Abstract
The recently discovered pathogen Bordetella holmesii has been isolated from the airways and blood of diseased humans. Genetic events contributing to the emergence of B. holmesii are not understood, and its phylogenetic position among the bordetellae remains unclear. To address these questions, B. holmesii strains were analyzed by comparative genomic hybridization (CGH) to a Bordetella pertussis microarray and by multilocus sequence typing. Both methods indicated substantial sequence divergence between B. pertussis and B. holmesii. However, CGH identified a putative pathogenicity island of 66 kb that is highly conserved between these species and contains several IS481 elements that may have been laterally transferred from B. pertussis to B. holmesii. This island contains, among other genes, a functional, iron-regulated locus encoding the biosynthesis, export, and uptake of the siderophore alcaligin. The acquisition of this genomic island by B. holmesii may have significantly contributed to its emergence as a human pathogen. Horizontal gene transfer between B. pertussis and B. holmesii may also explain the unusually high sequence identity of their 16S rRNA genes.
Bordetella holmesii is a recently described human pathogen first isolated in 1983 from the blood of a septicemic patient (48). Although most known systemic B. holmesii infections occur in immunocompromised individuals (41), serious systemic infection of a healthy adolescent has also been reported (36). Furthermore, although infrequently isolated, B. holmesii has been recovered from the nasopharynges of immunocompetent patients with pertussis-like symptoms (50), suggesting that B. holmesii may also cause respiratory disease. This bacterium is readily cultured on a variety of standard clinical microbiology media, so its discovery just a few decades ago suggests its recent emergence as a human pathogen (48).
In addition to B. holmesii, a number of other pathogenic species are found in the genus Bordetella. Best studied are the respiratory pathogens known as the classical or mammalian bordetellae: B. pertussis, human B. parapertussis, ovine B. parapertussis, and B. bronchiseptica. B. pertussis and human B. parapertussis are the causative agents of whooping cough, or pertussis, a disease which has reemerged despite widespread vaccination (8, 42). Ovine B. parapertussis infects the respiratory tracts of sheep, and B. bronchiseptica infects a wide range of mammalian species. The nonclassical Bordetella species include B. avium, B. hinzii, B. petrii, “B. ansorpii” (proposed name), and B. trematum, and these form a genetically diverse group that is clearly distinct from the mammalian bordetellae (16, 21).
Comparative analysis of their 16S rRNA sequences suggested that B. holmesii is very closely related to B. pertussis (48), a hypothesis supported by the discovery of the B. pertussis-specific insertion sequence element IS481 in B. holmesii (34). However, subsequent sequencing of housekeeping genes, analysis of cellular fatty acid composition, and characterization of the bvgAS locus suggested that B. holmesii may not be as closely related to B. pertussis as was first assumed (15, 48).
To identify genetic factors that may have contributed to the emergence of B. holmesii as a human pathogen and to clarify its relation to other members of the Bordetella genus, we used microarray-based comparative genome hybridization (CGH) and sequencing of housekeeping genes. Although gene sequencing indicated that B. holmesii is a uniform group that is more closely related to the nonclassical B. hinzii and B. avium than to the mammalian bordetellae, CGH detected a genomic region of approximately 66 kb that is highly conserved between B. pertussis and B. holmesii. This genomic island, encoding genes that could promote pathogenesis, may have been transferred from B. pertussis to B. holmesii, possibly contributing to the emergence of B. holmesii as a human pathogen.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Characteristics of the strains used in this study are listed in Table 1. For iron depletion tests, a previously described chemically defined medium (9) was adapted by adding 330 μM l-cysteine, 114 μM ascorbic acid, 33 μM niacin, and 325 μM reduced glutathione. Chelex-100 resin (Bio-Rad, Hercules, CA) was used for iron depletion, as described previously (9). Iron-replete medium was made by adding 66 μM ferrous sulfate to iron-depleted medium.
TABLE 1.
