Abstract
CbgA plays a role in cortex formation and the acquisition of a subset of stress resistance properties in Myxococcus xanthus spores. The cbgA mutant produces spores with thin or no cortex layers, and these spores are more sensitive to heat and sodium dodecyl sulfate than their wild-type counterparts.
In nature, biofilms formed by the soil bacterium Myxococcus xanthus feed on prey bacteria to obtain amino acids, which are used as sources of carbon, nitrogen, and energy (3, 5). When starved for amino acids, large groups of M. xanthus cells migrate to aggregation centers and build multicellular fruiting bodies. Rod-shaped cells inside these structures differentiate into spherical spores that are more resistant to sonication, heat, UV irradiation, and toxic substances than vegetatively growing cells (13, 20).
Expression studies suggest that over 500 proteins may be involved in endospore formation in Bacillus subtilis (6, 19, 24). Although it is likely that M. xanthus also uses a large number of proteins to construct a spore, relatively few proteins known to play roles specific to spore development within its fruiting bodies have been identified (8, 10, 11, 15, 16, 18).
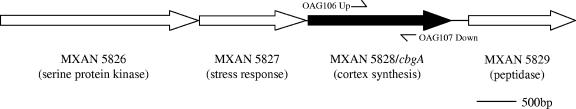
CbgA was first tagged as a potential M. xanthus sporulation protein based upon its sequence similarity (35% identity and 57% similarity) to SpoVR, a B. subtilis protein that plays an important role in the formation of the endospore cortex (2). To examine CbgA's role in sporulation, an insertion in the chromosomal copy of the cbgA gene in wild-type strain DK1622 was created as previously described (4). (Table 1 shows bacterial strains, plasmids, and primers used.) MXAN 5829 is the gene immediately downstream of cbgA (Fig. 1), and DNA sequencing data suggest that cbgA and MXAN 5829 are located in different operons. To confirm that the insertion in cbgA did not have a polar effect on the transcription of MXAN 5829, we used a real-time quantitative reverse transcription-PCR (qRT-PCR) protocol similar to that of Lancero et al. (14). We found that the expression levels of MXAN 5829 during growth in CTTYE (Casitone, Tris-HCl, yeast extract, KH2PO4, MgSO4) broth and development on TPM (Tris-HCl, KH2PO4, MgSO4) agar plates were similar for wild-type cells and the cbgA insertion mutant (data not shown), indicating that the insertion in cbgA did not affect MXAN 5829 expression.
TABLE 1.
Bacterial strains, plasmids and primers
| Strain, plasmid, or primer | Relevant characteristic(s) or sequence | Source or reference |
|---|---|---|
| Strains | ||
| AG840 | pAG331::cbgA (within cbgA) | This study |
| DK1622 | Wild-type motility and development | (12) |
| Plasmids | ||
| pCR2.1-TOPO | Kanr | Invitrogen |
| pAG331 | 621-bp fragment extending from bp 525 to bp 1146 of the cbgA gene | This study |
| Primers | ||
| OAG106 up | 5′-CCGAAGAAGGCCGAGGACGAG-3′ (amplicon size, 621 bp) | Invitrogen |
| OAG107 down | 5′-GAACTCGGGCGTCAGGAACGTG-3′ |
FIG. 1.
Organization of the cbgA locus. Arrows show the locations of the indicated genes and the predicted directions of gene transcription. The potential functions of the proteins encoded by these genes are shown in parentheses. OAG106 up and OAG106 down are the primers used to generate the 621-bp cbgA fragment in plasmid pAG331.
