Abstract
Yersinia enterocolitica causes human gastroenteritis, and many isolates have been classified as either “American” or “non-American” strains based on their geographic prevalence and virulence properties. In this study we describe identification of a transcriptional regulator that controls expression of the Y. enterocolitica ytxAB genes. The ytxAB genes have the potential to encode an ADP-ribosylating toxin with similarity to pertussis toxin. However, a ytxAB null mutation did not affect virulence in mice. Nevertheless, the ytxAB genes are conserved in many Y. enterocolitica strains. Interestingly, American and non-American strains have different ytxAB alleles encoding proteins that are only 50 to 60% identical. To obtain further insight into the ytxAB locus, we investigated whether it is regulated as part of a known or novel regulon. Transposon mutagenesis identified a LysR-like regulator, which we designated YtxR. Expression of ytxR from a nonnative promoter increased Φ(ytxA-lacZ) operon fusion expression up to 35-fold. YtxR also activated expression of its own promoter. DNase I footprinting showed that a His6-YtxR fusion protein directly interacted with the ytxA and ytxR control regions at similar distances upstream of their probable transcription initiation sites, identified by primer extension. Deletion analysis demonstrated that removal of the regions protected by His6-YtxR in vitro eliminated YtxR-dependent induction in vivo. The ytxAB locus is not present in most Yersinia species. In contrast, ytxR is conserved in multiple Yersinia species, as well as in the closely related organisms Photorhabdus luminescens and Photorhabdus asymbiotica. These observations suggest that YtxR may play a conserved role involving regulation of other genes besides ytxAB.
Three of the species that make up the genus Yersinia are widely accepted as organisms that are pathogenic to humans. Y. pestis is the etiological agent of plague, whereas Y. pseudotuberculosis and Y. enterocolitica usually cause intestinal disease. Y. enterocolitica is the species most frequently isolated from humans (6, 7), and infections are commonly acquired through ingestion of contaminated food or water (4). During a typical Y. enterocolitica infection the bacteria travel to the terminal ileum and penetrate the M cells overlaying the Peyer's patches. They multiply within the Peyer's patches before draining into and infecting the mesenteric lymph nodes. Disease usually manifests as self-limiting gastroenteritis and mesenteric lymphadenitis but can progress to septicemia, especially in patients with complicating conditions (6, 9).
Pathogenic Y. enterocolitica strains have been divided into two broad groups, based on serological typing and pathogenicity (7). The high-pathogenicity, so-called “American” strains are associated with large-scale outbreaks and more severe disease than their low-pathogenicity “non-American” counterparts (6). The variable pathogenicity of Y. enterocolitica is probably attributable to multiple factors, including the high-pathogenicity island that encodes an iron acquisition system unique to American serotypes (for a review, see reference 5).
An approximately 70-kb virulence plasmid is common to the three pathogenic Yersinia species (36). This plasmid encodes the Ysc type III secretion system and the Yop effector proteins that it exports, which disarm some features of the host innate immune response (8). This plasmid is necessary but not sufficient for virulence (7, 21). Chromosomal loci important for invasion of epithelial cells (48, 50), a stress response (11), and an additional type III secretion system (20) also play roles in virulence (for a review, see reference 37). There may be additional chromosomally encoded virulence factors that can be characterized.
Relatively common virulence factors of enteric pathogens are enterotoxins, which fall into two classes. Heat-stable enterotoxins are small peptides that induce fluid secretion from host cells (32). Heat-labile enterotoxins also play a role in inducing fluid secretion and are exemplified by cholera toxin of Vibrio cholerae and the heat-labile toxins of Escherichia coli (45). Each toxin consists of two different proteins associated in an A1B5 stoichiometry. The B pentamer binds to the host cell and triggers endocytic uptake of the complex. The A subunit is responsible for enzymatic modification of host cell proteins. The A subunits of both cholera toxin and the heat-labile toxins of E. coli are ADP-ribosyltransferases that modify the α subunit of a subset of heterotrimeric G proteins. This causes an increase in intracellular cyclic AMP levels, ultimately resulting in increased fluid secretion into the intestinal lumen (45).
Many bacterial genes are tightly regulated to ensure that they are expressed only in appropriate environments. This is especially true for virulence factors. For example, the V. cholerae ctxAB operon, which encodes cholera toxin, is subject to complex regulation in concert with several other members of overlapping regulons (for reviews, see references 35 and 43). In Yersinia species the Ysc-Yop regulon is also regulated by several proteins, some of which control many other genes (e.g., YmoA [23]).
Virulent Y. enterocolitica strains produce a heat-stable enterotoxin (Yst) that has been implicated as the cause of diarrhea in a rabbit model of infection (13, 14) but whose role in pathogenesis remains controversial (41). To date, genes with the potential to encode a heat-labile enterotoxin have not been described for Y. enterocolitica. Here we describe the ytxAB genes, which are conserved in several Y. enterocolitica strains and could encode a heat-labile enterotoxin. The role of these genes remains unknown, but we found that a previously uncharacterized member of the LysR family of transcriptional regulators, which we designated YtxR, positively regulates the ytxA promoter by direct interaction. The ytxR gene is conserved in many Yersinia species and in at least two members of the closely related genus Photorhabdus. In contrast, the ytxAB genes are not present in most Yersinia species or in any other genus. This suggests that YtxR regulates other genes besides ytxAB.
MATERIALS AND METHODS
Bacterial strains, plasmids, and routine growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Y. enterocolitica strains were routinely grown at 26°C in Luria-Bertani (LB) (Miller) broth or on LB agar plates (29). Antibiotics were used as described previously (27).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype or features | Source or reference |
|---|---|---|
| Escherichia coli B BL21-CodonPlus | ompT gal [dcm] [lon] hsdSB (rB− mB−) λDE3 lysogen pRIL | Stratagene |
| Yersinia enterocolitica 8081 strains (serogroup O:8, American strains)a | ||
| JB580v | ΔyenR (r− m+) pYV+ | 24 |
| YVM619 | Φ(ytxA-lacZYA) | This study |
| YVM707 | ΔytxAB::kan | This study |
| AJD239 | ΔytxR | This study |
| AJD199 | Φ(ytxA-lacZYA) Φ([TnMod-RKm′-lacIqtacp]-ytxR) | This study |
| AJD200 | Φ(ytxA-lacZYA) Φ([TnMod-RKm′-lacIqtacp]-ytxR) | This study |
| AJD254 | ΔytxR Φ(ytxA-lacZYA) | This study |
| AJD378 | ΔytxR Φ(ytxR-lacZYA) | This study |
| AJD1296 | ΔytxR ΔaraGFB::[Φ(ytxA-lacZY)] (Δ110 construct) | This study |
| AJD1297 | ΔytxR ΔaraGFB::[Φ(ytxA-lacZY)] (Δ52 construct) | This study |
| AJD1299 | ΔytxR ΔaraGFB::[Φ(ytxA-lacZY)] (Δ485 construct)b | This study |