B. holmesii strain characteristics
| Strain | Original name | Sourcea | Isolation site | Country | Region | Isolation yr |
|---|---|---|---|---|---|---|
| B0436 | 104394 | Institut Pasteur | —b | — | — | — |
| B0437 | 104395 | Institut Pasteur | — | — | — | — |
| B1850 | C690 | Pam Cassiday, CDC | Nasopharynx | United States | Massachusetts | 1994 |
| B1851 | C691 | Pam Cassiday, CDC | Blood | United States | Massachusetts | 1995 |
| B1852 | C692 | Pam Cassiday, CDC | Blood | United States | Ohio | — |
| B1853 | C693 | Pam Cassiday, CDC | Blood | United States | Colorado | — |
| B1854 | C694 | Pam Cassiday, CDC | Blood | United States | North Carolina | — |
| B1855 | USA 8/8/00 | Cathy Canthaboo, NIBSC | — | — | — | — |
| B2738 | BP244-01 | Norman Fry, University of Glasgow | Blood | United Kingdom | Isle of Wight | 2001 |
| B2739 | BP246-01 | Norman Fry, University of Glasgow | Blood | United Kingdom | London | 2001 |
| B2767 | RR 0000 0020 | Norman Fry, University of Glasgow | Blood | United Kingdom | — | 2003 |
| B2768 | HO 4290 0199 | Norman Fry, University of Glasgow | Blood | United Kingdom | Oxford | 2004 |
| Bho29 | G8350c | Robbin Weyant, CDC | Blood | Switzerland | — | — |
CDC, Centers for Disease Control and Prevention, United States; NIBSC, National Institute for Biological Standards and Control, United Kingdom.
—, unknown.
Isolation reported in reference 48.
Comparative genomic hybridization.
Experiments utilized a comprehensive classical Bordetella genome-wide DNA microarray, based on the microarray described in references 10 and 11. Genomic DNA was isolated using the Wizard genomic DNA purification kit (Promega, Madison, WI) by following the protocol for gram-negative bacteria. B. holmesii genomic DNA was labeled with Cy5 and hybridized to arrays with Cy3-labeled reference DNA (equimolar amounts of B. pertussis Tohama I, B. bronchiseptica RB50, and B. parapertussis 12822 genomic DNA) as described in reference 11. Arrays were scanned on a two-color GenePix 4000B scanner and analyzed with GenePix Pro 6.0 (Axon Instruments, Union City, CA).
For each spot on the array, the fluorescence intensity of each of the two dyes was calculated by subtracting the mean intensity of the background pixels from the median intensity of the spot pixels. The logarithm of the ratio of background-subtracted Cy5 intensity over background-subtracted Cy3 intensity [log2(Cy5/Cy3)] is near 0 when both samples hybridize equally well to the probe and significantly below 0 when the B. holmesii DNA hybridizes poorly compared to the reference DNA. Data normalization was achieved by calculating the mean log2(Cy5/Cy3) for 16 probes within the iron uptake island (IUI) that were verified by sequencing to be at least 99% identical to the B. holmesii genome (see Table S1 in the supplemental material) and then subtracting this value from the log2(Cy5/Cy3) for each probe on the array. Data from replicate spots for the same probe were averaged. A probe was considered to have a positive B. holmesii hybridization signal when its hybridization ratio exceeded the fifth percentile of ratios from a control hybridization of B. pertussis Tohama I. Further details are in Methods in the supplemental material.
Organization of the IUI.
The IUI was analyzed by amplifying overlapping 5- to 10-kb DNA fragments across the region. PCRs were analyzed by agarose gel electrophoresis to compare the fragment sizes from B. holmesii and B. pertussis Tohama I (see Table S2 in the supplemental material).
A total of 21 genes in the IUI were partially sequenced from three B. holmesii isolates. The nucleotide sequence of the 4.8-kb B. holmesii insert, absent from the classical bordetellae, was determined by genome walking inwards from the adjacent genes. In order to assess the organization of the BB3888 homolog, one of the open reading frames (ORFs) contained in the IUI, in B. pertussis and B. holmesii, we designed PCR primers in BB3888 that span the IS481 insertion and rearrangement point from B. pertussis Tohama I. Nucleotide sequence data adjacent to the left breakpoint of the IUI was obtained by PCR, using one primer inside the IUI, in BB0794, and one primer outside the IUI, in BB0795, which was not detected in B. holmesii by CGH. The sequence adjacent to the right breakpoint of the IUI was obtained using the TOPO walker kit (Invitrogen, Carlsbad, CA). Further details are in Methods in the supplemental material, and primers are listed in Table S3 in the supplemental material.
Phylogenetic analysis and broad-range PCR.
