Abstract
Bacillus anthracis, the spore-forming agent of anthrax, requires iron for growth and is capable of scavenging heme-iron during infection. We show here that the B. anthracis iron-regulated surface determinants (isd) locus encompasses isdC, specifying a heme-iron binding surface protein. Anchoring of IsdC to the cell wall envelopes of vegetative bacilli requires srtB, which encodes sortase B. Purified sortase B cleaves IsdC between the threonine and the glycine of its NPKTG motif sorting signal. B. anthracis variants lacking either isdC or srtB display defects in heme-iron scavenging, suggesting that IsdC binding to heme-iron in the cell wall envelope contributes to bacterial uptake of heme.
Bacillus anthracis is a spore-forming gram-positive bacterium pathogenic to both humans and animals (20). Depending on the route of entry, disease manifests as clinically distinct syndromes—cutaneous, gastrointestinal, or inhalational anthrax, all of which are rapidly fatal unless treated early with antimicrobials (18). One hallmark of anthrax disease is rapid multiplication of vegetative bacilli to a density of 1010 to 1011 CFU per gram of host tissue (20). To achieve such growth, B. anthracis must satisfy its nutritional requirements for iron (4, 42). Bioinformatic and genetic analyses suggest that B. anthracis can utilize diverse sources of iron that are sequestered within host iron storage and transport proteins, such as hemoglobin, lactoferrin, ferritin, and transferrin (7, 21, 32).
The ability to acquire iron during infection is an essential attribute of many bacterial pathogens of vertebrates (3, 43). Iron is required for numerous cellular processes that are fundamental to life, such as DNA replication, energy generation, and protection against reactive oxygen species (9). Most iron in vertebrates is sequestered within cells and stored as hemoproteins in red blood cells or is bound to transferrin or lactoferrin (6). In order to scavenge iron from transferrin or lactoferrin, bacteria secrete siderophores, molecules that capture iron with high affinity for subsequent receptor-mediated uptake by bacteria (10). The most abundant group of iron binding proteins in humans are hemoproteins, polypeptides that utilize heme as a cofactor (5, 9). Heme, a tetrapyrrole encircling an individual iron atom within the macrocyclic conjunction of the heme porphyrin ring, and hemoproteins represent approximately 80% of the iron in humans (9). In order to successfully mediate infection, most bacterial pathogens have developed systems capable of extracting heme from human hemoproteins and subsequently utilizing this cofactor as both a heme and an iron source (17, 34, 45).
The mechanism(s) whereby B. anthracis scavenges heme from host hemoproteins is not yet understood. Recent work with Staphylococcus aureus, another gram-positive pathogen, identified the isd (iron-regulated surface determinants) locus, which encodes proteins involved in heme-iron scavenging and transport (24, 25). Specifically, the locus codes for proteins with binding affinity for heme (IsdA and IsdC), hemoglobin (IsdB), and haptoglobin (IsdH/HarA) (12, 24, 25). Other genes provide heme-iron transport across bacterial membranes (IsdD, IsdE, and IsdF), in addition to IsdG oxygenase, which liberates iron from the tetrapyrrol scaffold (38, 39, 46). In S. aureus, two sortases, SrtA and SrtB, cleave the C-terminal sorting signals of surface proteins to form amide bonds between the amino groups of peptidoglycan cross bridges within cell wall envelopes and the C-terminal carboxyl group of surface proteins (22). Sortase A is responsible for anchoring IsdA, IsdB, and IsdH (HarA), whereas sortase B anchors IsdC in the cell wall envelope (23, 25). srtB, the structural gene for sortase B, but not that for sortase A, is located within the isd loci of S. aureus and B. anthracis (24).
The B. anthracis isdC homolog and srtB are present within isd; however, the locus lacks the surface protein genes isdA, isdB, and isdH found in S. aureus (Fig. 1A). Other isd genes encode a putative membrane transporter (isdE1, isdE2, and isdF) and an oxygenase (isdG) (37), while isdX1 and isdX2 specify two secreted proteins that may also contribute to heme-iron transport. To investigate a possible contribution of isd to heme-iron transport in B. anthracis, we examined the molecular properties of isdC and srtB.
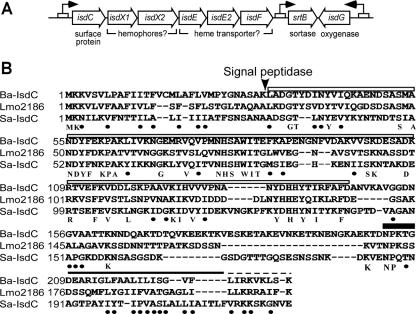
FIG. 1.
B. anthracis IsdC. (A) The Bacillus anthracis isd-like locus contains eight open reading frames: a sortase (srtB), isdC, secreted putative hemophores (isdX1 and isdX2), an iron transporter complex (isdE, -E2, and -F), and a monooxygenase (isdG). (B) Alignment of the amino acid sequences of B. anthracis isdC (BAS4444; Ba-IsdC), S. aureus IsdC (AAW38020; Sa-IsdC), and L. monocytogenes Lmo2186. The arrow indicates the predicted signal peptide cleavage site for B. anthracis IsdC. The closed bar, solid line, and dashed line indicate the proposed sortase cleavage motif (NPKTG for B. anthracis IsdC), hydrophobic domain, and positively charged tail of cell wall sorting signals, respectively. The highest degree of sequence similarity is within the NEAT domain (IsdC amino acids 32 to 146 [open bar]). Absolutely conserved amino acids are indicated below the alignment. The black dots indicate conservation of polarity, charge, or hydrophobicity.
MATERIALS AND METHODS
Bacterial strains, plasmids, and reagents.
The bacterial strains and plasmids used in this study are summarized in Table 1. For molecular biology studies, bacilli were grown in Luria broth (LB) or heart infusion broth (HIB) at 30°C. Antibiotics were used at the following concentrations: ampicillin (50 μg/ml for Escherichia coli), chloramphenicol (10 μg/ml for B. anthracis), erythromycin (5 μg/ml for B. anthracis), and kanamycin (20 μg/ml for E. coli/B. anthracis). The strains were stored at −80°C in the presence of 5% bovine serum albumin and 5% monosodium glutamate. Reagents were purchased from Sigma unless otherwise noted.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Property | Reference or source | ||
|---|---|---|---|---|
|
B. anthracis strains
|
||||
| Sterne 34F2 | B. anthracis strain Sterne 34F2 (pXO1+ pXO2−) | 40 | ||
| BAS2 | Δ(isdC) isdC (BAS4444) of strain Sterne replaced with erm | This study | ||
| BAS3 | Δ(srtB) srtB (BAS4437) of strain Sterne replaced with cat | This study | ||
| Plasmids
|
||||
| POS2 | Erm-resistant E. coli/B. anthracis shuttle vector | 35 | ||
| PBAS6 | pOS2 containing srtB and its Fur box/regulatory elements | This study | ||
| PLM5 | E. coli/B. anthracis shuttle vector with temperature-sensitive replicon | This study | ||
| PisdCH6 | pLM5 containing isdC with methionyl-six-histidyl codons upstream of the NPKTG motif | This study | ||
| PSrtB-IsdCH6 | pLM5 containing both srtB and isdCH6 | This study | ||
| Pgst-isdC | isdC32-212 inserted into pGEX2TK | This study | ||
| PsrtBΔN | srtBΔN inserted into pQE30 | This study | ||
B. anthracis srtB and isdC mutants were generated by previously established methods (16, 25, 37). Briefly, the srtB (BAS4437) and isdC (basK; BAS4444) genes of B. anthracis strain Sterne 34F2 were replaced by allelic exchange with chloramphenicol (cat) and erythromycin (erm) cassettes, respectively. B. anthracis template DNA was isolated using the Wizard Genomic DNA purification kit (Promega, Madison, WI). For the srtB mutant, 1,000 bp of 5′ and 3′ srtB-flanking sequences were PCR amplified from B. anthracis template DNA using the primer pairs srtB-PstI (restriction sites are underlined) (5′-GATCCTGCAGTTGAGTAGTGAAAAAGAACGG-3′) and srtB-XmaI (5′-GATCCCCGGGCACCTAATTCATAAA), as well as srtB-XmaI (5′-GATCCCCGGGCAGCAGGTGATCAAATTG-3′) and srtB-NotI (5′-GATCGCGGCCGCTTACTGTCTTTTTAC-3′). PCR products were cloned between the PstI/NotI sites of pLC28, and the cat cassette was inserted into the XmaI site (8). The srtB::cat cassette was excised with PstI and NotI, treated with Klenow enzyme, and cloned into the SmaI site of the temperature-sensitive shuttle vector pTS1 to generate the plasmid psrtB::cat (26). For the isdC mutant, 1,000-bp 5′ and 3′ isdC-flanking sequences were PCR amplified from B. anthracis template DNA using the primer pairs isdC-PstI (5′-GATCCTGCAGATGAAAAAGGTTTCTG-3′) and isdC-XmaI (5′-GATCCCCGGGCAGCTAGTTTTGCTG-3′), as well as isdC-XmaI (5′-GATCCCCGGGTAATCCAAAAACAGGCGATG-3′) and isdC-NotI (5′-GATCGCGGCCGCTTATTTACTCAATTTCAC-3′). PCR products were cloned into pLC28 and pTS1 as described above. Plasmid DNA was replicated in the dam mutant E. coli strain K1077 prior to electroporation into B. anthracis Sterne 34F2 (36). Allelic exchange was induced with a temperature shift to 43°C, followed by erythromycin or chloramphenicol selection.
