Abstract
The ability to form biofilms in the airways of people suffering from cystic fibrosis is a critical element of Pseudomonas aeruginosa pathogenesis. The 15-gene psl operon encodes a putative polysaccharide that plays an important role in biofilm initiation in nonmucoid P. aeruginosa strains. Biofilm initiation by a P. aeruginosa PAO1 strain with disruption of pslA and pslB (ΔpslAB) was severely compromised, indicating that psl has a role in cell-surface interactions. In this study, we investigated the adherence properties of this ΔpslAB mutant using biotic surfaces (epithelial cells and mucin-coated surfaces) and abiotic surfaces. Our results showed that psl is required for attachment to a variety of surfaces, independent of the carbon source. To study the potential roles of Psl apart from attachment, we generated a psl-inducible P. aeruginosa strain (Δpsl/pBAD-psl) by replacing the psl promoter region with araC-pBAD, so that expression of psl could be controlled by addition of arabinose. Analysis of biofilms formed by the Δpsl/pBAD-psl strain indicated that expression of the psl operon is required to maintain the biofilm structure at steps postattachment. Overproduction of the Psl polysaccharide led to enhanced cell-surface and intercellular adhesion of P. aeruginosa. This translated into significant changes in the architecture of the biofilm. We propose that Psl has an important role in P. aeruginosa adhesion, which is critical for initiation and maintenance of the biofilm structure.
Pseudomonas aeruginosa is an important opportunistic human pathogen that can cause life-threatening infections in cystic fibrosis (CF) patients and individuals with a compromised immune system. This environmental bacterium is capable of living planktonically or in surface-associated communities known as biofilms. P. aeruginosa biofilms can form on a variety of surfaces, including in mucus plugs of the CF lung and abiotic surfaces, such as contact lenses and catheters (8, 15, 30, 36). Bacteria growing in biofilms are less susceptible to antimicrobial agents and are protected from the host immune response, giving rise to chronic infections that are notoriously difficult to eradicate (9, 13, 27). Bacteria growing in biofilms produce one or more extracellular polymeric matrices that act as a scaffold, holding the cells of the biofilm community together. Polysaccharides are key components of the biofilm matrix, as they contribute to the overall biofilm architecture and to the resistance of bacteria in biofilms (5, 26, 29).
Biofilm development is a sequential process initiated by the attachment of planktonic cells to a surface, which is followed by formation of microcolonies and biofilm maturation in which individual bacteria, as well as the entire community, are embedded in a matrix composed of nucleic acid, protein, and polysaccharides (2, 5, 30, 34). Two potential polysaccharide biosynthetic loci, psl (PA2231 to PA2245) and pel (PA3058 to PA3064) of P. aeruginosa have been identified as loci that play important roles in biofilm initiation and formation in nonmucoid P. aeruginosa strains (11, 12, 20, 22, 29, 32, 41). The psl cluster contains 15 cotranscribed genes (pslA to pslO) encoding proteins predicted to be involved in polysaccharide biosynthesis. Overhage and colleagues recently mapped the psl operon promoter 41 bp upstream of the pslA start codon (32). Their data also suggested that psl expression was localized to the centers of microcolonies within biofilms (32). In addition, Kirisits et al. showed that the expression of psl and pel was elevated in variants isolated from aging P. aeruginosa PAO1 biofilms (22). It has recently been suggested that the mechanistic basis for psl and pel overproduction in these variants, as well as other autoaggregative variants, involves elevated levels of the intracellular signal cyclic diguanylate (c-diGMP). P. aeruginosa has several loci capable of modulating the c-diGMP level, including the wsp, LadS, and retS signal transduction systems (14, 18, 24, 42).
So far, it is not clear whether psl plays any role in biofilm formation beyond the initial adhesion. In the present study, we generated a psl-inducible strain by replacing the psl promoter with an araC-pBAD cassette and utilized this strain to address this issue. We also investigated the adherence properties of a ΔpslAB mutant on both biotic surfaces (epithelial cells and mucin-coated surfaces) and abiotic surfaces. Our results show that Psl polysaccharide plays an important role in P. aeruginosa adhesion by promoting cell-surface and intercellular interactions to initiate biofilms and maintain biofilm structure postattachment. Our results also indicate that the Psl polysaccharide is a critical component of the biofilm matrix, which functions as a scaffold, holding biofilm cells together.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. aeruginosa PAO1, its isogenic ΔpslAB mutant WFPA60 (20), the psl promoter deletion mutant WFPA800 (Δpsl), and the psl-inducible strain WFPA801 (Δpsl/pBAD-psl) were used in this study. For confocal laser scanning microscopy (CLSM), PAO1, WFPA60, WFPA800, and WFPA801 harboring pMRP9 (a plasmid expressing green fluorescent protein [GFP] constitutively) (8) were used. Plasmid pMRP9 was transferred to P. aeruginosa by transformation. Escherichia coli strain JM109 was used for all cloning, while SM10 was used to transfer plasmids to P. aeruginosa by conjugation. Unless otherwise indicated, E. coli strains were grown in Luria broth (LB), and P. aeruginosa was cultured in LB lacking sodium chloride or Jensen's medium, a chemically defined medium (21). Glucose was used as the carbon source in Jensen's medium. To obtain the data in Table 1, Jensen's medium was supplemented with a variety of carbon sources (0.4%, wt/vol). To prepare cultures for the rapid biofilm assay, overnight cultures in Jensen's media with the appropriate carbon sources were diluted 50-fold into similar Jensen's media, and the cultures were grown to the mid-log phase. Antibiotics were used at following concentrations: for E. coli, 100 μg/ml carbenicillin and 10 μg/ml gentamicin; and for P. aeruginosa, 300 μg/ml carbenicillin and 100 μg/ml gentamicin. Carbenicillin (150 μg/ml) was added to the media used for the flow cell system.
TABLE 1.
pslAB mutant WFPA60 is deficient in attachment to abiotic surfaces under a variety of conditions
| Carbon source | Crystal violet absorbance (A540)a
|
|
|---|---|---|
| PAO1 (wild type) | WFPA60 (ΔpslAB) | |
| Glucose | 0.142 ± 0.043 | 0.015 ± 0.002 |
| Mannose | 0.339 ± 0.069 | 0.038 ± 0.005 |
| Glycerol | 0.197 ± 0.056 | 0.016 ± 0.001 |
| Glutamate | 0.222 ± 0.005 | 0.015 ± 0.001 |
| Citrate | 0.144 ± 0.018 | 0.019 ± 0.002 |
Results of a rapid microtiter dish biofilm assay performed with cells grown in Jensen's chemically defined medium supplemented with different carbon sources.
Construction of WFPA800 and WFPA801.