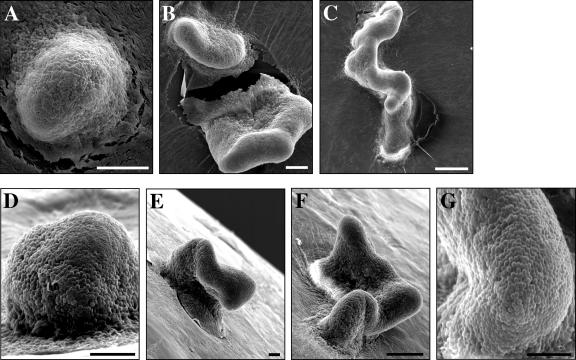
When cbgA and wild-type cells were spotted onto TPM starvation agar and their development was monitored using phase-contrast microscopy (4), we found that cbgA cells were capable of aggregating and forming fruiting bodies with about the same timing as wild-type cells (data not shown). However, scanning electron microscopy (SEM) analysis (22) revealed clear differences in the morphologies of wild-type and cbgA fruiting bodies (Fig. 2). For the most part, wild-type cells yielded the characteristic M. xanthus dome-shaped fruiting bodies (Fig. 2A and D). In contrast, the cbgA mutant formed towers of cells or elongated cell clusters that had snake-like appearances (Fig. 2B, C, E, and F). Thus, it appears that the cbgA mutant forms fruiting bodies with abnormal shapes.
FIG. 2.
High-resolution images of wild-type and cbgA fruiting bodies. SEM analysis was performed on wild-type (DK1622) fruiting bodies (A and D) and on cbgA mutant (AG840) fruiting bodies (B, C, E, F, and G) formed after 5 days of development on TPM agar. Black bars = 15 μm. White bars = 30 μm.
SEM (22) and transmission electron microscopy (25) analyses were used to compare the morphology of cbgA mutant spores to that of wild-type spores (Fig. 2 and 3). Surface-exposed spores in cbgA mutant fruiting bodies (Fig. 2G) were the same size and spherical shape as those found in wild-type fruiting bodies (Fig. 2D). Furthermore, phase-contrast microscopy showed that wild-type and cbgA fruiting bodies had similar numbers of spherical cells (data not shown), indicating that cells in the cbgA fruiting bodies were not undergoing lysis.
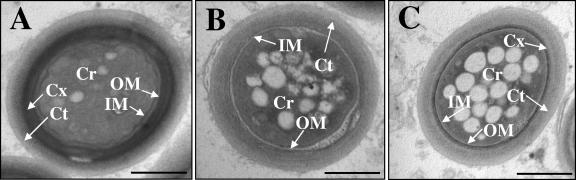
FIG. 3.
Ultrastucture of mature spores. Fruiting bodies were isolated after 5 days of development on TPM agar and analyzed using transmission electron microscopy. (A) A wild-type (DK1622) spore is shown with the core (Cr), inner membrane (IM), outer membrane (OM), cortex (Cx), and coat (Ct) labeled. Spores produced by the cbgA mutant (AG840) either lack the electron-dense cortex layer (B) or have relatively thin cortex layers (C). Bars = 500 nm.
Transmission electron micrographs of wild-type DK1622 spores revealed all of the structural features previously described for M. xanthus spores (10, 23). These structures include a spore core, an inner membrane and an outer membrane, a cortex, and an exterior spore coat (Fig. 3A). The cbgA mutant produced spores that lacked cortexes (Fig. 3B) or spores that had relatively thin cortex layers (Fig. 3C). The outer protective coat that normally surrounds the cortex, however, was indistinguishable in cbgA spores and wild-type spores.
The B. subtilis spore coat provides a protective barrier against bactericidal enzymes such as lysozyme (9), and mutations in cotE, spoVID, or yrbA, which affect coat assembly, render spores lysozyme sensitive (1, 21, 26). Presumably, the compromised spore coat in the mutant spores allows lysozyme to gain access to the peptidoglycan in the cortex. To confirm that the coats of the cbgA spores were intact, we exposed cbgA and wild-type spores to 250 μg/ml of lysozyme for 12 h as previously described (17, 21). The yield of viable, lysozyme-resistant cbgA mutant spores was similar to that of wild-type spores (data not shown). In addition, using the procedure of Inouye et al. (10), we were able to extract the major M. xanthus coat protein, protein S, from the surfaces of cbgA spores (data not shown). Taken together, our findings indicate that the coats of cbgA mutant spores are likely to be intact. These data also suggest that an intact cortex is not absolutely required for M. xanthus spores to maintain their characteristic shape, nor is it essential for synthesis of the spore coat.