Southern blotting was used to confirm the presence of srtB::cat and isdC::ermC alleles in B. anthracis. Briefly, chromosomal DNA was digested with SnaBI or MfeI. The restricted DNA was separated by electrophoresis, transferred to a nylon membrane, incubated in the presence of labeled probes, and detected by chemiluminescence. Probes were generated by PCR in the presence of digoxigenin-dUTP (DIG System; Roche Molecular Biochemicals, Germany) using the primers srtB-forward (5′-TTGAGTAGTGAAAAAGAACGG-3′) and srtB-reverse (5′-TTACTGTCTTTTTAC-3′), isdC-forward (5′-ATGAAAAAGGTTTCTG-3′) and isdC-reverse (5′-TTATTTACTCAATTTCAC-3′), ermC-forward (5′-TACACCTCCGGATAATAAA-3′) and ermC-reverse (5′-CACAAGACACTCTTTTTTC-3′), and cat-forward (5′-CGAGATTTTCAGGAGC-3′) and cat-reverse (5′-GATCCGGCGAATTTC-3′).
Protein purification.
Sortase B was purified as a recombinant protein with a replacement of amino acids 2 to 36 with six-histidyl (SrtBΔN) from E. coli lysates. Expression plasmid psrtBΔN was constructed by PCR amplification of srtB sequences from the B. anthracis genome using the primer pair srtB-forward-2 (5′-GTACGTACGGATCCATGGATTACTATGAAAATCG-3′) and srtB-reverse-2 (5′-GCTTTGATCATTTTTCTGTCATTCTCGAGCTAGCTAG-3′). Product DNA was digested with EcoRI/BamHI and cloned into pQE-30 restricted with the same enzymes. E. coli XL-1 Blue harboring psrtBΔN was propagated on LB agar with ampicillin overnight at 37°C. Bacteria were scraped from the agar plates and inoculated into 1 liter fresh LB and rotated at 250 rpm and 37°C. After 2 h of incubation, 1.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added, and the cultures were incubated for an additional 2 h. The bacteria were sedimented by centrifugation at 6,000 × g for 8 min and suspended in column buffer (50 mM Tris-HCl, pH 7.9, and 150 mM NaCl), and the cells were broken with two passes through a French press at 14,000 lb/in2. The bacterial lysate was centrifuged at 30,000 × g for 20 min, and soluble protein in the supernatants was passed through a 0.45-μm-pore-size cellulose filter. The filtrates were subjected to affinity chromatography on 1-ml nitrilotriacetic acid/Ni2+ agarose columns preequilibrated with column buffer. The columns were washed with 40 ml binding buffer, followed by 40 ml wash buffer (column buffer plus 10% glycerol and 5 mM imidazole). SrtBΔN was eluted with 2 ml column buffer with 0.25 M imidazole and 10% glycerol, dialyzed against 1 liter column buffer with 30% glycerol, and finally stored at −20°C.
Glutathione S-transferase (GST) and GST-IsdC were purified as recombinant proteins from E. coli. The expression plasmid pgst-isdC was constructed by PCR amplification of isdC sequences from the B. anthracis genome using the primer pair isdC-forward-2 (5′-GTACGTACGGATCCGCTGATGGTACTTACGATATTAA-3′) and isdC-reverse-2 (5′-GATCGATCGAATTCAATTGCTTCATCGCCTGTTTTTTGG-3′). The product DNA was digested with EcoRI/BamHI, cloned into pGEX2TK, and electroporated into E. coli XL-1 Blue. Bacterial lysates of strains harboring pGEX2TK or pgst-isdC were prepared as described above and loaded on glutathione Sepharose 4B. The columns were washed with 40 ml of column buffer, followed by 40 ml of wash buffer (column buffer plus 10% glycerol). Target proteins were eluted with 2 ml of binding buffer with 20 mM reduced glutathione and dialyzed against 1 liter column buffer with 30% glycerol in 50 mM Tris-HCl, pH 8.0. The purified GST-IsdC, as well as GST, was stored at −20°C.
Measuring sortase B activity.
Purified B. anthracis SrtAΔN, SrtBΔN, or SrtCΔN (10 μM) was incubated with a fluorescent resonance energy transfer peptide substrate (50 μM 2-aminobenzoyl-KTDNPKTGDEA-diaminopropionic acid-dinitrophenyl-NH2 [the sortase B recognition motif is underlined]) (Synpeptide, San Diego, CA) in 150 mM NaCl, 5 mM CaCl2, 50 mM Tris-HCl, pH 7.5, for 18 h at 37°C. Fluorescence was determined by measuring excitation at 320 nm and emission at 420 nm following incubation of the fluorescent resonance energy transfer substrate in the presence or absence of sortase, and arbitrary fluorescence units were recorded by fluorimetry. For SrtBΔN inhibition, 5 mM MTSET {2-(trimethylammonium)ethyl[methanethiosulfonate]} was added. Dithiothreitol (DTT) was added to 5 mM to MTSET-inhibited reactions. For analysis of sortase B cleavage products, 300 μl of sample was subjected to reversed-phase high-performance liquid chromatography (HPLC) on a C18 column (2 by 250 mm; C18 Hypersil; Keystone Scientific, Bellefonte, PA). The cleaved peptides were eluted with a linear gradient of acetonitrile in H2O from 1 to 100% (70 min; 1 fraction [500 μl]/min), and absorbance was monitored at 215 nm. The peak fractions were dried by vacuum centrifugation, suspended in 15 μl 0.1% trifluoroacetic acid (in H2O), mixed with H2O saturated with α-cyano-4-hydroxycinnamic acid, spotted onto a sample plate, and air dried. For matrix-assisted laser desorption ionization (MALDI)-time of flight experiments, samples were ionized by an N2 UV laser using a Reflectron time of flight spectrometer (Applied Biosystems, Foster City, CA) in reflectron mode. Two hundred laser shots were conducted at an accelerating voltage of 25,000 V and a laser intensity of 2,075 (repetition rate, 3 Hz). The instrument was calibrated using bovine serum albumin as an internal control, and scans were processed using Biosystems Voyager 6004 software.