WFPA800 and WFPA801 were constructed by using an unmarked, nonpolar deletion strategy described previously (19, 40). For construction of the psl promoter deletion mutant WFPA800, plasmid pMA8 was generated by cloning a 1.3-kb DNA fragment into the EcoRI and HindIII sites of pEX18Gm (19). The 1.3-kb fragment was obtained by overlapping PCR and ligation of a DNA fragment located at positions to −912 to −267 with a fragment at positions −54 to 591 relative to pslA. Unique SacI and KpnI sites were designed between these two fragments. Plasmid pMA8 was transferred to PAO1 by mating, and a double-crossover recombinant was isolated as previously described (19). This generated strain WFPA800, in which the promoter and 5′ regulatory sequences of the psl operon (positions −54 to −267 upstream of pslA) were deleted. To create the psl-inducible strain WFPA801, pMA9 was constructed by subcloning a fragment from pSW196 (3) containing araC-pBAD into the SacI and KpnI sites of pMA8. Plasmid pMA9 was transferred into PAO1, and a double-crossover recombinant was selected, resulting in WFPA801. In this derivative, the psl promoter and 5′ regulatory region were replaced by araC-pBAD so that Psl expression could be induced by addition of arabinose to the growth media. For both WFPA800 and WFPA801 the manipulations were validated by sequencing a fragment derived from PCR amplification of the DNA adjacent to pslA (data not shown).
Microtiter dish biofilm assay.
For the rapid attachment assays, 100 μl of a culture (optical density at 600 nm [OD600], ∼0.5) was added to wells of a microtiter dish (Falcon 3911). After incubation at room temperature for 30 min or 1 h, the planktonic and loosely adherent bacteria cells were washed off, and surface-attached cells were stained by addition of 0.1% crystal violet, solubilized in ethanol, and measured (A540) as described previously (20, 31). For the 8-h adherence assay, a 1/100 dilution of a saturated (overnight) culture was inoculated into wells of a microtiter dish; after 8 h of incubation at 30°C, the wells were washed, and surface-attached cells were stained and detected as described above.
Flow cell system and confocal laser scanning microscopy.
Unless otherwise indicated, biofilms were grown at room temperature in flow chambers with individual channel dimensions of 1 by 4 by 40 mm (Stovall Life Science, Inc.). The flow chambers were inoculated by injecting 200 μl of a mid-log-phase culture (OD600, 0.5; ∼107 cells) into each flow channel with a 1.0-ml syringe. After inoculation, the flow cells were left inverted for 1 h to allow bacterial cells to attach to the glass coverslips, and then each flow cell was turned upright and the flow was initiated (Jensen's medium; flow rate, 0.7 ml/min). All microscopic observations and image acquisition were performed with a Zeiss 510 CLSM (Carl Zeiss, Jena, Germany). Images were obtained using a 63×/1.3 water objective. The software included with the Zeiss LSM510 generated three-dimensional images and sections. CLSM-captured images were subjected to quantitative image analysis using the COMSTAT software (17).
Adherence of P. aeruginosa to mucin-coated surfaces and epithelial cells.
Respiratory mucin (Sigma) was covalently bound to glass coverslips as described previously (25). The presence and uniformity of the mucin on the coverslips were verified by atomic force microscopy. PAO1 and WFPA60 (ΔpslAB) were cultured in Jensen's medium (21) in a flowthrough biofilm culturing system at 30°C (6, 20, 25). For visualization, each strain was tagged with the GFP-containing plasmid pMRP9. Biofilm growth was assessed after 36 h with a Zeiss LSM 510 confocal laser scanning microscope. For adherence to epithelial cells, monolayers were cultured until they were confluent, washed twice with Dulbecco's phosphate-buffered saline (PBS) (Gibco), and then infected with ∼5.0 × 107 cells (A600, 0.7) for 1 h. One set of wells was washed five times with PBS, and then 0.1% Triton X-100 was added to disrupt epithelial cells. Triton X-100 (0.1%) was added to parallel infected monolayers to account for bacterial cell growth during the 1-h infection. The wells were scraped and vigorously mixed to disrupt aggregated cells, and the contents were serially diluted for viable cell counting. Each experiment was performed in triplicate, and the results are reported below as means of at least three experiments. A t test was used to determine the significance of the adhesion data. The equality-of-variance assumption was verified, eliminating the need for a nonparametric analysis of the data (38).
Congo red binding assay.
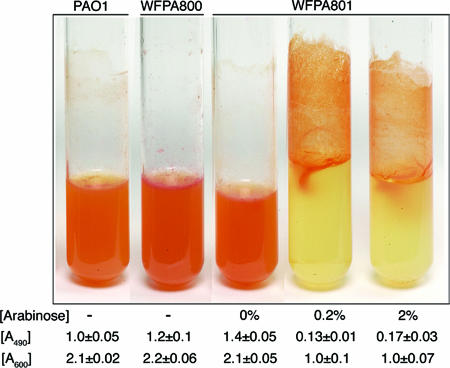
For liquid Congo red (CR) assays (see Fig. 3), 150 μl of a culture (OD600, ∼1.0) was inoculated into 5.0 ml Jensen's minimal medium with or without arabinose containing 40 μg/ml CR. The samples were incubated with agitation overnight at 37°C. The following day, images were obtained for all samples, and a 1.0-ml culture was removed in order to determine the OD600 before the bacterial cells were pelleted by centrifugation at 14,000 rpm. For quantification of CR binding, the A490 of the supernatant of each sample was determined. Each strain was assayed in triplicate to generate a mean and standard error.
FIG. 3.
Increased Psl production enhances Congo red staining and cell-surface interactions: CR binding assay of strains PAO1, WFPA800 (Δpsl), and WFPA801 (Δpsl/pBAD-psl) grown in liquid culture. The arabinose concentration, A600 of the culture, and A490 of the supernatant of the culture are indicated below each tube. The A490 values indicate the amount of CR dye retained in the supernatant following absorption to cells (i.e., the amount of CR dye that did not bind to the bacteria; lower numbers indicate more CR bound to the cells). The A600 values are the approximate numbers of cells remaining in suspension (not attached on the walls).
Isolation of total RNA and real-time RT-PCR.
Total RNA was purified from a 10.0-ml mid-log-phase culture grown in Jensen's medium using Ambion RNA isolation reagents (Ambion). TaqMan one-step reverse transcription (RT)-PCR Master Mix reagents (Roche) were used for PCR with 100 ng of total template RNA. Real-time PCR was performed with an ABI Prism 7000 sequence detection system (Applied Biosystem) using the following primers and probes: pslB forward primer, 5′-GCATGCCGAAACCCTTCA-3′; pslB reverse primer; 5′-GCGATACGCAGGAAGGTCTT-3′; and pslB probe, 5′-(DFAM)TGGCCGATGACCAGAGCCTGT(DTAM)-3′ (DFAM, 6-carboxyfluorescein; DTAM, tetramethylcarboxyrhodamide). Serially diluted genomic DNA was used as the template to obtain a relative standard curve. The constitutively expressed rpoD gene was used as an endogenous control to normalize quantification of the mRNA target. Real-time PCR data were analyzed, validated, and calculated according to the instructions of the manufacturer.