| AJD1295 | ΔytxR ΔaraGFB::[Φ(ytxR-lacZY)] (Δ150 construct) | This study |
| AJD1300 | ΔytxR ΔaraGFB::[Φ(ytxR-lacZY)] (Δ500 construct)b | This study |
| AJD1303 | ΔytxR ΔaraGFB::[Φ(ytxR-lacZY)] (Δ86 construct) | This study |
| AJD1304 | ΔytxR ΔaraGFB::[Φ(ytxR-lacZY)] (Δ21 construct) | This study |
| Y. enterocolitica CDC reference strainsc | ||
| 657-83 | Serogroup O:20, American strain | CDC |
| 658-83 | Serogroup O:21, American strain | CDC |
| 655-83 | Serogroup O:18, American strain | CDC |
| 634-83 | Serogroup O:4,32, American strain | CDC |
| 637-83 | Serogroup O:5,27, non-American strain | CDC |
| 661-83 | Serogroup O:27, non-American strain | CDC |
| Y. enterocolitica clinical isolates | ||
| MC5 | Biogroup 1, serogroup O:6,30, Crohn's disease | M. Cafferkey |
| MC7 | Biogroup 1, serogroup O:9, colitis with perforation | M. Cafferkey |
| MC8 | Biogroup 1, serogroup O:9, septicemia | M. Cafferkey |
| MC17 | Biogroup 1, serogroup O:3, acute diarrhea | M. Cafferkey |
| MC22 | Biogroup 3, serogroup O:3, acute appendicitis | M. Cafferkey |
| MC33 | Biogroup 3, serogroup O:3, acute colitis | M. Cafferkey |
| MC28 | Biogroup 4, serogroup O:3, acute diarrhea | M. Cafferkey |
| MC6 | Biogroup 4, serogroup O:3, mesenteric adenitis | M. Cafferkey |
| MC51 | Biogroup 4, serogroup O:3, acute terminal ileitis | M. Cafferkey |
| Other Yersinia strains | ||
| Y. pseudotuberculosis YPIII | pYV+ | 18 |
| Y. pseudotuberculosis K286 | Clinical isolate | 30 |
| Y. kristensenii | Walter Hill, FDAd | |
| Y. frederikensenii | Walter Hill, FDA | |
| Y. frederikensenii MC31 | Clinical isolate (chronic diarrhea, weight loss) | M. Cafferkey |
| Y. rohdei 3022-83 | Dog stool isolate | CDC |
| Y. rohdei 3435-85 | Human stool isolate | CDC |
| Y. aldovae 670-83 | Isolated from water | CDC |
| Y. intermedia | Walter Hill, FDA | |
| Plasmids | ||
| pFUSE | Cmr, mob+ (RP4), R6K ori, lacZYA+ operon fusion vector | 2 |
| pKN8 | BglII linker in SmaI site of pFUSE | 17 |
| pBAD18-Km | Kmr, araBp expression vector, Col E1 ori | 19 |
| pBAD33 | CmraraBp expression vector, p15A ori | 19 |
| pEP185.2 | Cmr, mob+ (RP4), R6K ori | 24 |
| pWSK129 | Kmr, low-copy-number cloning vector, pSC101 ori | 49 |
| pHG329 | Ampr, cloning vector, pBR322 ori | 46 |
| pQE30 | Apr, Col E1 ori, T5p expression vector for His6 fusion proteins | QIAGEN |
| pAJD213 | ΔytxAB::kan in pEP185.2 | This study |
| pAJD593 | araBp-ytxR in pBAD18-Km | This study |
| pAJD610 | ytxA full-length control region in pHG329 | This study |
| pAJD654 | araBp-ytxR in pBAD33 | This study |
| pAJD679 | araBp-His6-ytxR in pBAD18-Km | This study |
| pAJD905 | Cmr, R6K ori, mob+ (RP4), sacB1+, lacZY operon fusion vector | 28 |
| pAJD1060 | Δ110 ytxAp fragment in pAJD905 | This study |
| pAJD1061 | Δ53 ytxAp fragment in pAJD905 | This study |
| pAJD1062 | Δ485 (full length) ytxAp fragment in pAJD905 | This study |
| pAJD1065 | Δ500 (full length) ytxRp fragment in pAJD905 | This study |
| pAJD1057 | Δ150 ytxRp fragment in pAJD905 | This study |
| pAJD1058 | Δ110 ytxRp fragment in pAJD905 | This study |
| pAJD1059 | Δ21 ytxRp fragment in pAJD905 | This study |
| pAJD1252 | ytxR control region (positions −552 to 40) in pWSK129 | This study |
All Y. enterocolitica 8081 strains are derivatives of strain JB580v.
The Φ(ytxA-lacZY) Δ485 and Φ(ytxR-lacZY) Δ500 constructs represent the full-length control regions (all noncoding upstream DNA) upstream of the lacZY operon.
CDC, Centers for Disease Control and Prevention.
FDA, Food and Drug Administration.
Southern hybridization analysis.
Chromosomal DNA was digested with HindIII, resolved by electrophoresis on a 0.8% agarose gel, and transferred to nitrocellulose by the method of Southern (44). Approximately 300-bp “ytxA” probe fragments were generated by PCR using primers that annealed to the central region of ytxA from strains JB580v and MC22. Labeling, hybridization, and detection were done with the ECL direct nucleic acid labeling and detection system (GE Healthcare Life Sciences).
PCR amplification of the sapA-pspF intergenic region.
The following primers annealed to the 5′ end of sapA and the 3′ end of pspF, incorporating BamHI and XbaI sites, respectively (underlined): 5′-CGCGGATCCCCACTGACACAATAGACAAAACCGCGCTGAC (sapA primer) and 5′-GGCTCTAGAATTGGCTGCATAATAGTGAATATCAGATGCT (pspF primer).
The primers were used in PCRs with chromosomal DNA from various Y. enterocolitica strains. The products were cloned into plasmid pHG329, and their DNA sequences were determined.
Transposon mutagenesis.
Transposon mutagenesis of Y. enterocolitica strain YVM619 was performed exactly as described previously (27). Mutants with increased Φ(ytxA-lacZ) expression were identified as described in the Results. Southern blotting was done to ensure that each mutant contained a single transposon insertion, and the transposon-chromosome junctions were isolated and their DNA sequences were determined as described previously (27).
Strain and plasmid construction.
To construct a ytxAB deletion mutant, two DNA fragments were amplified from Y. enterocolitica strain JB580v chromosomal DNA by PCR. One fragment had a BglII site followed by the first 10 codons of ytxA and approximately 1 kb of upstream DNA. The other fragment had a BglII site followed by the last 22 codons of ytxB and approximately 1 kb of downstream DNA. These fragments were ligated at the BglII site and cloned into plasmid pEP185.2. The BamHI kanamycin resistance gene fragment from mini-Tn5 Km2 (15) was then cloned into the unique BglII site. The resulting plasmid, pAJD213, was integrated into the JB580v chromosome, and Kmr Cms exconjugants were isolated. The ΔytxAB::kan mutation was confirmed by Southern hybridization analysis (data not shown).
To construct Φ(ytxA-lacZ) and Φ(ytxR-lacZ) single-copy operon fusion strains, ytxA or ytxR control region fragments were amplified from strain JB580v chromosomal DNA by PCR. The fragments were cloned into plasmid pFUSE or pKN8 and integrated into the chromosome by homologous recombination (2) or were cloned into plasmid pAJD905 and integrated into the ara locus exactly as described previously (28).
A ΔytxR in-frame deletion mutant (AJD239) was constructed with the λ Red recombinase gene replacement system (12), adapted for use in Y. enterocolitica (27). Briefly, a ΔytxR::kan mutation was made using allelic exchange mediated by Red recombinase. The kanamycin resistance gene was removed by FLP recombinase-mediated excision, and the in-frame deletion was confirmed by Southern hybridization, colony PCR, and DNA sequencing (data not shown).
araBp-ytxR expression plasmids were constructed by amplifying fragments from Y. enterocolitica strain JB580v genomic DNA and cloning them into pBAD18-Km or pBAD33. To construct an araBp-His6-ytxR expression plasmid, a ytxR+ fragment was amplified by PCR and cloned into plasmid pQE30 (QIAGEN Inc.). It was then excised as an EcoRI-SalI fragment and cloned into pBAD18-Km to obtain plasmid pAJD679.
β-Galactosidase assays.
To determine the effect of transposon insertions on Φ(ytxA-lacZ) expression, saturated cultures were diluted into 4 ml of LB broth containing appropriate antibiotics in 18-mm-diameter test tubes so that the optical density at 600 nm was approximately 0.08. Cultures were grown on a roller drum at 26°C for 3 h. Then 1 mM (final concentration) isopropyl-β-d-thiogalactopyranoside (IPTG) was added, and the cultures were returned to the roller drum for an additional 2 h.