Anchoring of IsdCMH6 to the envelopes of bacilli.
pisdCMH6 was generated by a two-step PCR procedure using primers isdC-forward-3 (codons 1 to 202) (5′-GATCGATCGGTACCGATAACCAAGGAGGAAACAAG-3′) and isdC-reverse-3 (5′-CTAGCTAGGAATTCTTCAGCTTTTCCATTTTCATTTTTTG-3′), as well as isdC-forward-4 (codons 203 to 237) (5′-GATCGATCGAATTCATGCATCACCATCACCATCACAAAACAGATAATCCAAAAACA-3′) and isdC-reverse-4 (5′-CTAGCTAGGGTACCGATTTATTTACTCAATTTCACTTTAC-3′). Two PCR products, codons 1 to 202 and 203 to 237, were restricted with EcoRI, ligated, and subjected to PCR with isdC-forward-3 and isdC-reverse-4. The PCR product was cleaved with KpnI and ligated into KpnI-digested pLM5 to generate pIsdCH6. srtB was amplified from the B. anthracis genome with the primers srtB-forward-3 (5′-GATCGATCTCTAGACGTTTTGAGG TGATAATTTG-3′) and srtB-reverse-3 (5′-CTAGCTAGTCTAGATTACTGTCTTTTTACTAGTTTCG-3′). The PCR product was cleaved with XbaI and ligated into XbaI-restricted pIsdCH6 to generate pSrtB-IsdCH6. pIsdCH6, pSrtB-IsdCH6, or pLM5 was transformed into srtB::cat mutant bacilli, and the transformants were inoculated into 2 ml HIB and grown for 12 h at 30°C. The overnight cultures were inoculated 1:50 into 100 ml of HIB, and after 2 h of growth at 30°C, IPTG was added to 1.5 mM, followed by two additional hours of growth. The bacteria were sedimented by centrifugation at 10,000 × g; washed in 50 ml of 150 mM NaCl, 50 mM Tris-HCl, pH 7.5; suspended in 1 ml of UDS buffer (6 M urea, 5 mM DTT, 1% sodium dodecyl sulfate [SDS], 50 mM Tris-HCl, pH 8.0); and heated for 10 min at 100°C. Samples were divided into two 500-μl aliquots and centrifuged at 13,000 × g for 5 min. The supernatant was removed, and sediment containing murein sacculi was washed six times with 5 mM NaH2PO4, pH 6.0. The pellets were suspended in 50 μl of mutanolysin buffer (500 U/ml mutanolysin, 100 mM NaH2PO4, 1 mM phenylmethylsulfonyl fluoride, pH 6.0), and the cell wall was digested for 12 h at 37°C. Samples were centrifuged at 13,000 × g for 5 min, and the supernatants (digested cell wall) were isolated. Proteins in the medium fraction were precipitated by chloroform-methanol (44) and solubilized in 50 μl of SDS sample buffer (3 M urea, 4% SDS, 1% β-mercaptoethanol, 10% glycerol, 50 mM Tris-HCl, pH 7.5). Equivalent volumes (10 μl) of medium, lysate, and cell wall fractions were subjected to 15% SDS-polyacrylamide gel electrophoresis (PAGE), the proteins were transferred to polyvinylidene difluoride membranes, and the blots were probed with Ni-horseradish peroxidase (HRP), αSrtB, or αL6 rabbit antiserum (1:1,000), followed by mouse anti-rabbit HRP-linked antibody (1:10,000) and enhanced chemiluminescence (Pierce, Rockford, IL).
IsdC binding to heme.
GST-IsdC or GST (1 μM) was incubated with hemin chloride (0.1 to 25.6 μM) in 50 mM Tris-HCl, pH 7.5, for 5 min at 25°C, followed by spectrophotometric analysis of samples from 300 to 700 nm using a Varian Cary 50BIO instrument. Changes in absorbance at 412 nm were plotted for protein-hemin mixtures after subtraction of hemin absorbance derived from experimental determination in a reference cuvette. Spectral data (see Fig. 5B) were derived with 0.15 μM hemin. To remove hemin bound to GST-IsdC during purification from lysates of recombinant E. coli, GST-IsdC was dialyzed against 1 liter of 8 M urea (12 h), followed by dialysis with 50 mM Tris-HCl, pH 7.5 (12 h). About 80% of the hemin bound to GST-IsdC is removed by this procedure without affecting the ability of dialyzed GST-IsdC to bind hemin. Hemin chloride was solubilized in 0.1 M NH4OH.
FIG. 5.
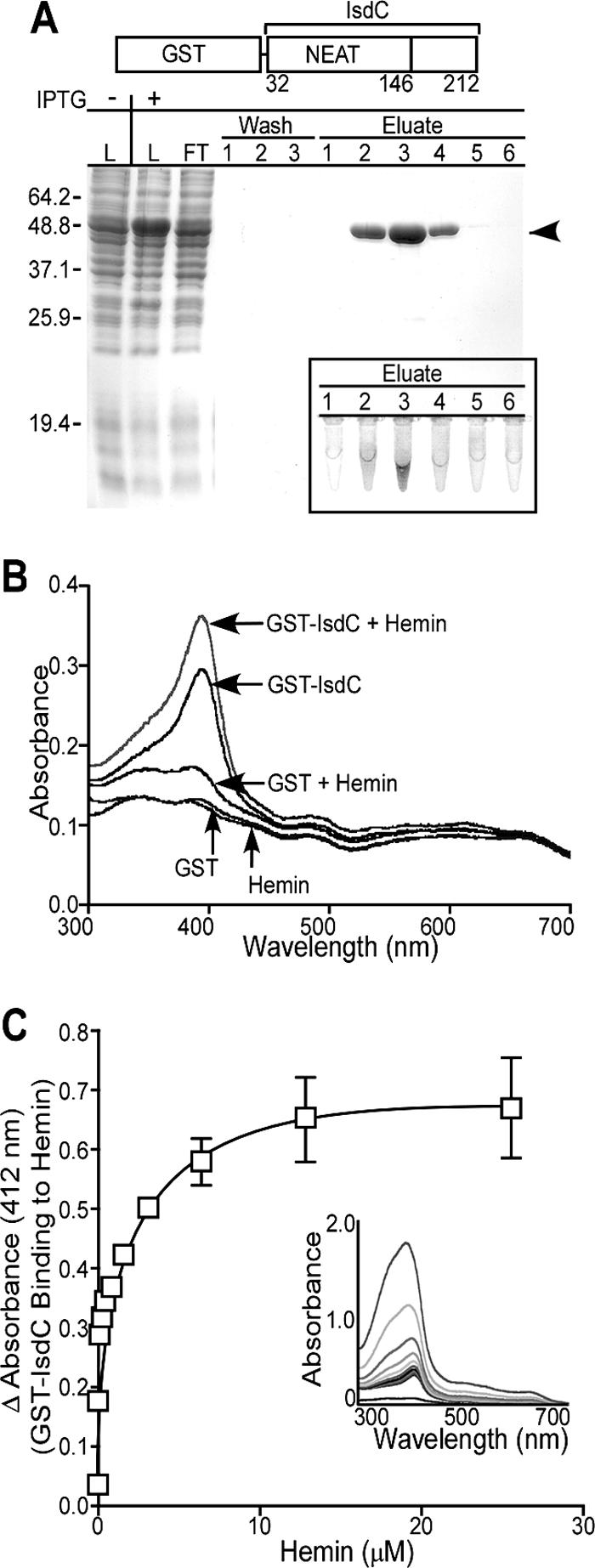
B. anthracis IsdC binds hemin. (A) Following IPTG-induced expression, GST-IsdC was purified from E. coli lysate (L) by affinity chromatography on glutathione-Sepharose. Flowthrough (FT), wash, and eluate fractions were sampled and analyzed by Coomassie-stained SDS-PAGE. The arrowhead denotes the electrophoretic mobility of GST-IsdC. The dark (brown) color of GST-IsdC peak elutions is characteristic of proteins that bind heme (inset) (24). (B) GST-IsdC or GST was incubated with or without hemin chloride, followed by spectrophotometric analysis of samples. The Soret peak at ∼412 nm is indicative of GST-IsdC binding to hemin. (C) Following dialysis to remove bound heme, GST-IsdC was incubated with increasing amounts of hemin, followed by spectrophotometry analysis. The inset displays the increase in absorbance at 412 nm of GST-IsdC plotted against the concentration of added hemin. The means and standard deviations of three independent determinations are displayed.
Bacterial growth in the presence of hemin.
