Abstract
The Mycobacterium tuberculosis genome encodes 12 alternative sigma factors, several of which regulate stress responses and are required for virulence in animal models of acute infection. In this work we investigated M. tuberculosis SigM, a member of the extracytoplasmic function subfamily of alternative sigma factors. This sigma factor is expressed at low levels in vitro and does not appear to function in stress response regulation. Instead, SigM positively regulates genes required for the synthesis of surface or secreted molecules. Among these are genes encoding two pairs of Esx secreted proteins, a multisubunit nonribosomal peptide synthetase operon, and genes encoding two members of the proline-proline-glutamate (PPE) family of proteins. Genes up regulated in a sigM mutant strain include a different PPE gene, as well as several genes involved in surface lipid synthesis. Among these are genes involved in synthesis of phthiocerol dimycocerosate (PDIM), a surface lipid critical for virulence during acute infection, and the kasA-kasB operon, which is required for mycolic acid synthesis. Analysis of surface lipids showed that PDIM synthesis is increased in a sigM-disrupted strain and is undetectable in a sigM overexpression strain. These findings demonstrate that SigM positively and negatively regulates cell surface and secreted molecules that are likely to function in host-pathogen interactions.
Mycobacterium tuberculosis remains a major human pathogen, causing widespread disease and mortality, particularly in the poorest countries of the world. The heightened susceptibility of human immunodeficiency virus-infected persons to M. tuberculosis and the emergence of increasingly drug-resistant strains underscore the urgency of implementing effective methods to control tuberculosis, as well as the challenges in doing so (10). A major factor in the difficulty in controlling tuberculosis is the 6- to 9-month course of treatment that is required to achieve cure of both active and latent tuberculosis. One reason that such prolonged treatment is thought to be needed is the lack of activity of current antituberculars against populations of nonreplicating persistent bacteria (53).
In contrast to many bacterial pathogens, where virulence mechanisms such as toxin production or secretion of other effector molecules cause host cell death and lead to acute disease following infection, primary M. tuberculosis infection typically causes minimal disease (26). Though in a small minority of persons initial infection may progress to clinically apparent disease, more commonly the infection is contained and infected individuals remain asymptomatic. In some infected persons, years to decades later, active bacterial replication resumes, causing disease that results both from extensive bacterial replication and a host inflammatory response that is unable to control the infection.
This disease pattern suggests the need for sophisticated bacterial mechanisms that are driven by the host environment, to allow initial active replication, long-term persistence, and the ability to resume active replication. While substantial progress has been made in identifying many genes required for initial replication and virulence in short-term infection models, these processes may be most relevant only for the early stages of M. tuberculosis infection. Adaptations associated with macrophage infection, starvation, and hypoxia have been identified (1, 31, 42, 44). Relatively little is known, however, about mechanisms that allow this organism to transition from active replication to a state of long-term persistence in the absence of extensive replication and then back again to active replication.
Most bacterial adaptations result from changes in specific gene expression. Our laboratory has focused on the regulation of M. tuberculosis transcription by alternative sigma factors as a mechanism by which this bacterium can achieve transcriptional regulation in response to specific stimuli. By controlling multiple genes and operons, alternative sigma factors can modify bacterial physiology and/or modulate the host-pathogen interaction in response to environmental signals. In past work, we and others have identified regulatory pathways that play critical roles in mycobacterial stress response regulation and in virulence in acute infection models in mice (16, 19, 22, 29, 30, 38, 40, 48, 52). In the current work, we have characterized the regulon of the M. tuberculosis alternative sigma factor SigM. Negative regulation of virulence-associated surface lipids and positive regulation of Esx family secreted protein and nonribosomal peptide synthetase genes by this sigma factor suggest that SigM activity may play a role in long-term in vivo adaptation to specific host environments, rather than in virulence early during the course of infection.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
M. tuberculosis H37Rv was used as the parental strain for generating sigM mutant and sigM overexpression strains and as the wild type (wt) for microarray experiments. Mycobacteria were grown in Middlebrook 7H9 medium (Difco) supplemented with ADC (0.5% albumin, 0.2% dextrose, and 0.085% sodium chloride), 0.2% glycerol, and 0.05% Tween 80. Liquid cultures were grown in vented flasks that were shaken at 100 rpm at 37°C. Solid medium was made using Middlebrook 7H9 supplemented with ADC, 0.2% glycerol, and 1.5% agar.
Escherichia coli strains DH5α (Life Technologies), TOP10 (Invitrogen), and XL1-Blue (Stratagene) were used as host strains for cloning experiments. Luria-Bertani (LB) broth or agar plates containing appropriate antibiotic were used for growing E. coli strains; 50 μg/ml kanamycin, 100 μg/ml ampicillin, and 100 μg/ml hygromycin were added to the culture medium as indicated, and 10% sucrose was added to the solid medium for counterselection of M. tuberculosis mutants.
Real time RT-PCR for mRNA quantitation.
RNA extracted from M. tuberculosis H37Rv starting at early log phase and ending at stationary phase was used to measure growth phase-dependent changes in expression of sigM. The reverse transcription and PCR were performed using the QuantiTech SYBR green reverse transcription-PCR (RT-PCR) kit (QIAGEN) and an ABI7000 sequence detection system. Duplicates of each reaction with 200 ng of templates were performed. Control reactions were performed using the sigA gene, which has been shown to be stably expressed during growth (28). Serial dilutions of genomic DNA were amplified using the sigM or sigA primers to generate standard curves of sigA or sigM DNA versus threshold cycle. The relative amounts of cDNA produced in the RT-PCRs were interpolated from these standard curves and expressed as the ratio of sigM to sigA.
Construction of M. tuberculosis sigM mutant and sigM-overexpressing strains and complementation.
The M. tuberculosis sigM mutant was constructed by targeted mutagenesis using a temperature-sensitive sacB delivery system as described previously (35, 38). Briefly, sigM was disrupted by insertion of a kanamycin resistance gene, and the resulting construct was cloned into pRH1351, which harbors the sacB and xylE genes (38). The resulting vector was electroporated into M. tuberculosis H37Rv, and transformants were selected at 30°C on kanamycin plates. Single colonies were grown in broth at 30°C and then plated on sucrose plates at 39°C. The resulting colonies were analyzed by PCR, and candidate sigM mutants were confirmed by Southern analysis.
An M. tuberculosis strain overexpressing sigM from the inducible acetamidase promoter was created as described previously (19). The sigM open reading frame was PCR amplified and cloned downstream of an inducible acetamidase promoter in the integrating vector pMV306 (47). The construct was sequenced and electroporated into wild-type M. tuberculosis H37Rv. The transformants were selected for kanamycin resistance. An extra copy of sigM at the L5 phage attB site was confirmed by PCR. Expression of sigM was measured by real-time PCR to confirm inducible expression.