RESULTS
Adherence of ΔpslAB mutant with abiotic and biotic surfaces.
Previous studies showed that biofilm initiation by nonmucoid P. aeruginosa strains with mutations in the psl genes was deficient, most likely due to the reduced ability of these mutants to interact with cell surfaces (12, 20, 29, 32). The attachment assays in these studies were performed with a rich medium, and it was unclear if Psl contributed to cell-surface interactions when bacteria were grown under other conditions. Since the carbon source (as an environmental signal) can regulate biofilm development (31) and the Psl polysaccharide is reportedly mannose rich (12), we compared the initial attachment of wild-type strain PAO1 and the initial attachment of strain WFPA60 (PAO1ΔpslAB) grown in Jensen's chemically defined medium supplemented with mannose or a variety of other carbon sources previously used to investigate effects on biofilm formation (31). In this assay, similar numbers of organisms derived from mid-log-phase cultures were inoculated into wells of a microtiter dish, the bacteria were allowed to attach for 30 min, the wells were washed, and the cells attached to the surface were stained with crystal violet (31). Although the levels of attachment varied for PAO1 when it was grown with different carbon sources, the adherence of WFPA60 was consistently approximately 10-fold less than the adherence of PAO1 (Table 1). These results indicated that the cell surface attachment defect of psl mutants was independent of the carbon source used. Previous studies demonstrated that PAO1 and WFPA60 have similar liquid culture growth rates, suggesting that the observed differences were not due to growth (20).
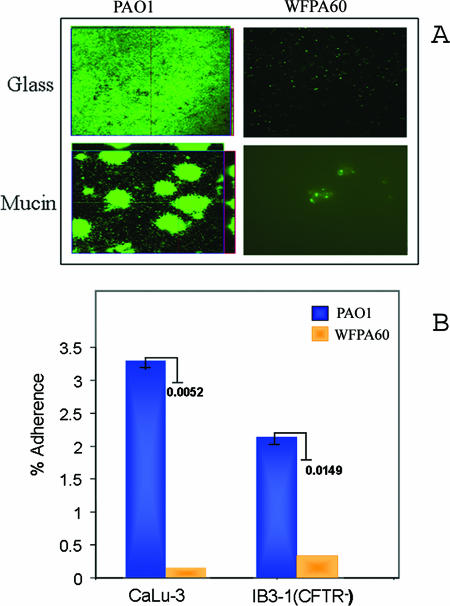
Since WFPA60 consistently exhibited an adherence defect on abiotic surfaces, we next tested whether this was true on biotic surfaces relevant to CF. Mucins, a large group of glycoproteins, are some of the major components of mucus, which covers the luminal surfaces of epithelial organs, including the lung (35). Therefore, mucin-coated surfaces are a physiologically and clinically relevant system with which to observe biofilm formation. We hypothesized that the initiation of biofilm formation by WFPA60 would be impaired on a mucin-coated surface. To examine this possibility, we utilized a system developed to observe biofilm formation in real time on mucin-coated glass coverslips (25). The mucin-coated coverslips were placed under continuous-flow culture conditions, and identical numbers of GFP-tagged cells of PAO1 and WFPA60 were inoculated into the system. The biofilms formed on the mucin-coated surfaces were visualized using CLSM. As a control, a non-mucin-coated coverslip was treated identically. As observed previously under these growth conditions, at 36 h PAO1 formed a dense but flat, uniform biofilm on the glass surface. On the mucin-coated coverslip, PAO1 formed highly structured mound-shaped cell clusters that were unevenly distributed on the surface (Fig. 1A). In contrast, there was minimal attachment of WFPA60 to either the glass or mucin-coated coverslips (Fig. 1A).
FIG. 1.
Adherence of ΔpslAB mutant to abiotic and biotic surfaces. (A) Qualitative analysis of PAO1 and WFPA60 (ΔpslAB) biofilm populations on glass and mucin-coated surface. Respiratory mucin was covalently bound to glass coverslips as previously described (25). Strains harboring pMRP9 were cultured on glass or mucin-coated glass coverslips in Jensen's medium in a flowthrough culture system. At 36 h postinoculation, images were acquired by CLSM. (B) Adherence of PAO1 and WFPA60 (ΔpslAB) to epithelial cells. Each strain was allowed to adhere to epithelial cell lines (multiplicity of infection, 10) for 1 h, and bacteria were recovered and enumerated by viable cell counting. The level of adherence was calculated by dividing the number of bacteria recovered by the initial number of bacteria used for adherence. The results are expressed as percentages of adherence to epithelial cells. P values are indicated next to the bars.
We also compared the attachment of PAO1 and the attachment of WFPA60 to respiratory epithelial cells. For these studies we utilized CaLu-3 cells, a serous mucin-overproducing tracheal epithelial cell line from a Caucasian male, and IB3-1 cells, an immortalized cell line created from a primary culture of bronchial epithelial cells from a CF patient (CFTR−). PAO1 and WFPA60 were allowed to adhere to the epithelial cells for 1 h (multiplicity of infection, 10). The cells were washed, and bacteria were recovered and enumerated by viable cell counting in order to quantify adherence. We found that with CaLu-3 cells, 3.3% of the initially deposited PAO1 population adhered, while the level of adherence of WFPA60 was more than 10-fold less than this level (0.3%) (P = 0.0052) (Fig. 1B). Although not quite as marked, a difference in surface adherence was also observed with the cftr-deficient IB3-1 cell line (Fig. 1B). These results were particularly striking because WFPA60 had an adherence defect similar to that of an rpoN mutant, a strain that lacks two recognized and well-studied P. aeruginosa adhesins (flagella and type IV pili) (data not shown).
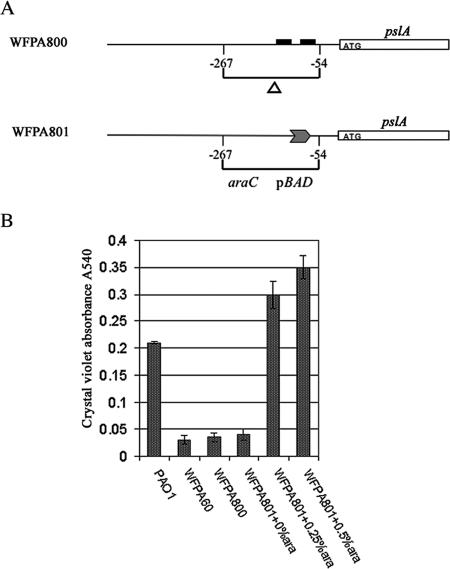
Generation of a P. aeruginosa psl-conditional strain.