To generate psrtB, the srtB gene plus 450 bp of upstream sequence was amplified from B. anthracis genomic DNA using forward (5′-GATCGATCGAATTCCGAAAGGGCTTTAGC-3′) and reverse-(5′-GCTTTGATCATTTTTCTGTCATTGCTAGCCTAGCTAG-3′) primers. The PCR products were restricted with EcoRI and NheI and ligated into pOS2 restricted with the same enzymes. Purified psrtB was electoporated into bacilli. Iron-free HIB, supplemented with 0.2% Tween 80, was generated by treatment with Chelex-100 for 12 h at 4°C (24). The medium was supplemented with Ca2+, Mg2+, Mn2+, and Zn2+ and passed through a 0.22-μM filter (iron-free HIB). B. anthracis strains were inoculated into iron-free HIB and incubated for 8 h at 37°C or 30°C, and the cultures at an optical density (OD) of ∼0.1 were diluted 1:100 in fresh iron-free HIB in the presence of 500 μM 2,2′-dipyridyl with or without hemin chloride (0.5 μM). After 16 h of incubation, growth was recorded as the OD reading at 600 nm (OD600), and the data were quantified as the growth index (growth in iron-free HIB with or without heme divided by growth in iron-rich HIB). All experiments were performed in the presence of antibiotics to maintain the appropriate plasmids.
RESULTS
B. anthracis IsdC.
BLAST searches with S. aureus isdC were used to identify the B. anthracis homolog, isdC (Fig. 1). A pfam search using cell wall sorting signals of surface proteins as queries identified the gene product of isdC as a potential sortase substrate (16) (isdC was originally designated basK, for B. anthracis surface protein K). Sequence alignment of B. anthracis IsdC, Listeria monocytogenes Lmo2186, and S. aureus IsdC revealed shared features (Fig. 1). All three proteins, which are synthesized as precursors with N-terminal signal peptides and C-terminal sorting signals, harbor a NEAT domain (near iron transporter; encompassed between residues 32 and 146 of B. anthracis IsdC) (Fig. 1) (1). Staphylococcal surface proteins harboring NEAT domains bind heme-iron (IsdA, IsdB, IsdC, and IsdH) and host hemoproteins, e.g., hemoglobin (IsdB) and haptoglobin (HarA/IsdH) (12, 24). Sortases recognize surface protein substrates by cleaving pentapeptide motif sequences within sorting signals (22). NPKTG, the sortase-motif sequence of B. anthracis IsdC, is similar to that of S. aureus IsdC (NPQTN) and Lmo2186 (NPKSS), each of which is recognized by sortase B in staphylococci and listeria, respectively (2, 16, 25, 30). These observations were integrated into a general hypothesis whereby IsdC may contribute to heme-iron scavenging of B. anthracis by binding heme-iron once the polypeptide is anchored to the cell wall envelope by sortase B.
Sortase B cleaves the NPKTG motif of IsdC.
Sortase B harbors an N-terminal signal peptide/membrane anchor and a C-terminal transpeptidase domain. SrtBΔN, with six-histidyl replacement of the SrtB signal peptide (residues 2 to 37), was purified from E. coli lysate by affinity chromatography (Fig. 2A). A fluorescence-based cleavage assay was used to measure SrtB activity. The fluorescence of 2-aminobenzyl-KTDNPKTGDEA-diaminopropionic acid-dinitrophenyl (a-KTDNPKTGDEA-d) is abolished due to close proximity of the fluorophore (amino benzyl) and quencher (dinitrophenyl). Incubation of a-KTDNPKTGDEA-d with SrtBΔN increased the sample fluorescence sevenfold above background, indicative of in vitro peptide cleavage (Fig. 2B). Incubation of SrtBΔN with MTSET abolished substrate cleavage (Fig. 2B), suggesting that MTSET may form a disulfide with the active-site cysteine of sortase B, a property observed for sortases of S. aureus (41). The activity of MTSET-treated SrtBΔN was restored upon incubation with the strong reductant DTT, indicating that the active-site sulfhydryl is essential for peptide cleavage. Purified SrtAΔN or SrtCΔN, two B. anthracis sortases involved in anchoring surface protein substrates with distinct sorting signals, did not cleave a-KTDNPKTGDEA-d, indicating that NPKTG motif sorting signals are specific substrates for SrtB (Fig. 2B).
FIG. 2.
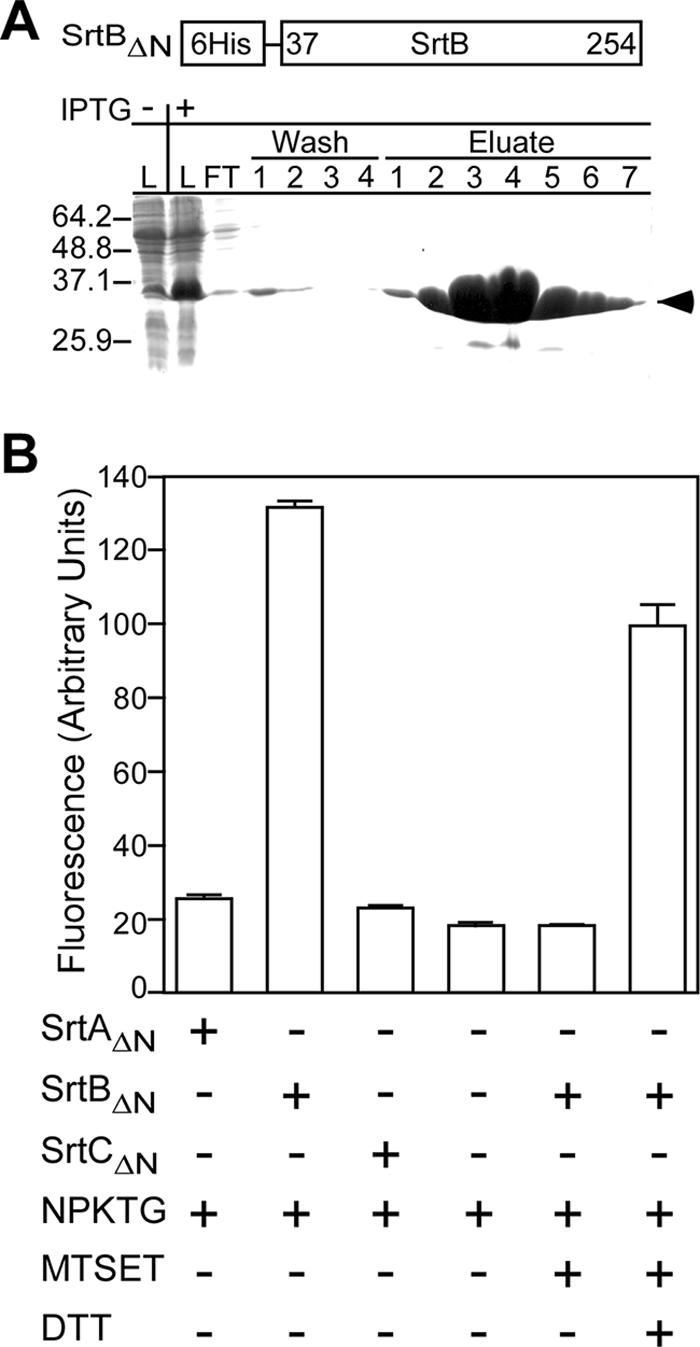
B. anthracis sortase B cleaves the IsdC sorting signal. (A) Following IPTG-induced expression, SrtBΔN was purified from E. coli lysate (L) by affinity chromatography on Ni-nitrilotriacetic acid Sepharose. Flowthrough (FT), wash, and eluate fractions were sampled and analyzed by Coomassie-stained SDS-PAGE. The arrowhead denotes the electrophoretic mobility of SrtBΔN. (B) Purified B. anthracis SrtAΔN, SrtBΔN, and SrtCΔN were incubated with the peptide a-KTDNPKTGDEA-d, and changes in fluorescence (excitation at 320 nm and emission at 420 nm) were quantified by fluorometry. MTSET-mediated inhibition of SrtBΔN cleavage of a-KTDNPKTGDEA-d was restored upon incubation with the reducing agent DTT. The error bars represent the mean and standard deviation derived from three independent experiments.