Complementation of the sigM mutant was performed as described previously (19). Briefly, sigM with the 5′ region containing the sigM promoter sequences was cloned into the integrating vector pMV306 containing a hygromycin resistance cassette. The construct was electroporated into the sigM mutant strain of M. tuberculosis, and the colonies were selected for hygromycin resistance. The resulting clones were screened by PCR.
Microarray experiments and gene identification.
Oligonucleotide microarrays of M. tuberculosis obtained from the Pathogen Functional Genomic Resource at The Institute for Genomic Research were used for microarray experiments to identify SigM-regulated genes. These microarrays represent all the annotated genes of M. tuberculosis H37Rv and additional genes from M. tuberculosis strain CDC1551. These additional genes either are not annotated or are differently annotated in strain H37Rv.
Two separate sets of microarray expression experiments were performed. In one set the wild type was compared to the sigM mutant, and in the second set the sigM overexpression strain of M. tuberculosis was compared to the sigM mutant. Six independent RNA samples were hybridized and analyzed for the comparison of wild type versus sigM mutant, and three independent samples were analyzed in the comparison of sigM overexpression strain versus mutant. In all experiments the test sample (wt or sigM overexpression strain) was labeled with Cy5 and the control (sigM mutant) was labeled with Cy3. In experiments involving the sigM overexpression strain, 0.2% acetamide was added to the cultures 1 hour prior to harvesting. RNA isolation, cDNA synthesis, labeling, and hybridization were performed as described previously (38).
The hybridized slides were scanned using an Axon 4000B scanner. Microarray spot intensities were defined and quantified using GENEPIX software (Axon, Union City, CA). The median value from the raw data was normalized by the LOWESS method (23). Statistical analysis was performed using BRB array tools (wt versus mutant arrays) (45) (http://linus.nci.nih.gov/BRB-ArrayTools.html) and the National Institute of Aging array analysis tool (overexpression versus mutant arrays) (43) (http://lgsun.grc.nia.nih.gov/ANOVA/). We used the most conservative error model, “the maximum of averaged and actual error variance,” in order to reduce false positives. P values are false detection rate values, i.e., the proportion of false positives expected among genes detected as significant.
RNA preparation and primer extension.
RNA isolation and primer extension experiments were performed as described previously (38). Total RNA was isolated from wild-type M. tuberculosis, the sigM mutant strain, and the sigM-overexpressing strain grown to mid-log phase (optical density at 600 nm = 0.4). Acetamide was added to each culture to a concentration of 0.2% at 1 hour prior to harvesting. For primer extension analysis, 0.5 pmol of γ-32P-labeled reverse primer was mixed with 8 μg of RNA in a 6-μl volume. Reverse transcription reactions were then performed as described, and the reaction products were analyzed on a sequencing gel. DNA sequencing reactions, performed with the same primer used for primer extension, were run in adjacent lanes to determine the sizes of the transcripts generated. Specific transcripts were quantified with a phosphorimager and normalized to a sigA transcript generated from the same RNA sample.
Lipid analysis.
Nonpolar surface lipids were extracted from late-log-phase cultures of wt, sigM insertional mutant, and sigM-overexpressing strains of M. tuberculosis. Each strain was grown in 50 ml of medium in a 250-ml flask to late log phase, and the wet weights of the cell pellets were determined. The pellets were resuspended in 1 ml of hexane and vortexed for 5 min. The cell suspension was centrifuged at 3,000 rpm for 5 minutes, and the hexane fraction was collected. Three milliliters of chloroform-methanol (1:1) was added to the hexane fraction. The extracted lipids in hexane-chloroform-methanol were dried under a stream of nitrogen and resuspended in 0.5 ml of chloroform-methanol (1:1).
The lipids were separated by silica thin-layer chromatography (TLC) with hexane and chloroform (9:1) as the solvent system. The lipids were visualized by spraying with acid (8% sulfuric acid and 3% cupric acetate) followed by heating at 110°C for 15 min. Pthiocerol dimycocerosate (PDIM) containing PIM A, PIM B, and PIM C from the laboratory of Gurdyal Besra was used as a standard.
For mass spectrometry (MS) analysis, hexane-extracted surface lipids from various strains and PDIM standards were loaded onto 250-μm silica-coated glass TLC plate (10 by 20 cm) and then developed in a hexane-chloroform (9:1) solvent system. The lipids were visualized by spraying the plate with water, and three spots comigrating with the PDIM standard were marked. After overnight air drying, silica at the marked spots was loaded into a nanospray tip, and lipids were dissolved in 10 μl of 10 mM ammonium acetate (NH4C2H3O2) in chloroform-methanol (2:1) for positive-mode electrospray ionization mass spectrometry (ThermoFinnigan LCQ Advantage). Selected ions were analyzed by collisionally induced dissociation (CID)-MS/MS. The MS data were compared to the PDIM molecular structure described by Camacho et al. (5).
RESULTS
Mutant construction, in vitro growth, and sigM expression.
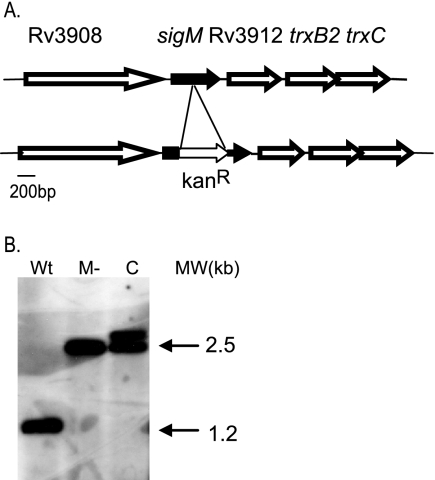
Genome sequence analysis indicates the presence of a homologue of M. tuberculosis sigM in Mycobacterium bovis, Mycobacterium smegmatis, and Mycobacterium avium paratuberculosis (18, 25, 50). In each of these organisms sigM is located 5′ of a thioredoxin reductase-thioredoxin operon, with a single gene of unknown function separating sigM from the thioredoxin reductase gene (trxB2) (Fig. 1A). In Mycobacterium leprae, remnants of a sigM homologue are present in the corresponding region, but this organism does not appear to contain a functional copy of sigM. Targeted inactivating mutations were made in sigM in both M. smegmatis and M. tuberculosis. Candidate mutants were screened by PCR and confirmed by Southern blot analysis (Fig. 1B shows results for M. tuberculosis). Complemented strains were prepared by integrating a single copy of sigM under the control of its own promoter into the L5 phage attB site of the sigM mutant (24). An inducible overexpression strain was constructed by introducing a copy of sigM under the control of the inducible acetamidase promoter (33) into wt H37Rv at the L5 attB site.