The results described above and previous work (12, 20, 29, 32) demonstrated that the Psl polysaccharide promotes attachment to both abiotic and biotic surfaces. We wanted to determine if Psl contributes to biofilm formation or architecture after surface adhesion and to evaluate the consequences of Psl overexpression. Two strains, WFPA800 and WFPA801, were constructed to address this question (Fig. 2A). Strain WFPA800 (Δpsl) was constructed by deleting psl regulatory sequences located at positions −54 to −267 relative to the pslA translation start codon (Fig. 2A). Since this deletion removed the mapped psl promoter and 5′ regulatory sequences (32), WFPA800 was expected to have negligible psl expression. WFPA801 (Δpsl/pBAD-psl) was a psl-inducible strain in which the pBAD promoter (16) replaced the promoter region of psl. In this strain, psl expression should have been induced by addition of arabinose to the growth medium. To evaluate WFPA800 and WFPA801, we examined them using a 30-min rapid attachment microtiter dish assay (Fig. 2B). In this assay, WFPA800 behaved like WFPA60 (ΔpslAB). In the absence of arabinose, the attachment of WFPA801 was severely compromised, similar to the results obtained for WFPA800 and WFPA60. However, after addition of arabinose, the attachment of WFPA801 exceeded the attachment observed with the wild-type PAO1 strain. The differences were not due to alterations in the growth rates since all strains exhibited similar growth kinetics in either rich media or Jensen's media with or without arabinose (data not shown). The differences were also not a result of changes in motility (data not shown). Since under psl-inducing conditions WFPA801 formed a biofilm similar to the biofilm formed by wild-type strain PAO1 (Fig. 2B) (see below), this indicates that the biofilm-deficient phenotype of a Δpsl strain can be fully restored by increased expression of psl.
FIG. 2.
Generation of a P. aeruginosa psl-conditional strain. (A) Schematic diagram showing construction of P. aeruginosa strains WFPA800 (Δpsl) and WFPA801 (Δpsl/pBAD-psl). The solid boxes indicate the putative −10 and −35 σ70 promoter elements. The transcription start site (position 1) is located 41 bp upstream of the pslA translation start site (32). (B) Biofilm formation by PAO1, WFPA60 (ΔpslAB), WFPA800 (Δpsl), and WFPA801 (Δpsl/pBAD-psl). The strains were assayed after 30 min of static incubation at room temperature in a microtiter dish. WFPA801 was cultured with the arabinose concentrations indicated below the bars (0%, 0.25%, and 0.5%). The values are the means and standard errors of three independent assays.
To quantify psl expression in these strains, we performed a real-time RT-PCR analysis of pslB transcription (Table 2). Expression of pslB in WFPA800 was reduced 50-fold compared with the PAO1 levels. When WFPA801 was grown without arabinose, the pslB expression was 10-fold lower than the pslB expression in PAO1. Addition of arabinose at a concentration of 0.05% led to levels of pslB expression in WFPA801 that were slightly elevated compared with the level in PAO1. Addition of 0.2% and 2% arabinose to cultures of WFPA801 led to levels of pslB transcription that were four- and sevenfold higher, respectively, than the levels in PAO1.
TABLE 2.
Real-time RT-PCR analysis of pslB transcription in WFPA800 (Δpsl) and WFPA801 (Δpsl/pBAD-psl) with different concentrations of arabinose
| Strain | Arabinose concn (%) | pslB expression relative to expression in PAO1a |
|---|---|---|
| PAO1 | 0 | 1 ± 0.1 |
| WFPA800 | 0 | 0.02 ± 0.002 |
| WFPA801 | 0 | 0.1 ± 0.01 |
| WFPA801 | 0.05 | 2 ± 0.17 |
| WFPA801 | 0.2 | 4 ± 0.25 |
| WFPA801 | 2 | 7 ± 0.39 |
The data are the means ± standard deviations for three replicate real-time RT-PCR assays.
Increased psl expression leads to enhanced cell-surface interactions, Congo red staining, and biofilm formation.
The psl cluster is predicted to encode a polysaccharide (20). We first examined differences in polysaccharide synthesis in the wild type, WFPA800 (Δpsl), and WFPA801 (psl-inducible strain) by using CR staining. CR is a dye that detects neutral polysaccharides or polysaccharides that contain either β-1,3- or β-1,4-glucopyranosyl units. CR was added to liquid growth media, and cells were grown overnight with shaking. When Psl was overproduced, we observed that WFPA801 cells stained by CR formed aggregates on the walls of tubes (Fig. 3). However, with PAO1, WFPA800, and uninduced WFPA801, no such aggregates were observed, and all staining was confined to cells in the medium (Fig. 3). This finding was verified by determining the OD600 of planktonic cells from the CR binding assay; approximately one-half of the WFPA801-induced cells remained attached to the culture tubes (Fig. 3). This showed that cell-surface and cell-cell interactions were enhanced when there was increased psl expression.
The CR binding was quantified by measuring the free CR left in the medium supernatant following centrifugation of the culture (A490). In this assay, increased synthesis of Psl led to a six- to eightfold reduction in the amount of CR in the culture supernatant, since the dye remained associated with the bacteria. As expected, the A490 values of WFPA800 and WFPA801 without arabinose were slightly higher than the A490 of PAO1 (Fig. 3). These results indicated that CR bound WFPA801 in a psl-dependent manner and suggested that proteins encoded by the psl operon were involved in synthesis of a neutral polysaccharide, possibly containing β-1,3- and/or β-1,4-glucopyranosyl units.
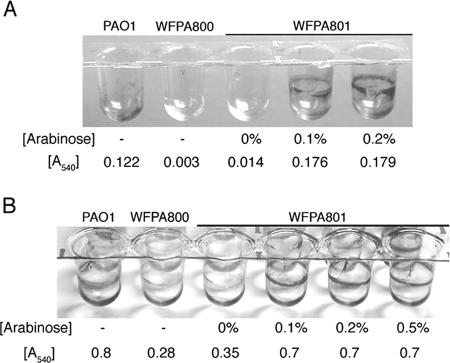
Our real-time RT-PCR data (Table 2) revealed that induction with 0.2% arabinose led to levels of pslB expression in WFPA801 that were fourfold higher than the wild-type levels. However, when WFPA801 was grown under these conditions, there was not a concomitant increase in biofilm formation. To reconcile this apparent discrepancy, we repeated the 30-min and 8-h microtiter dish attachment assays using a variety of psl expression levels (Fig. 4). In addition to measuring the soluble crystal violet released from attached cells (A540) (Fig. 4), the stained cells remaining on the walls were also visualized. At both times, we found that increased expression of psl led to significant retention of cells on the wall of a microtiter dish, even after ethanol solubilization (Fig. 4). This was not due to increased cell growth as identical numbers of cells were used in these assays and the growth rates of PAO1, WFPA800, and WFPA801 are similar. The failure to solubilize crystal violet with ethanol in induced WFPA801 cultures may have been due to increased adhesion of WFPA801 when Psl was overproduced, which resulted in a reduction in the number of bacteria detached from biofilms during the ethanol wash. Alternatively, it may have been due to the increased thickness of the biofilm matrix, which could have impeded diffusion of the dye. Nevertheless, our data clearly showed that elevated Psl polysaccharide synthesis resulted in increased biofilm formation and P. aeruginosa adhesion.
