMed] [Google Scholar]
- 11.Bonfil, D. J., M. Ronen-Tarazi, D. Sultemeyer, J. Lieman-Hurwitz, D. Schatz, and A. Kaplan. 1998. A putative HCO3− transporter in the cyanobacterium Synechococcus sp. strain PCC 7942. FEBS Lett. 430:236-240. [DOI] [PubMed] [Google Scholar]
- 12.Cannon, G. C., S. H. Baker, F. Soyer, D. R. Johnson, C. E. Bradburne, J. L. Mehlman, P. S. Davies, Q. L. Jiang, S. Heinhorst, and J. M. Shively. 2003. Organization of carboxysome genes in the thiobacilli. Curr. Microbiol. 46:115-119. [DOI] [PubMed] [Google Scholar]
- 13.Cannon, G. C., C. E. Bradburne, H. C. Aldrich, S. H. Baker, S. Heinhorst, and J. M. Shively. 2001. Microcompartments in prokaryotes: carboxysomes and related polyhedra. Appl. Environ. Microbiol. 67:5351-5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cannon, G. C., S. Heinhorst, C. E. Bradburne, and J. M. Shively. 2002. Carboxysome genomics: a status report. Funct. Plant Biol. 29:175-182. [DOI] [PubMed] [Google Scholar]
- 15.Cannon, G. C., and J. M. Shively. 1983. Characterization of a homogeneous preparation of carboxysomes from Thiobacillus neapolitanus. Arch. Microbiol. 134:52-59. [Google Scholar]
- 16.Dobrinski, K. P., D. L. Longo, and K. M. Scott. 2005. The carbon-concentrating mechanism of the hydrothermal vent chemolithoautotroph Thiomicrospira crunogena. J. Bacteriol. 187:5761-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Espie, G. S., and B. Colman. 2005. CO2-concentrating mechanisms in aquatic photosynthetic microorganisms. Can. J. Bot. 83:695-697. [Google Scholar]
- 18.Havemann, G. D., and T. A. Bobik. 2003. Protein content of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 185:5086-5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Havemann, G. D., E. M. Sampson, and T. A. Bobik. 2002. PduA is a shell protein of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 184:1253-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heinhorst, S., G. C. Cannon, and J. M. Shively. 2006. Carboxysomes and carboxysome-like inclusions, p. 141-165. Microbial monographs, vol. 2. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 21.Hiltonen, T., H. Bjoerkbacka, C. Forsman, A. K. Clarke, and G. Samuelsson. 1998. Intracellular beta-carbonic anhydrase of the unicellular green alga Coccomyxa. Plant Physiol. 117:1341-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holthuijzen, Y. A., E. F. M. VanDissel-Emiliani, J. G. Kuenen, and W. N. Konings. 1987. Energetic aspects of CO2 uptake in Thiobacillus neapolitanus. Arch. Microbiol. 147:285-290. [Google Scholar]
- 23.Kaji, E. H., and H. F. Lodish. 1993. In vitro unfolding of retinol-binding protein by dithiothreitol. J. Biol. Chem. 268:22195-22202. [PubMed] [Google Scholar]
- 24.Kaplan, A., and L. Reinhold. 1999. CO2 concentrating mechanisms in photosynthetic microorganisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50:539-570. [DOI] [PubMed] [Google Scholar]
- 25.Kerfeld, C. A., M. R. Sawaya, S. Tanaka, C. V. Nguyen, M. Phillips, M. Beeby, and T. O. Yeates. 2005. Protein structures forming the shell of primitive bacterial organelles. Science 309:936-938. [DOI] [PubMed] [Google Scholar]
- 26.Khalifah, R. G. 1971. The carbon hydroxide hydration activity of carbonic anhydrase. I. Stop-flow kinetic studies on the native human isozymes B and C. J. Biol. Chem. 246:2561-2573. [PubMed] [Google Scholar]
- 27.Larsson, S., H. Bjoerkbacka, C. Forsman, G. Samuelsson, and O. Olsson. 1997. Molecular cloning and biochemical characterization of carbonic anhydrase from Populus tremula × tremuloides. Plant Mol. Biol. 34:583-592. [DOI] [PubMed] [Google Scholar]
- 28.Li, Y. J., D. M. Rothwarf, and H. A. Scheraga. 1995. Mechanism of reductive protein unfolding. Nat. Struct. Biol. 2:489-494. [DOI] [PubMed] [Google Scholar]
- 29.McKay, R. M. L., S. P. Gibbs, and G. S. Espie. 1993. Effect of dissolved inorganic carbon on the expression of carboxysomes, localization of RubisCO and the mode of carbon transport in cells of the cyanobacterium Synechococcus UTEX 625. Arch. Microbiol. 159:21-29. [Google Scholar]
- 30.Mitra, M., S. M. Lato, R. A. Ynalvez, Y. Xiao, and J. V. Moroney. 2004. Identification of a new chloroplast carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol. 135:173-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitsuhashi, S., T. Mizushima, E. Yamashita, M. Yamamoto, T. Kumasaka, H. Moriyama, T. Ueki, S. Miyachi, and T. Tsukihara. 2000. X-ray structure of the beta-carbonic anhydrase from the red alga, Porphyridium purpureum, reveals a novel catalytic site for CO2 hydration. J. Biol. Chem. 275:5521-5526. [DOI] [PubMed] [Google Scholar]