To determine the effects of an araBp-ytxR+ plasmid, saturated cultures were diluted as described above into 4 ml of LB broth containing appropriate antibiotics. The cultures were grown on a roller drum at 26°C for 2 to 3 h (optical density at 600 nm, approximately 0.2 to 0.4), and then 0.2% (final concentration) arabinose was added. Cells were then grown for an additional 2 to 2.5 h at 26°C.
β-Galactosidase activity was determined at room temperature (approximately 22°C) using permeabilized cells (26). Activities were expressed in arbitrary units, which were determined using the formula described by Miller (29). Individual cultures were assayed in duplicate, and the activities reported below are the averages from three independent cultures.
RNA isolation and primer extension analysis.
Total RNA was isolated from Y. enterocolitica strains with a single-copy chromosomal Φ(ytxA-lacZY) operon fusion (AJD1299) or a Φ(ytxR-lacZY) operon fusion (AJD1300) and araBp-ytxR+ plasmid pAJD654. Cultures were grown as described above for the β-galactosidase assay experiments to determine the effect of an araBp-ytxR+ plasmid. RNA was isolated using an RNeasy mini kit (QIAGEN). End labeling of the oligonucleotide and primer extension reactions were done with the Primer Extension System avian myeloblastosis virus reverse transcriptase (Promega). The primer used was 5′-TCATCGGTTGTCGGATCGGA, which corresponds to a region in the template strand 60 bp downstream of the cloning site in the lacZ fusion plasmid pAJD905. The primer was labeled at the 5′ end with [γ-32P]ATP and used in extension reaction mixtures containing 5 μg of RNA. To generate size markers, the same primer was used in DNA sequencing reactions with the pAJD1062 (ytxAp) or pAJD1065 (ytxRp) template using the fmol DNA cycle sequencing system (Promega). Samples were resolved by denaturing 8% polyacrylamide-urea electrophoresis and visualized by autoradiography.
Purification of His6-YtxR.
A 1-liter culture of E. coli strain BL21-CodonPlus containing plasmid pAJD679 was grown at 30°C to an optical density at 600 nm of approximately 0.9. Arabinose (final concentration, 0.2%) was added, and the culture was incubated for an additional 3 h. Bacterial cells were collected by centrifugation, frozen at −20°C, and then resuspended in 20 ml of a solution containing 50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 5 mM β-mercaptoethanol, 0.1% Tween 20, and 5 mM MgCl2 (pH 7.5) containing 1× Complete protease inhibitor (Roche) and 1.25 mg/ml lysozyme. Cells were incubated on ice for 30 min and disrupted by sonication. The soluble and insoluble fractions were separated by centrifugation, and the soluble extract (supernatant) was mixed with 4 ml Ni-nitrilotriacetic acid-agarose (QIAGEN) for 1 h at 4°C and then poured into a column. The column was washed with 20 ml of a solution containing 50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, 5 mM β-mercaptoethanol, 0.1% Tween 20, and 5 mM MgCl2 (pH 7.5). His6-YtxR protein was eluted with 10 ml of a solution containing 50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, 5 mM β-mercaptoethanol, 0.1% Tween 20, and 5 mM MgCl2 (pH 7.5) and collected in 1-ml fractions, which were used directly in DNase I footprinting assays. Protein concentrations were estimated using NanoDrop ND-1000 spectrophotometer A280 measurement and a bovine serum albumin standard in the His6-YtxR elution buffer.
Preparation of probes for DNase I footprinting.
The ytxA control region fragment was generated from plasmid pAJD610 using M13 reverse primer and a primer that annealed approximately 450 bp upstream of ytxA position +1 and incorporated an EcoRI site. The product was digested with BamHI, which cleaved downstream of ytxA position 1, and was dephosphorylated with calf intestinal alkaline phosphatase (Promega). The bottom (template) strand was labeled at the 5′ end with [γ-32P]ATP using T4 polynucleotide kinase (Promega). Unincorporated [γ-32P]ATP was removed with the Promega Wizard SV gel and PCR cleanup system. To eliminate any label from the other end of the DNA fragment, the product was digested with EcoRI and cleaned again with the Promega Wizard SV gel and PCR cleanup system.
A ytxR control region fragment was generated from plasmid pAJD1252 using a primer that annealed to the cloning site of the plasmid (downstream of ytxRp position +1) and a primer that annealed approximately 550 bp upstream of ytxR position +1 and incorporated an XbaI site. The product was digested with BamHI, which cleaved downstream of ytxR position +1, and dephosphorylated. The bottom (template) strand was labeled as described above, except that an XbaI digest was used to eliminate any label from the other end of the DNA fragment.
DNase I footprinting assays.
Labeled ytxA or ytxR control region probes (approximately 2 nM) were mixed with His6-YtxR protein in a buffer containing 400 μg/ml salmon sperm DNA (Sigma-Aldrich), 100 mM HEPES (pH 7.6), 50 mM (NH4)2SO4, 5 mM dithiothreitol, 1% (vol/vol) Tween 20, and 150 mM KCl (total reaction volume, 50 μl). The reaction mixtures were incubated at 32°C for 15 min, and then 53 μl of a solution containing 5 mM CaCl2, 10 mM MgCl2, and 0.005 U/μl DNase I was added. Then the mixtures were incubated for 2 min, and digestion was stopped by adding 25 μl of a solution containing 2 M ammonium acetate, 250 mM EDTA, 100 μg/ml salmon sperm DNA, and 1 mg/ml glycogen. The DNA was precipitated with ethanol and resuspended in formamide loading dye. To generate a size marker, the pAJD610 (ytxAp) or pAJD1252 (ytxRp) plasmid was used in DNA sequencing reactions with the fmol DNA cycle sequencing system (Promega). The sequencing primers annealed downstream of the position +1 sites and had 5′ ends that corresponded exactly to the labeled ends of the fragments used in the footprint reactions. Samples were resolved by denaturing 8% polyacrylamide-urea electrophoresis and visualized by autoradiography.
Control region deletion analysis.
Truncated ytxA and ytxR control region fragments were generated by PCR using a common downstream primer that annealed within the 5′ ends of the coding regions and primers that annealed at various distances upstream. XbaI and BglII restriction sites were incorporated for the ytxAp fragments. XbaI and BamHI restriction digestion sites were incorporated for the ytxRp fragments. The fragments were cloned into pAJD905, and the DNA sequences were confirmed. The operon fusions were integrated into the ara locus and confirmed by colony PCR as described previously (28).
Nucleotide sequence accession numbers.
The nucleotide sequence data generated in this study have been assigned the following GenBank accession numbers: AY008264 for the ytxAB locus from Y. enterocolitica strain 8081(serotype O:8) and AY183120 for the ytxAB locus from Y. enterocolitica strain MC22 (serotype O:3).
RESULTS
Description of the ytxAB locus.
During characterization of the Y. enterocolitica phage shock protein (psp) locus (11) we identified two adjacent open reading frames (ytxAB) (Fig. 1). A BLASTP search revealed homology between YtxA and the catalytic subunit of pertussis toxin (PtxA; E = 1e-16). Notably, YtxA has the three conserved motifs characteristic of bacterial ADP-ribosylating toxins (ADPRT) (10, 16, 33), including the catalytic glutamic acid residue (Fig. 1). Other amino acids important for full catalytic activity of pertussis toxin (for a review, see reference 25) are also mostly conserved in YtxA (Fig. 1). Therefore, ytxA might encode an ADPRT. Two cysteine residues that form an important PtxA disulfide bond (31) are also present in YtxA, and YtxA is predicted to have a sec-dependent signal sequence.