Sortase B cleaves the NPKTG motif between the threonine and the glycine residues.
Reaction products of SrtBΔN or mock-treated peptide substrate was separated by reversed-phase HPLC, and elution was monitored by absorbance (Fig. 3). Mock-treated peptide eluted at 47 min (67% acetonitrile). MALDI-mass spectrometry (MS) of the eluted compound confirmed the substrate integrity (Table 2). Incubation of the substrate with SrtBΔN generated two product peaks eluting at 35 (49% acetonitrile) and 44 (61% acetonitrile) min. Mass spectrometry of the eluted products identified compounds with m/z of 922.46 (35 min) and 642.20 (44 min) (Fig. 3 and Table 2). These measurements are in agreement with the predicted masses of singly protonated compounds with the structure a-KTDNPKT (m/z 922.00) and GDEA-d (m/z 641.52). Thus, SrtBΔN cleaves the NPKTG signal motif of IsdC between its threonine and glycine residues.
FIG. 3.
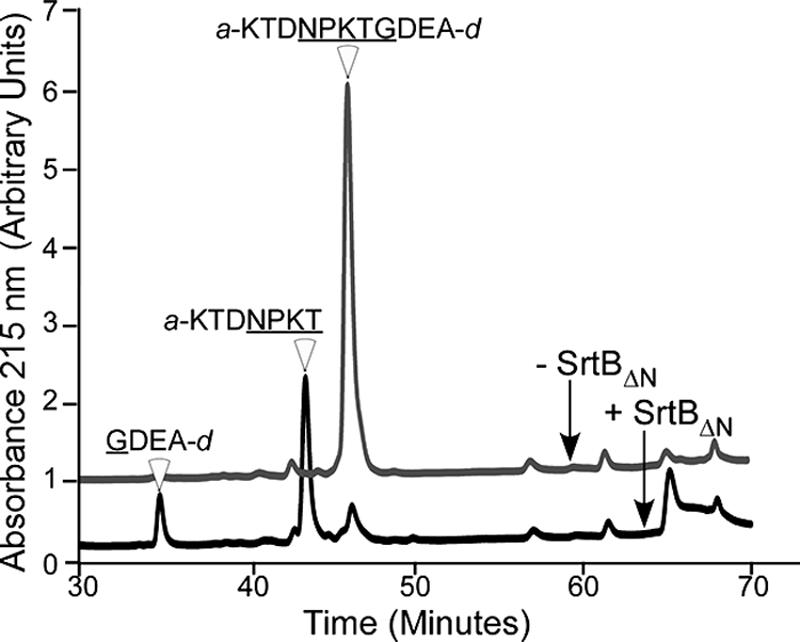
B. anthracis sortase B cleaves between the threonine and the glycine of the NPKTG sorting signal. IsdC substrate peptide, a-KTDNPKTGDEA-d, was incubated in the presence or absence of SrtBΔN; the reaction products were separated by reversed-phase HPLC; and the eluate was monitored via absorbance at 215 nm. The structures of the peptide substrate and product cleavage fragments were determined by MALDI-time of flight MS (Table 2). The data are representative of two independent determinations.
TABLE 2.
Mass spectrometry analysis of HPLC fractions 35, 44, and 47a
| Fraction (min) | Peptide structure | Expected m/z | Observed m/z |
|---|---|---|---|
| 47 | a-KTDNPKTGDEA-d | 1,545.01 | 1,545.62 |
| 44 | a-KTDNPKT | 922.0 | 922.46 |
| 35 | GDEA-d | 641.52 | 642.20 |
HPLC fractions 35, 44, and 47 (Fig. 3) were dried, suspended in 0.1% trifluoracetic acid in H2O, coupled to α-cyano-4-hydroxycinnamic acid, and subjected to MALDI-MS.
IsdC is anchored to the cell wall envelope in an SrtB-dependent manner.
The srtB gene of B. anthracis was replaced by homologous recombination with the antibiotic resistance determinant cat (chloramphenicol acetyltransferase). Southern blotting with srtB and cat DNA probes was used to verify the accuracy of the chromosomal mutation (see Fig. 6A). As expected, the isolated Δ(srtB) strain was resistant to chloramphenicol (data not shown). Plasmid pIsdCH6 provides IPTG-inducible expression of recombinant isdC carrying an in-frame insertion of methionine and six histidine codons (H6) upstream of the NPKTG motif sorting signal. pSrtB-IsdCH6 provides IPTG-inducible expression of srtB and isdCH6 (Fig. 4A). Both plasmids were transformed into the Δ(srtB) mutant strain BAS3. To isolate the B. anthracis cell wall, a new fractionation procedure was developed (Fig. 4B). Briefly, B. anthracis cultures were centrifuged, and the supernatant was removed (medium fraction). The bacterial sediments were boiled in UDS buffer (6 M urea, 5 mM DTT, 1% SDS, 50 mM Tris-HCl, pH 8.0) and centrifuged again. Sample supernatants, harboring SDS-soluble membrane and cytoplasmic proteins (ribosomal protein L6), were removed (lysate fraction). The sediment, containing murein sacculi, was solubilized by digestion with mutanolysin, an N-acetylmuramidase (cell wall fraction). IsdCH6 was found in the cell wall samples of bacilli expressing both srtB and isdCH6, but not in the cell wall of vector control samples or bacilli lacking srtB (Fig. 4C). Two SDS-soluble IsdCH6 species, 30 and 17 kDa, were identified in lysates of bacilli expressing srtB and isdCH6 (Fig. 4C). The 30-kDa species, present only in lysates of bacilli with srtB, migrated more slowly than anchored (mature) IsdCH6, i.e., a mobility of P2 precursors lacking an N-terminal signal peptide but harboring the NPKTG sorting signal at the C-terminal end of a 30-kDa sorting intermediate (the SDS-soluble precursor). IsdCH6 17 kDa is a product of protein degradation, as its abundance increased with sample incubation at room temperature (data not shown). Together, these data suggest that IsdC is anchored to the cell wall of B. anthracis by sortase B.
FIG. 6.