FIG. 1.
The M. tuberculosis sigM locus and mutant construction. A. sigM is located 5′ of the thioredoxin reductase-thioredoxin operon. Our sequence data indicate a 93-bp intergenic region between sigM and Rv3912 in H37Rv, identical to the CDC1551 genome sequence but distinct from the published H37Rv sequence (9, 17). B. Southern blot analysis demonstrates the insertional inactivation of sigM. Wt, H37Rv; M−, sigM mutant, C, complemented sigM mutant.
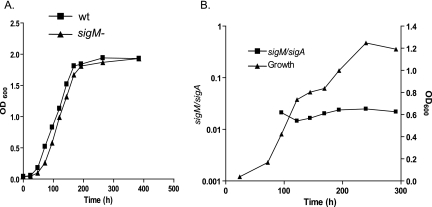
Comparison of growth in Middlebrook 7H9 liquid medium was performed for the wt and sigM mutant strains. These two strains had nearly identical growth rates throughout the growth cycle (Fig. 2A). Analysis of sigM gene expression was performed at serial time points during in vitro growth. The expression of M. tuberculosis sigM was shown to be markedly lower than that of the primary sigma factor gene, sigA, which was stably expressed at all stages of growth (Fig. 2B). Expression of sigM did not increase significantly at any growth stage, suggesting that sigM is not likely to play a major role in nutrient-replete growth or in stationary phase adaptation.
FIG. 2.
Growth of H37Rv versus that of the sigM mutant and expression of sigM in broth culture. A. Cultures were grown in Middlebrook 7H9 liquid medium, and the optical density at 600 nm (OD600)was measured at serial time points. B. RNA was extracted from H37Rv cultures at serial time points during growth and subjected to quantitative RT-PCR. Relative expression of sigM is shown as a ratio of sigM cDNA to sigA cDNA. Experiments were performed twice, with representative results of one experiment shown.
Based on its location adjacent to the thioredoxin reductase-thioredoxin locus in M. tuberculosis and M. smegmatis, we hypothesized that sigM might play a role in regulation of oxidative stress responses. Examination of survival following exposure to several types of oxidative stress (diamide, hydrogen peroxide, cumene hydroperoxide, and plumbagin) was performed, utilizing disc diffusion assays on agar plates and time-kill assays in broth culture. These experiments did not show a significant effect on viability of the M. smegmatis or M. tuberculosis sigM mutants relative to the parental wt strain (data not shown).
Identification of SigM-regulated genes and consensus SigM-dependent promoter elements.
In the absence of an apparent role in oxidative stress adaptation, we sought to determine the genes that are regulated by this sigma factor as a means to gain insight into SigM function. Genome-wide transcription profiling experiments were performed using oligonucleotide glass slide microarrays. Initial experiments compared gene expression between the wt and the sigM mutant during log-phase growth in broth culture. In these experiments, a small number of positively regulated genes were identified. The signal strength and the expression ratios of genes with increased expression in the wt versus the sigM mutant in these experiments were quite low (spot intensity of <1% of maximum). Primer extension experiments performed with five genes that showed increased expression in the wt versus the sigM mutant did not identify any promoters that were clearly SigM dependent. These microarray experiments also confirmed the low basal expression of sigM in wt H37Rv.
Several genes were determined to have significantly increased expression in the sigM mutant versus the wt strain (Table 1). Most striking was the markedly increased expression of PPE60 in the sigM mutant. This gene encodes a protein in the proline-glutamate (PE)_polymorphic GC-rich repetitive sequence (PGRS) family of over 100 homologous proteins. These proteins contain a PE or proline-proline-glutamate (PPE) motif near the amino-terminus, and the PE_PGRS proteins also encode peptides that rich in glycine and alanine. Some of these proteins have been shown to be surface associated and to be recognized by the host immune system during infection (3, 12). Also notable was the moderately increased expression of several lipid biosynthetic genes, including most genes from the first operon of the PDIM biosynthetic and transport locus, and the independently transcribed mas gene from this locus, but not the fadD28-mmpL7 operon, which is also required for PDIM synthesis and export (5). Also up regulated were genes in the kasA-kasB operon, which functions in mycolic acid synthesis, as well as other independently transcribed lipid biosynthesis genes. Expression of several genes in the region of the devR-devS-Rv3134c hypoxia-induced two-component system operon (44) was also increased in the sigM mutant.
TABLE 1.
Genes with increased expression in the sigM mutant strain versus H37Rva
| Gene | Ratio | P value | Gene product |
|---|---|---|---|
| Rv0237 | 1.7 | 0.007 | LpqI |
| Rv0885 | 1.8 | 0.007 | HPb |
| Rv1180 | 1.8 | 0.001 | Pks3 |
| Rv2245 | 1.6 | 0.002 | KasA |
| Rv2246 | 1.8 | <0.001 | KasB |
| Rv2247 | 1.9 | <0.001 | AccD6 |
| Rv2248 | 1.6 | 0.002 | HP |
| Rv2524c | 1.9 | 0.003 | Fas |
| Rv2590 | 1.6 | 0.002 | FadD9 |
| Rv2931 | 2.0 | 0.005 | PpsA |
| Rv2932 | 3.5 | 0.001 | PpsB |
| Rv2933 | 3.6 | <0.001 | PpsC |
| Rv2934 | 3.1 | <0.001 | PpsD |
| Rv2935 | 2.3 | 0.002 | PpsE |
| Rv2936 | 1.7 | <0.001 | DrrA |
| Rv2937 | 1.7 | 0.009 | DrrB |
| Rv2938 | 2.1 | 0.004 | DrrC |
| Rv2940c | 1.8 | 0.008 | Mas |
| Rv2950c | 1.7 | 0.007 | FadD29 |
| Rv3130c | 10.9 | 0.003 | HP |
| Rv3131 | 5.9 | 0.005 | HP |
| Rv3133c | 1.7 | 0.004 | DevR |
| Rv3134c | 2.2 | 0.004 | HP |
| Rv3135 | 2.2 | 0.001 | PPE50 |
| Rv3136 | 1.9 | <0.001 | PPE51 |
| Rv3137 | 2.0 | 0.001 | HP |
| Rv3138 | 1.6 | 0.009 | PflA |
| Rv3478 | 117.8 | <0.001 | PPE60 |
| Rv3616c | 2.0 | 0.009 | HP |
| Rv3689 | 1.7 | 0.009 | HP |
| Rv3825c | 1.8 | 0.002 | Pks2 |
Data are from six biological replicates. Genes with an expression ratio of ≥1.6 and a P value of < 0.01 are shown.