FIG. 1.
ytxAB locus encodes a putative ADP-ribosylating toxin. (A) Diagram of the arrangement of the ytxAB genes in the sapA-pspF intergenic region of Y. enterocolitica strain JB580v (8081). (B) CLUSTALW alignment of the YtxA protein with the catalytic subunit of pertussis toxin (PtxA; GenBank accession number P04947). The signal sequences (predicted for YtxA) are underlined. Identical, strongly similar, and similar residues are indicated by vertical lines, colons, and periods, respectively, between the sequences. Three conserved sequences characteristic of ADP-ribosyltransferases, including the catalytic glutamic acid residue, are indicated by boldface type (10, 16, 33). Residues important for full ADP-ribosyltransferase activity of PtxA are indicated by bullets below the sequence. Cysteine residues that form a disulfide bond in PtxA are indicated by arrows below the sequence.
A BLASTP search with YtxB revealed no significant homology (E < 10) to previously characterized proteins. YtxB is small, as are the B subunits of cholera toxin and the heat-labile enterotoxins of E. coli, and alignment of YtxB with the B subunit of cholera toxin revealed some similarities (data not shown). YtxB is also predicted to have a sec-dependent signal sequence.
A ytxAB deletion mutant was assessed using oral infection of 6- to 7-week-old female BALB/c mice essentially as described previously (34). The 50% lethal doses of ytxAB+ and ytxAB null strains were indistinguishable, as were the bacterial loads of these strains in different tissues over time (data not shown). From these experiments, we concluded that the ytxAB locus is not required for virulence in an adult mouse model of acute infection. This does not rule out a role for this putative toxin, perhaps a role that is limited to the intestinal stage of disease and/or is host species specific. We also attempted to overexpress the YtxA protein. However, the overexpressed protein was completely insoluble (data not shown), and we were unable to detect ADP-ribosyltransferase activity.
American and non-American strains have divergent ytxAB alleles.
The ytxAB locus might have been acquired recently because its G+C content (39%) is much lower than the average G+C content of the chromosome (47%) (http://www.sanger.ac.uk/Projects/Y_enterocolitica/). Therefore, to investigate ytxAB conservation, a Southern hybridization experiment was done using a probe that encoded the central region of the Y. enterocolitica strain JB580v ytxA gene.
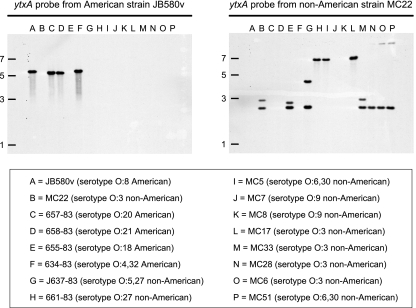
In addition to chromosomal DNA of the strain that it was derived from (serotype O:8), the ytxA probe hybridized to chromosomal DNA of Y. enterocolitica strains belonging to serotypes O:20, O:21, and O:4,32 (Fig. 2). These strains are all American strains (note that the probe did not hybridize to DNA from one American strain, a serotype O:18 strain). The probe did not hybridize to DNA from any non-American Y. enterocolitica strain (Fig. 2) or to DNA from any other Yersinia species listed in Table 1 (data not shown). Furthermore, a BLAST search of Y. pestis genomes, which were not included in this hybridization experiment, did not reveal any homology to ytxAB. Therefore, it appeared that the ytxA gene (and presumably ytxB) is present only in some American strains of Y. enterocolitica.
FIG. 2.
Southern hybridization analysis of ytxA conservation in Y. enterocolitica. Chromosomal DNA from various Y. enterocolitica strains was digested with HindIII, separated by electrophoresis on a 0.8% agarose gel, and transferred to nitrocellulose. In separate experiments the same nitrocellulose membrane was hybridized with a labeled ytxA fragment from American strain JB580v and with a labeled ytxA fragment from non-American strain MC22. The approximate positions (in kb) of size markers in the original agarose gel are indicated on the left. The lower panel shows the lane assignments for the chromosomal DNA samples.
To confirm the absence of ytxAB from non-American Y. enterocolitica strains, we amplified the sapA-pspF intergenic region (Fig. 1) of one of them by PCR. As a control, we also amplified an approximately 2.3-kb ytxAB+ fragment from the chromosome of Y. enterocolitica strain JB580v (data not shown). Unexpectedly, the non-American Y. enterocolitica strain (strain MC22 [Table 1]) produced a PCR product that was a similar size (data not shown). The DNA sequence of this fragment revealed genes that encoded proteins with 53% and 62% identity to the YtxA and YtxB proteins, respectively, of strain JB580v (data not shown). Despite the significant divergence of these ytxAB genes, residues predicted to be important for ADPRT activity of YtxA were conserved. Strikingly, although there was such a major difference between the ytxAB coding regions, the same was not true for the noncoding DNA sequences extending 200 bp upstream of the American and non-American ytxA initiation codons, which were 97% identical (data not shown).
The Southern hybridization experiment was repeated with a non-American strain ytxA probe. This probe hybridized to DNA from Y. enterocolitica strains belonging to serotypes O:5,27, O:27, O:6,30, and O:3 (Fig. 2), all of which were non-American strains (note that the probe did not hybridize to DNA from two non-American strains, both belonging to serotype O:9). The probe also hybridized to DNA from the only American Y. enterocolitica strain that did not hybridize to the ytxA probe from American strain JB580v (serotype O:18) (Fig. 2). The probe did not hybridize to DNA from any other Yersinia species listed in Table 1 (data not shown).
These data indicate that there are two versions of ytxAB in Y. enterocolitica. One is present only in American strains, and the other is present primarily in non-American strains.
DNA sequence analysis identified a ytxAB cassette.
Two clinical isolates belonging to non-American Y. enterocolitica serotype O:9 (strains MC7 and MC8 [Table 1]) are the only Y. enterocolitica strains tested that do not have a ytxAB locus. This conclusion was based on the failure of chromosomal DNA from these strains to hybridize to either probe (Fig. 2) and on the small size of their sapA-pspF intergenic region PCR fragments (data not shown).
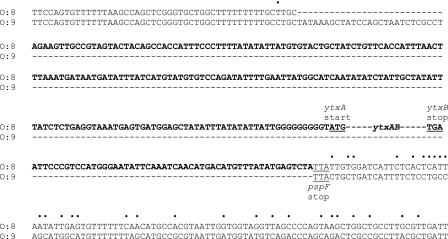
The DNA sequence of the relatively small sapA-pspF intergenic region PCR fragment from one of these Y. enterocolitica serotype O:9 strains was compared to that from the ytxAB+ strain JB580v (Fig. 3). This revealed the extent of the unique region present in the ytxAB+ strain. In addition to the ytxAB genes and the 48-bp intergenic region (not shown in Fig. 3), there are 207 bp of upstream DNA and 52 bp of downstream DNA. The non-American Y. enterocolitica strain MC22 has a region that is a similar length and has a similar sequence upstream of its ytxAB genes (data not shown). Therefore, strains with any version of the ytxAB locus probably contain a unique region that includes approximately 200 bp of noncoding upstream DNA. We hypothesized that this region probably contains the ytxA promoter and any important regulatory sequences. This hypothesis was investigated in the series of experiments described below.
FIG. 3.
Comparison of sapA-pspF intergenic region DNA sequences from Y. enterocolitica strains JB580v (serotype O:8, ytxAB+) and MC7 (serotype O:9, no ytxAB locus). Only part of each intergenic region is shown. The DNA sequence in boldface type is unique to strain JB580v (ytxAB gene sequences were omitted for clarity). Bullets above the sequences indicate differences between the regions conserved in both strains. The ytxA start codon, ytxB stop codon, and pspF stop codon (complementary strand) are labeled and underlined.