A mutation in the spoVR gene of B. subtilis, which affects production of the endospore cortex, results in a 3- to 10-fold decrease in heat resistance and resistance to toxic chemicals such as chloroform (2). To examine whether the stress resistance properties of cbgA mutant spores were compromised, cells that had developed on TPM agar plates for 5 days were harvested. Assays for resistance to sonication, heat, and UV irradiation were performed as previously described by Sudo and Dworkin (20), except that samples were sonicated before and after treatments (three 10-s bursts with 1-min cooling periods at room temperature between bursts) to disperse the cells. To determine spore resistance to sodium dodecyl sulfate (SDS), harvested cells were exposed to 1.0% SDS and incubated for 1 or 2 h at room temperature on an agitating orbital shaker. Counts of viable spores were determined as previously described (20).
The numbers of cbgA mutant spores that were viable after sonication or up to 10 min of exposure to UV irradiation were similar to those of wild-type spores (data not shown). However, cbgA mutant spores displayed a greater degree of sensitivity to temperatures of 50°C and 55°C than wild-type spores. As shown in Table 2, the number of cbgA spores that survived 50°C treatment was about 12-fold lower than that of wild-type spores, and the number of cbgA spores that survived 55°C heat treatment was about 30-fold lower than that of wild-type spores. The cbgA spores displayed a higher degree of SDS sensitivity than their wild-type counterparts; the number of cbgA spores that survived 1 and 2 h of exposure to SDS was about 50-fold lower than that of wild-type spores.
TABLE 2.
Resistance of wild-type and cbgA mutant spores to heat and SDS
| Strain | Viable spores followinga:
|
|||
|---|---|---|---|---|
| Heat treatment at:
|
SDS treatment at:
|
|||
| 50°C, 2 h | 55°C, 2 h | 1% SDS, 1 h | 1% SDS, 2 h | |
| DK1622 (wild type) | 100.0 ± 13.2 | 100.0 ± 16.9 | 100.0 ± 27.1 | 100.0 ± 16.6 |
| AG840 (cbgA) | 8.1 ± 4.1 | 3.4 ± 1.2 | 2.4 ± 1.1 | 2.0 ± 1.3 |
Medium recipes and procedures for developmental assays are described in the report of Caberoy et al. (4). The indicated spore resistance assays were performed at least three times for each strain. The mean values ± standard deviations for the assays are shown as percentages of the results for DK1622 (wild type). The number of wild-type spores that survived heat treatments ranged from 3 × 106 to 5 × 106, and the number that survived SDS treatments ranged from 6 × 105 to 1 × 106.
This study reports the finding that the cbgA gene product is necessary for proper cortex development in M. xanthus fruiting body spores. In addition, our results suggest that the cbgA gene product and an intact cortex are important for some, but not all, of the resistance properties normally associated with M. xanthus spores. The correlation between cortex defects and heat sensitivity was detected in previous studies of B. subtilis spore cortex mutants (reviewed in reference 7). Our results also suggest that the M. xanthus spore cortex plays an important role in resistance to SDS and perhaps other detergents.
Acknowledgments
We are thankful to Monsanto Company and TIGR for giving us access to the M. xanthus genome sequence prior to submission to GenBank (accession number CP000113).
This research was supported by National Science Foundation award MCB 031674 to J. Dahl and A. Garza.
Footnotes
Published ahead of print on 22 September 2006.