HP, hypothetical protein.
To attempt to more definitively identify SigM-regulated genes, additional microarray experiments were performed comparing RNA from an inducible sigM overexpression strain to RNA from the sigM mutant, also treated with inducer. Induction increases sigM expression approximately 60-fold in the overexpression strain, as determined by quantitative RT-PCR (data not shown), similar to the 78-fold induction observed in the microarray experiments. Several genes with highly increased expression in the sigM overexpression strain were identified, many of which clustered into apparent operons based on analysis of the M. tuberculosis genome sequence. The most highly regulated genes are shown in Table 2. The data for all significantly regulated genes from both sets of microarray experiments are available in the CIBEX database (accession no. CBX13).
TABLE 2.
Genes with increased expression in the sigM overexpression strain versus the sigM mutanta
| Gene | Ratio | P value | Gene product |
|---|---|---|---|
| MT2638 | 6.4 | <0.001 | HPb |
| Rv0098 | 10.9 | <0.001 | HP |
| Rv0099 | 10.9 | <0.001 | FadD10 |
| Rv0100 | 8.6 | <0.001 | HP |
| Rv1361 | 8.1 | 0.022 | PPE19 |
| Rv1457c | 7.3 | <0.001 | HP |
| Rv2514c | 7.0 | <0.001 | HP |
| Rv2515c | 9.2 | <0.001 | HP |
| Rv3396c | 6.5 | <0.001 | GuaA |
| Rv3422c | 6.1 | <0.001 | HP |
| Rv3439c | 735.7 | <0.001 | HP |
| Rv3440c | 321.5 | <0.001 | HP |
| Rv3443c | 6.5 | 0.012 | RplM |
| Rv3444c | 66.8 | <0.001 | EsxT |
| Rv3445c | 1,792.3 | <0.001 | EsxU |
| Rv3904c | 58.3 | <0.001 | EsxE |
| Rv3905c | 95.3 | <0.001 | EsxF |
| Rv3906c | 573.1 | <0.001 | HP |
| Rv3907c | 6.7 | <0.001 | PcnA |
| Rv3911 | 77.9 | <0.001 | SigM |
Data are from three biological replicates. Genes with an expression ratio of >6 and a P value of <0.05 are shown.
HP, hypothetical protein.
Among the most highly regulated genes were sigM itself, though not the adjacent 3′ gene Rv3912, and several genes predicted to be involved in the production of molecules that localize to the bacterial surface or are secreted. These include genes encoding the ESAT-6-like secreted proteins EsxU and EsxT, which are part of the Esx-4 putative protein secretion locus; a separate two-gene operon (Rv3440c-Rv3439c) of unknown function adjacent to this locus; and two other Esx family proteins (EsxE and EsxF) that are encoded in a different region of the chromosome. Also regulated by SigM were a two-gene operon carrying a gene of unknown function, PPE19, and an operon carrying PPE1 and a nonribosomal peptide synthetase (NRPS) gene together with three genes of unknown function and a putative fatty acid biosynthetic gene.
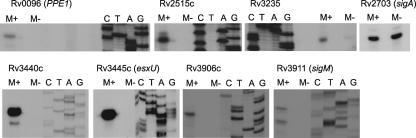
To identify candidate SigM-regulated promoters, primer extension analysis was performed on genes with the highest expression ratios. Where multiple genes from an apparent operon were up regulated in the sigM overexpression strain, primer extension analysis was performed on the first gene in the operon. These experiments identified several promoters that appear to be wholly dependent on SigM and one for which the majority of expression is SigM dependent (Fig. 3). The SigM-dependent esxU promoter identified in these experiments lies within the esxU coding region in the H37Rv and CDC1551 annotated genome sequences. This result indicates that the in vivo start codon of this gene is further 3′ than predicted. The potential initiation codons 3′ of the transcription start site would result in an EsxU protein of 105 or 92 amino acids (rather than the annotated 125 residues), which is more consistent with the predicted sizes of the other Esx proteins, which range in size from 90 to 119 residues.
FIG. 3.
Primer extension analysis of SigM-dependent promoters. RNA was isolated from the sigM overexpression strain (M+) and the sigM mutant (M−) and reverse transcribed with gene-specific antisense primers. A sequencing reaction was performed with the same primer, and reactions were resolved on sequencing gels to identify transcription start sites. Reverse transcription was performed using the same RNA with a sigA primer as a control in each experiment.
By aligning the transcription start sites of these promoters, consensus −35 and −10 elements were identified (Fig. 4), suggesting that these promoters are directly regulated by SigM. The −35 consensus sequence is similar to that of other sequences recognized by mycobacterial extracytoplasmic function (ECF) sigma factors. The SigM-dependent −35 sequence appears to be more highly conserved than the −10 element, consistent with the primary importance of the −35 element sequence in promoter recognition by many ECF subfamily sigma factors (27).
FIG. 4.
Alignment of SigM-dependent promoters. The regions 5′ of experimentally determined SigM-dependent transcription start sites were aligned. The conserved sequences in the −35 and −10 regions are boxed, with conserved bases shown in boldface. The transcription start site of each gene is underlined and shown in boldface.
Negative regulation of PDIM synthesis by SigM.
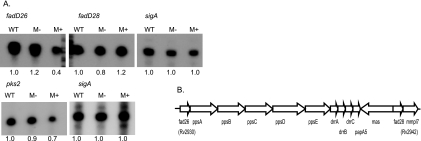
The microarray experiments indicated that SigM negatively regulates a number of lipid biosynthetic genes, including several genes involved in PDIM biosynthesis. To confirm this result, primer extension analyses were performed to examine specific transcript levels by using RNA from wt, sigM mutant, and sigM overexpression strains. Transcript signals were quantified for each strain and compared to signals from sigA transcripts generated using the same batches of RNA. These experiments confirmed moderately decreased expression of the transcript originating at the promoter of the first gene in the first PDIM operon, fadD26, in the sigM overexpression strain (Fig. 5). In contrast, fadD28 transcription, which was not found to be significantly different in the sigM mutant versus overexpression strains in the microarray analysis, was also not significantly altered in the sigM mutant or overexpression strains relative to the wt in the primer extension experiments. These experiments also confirmed decreased transcription in the sigM overexpression strain of pks2, a mas-like lipid biosynthetic gene whose product has been shown to synthesize precursors of sulfolipids (46) (Fig. 5).
FIG. 5.
sigM expression represses PDIM biosynthetic gene transcription. A. Quantitative primer extension analysis was performed using equal amount of total RNA from the H37Rv (wt), sigM mutant (M−), and sigM overexpression (M+) strains of M. tuberculosis. The numbers below each lane indicate quantification of the transcript relative to expression in the wt, normalized to sigA expression in the same experiment. Experiments were performed twice, and representative results are shown. B. Organization of the PDIM biosynthesis and transport locus. Arrows indicate the three transcripts previously identified in this region (5).