FIG. 4.
Increased Psl production enhances cell-surface interactions and biofilm formation: results of microtiter dish attachment assays with biofilms stained with crystal violet after 30 min of attachment (A) or after 8 h of attachment (B). The images show crystal violet-stained microtiter dish wells after ethanol elution, and the released soluble crystal violet was also evaluated by determining the A540 (the means of triplicate determinations are indicated below the wells). The arabinose concentrations used for Psl induction during growth of WFPA801 are indicated below the wells.
psl contributes to maintenance of the mature biofilm structure.
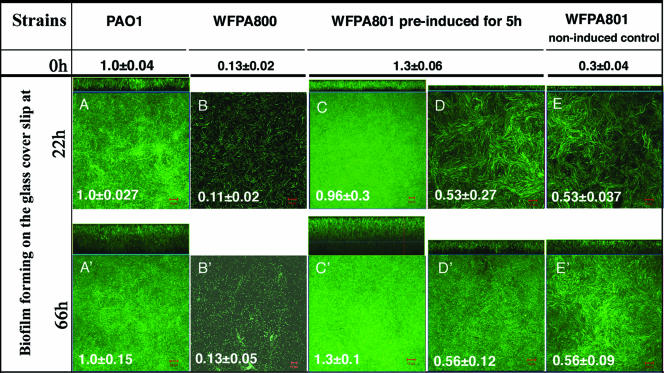
P. aeruginosa undergoes distinct stages of biofilm development, including attachment, maturation, and dispersion (30, 34, 39). Since the psl mutants did not attach efficiently, it was difficult to evaluate any potential role for Psl at developmental stages beyond attachment. However, use of the psl-conditional strain WFPA801 allowed us to selectively turn on and off psl expression during different stages of biofilm development. Two experiments were performed to determine if psl contributes to biofilm formation or structure at steps postattachment. First, WFPA801 was induced for 5 h with arabinose prior to injection into the flow cell. This ensured that wild-type adherence occurred in the initial attachment step, which was verified by a rapid attachment assay performed with cultures identical to those used to inoculate the flow cell reactors (Fig. 5). The bacteria were allowed to attach for 1 h in the flow cell chambers without medium flow, after which medium flow was initiated with or without arabinose for either 22 or 66 h. After these times, images of biofilms were acquired by CLSM. The biofilm thickness of each strain was calculated by averaging the maximum thicknesses of nine independent image stacks, and the values were normalized to the values for wild-type strain PAO1 (18 μm at 22 h and 34 μm at 66 h) (Fig. 5A). A representative image of each strain is also shown for 22 h and 66 h in Fig. 5.
FIG. 5.
psl contributes to biofilm formation and structure following initial attachment. Biofilms of PAO1, WFPA800 (Δpsl), and WFPA801 (Δpsl/pBAD-psl) were analyzed in a continuous-culture flow cell system. Cultures of each strain were grown in Jensen's medium (OD600, ∼0.5) with or without arabinose and were injected into a flow cell device. After 1 h of incubation without flow to allow the cells to attach, a continuous flow of Jensen's medium was started. The values at the top (0 h) indicate the initial attachment values obtained in a 1-h attachment microtiter dish assay of the cultures used in the flow cells. These values were normalized to the results for PAO1 (A540, 0.295). The images of the biofilms that formed on the glass coverslips were acquired by CLSM after 22 and 66 h under flow conditions. For each strain, biofilm images were obtained for nine areas covering 2.0 × 105 μm2 along the flow cell. The biofilm thickness of each sample was calculated by averaging the maximum thicknesses of the nine image stacks. A representative biofilm image for each sample is shown. The large square image is a horizontal view of the biofilm, and the rectangular image is a side view of the biofilm. The values for average biofilm thickness on the images were normalized to the PAO1 values (18 μm at 22 h and 34 μm at 66 h). Scale bar = 10 μm. The following strains and growth condition were used: (A and A′) PAO1 with no arabinose-containing medium flow; (B and B′) WFPA800 with no arabinose-containing medium flow; (C and C′) WFPA801 preinduced with 0.2% arabinose for 5 h before injection and then continuous addition of 0.2% arabinose to the medium; (D and D′) WFPA801 preinduced with 0.2% arabinose for 5 h before injection and then continuous addition of medium without arabinose; (E and E′) noninduced control WFPA801 continually grown without arabinose.
When WFPA801 was grown under continuous psl-inducing conditions, it formed a biofilm comparable to the biofilm formed by wild-type strain PAO1 (Fig. 5C). However, when WFPA801 was allowed to attach under psl-permissive conditions but subsequently grown under psl-nonpermissive conditions, the biofilm thickness was one-half the biofilm thickness obtained for the wild type or WFPA801 grown in the continual psl-permissive conditions (Fig. 5A, C, and D). The difference was reproducible and statistically significant (P = 0.001, as determined by a two-tailed t test). Biofilms formed by WFPA801 grown with no arabinose were clearly thinner than induced WFPA801 or wild-type biofilms (Fig. 5A, C, and E), but the thickness was not identical to the thickness of biofilms formed by WFPA800 (Fig. 5B). For Δpsl strain WFPA800 only a single layer of bacteria was attached on the coverslip after 22 h of growth in the flow cell, and a few microcolonies had formed by 66 h (Fig. 5B). Similar results were obtained previously with WFPA60 (ΔpslAB) (20). Compared with the wild type, the reduction in biofilm development exhibited by WFPA801 under psl-nonpermissive conditions (Fig. 5E) was identical to the reduction observed when WFPA801 was precultured in psl-permissive conditions (Fig. 5D). This was probably due to leaky expression from the pBAD promoter driving psl, since there is no catabolite control system in P. aeruginosa that represses pBAD, as there is in E. coli. Further support for low-level psl expression even in the absence of arabinose was evident when the results for WFPA800 and uninduced WFPA801 obtained with the real-time RT-PCR assay were compared (Table 2).