PCR primers for phylogenetic analysis were designed to hybridize to conserved regions of the atpD, rpoB, tuf, and rnpB genes (see Table S3 in the supplemental material). PCR products were directly sequenced. Concatenated sequences of these four genes were aligned for the construction of neighbor-joining trees using PHYLIP software (J. Felenstein. 2005. Phylogeny Inference Package, version 3.6 [distributed by the author]. Department of Genomic Sciences, University of Washington, Seattle). Details are in Methods in the supplemental material.
B. holmesii 16S rRNA genes were amplified by broad-range PCR using the 8F and 806R primers (35), and 23S rRNA genes were amplified using the MS37 and MS38 primers (22). PCR products were cloned using the Topo TA cloning kit (Invitrogen). Twelve clones were picked for each strain, and plasmids were purified using the Wizard miniprep kit (Promega) and sequenced using M13 forward and reverse primers.
Southern blot hybridization.
The genomic DNA of B. holmesii, B. pertussis, and B. avium isolates was digested with the restriction enzymes ClaI and NcoI. Southern blotting and hybridization with biotin-labeled oligonucleotide probes were performed essentially as described by Schouls et al. (37). The same blot was used for both 16S rRNA gene probe hybridizations, with a stripping step between experiments. Table S3 in the supplemental material lists the probes that were hybridized to the genomes of the Bordetella isolates.
Transcription of alcaligin genes.
B. holmesii strains were grown to mid-logarithmic phase in iron-depleted or iron-replete medium. Total RNA was isolated using the QIAGEN RNeasy kit (QIAGEN Inc, Valencia, CA). Real-time PCR experiments were performed with a LightCycler (Roche Diagnostics, Indianapolis, IN) using the LightCycler DNA master hybridization probe kit (Roche Diagnostics). alcA and fauA transcript levels were compared to that of rpoB, which served as the internal reference. Further details are in Methods in the supplemental material.
MS.
Culture supernatant samples were analyzed by nanoscale reversed-phase liquid chromatography (LC) on a model HP1100 LC system (Hewlett Packard GmbH, Waldbronn, Germany) coupled to an electrospray mass spectrometer (MS) (liquid chromatography quadrupole classic ion trap), essentially as described by Meiring et al. (25). Technical details are in Methods in the supplemental material.
Microarray data accession number.
Raw microarray data have been deposited at ArrayExpress under accession number E-TABM-55.
Nucleotide sequence accession numbers.
GenBank accession numbers DQ402382 to DQ402420 and DQ420056 to DQ420164 are published for the first time.
RESULTS
Comparative genome hybridization.
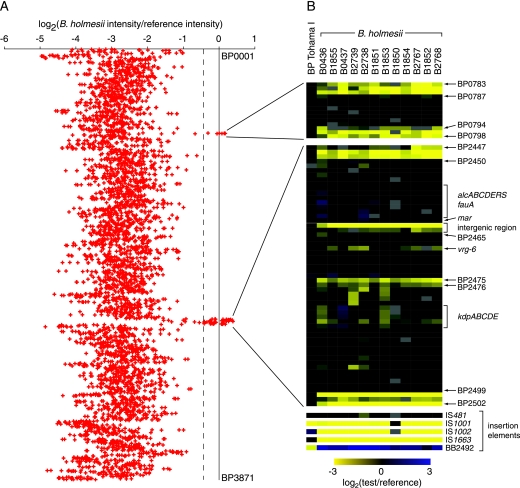
The genomes of 12 B. holmesii strains independently isolated in diverse locations during different years (Table 1) were each hybridized to a microarray representing the ORFs of B. pertussis Tohama I, B. parapertussis 12822, and B. bronchiseptica RB50 (11). Although most of the 5,515 microarray probes failed to hybridize significantly to B. holmesii genomic DNA, an average of 157 probes (ranging from 59 to 311) hybridized to the B. holmesii genomes as strongly as they did to B. pertussis genomic DNA (Fig. 1A; see normalized microarray data available as Table S4 in the supplemental material). On average, 41 of these positive probes mapped to two contiguous regions in the Tohama I genome: BP0787-BP0794 and BP2450-BP2499 (Fig. 1B). The remaining probes were scattered across the genome and usually represented genes with relatively high sequence conservation across the Bordetella genus (e.g., genes encoding ribosomal proteins [data not shown]). The hybridization of B. holmesii genomic DNA to a probe for IS481 was consistent with previous work that indicated the presence of eight IS481 copies in the B. holmesii genome (34).
FIG. 1.