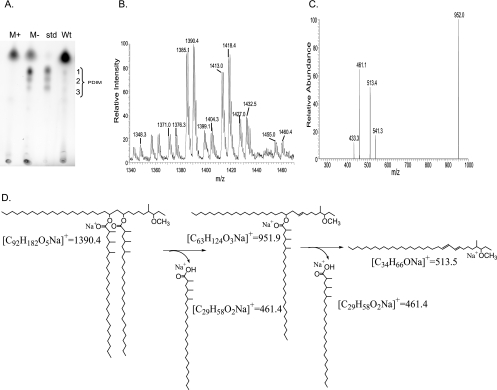
To determine whether the effects of sigM expression on transcription of PDIM biosynthesis genes resulted in altered PDIM production, nonpolar surface lipids were extracted from wt, sigM mutant, and sigM overexpression strains and analyzed by TLC (Fig. 6A). Three faintly visible spots from the wild-type extracts comigrated with a standard containing a mixture of PDIM A, PDIM B, and PDIM C. Markedly stronger spots were consistently present in the sigM-disrupted strain. In contrast, no spots were visible in this region in the sigM overexpression strain (Fig. 6A). To verify the identity of these lipids that were negatively regulated by SigM, preparative TLC was performed and lipids comigrating with PDIM standards were directly transferred to nanospray tips, extracted from the matrix, and analyzed by mass spectrometry. Initial analysis showed ions from all three TLC spots with mass/charge (m/z) ratios similar or identical to m/z ratios observed in the PDIM standards (Fig. 6B). The top spot in the triad was further analyzed, and two series of ions were observed. One series of monovalent ions (m/z 1376, 1390, 1404, 1418, and 1432) corresponded to the sodium adducts (M + Na)+ of PDIM A containing 91 to 95 carbon atoms (5). Another ion series corresponded to the same set of molecules detected as ammonium adducts (M + NH4)+ (Fig. 6B). CID-MS/MS of the ion at m/z 1390.4 generated the product ion at m/z 951.9 ([M + Na]+) corresponding to the loss of one mycocerosic acid, and the mycocerosic acid could be detected directly as well ([M + Na]+ = 461). Fragmentation of the ion of m/z 951.9 generated a product ion corresponding to that expected from loss of the second mycocerosic acid (Fig. 6C and D). These results confirm the identity of the SigM down-regulated lipids as PDIM.
FIG. 6.
sigM expression inhibits PDIM synthesis. A. Nonpolar surface lipids were extracted and subjected to silica TLC in comparison with a PDIM standard B. Mass spectrometry analysis of lipids corresponding to the uppermost spot in the PDIM triad, which were directly scraped onto nanospray tips, extracted by solvent, and analyzed by nanospray electrospray ionization-MS, yielding ions that correspond to those expected from an alkane series of ammonium (m/z 1371, 1385, 1399, 1413, and 1427) and sodium (m/z 1376, 1390, 1404, 1418, and 1432) adducts of PDIM. C and D. CID mass spectra of the ion at m/z 1390.4 yielded fragment ions corresponding to sodium adducts of products that have lost one (m/z 951.9) or two (m/z 513.5) mycocerosic acids (m/z 461).
DISCUSSION
This work defines a SigM regulon that includes genes encoding two pairs of Esx family secreted proteins, two additional genes adjacent to the Esx-4 secretion locus, a nonribosomal-peptide synthesis operon, two PPE genes, and several genes of unknown function. The low-level expression of this regulon in vitro during vegetative growth suggests that sigM expression is likely to be induced in response to specific stimuli during infection. In contrast to these positively regulated genes, our data show that SigM negatively regulates PPE60 expression and the expression of several genes involved in surface lipid biosynthesis and transport.
The function of the PPE proteins and the related PE and PE_PGRS proteins is not known. Different members of this family have been shown to be induced or repressed in response to different conditions in vitro, and some of these proteins have been shown to be antigenic during human or animal infection or to be required for virulence (7, 8, 32, 37, 51). These data suggest that these proteins function to modulate immunity or other aspects of host-pathogen interactions. The positive regulation by SigM of PPE1 and PPE19, versus negative regulation of PPE60 by SigM, suggests distinct functions for these PPE proteins in vivo and is consistent with the recently described opposite regulation in vivo of PE_PGRS16 versus PE_PGRS26 (14).
Among the lipid genes negatively regulated by SigM are the kasA-kasB operon, required for mycolic acid synthesis; the type I fatty acid synthase gene fas; pks2 and pks3; and several genes involved in the synthesis and transport of PDIMs. These data suggest that sigM expression may have extensive effects on regulating the content and/or quantity of M. tuberculosis surface lipids. Consistent with the gene expression data, we observed striking derepression of PDIM surface lipids in the M. tuberculosis sigM deletion strain. PDIMs have been shown to be important virulence determinants required for efficient replication in the lung during short-term infection of mice (6, 11). The negative regulation by SigM of these lipid virulence determinants suggests that SigM is not required for virulence early during the course of infection. In support of this interpretation, microarray experiments examining gene expression following acute macrophage infection did not demonstrate induction of sigM expression, nor was sigM expression significantly increased during short-term mouse infection (42, 49). Also consistent with this inference, a recent study in which sigma factor mutant strains were examined in the guinea pig, a highly susceptible animal model of tuberculosis, found that a sigM-deleted strain was not attenuated (21). Strikingly, however, and consistent with our data showing derepression of PDIM synthesis, the sigM mutant strain appeared to be hypervirulent at the early time points in this model, with greater numbers of granulomas and increased necrosis compared to the wild type.
Our transcription and lipid synthesis data, together with these infection data, suggest the hypothesis that regulation of gene expression by SigM may play a role in bacterial adaptation later in the course of in vivo infection or in specific host tissues. In this model, SigM activity, induced in response to specific host stimuli, would result in adaptation not only via increased expression of SigM-dependent genes but also by down-regulating surface lipids and other factors that are important in acute infection. Microarray transcription analysis of in vitro models of persistence that may be relevant to latency, i.e., starvation and hypoxia, does not show increased expression of sigM or the genes that it regulates under these conditions (1, 44). Similarly, our data do not show increased sigM transcription on entry into stationary phase. If SigM does play a role in adaptation to specific host environments, these data indicate that the host signals required to induce sigM expression are not modeled by these in vitro conditions.