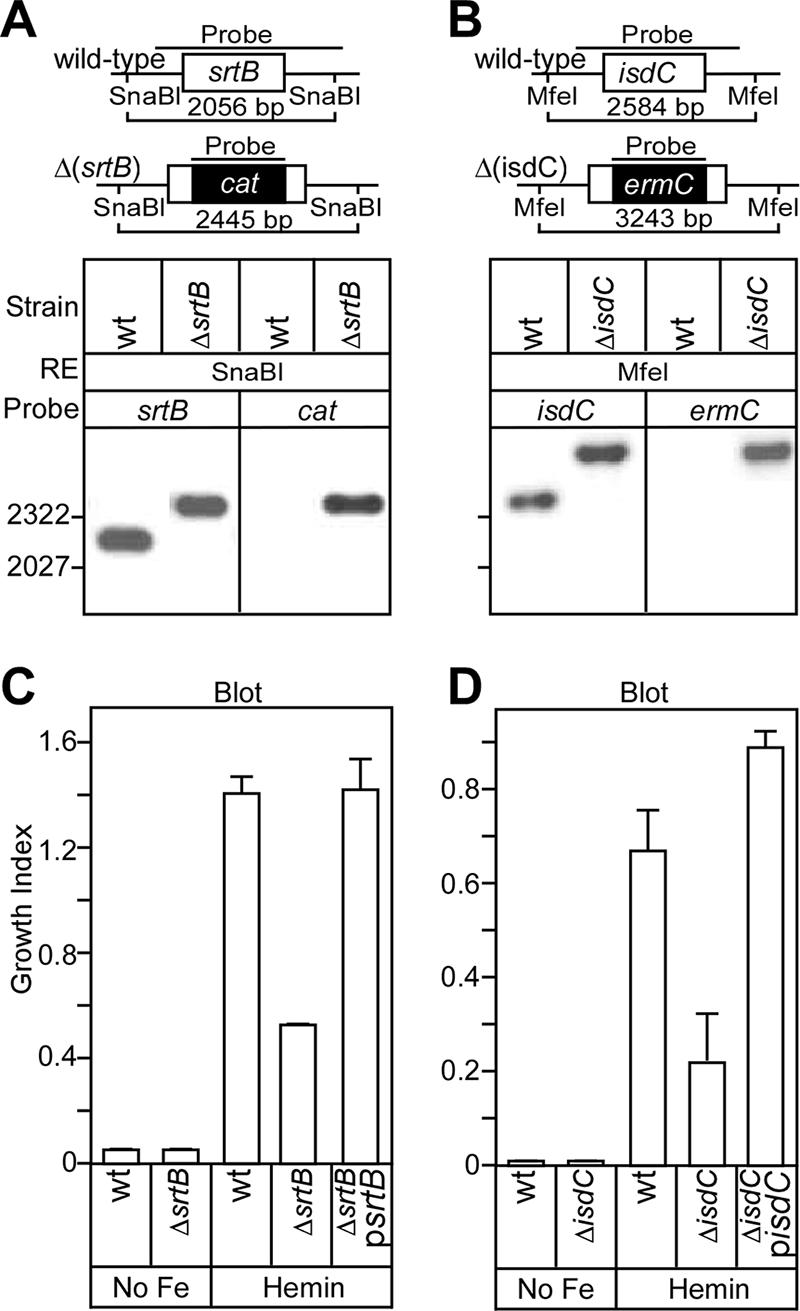
Sortase B and IsdC are required for efficient heme utilization. B. anthracis srtB (A) and isdC (B) were replaced with chloramphenicol (cat) and erythromycin (erm) cassettes, respectively. Following SnaB1 [Δ(srtB)] or MfeI [Δ(isdC)] restriction, Southern blot analysis of wild-type (wt) and mutant [Δ(srtB) and Δ(isdC)] DNAs, as well as antibiotic resistance cassettes (cat and erm), provided confirmation of the deleted genes. (C and D) B. anthracis strains were grown in HIB containing 500 μM 2,2′-dipyridyl with or without hemin (0.5 μM) for 16 h. The OD600 was recorded, and the growth index was quantified (growth in iron-free HIB with or without heme divided by growth in iron-rich HIB). The bars represent the means and standard deviations of three independent determinations.
FIG. 4.
Sortase B-mediated anchoring of IsdC to the cell wall envelope of B. anthracis. (A) Schematic representation of plasmids pIsdCH6, pSrtB-IsdCH6, and pLM5 (parent vector). Pspac indicates the Pspac promoter and kan the kanamycin resistance gene. (B) Plasmids were electroporated into B. anthracis mutants lacking srtB, and cultures of the transformants were fractionated into medium, lysate, and cell wall fractions. (C) Immunoreactive signals for IsdCH6, SrtB, and a cytoplasmic control protein (L6) were generated with antibodies against six-histidyl (Ni-HRP), sortase B (αSrtB), and L6 (αL6), respectively. The electrophoretic mobilities of molecular mass marker proteins, as well as IsdC precursor (P) and anchored (M; mature) species, are indicated. The numbers above the lanes refer to plasmid constructs displayed in panel A. The asterisk marks a degradation product of IsdC. The data are representative of three independent determinations.
IsdC binds heme-iron.
The mature portion of IsdC, residues 32 to 212, including its NEAT domain, was fused to the C-terminal end of GST. GST-IsdC was expressed in E. coli, purified by affinity chromatography using glutathione-Sepharose, and eluted with glutathione (Fig. 5A). Eluate fractions containing abundant amounts of protein were colored red-brown, consistent with the hypothesis that GST-IsdC copurifies with iron-containing compounds (Fig. 5A, inset). Spectral-absorbance profiles of GST-IsdC revealed a Soret band at 412 nm, indicative of protein binding to heme-iron (Fig. 5B) (38). Further, the intensity of GST-IsdC Soret absorbance increased upon addition of hemin. As a control, the spectral properties of purified GST did not reveal a Soret band even when an equivalent amount of hemin was added to the sample. The association of GST-IsdC with hemin was quantified by spectrophotometry. Purified GST-IsdC was incubated with increasing concentrations of hemin (0.1 to 25.6 μM), and the spectral absorbance at 412 nm was recorded (Fig. 5C). GST-IsdC bound hemin in a concentration-dependent manner with a Kd of 3.1 μM (±0.42 μM); saturation of binding occurred at 25 μM hemin. As a control, purified GST did not bind to hemin (data not shown). Together, these observations indicate that B. anthracis IsdC binds heme-iron.
SrtB and IsdC are required for efficient heme-iron acquisition.
The isdC gene of B. anthracis was replaced by homologous recombination with the antibiotic resistance determinant ermC. Southern blotting with isdC and ermC DNA probes was used to verify the accuracy of the chromosomal mutation (Fig. 6B). As expected, the isolated Δ(isdC) strain BAS2 was resistant to erythromycin (data not shown). The B. anthracis wild-type parent, strain Sterne 34F2, and its isogenic variants lacking either srtB or isdC were inoculated into Chelex-treated iron-free HIB with or without 0.5 μM hemin as the sole source of iron. The OD600s of culture samples after 16 h of incubation were indicative of bacterial growth and iron uptake (Fig. 6C and D). Chelex-treated, iron-free HIB did not support the growth of wild-type B. anthracis or the Δ(srtB) or Δ(isdC) strain, corroborating the general hypothesis that B. anthracis absolutely requires iron as a nutrient for growth. The B. anthracis wild-type strain Sterne grew well in iron-free HIB that had been supplemented with hemin, indicating that iron sequestered within heme tetrapyrrol can serve as a source of nutrients and support growth. Isogenic variants lacking either srtB or isdC displayed significant growth defects in iron-free HIB supplemented with hemin. These phenotypes were restored to wild-type levels of bacterial growth upon transformation of the mutant strains with plasmid-borne wild-type alleles of either srtB or isdC (Fig. 6C and D). Collectively, these data suggest that SrtB anchors the heme-binding protein IsdC to the cell wall envelope, a process that is necessary for the acquisition of heme and efficient growth of B. anthracis under iron-limiting conditions.
DISCUSSION
Elemental iron is an essential nutrient for the establishment of many bacterial infections, with bacterial iron uptake proceeding through two types of mechanisms (42). First, receptor proteins capture host factors with iron cargo on the bacterial surface, which is followed by removal and transport of iron across the bacterial envelope. A second strategy involves the bacterial secretion of siderophores, molecules with high affinity for iron (association constants up to 1050), designed for the sole purpose of capturing the nutrient in host tissues. Upon binding to specific transporters, iron-siderophore complexes are eventually imported back into bacterial cells (13, 31). B. anthracis most likely employs both strategies to acquire iron during infection, as its genome encodes several iron acquisition systems, such as two type III hemolysins (32), 15 ferrichrome-type ABC transporters (7, 14, 32), three Fur-type transcriptional regulators (32), two ferritins, and several other iron-binding proteins (28). Petrobactin, a catacholate, is the primary siderophore secreted by B. anthracis during iron starvation, whereas the siderophore bacillibactin represents a minor component (14). Finally, the B. anthracis asb (anthrax siderophore biosynthesis) system secretes anthrachelin, a siderophore that is known to be required for anthrax pathogenesis (7, 15).
B. anthracis isd displays features that are both shared by S. aureus isd and unique. Features shared by B. anthracis isd and S. aureus isd include a sortase (srtB), a ferrichrome transporter (isdD, isdE, and isdF), and a heme monooxygenase (isdG). Fur box consensus sequences indicate ferric uptake repressor-mediated gene regulation (24, 37-39). S. aureus isdA, isdB, and isdH (harA) encode sortase A-anchored surface proteins that bind hemoglobin, heme, or haptoglobin (24, 29); however, these genes are not found in the genome of B. anthracis. Instead, B. anthracis (but not S. aureus) isd encompasses isdX1 and isdX2, whose products contain signal peptides but lack obvious sorting motifs and which may function as secreted polypeptides that capture heme-iron.