- 32.Orus, M. I., M. L. Rodriguez-Buey, E. Marco, and E. Fernandez-Valiente. 2001. Changes in carboxysome structure and grouping and in photosynthetic affinity for inorganic carbon in Anabaena strain PCC 7119 (Cyanophyta) in response to modification of CO2 and Na+ supply. Plant Cell Physiol. 42:46-53. [DOI] [PubMed] [Google Scholar]
- 33.Penrod, J. T., and J. R. Roth. 2006. Conserving a volatile metabolite: a role for carboxysome-like organelles in Salmonella enterica. J. Bacteriol. 188:2865-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price, G. D., and M. R. Badger. 1991. Evidence for the role of carboxysomes in the cyanobacterial CO2-concentrating mechanism. Can. J. Bot. 69:963-973. [Google Scholar]
- 35.Price, G. D., J. R. Coleman, and M. R. Badger. 1992. Association of carbonic anhydrase activity with carboxysomes isolated from the cyanobacterium Synechococcus PCC7942. Plant Physiol. 100:784-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price, G. D., D. Sultemeyer, B. Klughammer, M. Ludwig, and M. R. Badger. 1998. The functioning of the CO2 concentrating mechanism in several cyanobacterial strains: a review of general physiological characteristics, genes, proteins and recent advances. Can. J. Bot. 76:973-1002. [Google Scholar]
- 37.Reinhold, L., M. Zviman, and A. Kaplan. 1987. Inorganic carbon fluxes and photosynthesis in cyanobacteria. A quantitative model, p. 6.289-6.296. In J. Biggens (ed.), Progress in photosynthetic research, vol. 4. Martinus Nijhoff, Dordrecht, The Netherlands. [Google Scholar]
- 38.Reinhold, L., M. Zviman, and A. Kaplan. 1989. A quantitative model for carbon fluxes and photosynthesis in cyanobacteria. Plant Physiol. Biochem. 27:945-954. [Google Scholar]
- 39.Sawaya, M. R., G. C. Cannon, S. Heinhorst, S. Tanaka, E. B. Williams, T. O. Yeates, and C. A. Kerfeld. 2006. The structure of beta-carbonic anhydrase from the carboxysomal shell reveals a distinct subclass with one active site for the price of two. J. Biol. Chem. 281:7546-7555. [DOI] [PubMed] [Google Scholar]
- 40.Shively, J. M., F. Ball, D. H. Brown, and R. E. Saunders. 1973. Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) in Thiobacillus neapolitanus. Science 182:584-586. [DOI] [PubMed] [Google Scholar]
- 41.Shively, J. M., and R. S. English. 1991. The carboxysome, a prokaryotic organelle: a mini review. Can. J. Bot. 69:957-962. [Google Scholar]
- 42.Smith, K. S., N. J. Cosper, C. Stalhandske, R. A. Scott, and J. G. Ferry. 2000. Structural and kinetic characterization of an archaeal beta-class carbonic anhydrase. J. Bacteriol. 182:6605-6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith, K. S., and J. G. Ferry. 1999. A plant-type (beta-class) carbonic anhydrase in the thermophilic methanoarchaeon Methanobacterium thermoautotrophicum. J. Bacteriol. 181:6247-6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith, K. S., and J. G. Ferry. 2000. Prokaryotic carbonic anhydrases. FEMS Microbiol. Rev. 24:335-366. [DOI] [PubMed] [Google Scholar]
- 45.Smith, K. S., C. Jakubzick, T. S. Whittam, and J. G. Ferry. 1999. Carbonic anhydrase is an ancient enzyme widespread in prokaryotes. Proc. Natl. Acad. Sci. USA 96:15184-15189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Snead, R. M., and J. M. Shively. 1978. d-Ribulose-1,5-carboxylase from Thiobacillus neapolitanus. Curr. Microbiol. 1:309-314. [Google Scholar]
- 47.So, A. K., G. S. Espie, E. B. Williams, J. M. Shively, S. Heinhorst, and G. C. Cannon. 2004. A novel evolutionary lineage of carbonic anhydrase (epsilon class) is a component of the carboxysome shell. J. Bacteriol. 186:623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.So, A. K., M. John-McKay, and G. S. Espie. 2002. Characterization of a mutant lacking carboxysomal carbonic anhydrase from the cyanobacterium Synechocystis PCC6803. Planta 214:456-467. [DOI] [PubMed] [Google Scholar]
- 49.So, A. K.-C., and G. S. Espie. 2005. Cyanobacterial carbonic anhydrases. Can. J. Bot. 83:721-734. [Google Scholar]
- 50.Tripp, B. C., K. S. Smith, and J. G. Ferry. 2001. Carbonic anhydrase: new insights for an ancient enzyme. J. Biol. Chem. 276:48615-48618. [DOI] [PubMed] [Google Scholar]
- 51.Yagawa, Y., S. Muto, and S. Miyachi. 1987. Carbonic anhydrase of a unicellular red alga Porphyridium cruentum R-1. I. Purification and properties of the enzyme. Plant Cell Physiol. 28:1258-1262. [Google Scholar]
- 52.Yoshizawa, Y., K. Toyoda, H. Arai, M. Ishii, and Y. Igarashi. 2004. CO2-responsive expression and gene organization of three ribulose-1,5-bisphosphate carboxylase/oxygenase enzymes and carboxysomes in Hydrogenovibrio marinus strain MH-110. J. Bacteriol. 186:5685-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]