SigM also regulates the Esx proteins EsxT and EsxU, present in the Esx-4 putative secretion locus, and two other Esx proteins, EsxE and EsxF, that are not physically linked to genes encoding a putative secretion apparatus. ESAT-6 and Cfp-10, the first-described members of the Esx family of small secreted proteins, have been extensively characterized as potent T-cell antigens and important virulence factors (reviewed in reference 4). Of the other 21 M. tuberculosis Esx proteins, EsxG and EsxH have been shown to be regulated by IdeR in response to iron depletion, and a number of Esx proteins have been shown to be recognized by immune T cells in humans or animals infected with M. tuberculosis (4, 41). Based on the data discussed above, in contrast to ESAT-6 and Cfp-10, which are important in the pathogenesis of acute infection, we hypothesize that these SigM-regulated Esx proteins may function in bacterial adaptation and/or host modulation at later stages of infection.
The SigM-dependent operon encoding an NRPS (Rv0096-Rv0101) is also likely to function in modulation of the host-bacterium interaction. In Streptomyces spp., where they have been extensively studied, NRPSs synthesize a wide range of bioactive molecules, including several antibiotics, many of which are secreted. A separate M. tuberculosis gene cluster that includes genes encoding NRPSs and polyketide synthases has been shown to synthesize mycobactins, secreted iron-chelating siderophores that are required for bacterial replication in macrophages (13, 36). The SigM-dependent NRPS-encoding operon is highly conserved in M. bovis and M. leprae but not in M. smegmatis, although there is significant similarity between an M. smegmatis NRPS-encoding gene and the M. tuberculosis nrp gene. Disruption of this M. smegmatis locus, which contains four NRPS modules, resulted in absence of glycopeptidolipid biosynthesis, suggesting that these genes may synthesize the peptide core of the glycopeptidolipids in this species (2). Though only Rv0101 is annotated as encoding an NRPS in the M. tuberculosis H37Rv genome, using sequence- and structure-based motif analysis methods (15), we have identified NRPS motifs in both Rv0099 and Rv0100. This analysis indicates that Rv0099 (annotated as fadD10) encodes an adenylation/activation domain-containing protein and that Rv0100 encodes a peptidyl carrier protein. In addition, our analysis indicates that the amino terminus of Rv0101 (nrp) contains an unannotated NRPS condensation domain. These analyses indicate that this operon encodes an NRPS composed of three modules, each of which contains activation, peptidyl carrier, and condensation domains, with the first two domains of the first module encoded in Rv0099 and Rv0100.
This NRPS-encoding operon has previously been shown to be up regulated in an M. tuberculosis mutant strain in which the gene encoding the response regulator of the SenX3-RegX3 two-component system was deleted. This mutant was attenuated for growth in macrophages and mildly attenuated for replication in vivo in mice, suggesting that the products of the PPE1-NRPS operon are not required for macrophage survival or short-term infection in mice (34), which is consistent with the hypothesis that SigM-regulated genes are not required for acute infection. This locus was also analyzed in a transposon mutant of M. bovis in which Mb100, the homologue of Rv0097, was disrupted (20). These authors observed that this strain did not produce PDIM or glycosylphenol-PDIM, and they hypothesized that this locus is directly involved in PDIM biosynthesis. They also demonstrated marked up regulation of transcription 5′ of the insertion and decreased gene expression 3′ of the insertion (including all genes containing NRPS motifs). Our data suggest a mechanism for the increased expression of the 5′ genes of this operon and an alternative explanation for the absence of PDIMs in the Mb100-disrupted strain. Our data suggest a model in which sigM is repressed by the products of the nrp locus. Expression of sigM is derepressed in the Mb100-disrupted strain because of decreased expression of NRPS-encoding genes 3′ of the insertion. Increased sigM expression leads to increased expression from the SigM-dependent promoter 5′ of PPE1, the first gene of this operon. The absence of PDIMs in this strain would result from repressed transcription of the known PDIM biosynthesis genes that we have shown to be negatively regulated by SigM.
This work also identified clear SigM-dependent consensus promoter elements, particularly in the −35 region. Consensus promoters recognized by four other ECF sigma factors, SigD, SigE, SigH, and SigL, have been identified experimentally (19, 38-40). When the SigM-dependent consensus is compared to these promoters, a core five-base sequence, GGAAC, is found in the −35 element, with T in place of the first G in the SigL-dependent consensus and the T in place of the second G in the SigD-dependent consensus. There is more variation among the consensus sequences at the positions immediately 5′ and 3′ to this core sequence. The −10 element consensus sequences are more divergent than the −35 element. The SigM consensus is most similar to the −35 region of the promoters recognized by SigE and SigH, but it lacks the conserved −10 GTT sequence of these promoters. Reviews of our and other groups' published microarray data do not suggest that the SigM-regulated genes identified here are also significantly regulated by any of these other ECF sigma factors (19, 22, 30, 38). These findings suggest that the core −35 region is required for efficient promoter binding and/or transcription initiation of all of these sigma factors and that binding specificity results from a limited number of nucleotides at either end of the −35 element and from differences in the −10 region sequences.
This work demonstrates the regulation by SigM of genes that are likely to modulate the host-pathogen interaction. Changes in SigM activity provide a mechanism by which the production of surface-associated and secreted molecules can be altered during infection. The absence of induction of sigM following macrophage or short-term mouse infection, the negative regulation by SigM of PDIM synthesis, the increased virulence of a sigM mutant in guinea pigs, and the attenuation in macrophages and short-term mouse infection of a mutant in which the SigM-dependent PPE1-nrp operon is up regulated indicate that the SigM regulon does not play a role in M. tuberculosis virulence early during infection. Rather, we hypothesize that SigM may function in M. tuberculosis pathogenesis at later stages of infection, e.g., during latency or in specific host tissue environments. Elucidation of the function of SigM-dependent genes and analysis of the expression of SigM-regulated genes during the course of acute and chronic infection will be essential to gain insight into the role of the SigM regulon in the complex pathogenesis of tuberculosis.
Acknowledgments
This work was supported by Public Health Service grant AI37901 from the National Institute of Allergy and Infectious Diseases (NIAID) to R.N.H. and by grants AI07155 and AI049313 from NIAID and a grant from the Pew Foundation to D.B.M.
Microarrays were provided by the NIAID-sponsored Pathogen Functional Genomic Resource at TIGR. We thank Gurdyal S. Besra for the gift of the PDIM standard, Seby L. Edassery for assistance with microarray analysis, Neal Padte for construction of the M. smegmatis sigM mutant, and Simon Dove for review of the manuscript.
Footnotes
Published ahead of print on 6 October 2006.