A LysR-like transcriptional regulator induces ytxAB expression.
Understanding whether ytxAB expression is regulated and, if it is, the underlying mechanism(s) might provide insight into its role. To begin to investigate this, a single-copy Φ(ytxA-lacZ) operon fusion strain was constructed. The level of β-galactosidase activity expressed from this fusion was relatively low at 26°C or 37°C (less than 100 Miller units), suggesting that ytxA was poorly expressed under standard laboratory conditions. We hypothesized that ytxAB might be expressed only under specific conditions and that control is mediated by a regulatory protein interacting with the unique region upstream of ytxA (Fig. 3). Therefore, a genetic screen was devised to identify regulatory proteins, even if they were poorly expressed in our standard growth conditions.
The screen relied on a transposon encoding the E. coli lac repressor (lacI) and an outward-facing tac promoter (27). This transposon causes null mutations by insertion (e.g., a ytxA repressor) and/or IPTG-dependent overexpression of downstream genes (e.g., a ytxA activator). Approximately 40,000 transposon mutants of a Φ(ytxA-lacZ) operon fusion strain were screened after growth at 26°C in the presence of IPTG on LB indicator agar containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). Mutants with increased Φ(ytxA-lacZ) expression were identified as dark blue colonies and were later confirmed by β-galactosidase assays (data not shown). Any mutants with transposon insertions immediately upstream of Φ(ytxA-lacZ) were identified by Southern hybridization analysis and eliminated from further analysis.
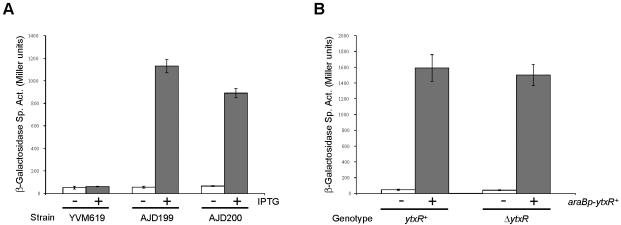
The screen did not identify any mutants with an IPTG-independent increase in Φ(ytxA-lacZ) expression. However, we identified six mutants that exhibited severalfold IPTG-dependent increases in Φ(ytxA-lacZ) expression, suggesting that this expression was caused by overexpression of a gene downstream of the transposon. Southern hybridization analysis indicated that all six mutants had a single transposon insertion in the same chromosomal region (data not shown). Two of these mutants (designated strains AJD199 and AJD200) were randomly selected for further analysis. These mutants exhibited 14- to 20-fold IPTG-dependent induction of Φ(ytxA-lacZ) expression (Fig. 4).
FIG. 4.
Increased ytxR expression induces a Φ(ytxA-lacZ) operon fusion. (A) Expression of Φ(ytxA-lacZ) in the presence (+) or absence (−) of IPTG in a strain without a transposon (YVM619) or two mutants with tacp transposon insertions upstream of ytxR (AJD199 and AJD200). (B) Expression of Φ(ytxA-lacZ) in ytxR+ (YVM619) or ΔytxR null (AJD254) strains with either araBp-ytxR+ expression plasmid pAJD593 (+) or the pBAD18-Km control vector (−). Cultures were grown and β-galactosidase activities were determined as described in Materials and Methods. The data are averages from three independent cultures, and the error bars indicate the standard deviations from the means. Sp. Act., specific activity.
The DNA sequences of the transposon-chromosome junctions from the two mutants revealed that the transposon had inserted 186 bp (AJD199) and 156 bp (AJD200) upstream of the same open reading frame (YE2253), in an orientation that would direct its expression from the tac promoter of the transposon. YE2253 is located on the complementary strand between nucleotides 2461446 and 2462306 of the Y. enterocolitica chromosome (http://www.sanger.ac.uk/Projects/Y_enterocolitica/). It is predicted to encode an uncharacterized member of the family of LysR-type transcriptional regulators (LTTRs) (for a review, see reference 40). We designated YE2253 the ytxR gene.
Finally, we checked whether ytxR overexpression alone was responsible for increasing Φ(ytxA-lacZ) expression. The ytxR gene was cloned into the araBp expression plasmid, pBAD18-Km. Expression of ytxR from this plasmid induced Φ(ytxA-lacZ) expression approximately 35-fold (Fig. 4). This did not occur for unrelated lacZ operon fusions studied in our laboratory (data not shown). A ytxR in-frame deletion mutant was also constructed. However, there was no difference in Φ(ytxA-lacZ) expression between ytxR+ and ytxR null strains (Fig. 4). This suggests that the ytxR gene is not significantly expressed from its own promoter under standard laboratory conditions. Taken together, all of these data show that expression of ytxR from a nonnative promoter is sufficient to induce Φ(ytxA-lacZ) expression.
YtxR is an autoregulator.
Most LTTRs act as autoregulators, enhancing or repressing their own transcription (40). To test whether this is the case for YtxR, a single-copy Φ(ytxR-lacZ) operon fusion was constructed in a strain with a ΔytxR mutation. When ytxR was expressed from araBp expression plasmid pAJD593, it induced Φ(ytxR-lacZ) expression approximately 100-fold (data not shown). This indicates that YtxR is a positive autoregulator.
Determination of ytxA and ytxR 5′ mRNA ends.
Next we wanted to characterize the ytxA and ytxR promoters and their control by YtxR at the molecular level. An important first step was to locate the 5′ ends of the ytxA and ytxR mRNAs. Therefore, RNA was isolated separately from Φ(ytxA-lacZ) and Φ(ytxR-lacZ) strains containing an araBp-ytxR+ expression plasmid and analyzed by primer extension (see Materials and Methods).
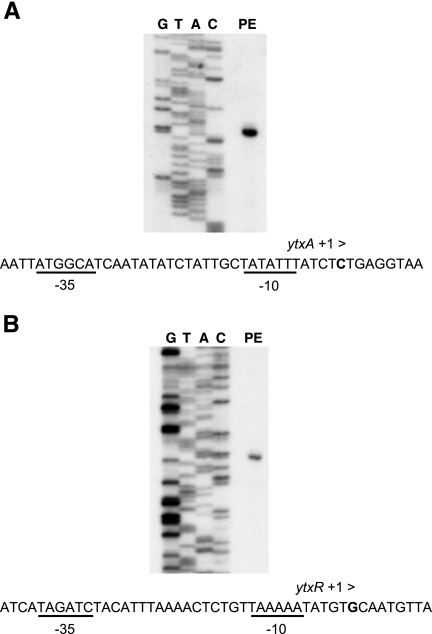
A single ytxA 5′ end was detected that corresponded to 50 nucleotides upstream of the probable ytxA ATG start codon (Fig. 5 and data not shown). This result was confirmed by 5′ rapid amplification of cDNA ends using a different ytxA template (data not shown). This 5′ mRNA end may have originated from a σ70-dependent promoter because putative −10 and −35 sequences were identified upstream (Fig. 5).
FIG. 5.
Primer extension analyses of the ytxA and ytxR control regions. RNA was extracted from Y. enterocolitica strains with either Φ(ytxA-lacZ) (A) or Φ(ytxR-lacZ) (B) single-copy operon fusions and the arap-ytxR+ expression plasmid pAJD654. Lane PE, primer extension reaction; lanes G, T, A, and C, DNA sequencing reactions. The nucleotide sequences surrounding the putative transcription initiation sites are shown below the panels. Nucleotides corresponding to the 5′ mRNA end sites are indicated by boldface type and labeled “+1 >.” Putative −10 and −35 elements are underlined and labeled.
In the case of ytxR the 5′ mRNA end corresponded to 237 nucleotides upstream of the probable ATG start codon (Fig. 5). No other smaller products of the primer extension reaction were detected (Fig. 5 and data not shown). Therefore, ytxR has an unusually long 5′ untranslated region. However, this is not unprecedented, even for genes that encode LTTRs (39). Once again, sequences with some similarity to −10 and −35 elements were detected upstream of the position corresponding to the 5′ mRNA end.