To further investigate the role of Psl postattachment, a second assay was performed, in which WFPA801 was cultured under psl-permissive conditions for 24 h, analyzed, and then cultured for an additional 68 h under psl-nonpermissive conditions. Twenty-four hours of development was chosen since the thicknesses of biofilms in our flow cell system at this point were >10 μm, which suggested that biofilm maturation had occurred. Biofilms formed by PAO1, WFPA800, and WFPA801 were analyzed in the same way. Images were obtained for nine areas covering 2.0 × 105 μm2 along the flow cell apparatus and were analyzed with the COMSTAT software to determine the biomass (Fig. 6A) and average biofilm thickness (Fig. 6B). Following growth of WFPA801 for 24 h under psl-permissive conditions, the biofilm biomass and thickness were slightly greater (∼1.5-fold) than the biofilm biomass and thickness observed for PAO1 (P = 0.25), yet WFPA800 biofilm development was severely compromised (∼5% of the wild type) (Fig. 6). However, when WFPA801 was subsequently grown for 68 h under psl-nonpermissive conditions, the biofilm biomass and average thickness were 80% of the wild-type biofilm biomass and average thickness (Fig. 6). These data were reproducible and statistically significant (P = 0.002 for biomass; P = 0.001 for average thickness). The biofilm biomass and thickness for WFPA800 at this time were 24% and 32% of the wild-type biofilm biomass and thickness, respectively (Fig. 6). Collectively, the data in Fig. 5 and 6 show that elimination of psl expression postattachment resulted in reductions in biofilm biomass and thickness, indicating that continuous Psl synthesis is necessary to maintain normal biofilm structure.
FIG. 6.
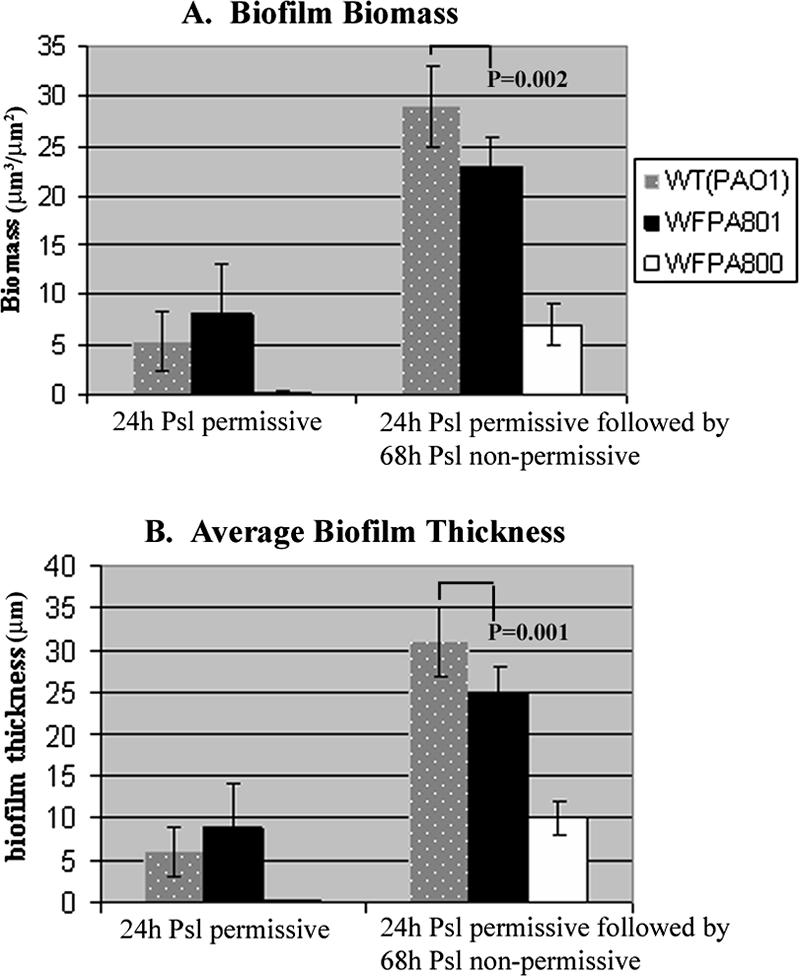
psl contributes to biofilm formation and structure following maturation: comparison of biofilms formed in flow cells by strains PAO1, WFPA801 (Δpsl/pBAD-psl), and WFPA800 (Δpsl). Each flow cell chamber was inoculated with 200 μl of a strain cultured with 0.2% arabinose (OD600, ∼0.5). The images of biofilms were acquired by CLSM after 24 h of growth in Jensen's minimal medium with 0.2% arabinose (Psl permissive). Following this, biofilms were allowed to grow for an additional 68 h in the absence of arabinose (Psl nonpermissive), and another series of images was obtained. For each condition, images of biofilms were obtained from nine areas covering 2.0 × 105 μm2 along the flow cell. All images generated by CLSM were analyzed by the COMSTAT software to determine biofilm biomass and average biofilm thickness. P values were calculated by using a two-tailed t test.
Overproduction of Psl results in biofilm architecture changes.
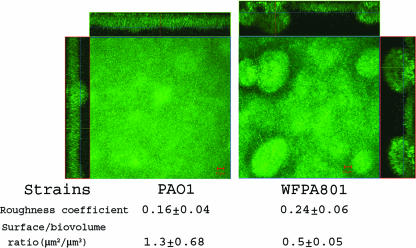
Finally, we examined the effect of Psl overproduction on biofilm architecture in a flow cell chamber. WFPA801 biofilms were formed with medium containing 2.0% arabinose for 48 h. Under these conditions, psl transcription was sevenfold higher than the psl transcription in PAO1 (Table 2). Compared with PAO1, WFPA801 formed biofilms with distinct three-dimensional mushroom-like structures (Fig. 7). The biofilm images were analyzed using the COMSTAT software. Induced WFPA801 biofilms had a roughness coefficient higher than that of PAO1 biofilms (P = 0.04, as determined by a one-tailed t test), which indicated that there was increased three-dimensional structure (Fig. 7). The ratio of surface to biovolume for PAO1 biofilms was 2.6-fold higher than the ratio for induced WFPA801 biofilms (P = 0.01, as determined by a one-tailed t test). This indicated that PAO1 formed flat, undifferentiated biofilms that covered 2.6 times more surface (with the same amount of biomass) than WFPA801 biofilms overproducing Psl covered. These data indicate that Psl overproduction resulted in a hyper-biofilm structure and architecture. The hyper-biofilm-forming phenotype is similar to that of variants isolated from aging P. aeruginosa PAO1 biofilms, in which both psl and pel levels are elevated (22). Collectively, the data in Fig. 4 to 7 provide evidence that the Psl polysaccharide functions as a scaffold, holding biofilm cells together. We propose that this is independent of, but physiologically linked to, the role of Psl in adhesion.
FIG. 7.
Overproduction of Psl causes biofilm architectural changes: biofilms of strains PAO1 and WFPA801 (Δpsl/pBAD-psl) in flow cells with a continuous flow of medium containing 2% arabinose. The images were acquired by CLSM 48 h after flow initiation. For each strain, biofilms were observed, and five image stacks were obtained from five areas covering 1.0 × 105 μm2 along the flow cell. A representative biofilm image for each sample is shown. CLSM-captured images were subjected to quantitative image analysis for roughness and surface/biovolume using the COMSTAT software, and the results are shown below the images.