REFERENCES
- 1.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. [DOI] [PubMed] [Google Scholar]
- 2.Billman-Jacobe, H., M. J. McConville, R. E. Haites, S. Kovacevic, and R. L. Coppel. 1999. Identification of a peptide synthetase involved in the biosynthesis of glycopeptidolipids of Mycobacterium smegmatis. Mol. Microbiol. 33:1244-1253. [DOI] [PubMed] [Google Scholar]
- 3.Brennan, M. J., and G. Delogu. 2002. The PE multigene family: a ‘molecular mantra’ for mycobacteria. Trends Microbiol. 10:246-249. [DOI] [PubMed] [Google Scholar]
- 4.Brodin, P., I. Rosenkrands, P. Andersen, S. T. Cole, and R. Brosch. 2004. ESAT-6 proteins: protective antigens and virulence factors? Trends Microbiol. 12:500-508. [DOI] [PubMed] [Google Scholar]
- 5.Camacho, L. R., P. Constant, C. Raynaud, M. A. Laneelle, J. A. Triccas, B. Gicquel, M. Daffe, and C. Guilhot. 2001. Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis. Evidence that this lipid is involved in the cell wall permeability barrier. J. Biol. Chem. 276:19845-19854. [DOI] [PubMed] [Google Scholar]
- 6.Camacho, L. R., D. Ensergueix, E. Perez, B. Gicquel, and C. Guilhot. 1999. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol. Microbiol. 34:257-267. [DOI] [PubMed] [Google Scholar]
- 7.Choudhary, R. K., S. Mukhopadhyay, P. Chakhaiyar, N. Sharma, K. J. Murthy, V. M. Katoch, and S. E. Hasnain. 2003. PPE antigen Rv2430c of Mycobacterium tuberculosis induces a strong B-cell response. Infect. Immun. 71:6338-6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choudhary, R. K., R. Pullakhandam, N. Z. Ehtesham, and S. E. Hasnain. 2004. Expression and characterization of Rv2430c, a novel immunodominant antigen of Mycobacterium tuberculosis. Protein Expr. Purif. 36:249-253. [DOI] [PubMed] [Google Scholar]
- 9.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 10.Corbett, E. L., C. J. Watt, N. Walker, D. Maher, B. G. Williams, M. C. Raviglione, and C. Dye. 2003. The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch. Intern. Med. 163:1009-1021. [DOI] [PubMed] [Google Scholar]
- 11.Cox, J. S., B. Chen, M. McNeil, and W. R. Jacobs, Jr. 1999. Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 402:79-83. [DOI] [PubMed] [Google Scholar]
- 12.Delogu, G., C. Pusceddu, A. Bua, G. Fadda, M. J. Brennan, and S. Zanetti. 2004. Rv1818c-encoded PE_PGRS protein of Mycobacterium tuberculosis is surface exposed and influences bacterial cell structure. Mol. Microbiol. 52:725-733. [DOI] [PubMed] [Google Scholar]
- 13.De Voss, J. J., K. Rutter, B. G. Schroeder, H. Su, Y. Zhu, and C. E. Barry III. 2000. The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc. Natl. Acad. Sci. USA 97:1252-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dheenadhayalan, V., G. Delogu, M. Sanguinetti, G. Fadda, and M. J. Brennan. 2006. Variable expression patterns of Mycobacterium tuberculosis PE-PGRS genes: evidence that PE_PGRS16 and PE_PGRS26 are inversely regulated in vivo. J. Bacteriol. 188:3721-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.EMBL. 15 May 2006, accession date. Interpro Scan. http://www.ac.uk/InterProScan/index.html.
- 16.Fernandes, N., Q.-L. Wu, D. Kong, X. Puyang, S. Garg, and R. Husson. 1999. A mycobacterial extracytoplasmic function sigma factor involved in survival following heat shock and oxidative stress. J. Bacteriol. 181:4266-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleischmann, R. D., D. Alland, J. A. Eisen, L. Carpenter, O. White, J. Peterson, R. DeBoy, R. Dodson, M. Gwinn, D. Haft, E. Hickey, J. F. Kolonay, W. C. Nelson, L. A. Umayam, M. Ermolaeva, S. L. Salzberg, A. Delcher, T. Utterback, J. Weidman, H. Khouri, J. Gill, A. Mikula, W. Bishai, W. R. Jacobs, Jr., J. C. Venter, and C. M. Fraser. 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 184:5479-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garnier, T., K. Eiglmeier, J. C. Camus, N. Medina, H. Mansoor, M. Pryor, S. Duthoy, S. Grondin, C. Lacroix, C. Monsempe, S. Simon, B. Harris, R. Atkin, J. Doggett, R. Mayes, L. Keating, P. R. Wheeler, J. Parkhill, B. G. Barrell, S. T. Cole, S. V. Gordon, and R. G. Hewinson. 2003. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. USA 100:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hahn, M. Y., S. Raman, M. Anaya, and R. N. Husson. 2005. The Mycobacterium tuberculosis ECF sigma factor SigL regulates polyketide synthases and membrane/secreted proteins, and is required for virulence. J. Bacteriol. 187:7062-7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hotter, G. S., B. J. Wards, P. Mouat, G. S. Besra, J. Gomes, M. Singh, S. Bassett, P. Kawakami, P. R. Wheeler, G. W. de Lisle, and D. M. Collins. 2005. Transposon mutagenesis of Mb0100 at the ppe1-nrp locus in Mycobacterium bovis disrupts phthiocerol dimycocerosate (PDIM) and glycosylphenol-PDIM biosynthesis, producing an avirulent strain with vaccine properties at least equal to those of M. bovis BCG. J Bacteriol. 187:2267-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karls, R. K., J. Guarner, D. N. McMurray, K. A. Birkness, and F. D. Quinn. 2006. Examination of Mycobacterium tuberculosis sigma factor mutants using low-dose aerosol infection of guinea pigs suggests a role for SigC in pathogenesis. Microbiology 152:1591-1600. [DOI] [PubMed] [Google Scholar]
- 22.Kaushal, D., B. G. Schroeder, S. Tyagi, T. Yoshimatsu, C. Scott, C. Ko, L. Carpenter, J. Mehrotra, Y. C. Manabe, R. D. Fleischmann, and W. R. Bishai. 2002. Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative sigma factor, SigH. Proc. Natl. Acad. Sci. USA 99:8330-8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kerr, M. K., M. Martin, and G. A. Churchill. 2000. Analysis of variance for gene expression microarray data. J. Comput. Biol. 7:819-837. [DOI] [PubMed] [Google Scholar]
- 24.Lee, M., L. Pascopella, W. Jacobs, Jr., and G. Hatfull. 1991. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis and bacille Calmette-Guerin. Proc. Natl. Acad. Sci. USA 88:3111-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, L., J. P. Bannantine, Q. Zhang, A. Amonsin, B. J. May, D. Alt, N. Banerji, S. Kanjilal, and V. Kapur. 2005. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc. Natl. Acad. Sci. USA 102:12344-12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lincoln, E., and E. Sewell. 1963. Tuberculosis in children, p. 71-132. McGraw-Hill Book Co., New York, N.Y.