DNase I footprint analysis of His6-YtxR interaction with the ytxA and ytxR control regions.
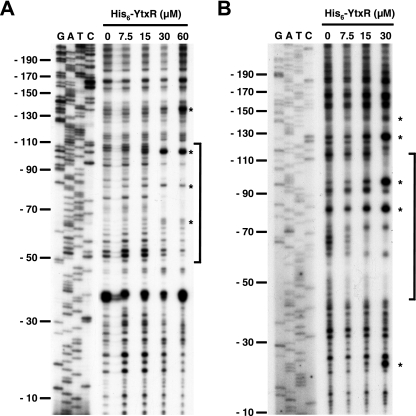
The simplest hypothesis to explain how ytxR overexpression induces Φ(ytxA-lacZ) and Φ(ytxR-lacZ) expression is that YtxR directly binds to the ytxA/ytxR control regions. To test this hypothesis, a His6-YtxR fusion protein was purified [we first confirmed that production of His6-YtxR was able to induce Φ(ytxA-lacZ) in vivo (data not shown)]. His6-YtxR protected nucleotides in both the ytxA and ytxR control regions from DNase I cleavage (Fig. 6). In both cases the protected regions were centered at approximately position −75. This is in good agreement with the binding site locations of other members of the LTTR family (40). An alignment of the protected regions revealed significant sequence similarity (Fig. 7). Similar concentrations of the His6-YtxR protein produced clearly observable DNase I footprints of the ytxA and ytxR control regions, suggesting that the binding affinities were comparable. These data demonstrate that His6-YtxR interacts with defined regions upstream of ytxA and ytxR in vitro. Therefore, YtxR probably activates ytxA and ytxR transcription directly in vivo. Further support for this conclusion came from the set of experiments described below.
FIG. 6.
DNase I footprint analysis of the ytxA and ytxR control regions. Labeled ytxA (A) or ytxR (B) control region fragments were incubated with different concentrations of His6-YtxR protein as indicated above the lanes and then treated with DNase I. Lanes G, A, T, and C show the results for sequencing reactions for each control region and are calibrated with respect to the number of base pairs from the transcription start site. Brackets indicate the approximate region of DNase I protection. Asterisks indicate sites hypersensitive to DNase I cleavage in the presence of His6-YtxR.
FIG. 7.
Important features of the ytxA and ytxR control regions. (A) ytxA control region. (B) ytxR control region. The numbering is relative to the putative transcriptional start sites, indicated by boldface type and labeled “+1>.” Putative −10 and −35 elements are underlined and labeled. Regions protected from DNase I cleavage in the presence of His6-YtxR are double underlined. Deletion endpoints of constructs used in this study are indicated. (C) CLUSTALW alignment of the protected areas of both control regions. Asterisks indicate identical nucleotides.
5′ Deletion analysis of the ytxA and ytxR control regions.
The final series of experiments was designed to test whether the regions protected by His6-YtxR in vitro were required for YtxR-dependent regulation in vivo. A set of single-copy Φ(ytxA-lacZ) and Φ(ytxR-lacZ) operon fusion strains was constructed with progressive 5′ deletions of their control regions (Fig. 7). These strains were grown with or without ytxR expression from an arabinose-inducible plasmid, and β-galactosidase activities were determined (Table 2).
TABLE 2.
Effects of control region deletions on Φ(ytxA-lacZ) and Φ(ytxR-lacZ) expression
| Control regiona | β-Galactosidase sp actb
|
|
|---|---|---|
| Without YtxRc | With YtxR | |
| Φ(ytxA-lacZ) | ||
| Δ485 | 32 ± 5 | 950 ± 80 |
| Δ110 | 40 ± 4 | 2,120 ± 320 |
| Δ52 | 47 ± 4 | 48 ± 6 |
| Φ(ytxR-lacZ) | ||
| Δ500 | 23 ± 6 | 1,030 ± 340 |
| Δ150 | 310 ± 120 | 2,140 ± 560 |
| Δ86 | 510 ± 60 | 1,810 ± 610 |
| Δ21 | 420 ± 32 | 400 ± 100 |
Each strain has a Φ(ytxA-lacZY) or Φ(ytxR-lacZY) fusion integrated on the chromosome with a different amount of DNA upstream of the transcription initiation site, as shown in Fig. 7.
β-Galactosidase specific activity was determined as described in Materials and Methods and is expressed in arbitrary (Miller) units. The values are averages± standard deviations.
Strains with the araBp vector pBAD33 (without YtxR) or the araBp-ytxR+ expression plasmid pAJD654 (with YtxR) were grown in the presence of 0.2% arabinose as described in Materials and Methods.
Deletion of sequences upstream of position −110 did not affect YtxR-dependent induction of Φ(ytxA-lacZ) expression (Table 2). However, deletion to position −52 completely eliminated YtxR-induced activity without affecting the basal (YtxR-independent) activity. Therefore, the region between positions −110 and −52 is essential for YtxR-dependent induction in vivo. This is in agreement with the region identified by DNase I footprinting in vitro (Fig. 6 and 7).
In the Φ(ytxR-lacZ) deletion analysis, two different phenomena were observed. First, deletion of the region from position −500 to position −150 significantly elevated both YtxR-independent expression and YtxR-dependent expression, while the ability of YtxR to activate expression was maintained (Table 2). The next deletion, from position −150 to position −86, did not have any additional effect on Φ(ytxR-lacZ) expression. However, deletion to position −21 eliminated YtxR-dependent induction. Once again, these results are in agreement with the region identified by DNase I footprint analysis.
Conservation of ytxR.
Southern hybridization analysis (Fig. 2) and BLASTP searches (data not shown) indicated that the ytxAB genes are not present in most Yersinia species and in all other genera. The ytxR gene (YE2253) is not linked to ytxAB (YE2124 and YE2123), and so we were interested in investigating ytxR conservation. We performed BLASTP searches with the predicted YtxR protein sequence. This analysis revealed that ytxR is intact and conserved (more than 90% amino acid identity) in the seven Yersinia species whose genome sequences are available (http://www.ncbi.nlm.nih.gov), including several different Y. pestis genomes (e.g., YPO2169 in Y. pestis CO92 [data not shown]). We confirmed this conservation by successfully amplifying an internal “ytxR” fragment from the chromosomes of all the strains tested in the ytxA hybridization analysis (Fig. 2) except Y. aldovae 670-83 (data not shown). As a negative control, the PCR failed to amplify a fragment from the ΔytxR strain AJD239 (Table 1). Besides Yersinia, BLASTP searches also revealed that ytxR is conserved in the insect pathogen Photorhabdus luminescens and also in Photorhabdus asymbiotica (more than 60% amino acid identity and the same chromosomal context). These observations suggest that YtxR probably regulates genes besides ytxAB and may play an important conserved role in the closely related genera Yersinia and Photorhabdus.
DISCUSSION
Multiple Y. enterocolitica strains have genes (ytxAB) that have the potential to encode an ADPRT. YtxA is a member of a large family of proven and putative bacterial ADPRTs (33), but a ytxAB null mutant is virulent in an adult mouse model of acute infection. However, YtxAB could play a role limited to the intestinal stage of disease and/or be host specific. For example, the Y. enterocolitica heat-stable enterotoxin Yst had no detectable role in mice (41) or gnotobiotic piglets (38) but did affect diarrhea, weight loss, and death in young rabbits (13). In an attempt to obtain more insight into the ytxAB locus, we have begun to investigate the regulation of its expression. Here we report identification of YtxR, an LTTR that induces expression of ytxAB and also of its own gene. This regulation is mediated by direct interaction of YtxR with the ytxA and ytxR control regions.