Sortase B enzymes from B. anthracis and S. aureus have been crystallized, and their three-dimensional structures have been solved (47). Cys233 of B. anthracis SrtB assumes the same role as Cys194 of S. aureus SrtA—the active-site thiol performs a nucleophilic attack on the amide bond between the threonine (T) and the glycine (G) residues of NPKTG (this work) or NPQTN (25) motif sorting signals. In staphylococci, sortase B-mediated anchoring of IsdC immobilizes the polypeptide within the cell wall envelope (23), a position that may be critical for heme-iron delivery to the membrane transporters, IsdD, IsdE, and IsdF. In B. anthracis, IsdC is anchored to the cell wall by SrtB and purified IsdC binds heme. Further, Δ(srtB) and Δ(isdC) B. anthracis displayed defects in the utilization of heme as a source of iron. Together, these data support a general model whereby cell wall-anchored IsdC is required for B. anthracis scavenging of the essential nutrient iron from the bacterial surface. Consistent with the notion that heme-iron transport is important for pathogenesis in B. anthracis, Zink and Burns reported that srtB mutant bacilli display defects in intracellular replication in macrophages (48).
Genes with homology to isd components have also been identified in Listeria monocytogenes (2, 27). L. monocytogenes isd contains eight genes, including sortase B (srtB; lmo2181) and two presumed sortase B substrate genes, lmo2186 (isdC) and svpA (lmo2185) (27). Sortase B-dependent surface display of SvpA is regulated by Fur (2, 27). Following intravenous infections, L. monocytogenes mutants lacking srtB or svpA display no defect in the ability to either cause lethal infections in mice or replicate in the cytosol of tissue culture cells invaded by listeria (2, 27). However, some isd mutants displayed organ-specific replication defects during murine infections via the oral route, suggesting that a precise function of isd in listerial pathogenesis has not yet been established (27). Similar to S. aureus IsdC (12, 24, 25) and B. anthracis IsdC (this report), L. monocytogenes SvpA also binds hemin. Molecular interactions between L. monocytogenes IsdC and hemin have not yet been reported; however, it seems likely that this polypeptide may also bind hemin. Surprisingly, mutant listeria lacking srtB, srtA, lmo2186, or svpA were not required for iron scavenging when heme or hemoglobin were the sole iron sources in disk diffusion assays on agar media (19, 27). Thus, the physiological role of SvpA binding to hemin requires further investigation.
L. monocytogenes sortases A and B have not yet been purified, and their enzymatic activities remain unknown. Using a proteomic approach, 13 LPXTG motif-type surface proteins were identified in the cell wall compartment of L. monocytogenes EGD-e that were absent from the cell wall compartments of isogenic variants lacking the srtA gene (30). Lmo2186 (IsdC) and Lmo2185 (SvpA) were also identified in the cell wall compartment of wild-type listeria, but not in the cell walls of isogenic srtB mutants (30). Lmo2186 (IsdC) harbors two putative motif sequences (NKVTN and NPKSS), whereas Lmo2185 (SvpA) carries only one such motif sequence (NAKTN). Considering the recognition of specific motif sequences by staphylococcal sortases (25, 41), it seems unlikely that listerial sortase B recognizes more than one sorting signal motif sequence. These observations also suggest that the molecular features of a recognition motif for listerial sortase B and its substrates, cleavage site, and surface protein anchor structure still need to be determined. Further, molecular analysis of an involvement of sortase B in listerial hemin transport may also require knowledge of its polypeptide substrates.
A general hypothesis for the acquisition of heme through the isd-like locus in B. anthracis can be derived from this work. Under iron-limiting conditions, B. anthracis initiates the production of Isd proteins, which presumably constitute a relay system for heme import into bacilli. IsdX1 and IsdX2, NEAT domain-containing proteins with signal peptides, are either secreted or associated with the envelope and may be the most distal components in this relay system. SrtB anchors IsdC to the cell wall via the carboxyl of threonine (NPKTG) and most likely to the free amino group of meso-diaminopimelic acid cross bridges (11, 33). Heme, bound by IsdC, may then be transferred to the iron permease system (IsdD, IsdE, and IsdF), followed by transport across the bacterial membrane. Finally, a monooxygenase, IsdG, degrades heme to liberate iron for use by B. anthracis (37).
Acknowledgments
We thank Christina Tam and Andy Gaspar for plasmids and help with mutagenesis experiments and members of our laboratory for technical advice and discussions. Mass spectrometry was performed in the Mass Spectrometry Core at the University of Chicago.
This work was supported by grants from the National Institutes of Health (AI38897 and AI52474). A.W.M. and O. S. acknowledge membership in and support from the Region V “Great Lakes” Regional Center of Excellence in Biodefense and Emerging Infectious Diseases Consortium (GLRCE; National Institute of Allergy and Infectious Diseases Award 1-U54-AI-057153).
Footnotes
Published ahead of print on 29 September 2006.
REFERENCES
- 1.Andrade, M. A., F. D. Ciccarelli, C. Perez-Iratxeta, and P. Bork. 2002. NEAT: a domain duplicated in genes near the components of a putative Fe3+ siderophore transporter from Gram-positive pathogenic bacteria. Genome Biol. 3:RESEARCH0047. [Online.] http://genomebiology.com/2002/3/9/RESEARCH/0047. [DOI] [PMC free article] [PubMed]
- 2.Bierne, H., C. Garandeau, M. G. Pucciarelli, C. Sabet, S. Newton, F. Garcia-del Portillo, P. Cossart, and A. Charbit. 2004. Sortase B, a new class of sortase in Listeria monocytogenes. J. Bacteriol. 186:1972-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun, V. 2001. Iron uptake mechanisms and their regulation in pathogenic bacteria. Int. J. Med. Microbiol. 291:67-79. [DOI] [PubMed] [Google Scholar]
- 4.Brown, J. S., and D. W. Holden. 2002. Iron acquisition by Gram-positive bacterial pathogens. Microbes Infect. 4:1149-1156. [DOI] [PubMed] [Google Scholar]
- 5.Bullen, J. J. 1981. The significance of iron in infection. Rev. Infect. Dis. 3:1127-1138. [DOI] [PubMed] [Google Scholar]
- 6.Bullen, J. J., and E. Griffiths. 1999. Iron and infection: molecular, physiological and clinical aspects. John Wiley and Sons, New York, N.Y.
- 7.Cendrowski, S., W. MacArthur, and P. Hanna. 2004. Bacillus anthracis requires siderophore biosynthesis for growth in macrophages and mouse virulence. Mol. Microbiol. 51:407-417. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, L. W., D. M. Anderson, and O. Schneewind. 1997. Two independent type III secretion mechanisms for YopE in Yersinia enterocolitica. Mol. Microbiol. 24:757-765. [DOI] [PubMed] [Google Scholar]
- 9.Crichton, R. 2001. Inorganic biochemistry of iron metabolism: from molecular mechanisms to clinical consequences, vol. 2. John Wiley & Sons, Ltd., Chichester, West Sussex, England.