- 27.Lonetto, M., K. Brown, K. Rudd, and M. Buttner. 1994. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase σ factors involved in the regulation of extracytoplasmic functions. Proc. Natl. Acad. Sci. USA 91:7573-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manganelli, R., E. Dubnau, S. Tyagi, F. R. Kramer, and I. Smith. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol. Microbiol. 31:715-724. [DOI] [PubMed] [Google Scholar]
- 29.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, E. Dubnau, M. Gomez, and I. Smith. 2002. Role of the extracytoplasmic-function sigma factor sigmaH in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 45:365-374. [DOI] [PubMed] [Google Scholar]
- 30.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, and I. Smith. 2001. The Mycobacterium tuberculosis ECF sigma factor sigmaE: role in global gene expression and survival in macrophages. Mol. Microbiol. 41:423-437. [DOI] [PubMed] [Google Scholar]
- 31.McKinney, J. D., K. Honer zu Bentrup, E. J. Munoz-Elias, A. Miczak, B. Chen, W. T. Chan, D. Swenson, J. C. Sacchettini, W. R. Jacobs, Jr., and D. G. Russell. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735-738. [DOI] [PubMed] [Google Scholar]
- 32.Okkels, L. M., I. Brock, F. Follmann, E. M. Agger, S. M. Arend, T. H. Ottenhoff, F. Oftung, I. Rosenkrands, and P. Andersen. 2003. PPE protein (Rv3873) from DNA segment RD1 of Mycobacterium tuberculosis: strong recognition of both specific T-cell epitopes and epitopes conserved within the PPE family. Infect. Immun. 71:6116-6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parish, T., E. Mahenthiralingam, P. Draper, E. O. Davis, and M. J. Colston. 1997. Regulation of the inducible acetamidase gene of Mycobacterium smegmatis. Microbiology 143:2267-2276. [DOI] [PubMed] [Google Scholar]
- 34.Parish, T., D. A. Smith, G. Roberts, J. Betts, and N. G. Stoker. 2003. The senX3-regX3 two-component regulatory system of Mycobacterium tuberculosis is required for virulence. Microbiology 149:1423-1435. [DOI] [PubMed] [Google Scholar]
- 35.Pelicic, V., M. Jackson, J.-M. Reyrat, W. J. Jacobs, B. Gicquel, and C. Guilhot. 1997. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 94:10955-10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quadri, L. E., J. Sello, T. A. Keating, P. H. Weinreb, and C. T. Walsh. 1998. Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chem. Biol. 5:631-645. [DOI] [PubMed] [Google Scholar]
- 37.Ramakrishnan, L., N. A. Federspiel, and S. Falkow. 2000. Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science 288:1436-1439. [DOI] [PubMed] [Google Scholar]
- 38.Raman, S., R. Hazra, C. C. Dascher, and R. N. Husson. 2004. Transcription regulation by the Mycobacterium tuberculosis alternative sigma factor SigD and its role in virulence. J. Bacteriol. 186:6605-6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raman, S., and R. N. Husson. 2006. Unpublished data.
- 40.Raman, S., T. Song, X. Puyang, S. Bardarov, W. Jacobs, Jr., and R. Husson. 2001. The alternative sigma factor SigH regulates major components of the oxidative and heat stress responses in Mycobacterium tuberculosis. J. Bacteriol. 183:6119-6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodriguez, G. M., M. I. Voskuil, B. Gold, G. K. Schoolnik, and I. Smith. 2002. ideR, an essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 70:3371-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schnappinger, D., S. Ehrt, M. I. Voskuil, Y. Liu, J. A. Mangan, I. M. Monahan, G. Dolganov, B. Efron, P. D. Butcher, C. Nathan, and G. K. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharov, A. A., D. B. Dudekula, and M. S. Ko. 2005. A web-based tool for principal component and significance analysis of microarray data. Bioinformatics 21:2548-2549. [DOI] [PubMed] [Google Scholar]
- 44.Sherman, D. R., M. Voskuil, D. Schnappinger, R. Liao, M. I. Harrell, and G. K. Schoolnik. 2001. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha-crystallin. Proc. Natl. Acad. Sci. USA 98:7534-7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simon, R., and A. Lam. 2005. BRB array tools, version 3.0. NIH, Bethesda, Md.
- 46.Sirakova, T. D., A. K. Thirumala, V. S. Dubey, H. Sprecher, and P. E. Kolattukudy. 2001. The Mycobacterium tuberculosis pks2 gene encodes the synthase for the hepta- and octamethyl-branched fatty acids required for sulfolipid synthesis. J. Biol. Chem. 276:16833-16839. [DOI] [PubMed] [Google Scholar]
- 47.Stover, C., V. de la Cruz, T. Fuerst, J. Burlein, L. Benson, L. Bennett, G. Bansal, J. Young, M. Lee, G. Hatfull, S. Snapper, R. Barletta, W. Jacobs, Jr., and B. Bloom. 1991. New use of BCG for recombinant vaccines. Nature 351:456-460. [DOI] [PubMed] [Google Scholar]
- 48.Sun, R., P. J. Converse, C. Ko, S. Tyagi, N. E. Morrison, and W. R. Bishai. 2004. Mycobacterium tuberculosis ECF sigma factor sigC is required for lethality in mice and for the conditional expression of a defined gene set. Mol. Microbiol. 52:25-38. [DOI] [PubMed] [Google Scholar]
- 49.Talaat, A. M., R. Lyons, S. T. Howard, and S. A. Johnston. 2004. The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc. Natl. Acad. Sci. USA 101:4602-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.TIGR. 2006. Mycobacterium smegmatis MC2 genome sequence. TIGR, Rockville, Md.
- 51.Voskuil, M. I., D. Schnappinger, R. Rutherford, Y. Liu, and G. K. Schoolnik. 2004. Regulation of the Mycobacterium tuberculosis PE/PPE genes. Tuberculosis (Edinburgh) 84:256-262. [DOI] [PubMed] [Google Scholar]
- 52.Wu, Q.-L., D. Kong, K. Lam, and R. Husson. 1997. A mycobacterial extracytoplasmic function sigma factor involved in survival following stress. J. Bacteriol. 179:2922-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, Y., K. Post-Martens, and S. Denkin. 2006. New drug candidates and therapeutic targets for tuberculosis therapy. Drug Discov. Today 11:21-27. [DOI] [PubMed] [Google Scholar]