We discovered two different versions of the ytxAB locus in Y. enterocolitica. One version is specific to American strains, and the other is specific to non-American strains. An exception is American serotype O:18, which has the non-American version of ytxAB. Hybridization analysis with fragments of the ail gene also distinguished between American and non-American strains (30). However, the ail hybridization pattern of the same O:18 serotype strain placed it with the other American strains. We also found a third version of the ytxAB locus in a Y. intermedia isolate provided by the Food and Drug Administration (Darwin and Miller, unpublished data). The genome sequence of an American Type Culture Collection Y. intermedia strain is also now available (http://www.ncbi.nlm.nih.gov), and the genome contains genes similar to ytxAB in the sapA-pspF intergenic region. Y. intermedia, like several other Yersinia species, is considered nonpathogenic. However, it has been suggested that some of these species may sometimes cause disease by using uncharacterized virulence proteins (47).
The ytxAB locus has a G+C content of 39%, which is much lower than the average G+C content of the chromosome (47%). Perhaps the ytxAB locus was acquired by horizontal transfer. Furthermore, genome sequence analysis revealed that in Y. pestis CO92 there is an insertion element instead of ytxAB between sapA and pspF. In Y. pseudotuberculosis and some other Yersinia species there do not appear to be any coding regions between sapA and pspF. Strikingly, like ytxAB, the unlinked ytxR gene encoding their regulator also has an extremely low G+C content (33%).
The divergence of the two different versions of ytxAB in Y. enterocolitica is surprising. The two YtxA versions and two YtxB versions are only 53% and 62% identical, respectively. This contrasts with the 95% amino acid identity between Ail proteins from American and non-American Y. enterocolitica strains (3). However, despite the significant dissimilarity between the ytxA coding regions of the two American and non-American strains that we studied in detail, the 200 bp of noncoding DNA upstream of their ytxA start codons is 97% identical. This suggests that genetic drift may not explain the divergence and that different ytxAB cassettes might have been introduced two or more times into the genus Yersinia. Of course, we cannot rule out the possibility that a single ytxAB locus was acquired by an ancestral strain and strong selective pressures resulted in marked divergence of only the coding sequences.
Many bacterial genes are expressed only weakly in normal laboratory growth conditions. For example, the cholera toxin genes of V. cholerae El Tor require highly specialized conditions for expression outside the host (22). Similarly, a Φ(ytxA-lacZ) operon fusion was expressed only weakly in the laboratory, which led to the screen that identified ytxR. Like the majority of LTTRs, YtxR is an autoregulator. Most LTTRs are negative autoregulators (40), but YtxR falls into a smaller group whose members activate their own expression (for example, see reference 1). Another common feature of most (but not all) LTTRs is regulation of a gene divergently transcribed a short distance immediately upstream (40). The divergently transcribed xthA gene (YE2254) is located upstream of ytxR. However, it is separated from ytxR by almost 800 bp, and a Φ(xthA-lacZ) operon fusion is not regulated by YtxR (Axler-DiPerte and Darwin, unpublished data).
Most LTTRs are activated by an interaction with a small coinducer molecule (40). However, our experiments suggested that a coinducer may not be required for YtxR because increased expression of ytxR is sufficient to activate its target promoters. The Nac protein of Klebsiella aerogenes is an example of an LTTR that does not require a coinducer molecule. Like expression of YtxR, increased expression of nac from an IPTG-inducible promoter is sufficient to allow it to regulate target promoters (42). If YtxR does not need a coinducer, then activation of the YtxR regulon might be initiated by upregulation of ytxR expression. We do not yet know the environmental conditions that allow this to occur. However, we are beginning to obtain some clues about a possible mechanism. First, deletion analysis has suggested that a region far upstream of the ytxR promoter negatively regulates its expression (Table 2). Primer extension analysis also revealed that ytxR has a long 5′ untranslated region. Therefore, we speculated that ytxR regulation is complex and that the untranslated region may play a pivotal role in activation of the YtxR regulon.
The ytxR gene is conserved in all Yersinia and Photorhabdus species that have been sequenced, most of which do not have the ytxAB genes. This strongly suggests that there are probably other unidentified YtxR target promoters. However, we cannot yet ascribe a function to the YtxR regulon. We have not discovered a robust phenotype for a Y. enterocolitica ytxR null mutant, including in a mouse model of acute infection (Axler-DiPerte and Darwin, unpublished data). Our favored working hypothesis is that the regulon might be activated in an environment outside the host and then deactivated soon after infection. Therefore, even if the regulon primes Y. enterocolitica for stages early in infection, a phenotype in animals would not be apparent if bacteria were not grown in this YtxR-activating environment prior to infection. Another equally plausible possibility is that activation of the YtxR regulon occurs during infection but that it is host specific. Finally, YtxR may play an environmental role unrelated to host interaction. Uncovering environmental conditions that activate the YtxR regulon, and especially identifying all of the YtxR target promoters, should considerably increase our understanding of the role of the regulon in Y. enterocolitica physiology. Addressing these questions will be the major goal of our future investigations. Answering them could provide significant insight into the two very important and well-studied genera Yersinia and Photorhabdus.
Acknowledgments
This study was supported by institutional startup funds from NYU School of Medicine and by NIH grant AI01230 awarded to V.L.M. G.L.A. was supported by grant T32 AI007180 from the NIH.
We thank Joe Barbieri for many helpful discussions and Heran Darwin for a critical review of the manuscript.
Footnotes
Published ahead of print on 22 September 2006.