DISCUSSION
Infection by P. aeruginosa is the main cause of mortality in CF patients. A critical factor in the pathogenesis is the ability to form biofilms in the lungs. Bacteria in biofilms are surrounded by an extracellular polymeric matrix, which provides protection from biocides and the host immune system. The psl locus encodes a putative polysaccharide that plays an important role in biofilm formation in nonmucoid P. aeruginosa strains (12, 20, 29, 32). In this paper, we show that Psl is required for cell-surface interactions on both biotic and abiotic surfaces. We also provide data showing that Psl has a role at stages of biofilm development beyond surface attachment. Finally, we show that overproduction of the Psl polysaccharide results in enhanced CR staining and adhesion of P. aeruginosa, which translates into significant changes in the architecture and physical properties of biofilms.
Biofilm development is a coordinated series of events beginning with surface attachment by planktonic bacteria. The data presented here and elsewhere (12, 20, 29, 32) indicate that psl plays a prominent role during biofilm development by promoting cell-surface and intercellular interactions. Since psl mutant strains are as deficient in binding to airway epithelial cells as strains lacking RpoN-dependent adhesins (e.g., flagella and pili) (Fig. 1) are, Psl must be given consideration as an important component of P. aeruginosa adhesion. This is not without precedent, as polysaccharides such as Staphylococcus polysaccharide intercellular adhesin/poly-N-acetylglucosamine (10, 23), Escherichia coli PGA (poly-β-1,6-N-acetylglucosamine) (1), N-acetylglucosamine of the Caulobacter crescentus holdfast organelle (37), P. aeruginosa alginate (28, 33), and lipopolysaccharides or lipooligosaccharides, contribute significantly to the adhesion of bacteria. Given that the psl operon is highly conserved among clinical and environmental nonmucoid isolates of P. aeruginosa (43), Psl is an attractive vaccine target for preventing colonization and perhaps subsequent chronic infection.
Our data indicate that psl is required to maintain biofilm structure postattachment. This was determined when we eliminated Psl expression immediately following attachment (Fig. 5) or at later stages (Fig. 6). When Psl synthesis was shut down following the initial attachment, the thickness of the biofilms was reduced significantly compared with the thickness of biofilms continually expressing Psl. Likewise, when biofilms were grown under Psl-permissive conditions for 24 h and then cultured for an additional 68 h under Psl-nonpermissive conditions, the biofilm biomass and thickness were reduced. These studies revealed a subtle but nonetheless important role for Psl postattachment. Our results probably underestimated the actual importance of Psl postattachment, since the Psl enzymes and the polysaccharide may be relatively stable and continue to function for some time even in the absence of psl gene expression. This effect was even more pronounced since the Δpsl/pBAD-psl strain, WFPA801, exhibited some leaky expression from the pBAD promoter under noninduced conditions. Taken together, these data suggest that Psl promotes intercellular interactions necessary for maintaining the biofilm architecture and that continuous Psl synthesis is necessary for proper biofilm maturation. The data further suggest that Psl is a primary scaffolding component of the extracellular matrix of P. aeruginosa biofilms and that therapies targeting Psl may facilitate biofilm dissolution.
We also discovered that overproduction of Psl led to enhanced CR binding and a “hyper-biofilm” phenotype. Workers in several laboratories have recently described colony morphology variants obtained from extended growth of biofilm cultures or from mutagenesis (4, 7, 14, 18, 22). The phenotypes of these mutants or phenotypic variants are remarkably similar to the phenotypes observed when Psl is overproduced. Indeed, subsequent analyses of some of the variants described above revealed that they exhibit elevated psl and pel expression (14, 18, 22, 42). Although the details regarding the complex regulatory pathways triggering psl and pel expression remain to be firmly established, at least three independent signal transduction networks mediated by RetS, GacS, and LadR control the activity of an RNA-protein complex (RsmA-RsmZ) that ultimately modulates psl and pel levels in P. aeruginosa (42). Additionally, the intracellular signaling molecule c-diGMP is necessary for psl and pel expression (7, 18). c-diGMP levels are modulated in part by the WspF-WspR chemosensory system (7, 18). How c-diGMP and the signal transduction pathways mentioned above (RetS, GacS, LadR, RsmAZ) converge to coordinately control psl and pel levels, as well as other critical P. aeruginosa phenotypes (14, 42, 45), is not known. Nonetheless, our data indicate that overexpression of Psl is sufficient to convert the P. aeruginosa phenotype to a phenotype that resembles the small, rough, strongly cohesive, autoaggregative variants mentioned above.
The psl cluster is predicted to synthesize a polysaccharide since proteins encoded by this operon exhibit homology to proteins involved in polysaccharide biosynthesis, modification, and transport from other bacteria. Previous data suggest that psl is involved in the production of a mannose-rich matrix material (12). Our CR binding data suggest that psl synthesizes a neutral polysaccharide and/or a polysaccharide with β-1,3- and β-1,4-glucopyranosyl units. This is consistent with our previous studies, which revealed that mannose, rhamnose, and glucose are the primary carbohydrate components of nonmucoid P. aeruginosa biofilms (44). The psl-inducible strain WFPA801 should be very useful for planned future studies to define the structure of the Psl polysaccharide, as well as to develop reagents (e.g., antiserum and lectins) to probe Psl expression within biofilms and in vivo. This should allow us to evaluate the potential contributions and interactions of the Psl, Pel, and alginate polysaccharides in acute and chronic P. aeruginosa infections.
Acknowledgments
We acknowledge Ken Grant, Micromed at WFUHS, for his assistance with microscopy and Cameron Dennis for photography. We also acknowledge Yolanda Sanchez and Laura Cobb for their assistance with the epithelial cell adhesion experiments.
This work was supported by Public Health Service grants AI061396 and HL58334 to D.J.W.
Footnotes
Published ahead of print on 15 September 2006.