REFERENCES
- 1.Beall, B., A. Driks, R. Losick, and C. P. Moran, Jr. 1993. Cloning and characterization of a gene required for assembly of the Bacillus subtilis spore coat. J. Bacteriol. 175:1705-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beall, B., and C. P. Moran, Jr. 1994. Cloning and characterization of spoVR, a gene from Bacillus subtilis involved in spore cortex formation. J. Bacteriol. 176:2003-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bretscher, A. P., and D. Kaiser. 1978. Nutrition of Myxococcus xanthus, a fruiting myxobacterium. J. Bacteriol. 133:763-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caberoy, N. B., R. D. Welch, J. S. Jakobsen, S. C. Slater, and A. G. Garza. 2003. Global mutational analysis of NtrC-like activators in Myxococcus xanthus: identifying activator mutants defective for motility and fruiting body development. J. Bacteriol. 185:6083-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dworkin, M. 1962. Nutritional requirements for vegetative growth of Myxococcus xanthus. J. Bacteriol. 84:250-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eichenberger, P., M. Fujita, S. T. Jensen, E. M. Conlon, D. Z. Rudner, S. T. Wang, C. Fergusun, K. Haga, T. Sato, J. S. Liu, and R. Losick. 2004. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2:1664-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellar, D. J. 1978. Spore specific structures and their function. Symp. Soc. Gen. Microbiol. 28:295-334. [Google Scholar]
- 8.Gollop, R., M. Inouye, and S. Inouye. 1991. Protein U, a late-developmental spore coat protein of Myxococcus xanthus, is a secretory protein. J. Bacteriol. 173:3597-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gould, G. W. 1983. Mechanisms of resistance and dormancy, p. 173-210. In A. Hurst and G. W. Gould (ed.), The bacterial spore, vol. 2. Academic Press, London, United Kingdom. [Google Scholar]
- 10.Inouye, M., S. Inouye, and D. R. Zusman. 1979. Biosynthesis and self-assembly of protein S, a development specific protein of Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 76:209-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inouye, S., Y. Ike, and M. Inouye. 1983. Tandem repeat of the genes for protein S, a development-specific protein of Myxococcus xanthus. J. Biol. Chem. 258:38-40. [PubMed] [Google Scholar]
- 12.Kaiser, D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 76:5952-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kottel, R. H., K. Bacon, D. Clutter, and D. White. 1975. Coats from Myxococcus xanthus: characterization and synthesis during myxospore differentiation. J. Bacteriol. 124:550-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lancero, H., N. B. Caberoy, S. Casteneda, Y. Li, A. Lu, D. Dutton, X. Y. Duan, H. B. Kaplan, W. Shi, and A. G. Garza. 2004. Characterization of a Myxococcus xanthus mutant that is defective for adventurous motility and social motility. Microbiology 150:4085-4093. [DOI] [PubMed] [Google Scholar]
- 15.Licking, E., L. Gorski, and D. Kaiser. 2000. A common step for changing cell shape in fruiting body and starvation-independent sporulation of Myxococcus xanthus. J. Bacteriol. 182:3553-3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCleary, W. R., B. Esmon, and D. R. Zusman. 1991. Myxococcus xanthus protein C is a major spore surface protein. J. Bacteriol. 173:2141-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 18.Otani, M., K. Satoshi, X. Chunying, C. Umezawa, K. Sano, and S. Inouye. 1998. Protein W, a spore-specific protein in Myxococcus xanthus, formation of a large electron-dense particle in a spore. Mol. Microbiol. 30:57-66. [DOI] [PubMed] [Google Scholar]
- 19.Steil, L., M. Serrano, A. O. Henriques, and U. Volker. 2005. Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis. Microbiology 151:399-420. [DOI] [PubMed] [Google Scholar]
- 20.Sudo, S. Z., and M. Dworkin. 1969. Resistance of vegetative cells and microcysts of Myxococcus xanthus. J. Bacteriol. 98:883-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takamatsu, H., T. Kodama, T. Nakayama, and K. Watabe. 1999. Characterization of the yrbA gene of Bacillus subtilis, involved in resistance and germination of spores. J. Bacteriol. 181:4986-4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vasquez, G. M., F. Qualls, and D. White. 1985. Morphogesis of Stigmatella aurantiaca fruiting bodies. J. Bacteriol. 163:515-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voelz, H., and M. Dworkin. 1962. Fine structure of Myxococcus xanthus during morphogenesis. J. Bacteriol. 84:943-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang, S. T., B. Setlow, E. M. Conlon, J. L. Lyon, D. Imamura, T. Sato, P. Setlow, R. Losick, and P. Eichenberger. 2006. The forespore line of gene expression in Bacillus subtilis. J. Mol. Biol. 358:16-37. [DOI] [PubMed] [Google Scholar]
- 25.White, D. J., and P. L. Hartzell. 2000. AglU, a protein required for gliding motility and spore maturation of Myxococcus xanthus, is related to WD-repeat proteins. Mol. Microbiol. 36:662-678. [DOI] [PubMed] [Google Scholar]
- 26.Zheng, L. B., W. P. Donovan, P. C. Fitz-James, and R. Losick. 1988. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev. 2:1047-1054. [DOI] [PubMed] [Google Scholar]