REFERENCES
- 1.Abe, A., H. Matsui, H. Danbara, K. Tanaka, H. Takahashi, and K. Kawahara. 1994. Regulation of spvR gene expression of Salmonella virulence plasmid pKDSC50 in Salmonella choleraesuis serovar Choleraesuis. Mol. Microbiol. 12:779-787. [DOI] [PubMed] [Google Scholar]
- 2.Bäumler, A. J., R. M. Tsolis, A. W. van der Velden, I. Stojiljkovic, S. Anic, and F. Heffron. 1996. Identification of a new iron regulated locus of Salmonella typhi. Gene 183:207-213. [DOI] [PubMed] [Google Scholar]
- 3.Beer, K. B., and V. L. Miller. 1992. Amino acid substitutions in naturally occurring variants of Ail result in altered invasion activity. J. Bacteriol. 174:1360-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bottone, E. J. 1999. Yersinia enterocolitica: overview and epidemiologic correlates. Microbes Infect. 1:323-333. [DOI] [PubMed] [Google Scholar]
- 5.Carniel, E. 2001. The Yersinia high-pathogenicity island: an iron-uptake island. Microbes Infect. 3:561-569. [DOI] [PubMed] [Google Scholar]
- 6.Carniel, E., and H. H. Mollaret. 1990. Yersiniosis. Comp. Immunol. Microbiol. Infect. Dis. 13:51-58. [DOI] [PubMed] [Google Scholar]
- 7.Cornelis, G., Y. Laroche, G. Balligand, M. P. Sory, and G. Wauters. 1987. Yersinia enterocolitica, a primary model for bacterial invasiveness. Rev. Infect. Dis. 9:64-87. [DOI] [PubMed] [Google Scholar]
- 8.Cornelis, G. R. 2002. The Yersinia Ysc-Yop ‘type III’ weaponry. Nat. Rev. Mol. Cell. Biol. 3:742-752. [DOI] [PubMed] [Google Scholar]
- 9.Cover, T. L., and R. C. Aber. 1989. Yersinia enterocolitica. N. Engl. J. Med. 321:16-24. [DOI] [PubMed] [Google Scholar]
- 10.Coye, L. H., and C. M. Collins. 2004. Identification of SpyA, a novel ADP-ribosyltransferase of Streptococcus pyogenes. Mol. Microbiol. 54:89-98. [DOI] [PubMed] [Google Scholar]
- 11.Darwin, A. J., and V. L. Miller. 2001. The psp locus of Yersinia enterocolitica is required for virulence and for growth in vitro when the Ysc type III secretion system is produced. Mol. Microbiol. 39:429-444. [DOI] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delor, I., and G. R. Cornelis. 1992. Role of Yersinia enterocolitica Yst toxin in experimental infection of young rabbits. Infect. Immun. 60:4269-4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delor, I., A. Kaeckenbeeck, G. Wauters, and G. R. Cornelis. 1990. Nucleotide sequence of yst, the Yersinia enterocolitica gene encoding the heat-stable enterotoxin, and prevalence of the gene among pathogenic and nonpathogenic yersiniae. Infect. Immun. 58:2983-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in Gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Domenighini, M., and R. Rappuoli. 1996. Three conserved consensus sequences identify the NAD-binding site of ADP-ribosylating enzymes, expressed by eukaryotes, bacteria and T-even bacteriophages. Mol. Microbiol. 21:667-674. [DOI] [PubMed] [Google Scholar]
- 17.Ellison, D. W., B. Young, K. Nelson, and V. L. Miller. 2003. YmoA negatively regulates expression of invasin from Yersinia enterocolitica. J. Bacteriol. 185:7153-7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gemski, P., J. R. Lazere, T. Casey, and J. A. Wohlieter. 1980. Presence of a virulence-associated plasmid in Yersinia pseudotuberculosis. Infect. Immun. 28:1044-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haller, J. C., S. Carlson, K. J. Pederson, and D. E. Pierson. 2000. A chromosomally encoded type III secretion pathway in Yersinia enterocolitica is important in virulence. Mol. Microbiol. 36:1436-1446. [DOI] [PubMed] [Google Scholar]
- 21.Heesemann, J., B. Algermissen, and R. Laufs. 1984. Genetically manipulated virulence of Yersinia enterocolitica. Infect. Immun. 46:105-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwanaga, M., and T. Kuyyakanond. 1987. Large production of cholera toxin by Vibrio cholerae O1 in yeast extract peptone water. J. Clin. Microbiol. 25:2314-2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson, M. W., E. Silva-Herzog, and G. V. Plano. 2004. The ATP-dependent ClpXP and Lon proteases regulate expression of the Yersinia pestis type III secretion system via regulated proteolysis of YmoA, a small histone-like protein. Mol. Microbiol. 54:1364-1378. [DOI] [PubMed] [Google Scholar]
- 24.Kinder, S. A., J. L. Badger, G. O. Bryant, J. C. Pepe, and V. L. Miller. 1993. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O8 and construction of a transformable R−M+ mutant. Gene 136:271-275. [DOI] [PubMed] [Google Scholar]
- 25.Krueger, K. M., and J. T. Barbieri. 1995. The family of bacterial ADP-ribosylating exotoxins. Clin. Microbiol. Rev. 8:34-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maloy, S. R., V. J. Stewart, and R. K. Taylor. 1996. Genetic analysis of pathogenic bacteria. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 27.Maxson, M. E., and A. J. Darwin. 2004. Identification of inducers of the Yersinia enterocolitica phage shock protein system and comparison to the regulation of the RpoE and Cpx extracytoplasmic stress responses. J. Bacteriol. 186:4199-4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maxson, M. E., and A. J. Darwin. 2005. Improved system for construction and analysis of single-copy β-galactosidase operon fusions in Yersinia enterocolitica. Appl. Environ. Microbiol. 71:5614-5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 30.Miller, V. L., J. J. Farmer III, W. E. Hill, and S. Falkow. 1989. The ail locus is found uniquely in Yersinia enterocolitica serotypes commonly associated with disease. Infect. Immun. 57:121-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moss, J., S. J. Stanley, D. L. Buns, J. A. Hsia, D. A. Yost, G. A. Myers, and E. L. Hewlett. 1983. Activation by thiol of the latent NAD glycohydrolase and ADP-ribosyltransferase activities of Bordetella pertussis toxin (islet-activating protein). J. Biol. Chem. 258:11879-11882. [PubMed] [Google Scholar]
- 32.Nair, G. B., and Y. Takeda. 1998. The heat-stable enterotoxins. Microb. Pathog. 24:123-131. [DOI] [PubMed] [Google Scholar]
- 33.Pallen, M. J., A. C. Lam, N. J. Loman, and A. McBride. 2001. An abundance of bacterial ADP-ribosyltransferases—implications for the origin of exotoxins and their human homologues. Trends Microbiol. 9:302-307. [DOI] [PubMed] [Google Scholar]
- 34.Pepe, J. C., and V. L. Miller. 1993. Yersinia enterocolitica invasin: a primary role in the initiation of infection. Proc. Natl. Acad. Sci. USA 90:6473-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peterson, K. M. 2002. Expression of Vibrio cholerae virulence genes in response to environmental signals. Curr. Issues Intest. Microbiol. 3:29-38. [PubMed] [Google Scholar]
- 36.Portnoy, D. A., S. L. Moseley, and S. Falkow. 1981. Characterization of plasmids and plasmid-associated determinants of Yersinia enterocolitica pathogenesis. Infect. Immun. 31:775-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Revell, P. A., and V. L. Miller. 2001. Yersinia virulence: more than a plasmid. FEMS Microbiol. Lett. 205:159-164. [DOI] [PubMed] [Google Scholar]
- 38.Robins-Browne, R. M., S. Tzipori, G. Gonis, J. Hayes, M. Withers, and J. K. Prpic. 1985. The pathogenesis of Yersinia enterocolitica infection in gnotobiotic piglets. J. Med. Microbiol. 19:297-308. [DOI] [PubMed] [Google Scholar]
- 39.Russell, D. A., G. A. Byrne, E. P. O'Connell, C. A. Boland, and W. G. Meijer. 2004. The LysR-type transcriptional regulator VirR is required for expression of the virulence gene vapA of Rhodococcus equi ATCC 33701. J. Bacteriol. 186:5576-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 41.Schiemann, D. A. 1981. An enterotoxin-negative strain of Yersinia enterocolitica serotype O:3 is capable of producing diarrhea in mice. Infect. Immun. 32:571-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwacha, A., and R. A. Bender. 1993. The product of the Klebsiella aerogenes nac (nitrogen assimilation control) gene is sufficient for activation of the hut operons and repression of the gdh operon. J. Bacteriol. 175:2116-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skorupski, K., and R. K. Taylor. 1997. Control of the ToxR virulence regulon in Vibrio cholerae by environmental stimuli. Mol. Microbiol. 25:1003-1009. [DOI] [PubMed] [Google Scholar]
- 44.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 45.Spangler, B. D. 1992. Structure and function of cholera toxin and the related Escherichia coli heat-labile enterotoxin. Microbiol. Rev. 56:622-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stewart, G. S., S. Lubinsky-Mink, C. G. Jackson, A. Cassel, and J. Kuhn. 1986. pHG165: a pBR322 copy number derivative of pUC8 for cloning and expression. Plasmid 15:172-181. [DOI] [PubMed] [Google Scholar]
- 47.Sulakvelidze, A. 2000. Yersiniae other than Y. enterocolitica, Y. pseudotuberculosis, and Y. pestis: the ignored species. Microbes Infect. 2:497-513. [DOI] [PubMed] [Google Scholar]
- 48.Wachtel, M. R., and V. L. Miller. 1995. In vitro and in vivo characterization of an ail mutant of Yersinia enterocolitica. Infect. Immun. 63:2541-2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 50.Young, V. B., V. L. Miller, S. Falkow, and G. K. Schoolnik. 1990. Sequence, localization and function of the invasin protein of Yersinia enterocolitica. Mol. Microbiol. 4:1119-1128. [DOI] [PubMed] [Google Scholar]