REFERENCES
- 1.Agladze, K., X. Wang, and T. Romeo. 2005. Spatial periodicity of Escherichia coli K-12 biofilm microstructure initiates during a reversible, polar attachment phase of development and requires the polysaccharide adhesin PGA. J. Bacteriol. 187:8237-8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allesen-Holm, M., K. B. Barken, L. Yang, M. Klausen, J. S. Webb, S. Kjelleberg, S. Molin, M. Givskov, and T. Tolker-Nielsen. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 59:1114-1128. [DOI] [PubMed] [Google Scholar]
- 3.Baynham, P. J., D. M. Ramsey, B. V. Gvozdyev, E. M. Cordonnier, and D. J. Wozniak. 2006. The Pseudomonas aeruginosa ribbon-helix-helix DNA-binding protein AlgZ (AmrZ) controls twitching motility and biogenesis of type IV pili. J. Bacteriol. 188:132-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boles, B. R., M. Thoendel, and P. K. Singh. 2004. Self-generated diversity produces “insurance effects” in biofilm communities. Proc. Natl. Acad. Sci. USA 101:16630-16635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Branda, S. S., A. Vik, L. Friedman, and R. Kolter. 2005. Biofilms: the matrix revisited. Trends Microbiol. 13:20-26. [DOI] [PubMed] [Google Scholar]
- 6.Christensen, B., C. Sternberg, J. B. Andersen, R. J. Palmer, A. T. Nielsen, M. Givskov, and S. Molin. 1999. Molecular tools to study biofilm physiology. Methods Enzymol. 310:20-42. [DOI] [PubMed] [Google Scholar]
- 7.D'Argenio, D. A., M. W. Calfee, P. B. Rainey, and E. C. Pesci. 2002. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J. Bacteriol. 184:6481-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies, D. J., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell to cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 9.Drenkard, E. 2003. Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microb. Infect. 5:1213-1219. [DOI] [PubMed] [Google Scholar]
- 10.Fluckiger, U., M. Ulrich, A. Steinhuber, G. Doring, D. Mack, R. Landmann, C. Goerke, and C. Wolz. 2005. Biofilm formation, icaADBC transcription, and polysaccharide intercellular adhesin synthesis by staphylococci in a device-related infection model. Infect. Immun. 73:1811-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedman, L., and R. Kolter. 2004. Genes involved in matrix formation of Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51:675-690. [DOI] [PubMed] [Google Scholar]
- 12.Friedman, L., and R. Kolter. 2004. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 186:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fux, C. A., J. W. Costerton, P. S. Stewart, and P. Stoodley. 2005. Survival strategies of infectious biofilms. Trends Microbiol. 13:34-40. [DOI] [PubMed] [Google Scholar]
- 14.Goodman, A. L., B. Kulasekara, A. Rietsch, D. Boyd, R. S. Smith, and S. Lory. 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 7:745-754. [DOI] [PubMed] [Google Scholar]
- 15.Govan, J. R., and V. Deretic. 1996. Microbiol. pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guzman, L., D. Belin, M. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heydorn, A. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 18.Hickman, J. W., D. F. Tifrea, and C. S. Harwood. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. USA 102:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. Schweizer. 1998. A broad-host range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: applications for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 20.Jackson, K. D., M. Starkey, S. Kremer, M. R. Parsek, and D. J. Wozniak. 2004. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J. Bacteriol. 186:4466-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jensen, S. E., I. T. Facycz, and J. N. Campbell. 1980. Nutritional factors controlling exocellular protease production by Pseudomonas aeruginosa. J. Bacteriol. 144:844-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirisits, M. J., L. Prost, M. Starkey, and M. R. Parsek. 2005. Characterization of colony morphology variants isolated from Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 71:4809-4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kropec, A., T. Maira-Litran, K. K. Jefferson, M. Grout, S. E. Cramton, F. Gotz, D. A. Goldmann, and G. B. Pier. 2005. Poly-N-acetylglucosamine production in Staphylococcus aureus is essential for virulence in murine models of systemic infection. Infect. Immun. 73:6868-6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kulasakara, H., V. Lee, A. Brencic, N. Liberati, J. Urbach, S. Miyata, D. G. Lee, A. N. Neely, M. Hyodo, Y. Hayakawa, F. M. Ausubel, and S. Lory. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc. Natl. Acad. Sci. USA 103:2839-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landry, R. M., D. An, J. T. Hupp, P. K. Singh, and M. R. Parsek. 2006. Mucin-Pseudomonas aeruginosa interactions promote biofilm formation and antibiotic resistance. Mol. Microbiol. 59:142-151. [DOI] [PubMed] [Google Scholar]
- 26.Mah, T.-F., B. Pitts, B. Pellock, G. C. Walker, P. S. Stewart, and G. A. O'Toole. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306-310. [DOI] [PubMed] [Google Scholar]
- 27.Mah, T. C., and G. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34-39. [DOI] [PubMed] [Google Scholar]
- 28.Marcus, H., and N. R. Baker. 1985. Quantitation of adherence of mucoid and nonmucoid Pseudomonas aeruginosa to hamster tracheal epithelium. Infect. Immun. 47:723-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsukawa, M., and E. P. Greenberg. 2004. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J. Bacteriol. 186:4449-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 31.O'Toole, G. A., and R. Kolter. 1998. The initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 32.Overhage, J., M. Schemionek, J. S. Webb, and B. H. A. Rehm. 2005. Expression of the psl operon in Pseudomonas aeruginosa PAO1 biofilms: PslA performs an essential function in biofilm formation. Appl. Environ. Microbiol. 71:4407-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramphal, R., and G. B. Pier. 1985. Role of Pseudomonas aeruginosa mucoid exopolysaccharide in adherence to tracheal cells. Infect. Immun. 47:1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. J. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi, L., R. Ardehali, K. D. Caldwell, and P. Valint. 2000. Mucin coating on polymeric material surfaces to suppress bacterial adhesion. Colloids Surf. 17:229-239. [Google Scholar]
- 36.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 37.Smith, C. S., A. Hinz, D. Bodenmiller, D. E. Larson, and Y. V. Brun. 2003. Identification of genes required for synthesis of the adhesive holdfast in Caulobacter crescentus. J. Bacteriol. 185:1432-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.StatSoft. 2004. Electronic statistics textbook. StatSoft, Tulsa, OK.
- 39.Stoodley, P., K. Sauer, D. G. Davies, and J. W. Costerton. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187-209. [DOI] [PubMed] [Google Scholar]
- 40.Sundin, C., M. C. Wolfgang, S. Lory, A. Forsberg, and E. Frithz-Lindsten. 2002. Type IV pili are not specifically required for contact dependent translocation of exoenzymes by Pseudomonas aeruginosa. Microb. Pathog. 33:265-277. [DOI] [PubMed] [Google Scholar]
- 41.Vasseur, P., I. Vallet-Gely, C. Soscia, S. Genin, and A. Filloux. 2005. The pel genes of the Pseudomonas aeruginosa PAK strain are involved at early and late stages of biofilm formation. Microbiology 151:985-997. [DOI] [PubMed] [Google Scholar]
- 42.Ventre, I., A. L. Goodman, I. Vallet-Gely, P. Vasseur, C. Soscia, S. Molin, S. Bleves, A. Lazdunski, S. Lory, and A. Filloux. 2006. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Natl. Acad. Sci. USA 103:171-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolfgang, M. C., B. R. Kulasekara, X. Liang, D. Boyd, K. Wu, Q. Yang, C. G. Miyada, and S. Lory. 2003. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 100:8484-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wozniak, D. J., T. J. Wyckoff, M. Starkey, R. Keyser, P. Azadi, G. O'Toole, and M. R. Parsek. 2003. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc. Natl. Acad. Sci. USA 100:7907-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yahr, T., and E. P. Greenberg. 2004. The genetic basis for the commitment to chronic versus acute infection in Pseudomonas aeruginosa. Mol. Cell 16:497-503. [DOI] [PubMed] [Google Scholar]