- 10.Crosa, J. H., and C. T. Walsh. 2002. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 66:223-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dhar, G., K. F. Faull, and O. Schneewind. 2000. Anchor structure of cell wall surface proteins in Listeria monocytogenes. Biochemistry 39:3725-3733. [DOI] [PubMed] [Google Scholar]
- 12.Dryla, A., D. Gelbmann, A. von Gabain, and E. Nagy. 2003. Identification of a novel iron regulated staphylococcal surface protein with haptoglobin-haemoglobin binding activity. Mol. Microbiol. 49:37-53. [DOI] [PubMed] [Google Scholar]
- 13.Faraldo-Gomez, J. D., and M. S. Sansom. 2003. Acquisition of siderophores in gram-negative bacteria. Nat. Rev. Mol. Cell. Biol. 4:105-116. [DOI] [PubMed] [Google Scholar]
- 14.Francis, A. W., C. E. Ruggiero, A. T. Koppisch, J. Dong, J. Song, T. Brettin, and S. Iyer. 2005. Proteomic analysis of Bacillus anthracis Sterne vegetative cells. Biochim. Biophys. Acta 1748:191-200. [DOI] [PubMed] [Google Scholar]
- 15.Garner, B. L., J. E. Arceneaux, and B. R. Byers. 2004. Temperature control of a 3,4-dihydroxybenzoate (protocatechuate)-based siderophore in Bacillus anthracis. Curr. Microbiol. 49:89-94. [DOI] [PubMed] [Google Scholar]
- 16.Gaspar, A. H., L. A. Marraffini, E. M. Glass, K. L. DeBord, H. Ton-That, and O. Schneewind. 2005. Bacillus anthracis sortase A (SrtA) anchors LPXTG motif-containing surface proteins to the cell wall envelope. J. Bacteriol. 187:4646-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Genco, C. A., and D. W. Dixon. 2001. Emerging strategies in microbial haeme capture. Mol. Microbiol. 39:1-11. [DOI] [PubMed] [Google Scholar]
- 18.Inglesby, T. V., D. A. Henderson, J. G. Bartlett, M. S. Ascher, E. Eitzen, P. B. Jahrling, J. Hauer, M. Layton, J. McDade, M. T. Osterholm, T. O'Toole, G. Parker, T. Perl, P. K. Russel, and K. Tonat. 1999. Anthrax as a biological weapon: medical and public health management. JAMA 281:2127-2137. [DOI] [PubMed] [Google Scholar]
- 19.Jin, B., S. M. Newton, Y. Shao, X. Jiang, A. Charbit, and P. E. Klebba. 2006. Iron acquisition systems for ferric hydroxamates, haemin and haemoglobin in Listeria monocytogenes. Mol. Microbiol. 59:1185-1198. [DOI] [PubMed] [Google Scholar]
- 20.Koch, R. 1876. Die Átiologie der Milzbrand-Krankheit, begründet auf die Entwicklungsgeschichte des Bacillus anthracis. Beitr. Biol. Pflanz. 2:277-310. [Google Scholar]
- 21.Koppisch, A. T., C. C. Browder, A. L. Moe, J. T. Shelley, B. A. Kinkel, L. E. Hersman, S. Iyer, and C. E. Ruggiero. 2005. Petrobactin is the primary siderophore synthesized by Bacillus anthracis str. Sterne under conditions of iron starvation. Biometals 18:577-585. [DOI] [PubMed] [Google Scholar]
- 22.Marraffini, L. A., A. C. DeDent, and O. Schneewind. 2006. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol. Mol. Biol. Rev. 70:192-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marraffini, L. A., and O. Schneewind. 2005. Anchor structure of staphylococcal surface proteins. V. Anchor structure of the sortase B substrate IsdC. J. Biol. Chem. 280:16263-16271. [DOI] [PubMed] [Google Scholar]
- 24.Mazmanian, S. K., E. P. Skaar, A. H. Gasper, M. Humayun, P. Gornicki, J. Jelenska, A. Joachimiak, D. M. Missiakas, and O. Schneewind. 2003. Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299:906-909. [DOI] [PubMed] [Google Scholar]
- 25.Mazmanian, S. K., H. Ton-That, K. Su, and O. Schneewind. 2002. An iron-regulated sortase enzyme anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc. Natl. Acad. Sci. USA 99:2293-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDevitt, D., P. Francois, P. Vaudaux, and T. J. Foster. 1994. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol. Microbiol. 11:237-248. [DOI] [PubMed] [Google Scholar]
- 27.Newton, S. M., P. E. Klebba, C. Raynaud, Y. Shao, X. Jiang, I. Dubail, C. Archer, C. Frehel, and A. Charbit. 2005. The svpA-srtB locus of Listeria monocytogenes: fur-mediated iron regulation and effect on virulence. Mol. Microbiol. 55:927-940. [DOI] [PubMed] [Google Scholar]
- 28.Papinutto, E., W. G. Dundon, N. Pitulis, R. Battistutta, C. Montecucco, and G. Zanotti. 2002. Structure of two iron-binding proteins from Bacillus anthracis. J. Biol. Chem. 277:15093-15098. [DOI] [PubMed] [Google Scholar]
- 29.Pilpa, R. M., E. A. Fadeev, V. A. Villareal, M. L. Wong, M. Phillips, and R. T. Clubb. 2006. Solution structure of the NEAT (NEAr Transporter) domain from IsdH/HarA: the human hemoglobin receptor in Staphylococcus aureus. J. Mol. Biol. 360:435-447. [DOI] [PubMed] [Google Scholar]
- 30.Pucciarelli, M. G., E. Calvo, C. Sabet, H. Bierne, P. Cossart, and F. Garcia-del Portillo. 2005. Identification of substrates of the Listeria monocytogenes sortases A and B by a non-gel proteomic analysis. Proteomics 5:4808-4817. [DOI] [PubMed] [Google Scholar]
- 31.Ratledge, C., and L. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]
- 32.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baille, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. T. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolsto, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81-86. [DOI] [PubMed] [Google Scholar]
- 33.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmitt, M. P., and E. S. Drazek. 2001. Construction and consequences of directed mutations affecting the hemin receptor in pathogenic Corynebacterium species. J. Bacteriol. 183:1476-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneewind, O., D. Mihaylova-Petkov, and P. Model. 1993. Cell wall sorting signals in surface protein of Gram-positive bacteria. EMBO J. 12:4803-4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schurter, W., M. Geiser, and D. Mathe. 1989. Efficient transformation of Bacillus thuringiensis and B. cereus via electroporation: transformation of acrystalliferous strains with a cloned delta-endotoxin gene. Mol. Gen. Genet. 218:177-181. [DOI] [PubMed] [Google Scholar]
- 37.Skaar, E. P., A. H. Gaspar, and O. Schneewind. 2006. Bacillus anthracis IsdG, a heme degrading monooxygenase. J. Bacteriol. 188:1071-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skaar, E. P., A. H. Gaspar, and O. Schneewind. 2004. IsdG and IsdI, heme degrading enzymes in the cytoplasm of Staphylococcus aureus. J. Biol. Chem. 279:436-443. [DOI] [PubMed] [Google Scholar]
- 39.Skaar, E. P., and O. Schneewind. 2004. Iron-regulated surface determinants (Isd) of Staphylococcus aureus: stealing iron from heme. Microbes Infect. 6:390-397. [DOI] [PubMed] [Google Scholar]
- 40.Sterne, M. 1937. Avirulent anthrax vaccine. Onderstepoort J. Vet. Sci. Anim. Ind. 21:41-43. [PubMed] [Google Scholar]
- 41.Ton-That, H., G. Liu, S. K. Mazmanian, K. F. Faull, and O. Schneewind. 1999. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc. Natl. Acad. Sci. USA 96:12424-12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wandersman, C., and P. Delepelaire. 2004. Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol. 58:611-647. [DOI] [PubMed] [Google Scholar]
- 43.Wandersman, C., and I. Stojiljkovic. 2000. Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores. Curr. Opin. Microbiol. 3:215-220. [DOI] [PubMed] [Google Scholar]
- 44.Wessel, D., and U. I. Flugge. 1984. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138:141-143. [DOI] [PubMed] [Google Scholar]
- 45.Wilks, A. 2002. Heme oxygenase: evolution, structure, and mechanism. Antioxid. Redox Signal. 4:603-614. [DOI] [PubMed] [Google Scholar]
- 46.Wu, R., E. P. Skaar, R. Zhang, G. Joachimiak, P. Gornicki, O. Schneewind, and A. Joachimiak. 2005. Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases. J. Biol. Chem. 280:2840-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, R., R. Wu, G. Joachimiak, S. K. Mazmanian, D. M. Missiakas, P. Gornicki, O. Schneewind, and A. Joachimiak. 2004. Structures of sortase B from Staphylococcus aureus and Bacillus anthracis reveal catalytic amino acid triad in the active site. Structure 12:1147-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zink, S. D., and D. L. Burns. 2005. Importance of srtA and srtB for growth of Bacillus anthracis in macrophages. Infect. Immun. 73:5222-5228. [DOI] [PMC free article] [PubMed] [Google Scholar]