Comparative genome hybridization of B. holmesii strains to a classical Bordetella DNA microarray. (A) CGH of 12 B. holmesii isolates to a microarray comprising the genomes of B. pertussis Tohama I, B. parapertussis 12822, and B. bronchiseptica RB50. The moving average (with a sliding window of three adjacent values) of the mean log2(Cy5/Cy3) of 12 B. holmesii genomes is plotted on the x axis. Microarray probes that are represented in the B. pertussis Tohama I genome are arranged on the y axis in B. pertussis Tohama I genome order. The dashed line indicates the fifth percentile of B. pertussis Tohama I hybridization ratios; ratios below the fifth percentile are considered to indicate the lack of B. holmesii hybridization. (B) Probes that hybridized to the B. holmesii genome with a strength comparable to that of the reference and adjacent nonhybridizing probes are shown in detail for individual B. holmesii strains and for B. pertussis Tohama I. ORF and gene designations for selected loci are provided as landmarks. Probes representing insertion sequence elements are also shown. BB2492 is a putative IS3 family transposase found in B. bronchiseptica RB50 but not in B. pertussis. Strain numbers are indicated above the columns. Each row represents one probe in B. pertussis Tohama I gene order. The logarithm of the hybridization ratio [log2(Cy5/Cy3)] is indicated by the yellow-black-blue color scale. Missing data are represented in gray. The data represented in this figure are available as Table S4 in the supplemental material.
The two B. pertussis genomic regions shared by the classical bordetellae and B. holmesii contained, among a number of other genes, a locus comprising alcABCDERS, encoding the alcaligin biosynthetic pathway, the exporter for alcaligin (AlcS), and a transcriptional activator of the locus (AlcR), and fauA, encoding the alcaligin uptake receptor (3, 6, 7, 19, 31). Alcaligin is a siderophore produced by B. pertussis, B. bronchiseptica, and Alcaligenes denitrificans (27) that scavenges free iron from the extracellular milieu. Accordingly, we designated this genomic fragment the B. holmesii IUI.
Molecular characterization of the B. holmesii IUI.
Partial nucleotide sequences of 21 genes in the IUI, indicated to be present by CGH, were obtained from B. holmesii isolates B0436, B1850, and B1852 after PCR amplification using B. pertussis primer pairs (GenBank accession numbers DQ402382 to DQ402418). All of these sequences were at least 99.4% identical to the B. pertussis sequence, with the exception of BP0794 (94.1%), located at the putative right end of the IUI (see below; see also Table S1 in the supplemental material).
The GC content of the IUI (66.0%) is substantially higher than the average GC content of B. holmesii genes (61.5 to 62.3%) (48; see below) but comparable to that of the B. pertussis genome (67.7%) (30).
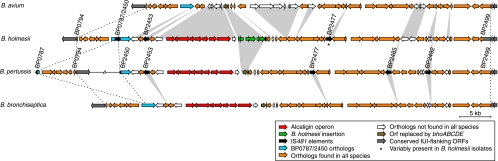
In B. bronchiseptica and B. parapertussis, but not in B. pertussis, the two genomic regions identified by CGH constitute a single continuous chromosomal fragment. The presence of IS481 sequences at the putative rearrangement breakpoint in BP0787 and BP2450, which are the two halves of a single ORF in B. bronchiseptica and B. parapertussis, suggests that this region has been rearranged in the B. pertussis lineage by recombination between transposon copies (Fig. 2). PCR was performed using a primer pair that spans the breakpoint (BB3888-480f and BB3888-720r; see also Table S3 in the supplemental material). This reaction yielded a 240-bp fragment from B. bronchiseptica RB50 but failed to amplify a product from B. pertussis Tohama I due to the chromosomal rearrangement between these primers.
FIG. 2.
Comparison of IUI genomic organizations in B. holmesii, B. pertussis, B. bronchiseptica, and B. avium. Deletions and insertions between species are indicated by gray surfaces. The dashed lines connect the ORFs at the borders of the orthologous sequences. The ORFs are color coded as described in the key. The ORF composition of B. holmesii was deduced from PCR and CGH data, while the representations of B. avium 197N, B. pertussis Tohama I, and B. bronchiseptica RB50 ORFs were based on published genome sequences.
PCR amplification of DNA from all B. holmesii strains using this primer pair yielded a product of approximately 1.3 kb, indicating the chromosomal proximity of the primer-binding sites but suggesting the presence of a 1-kb insertion, which is the approximate size of an IS481 element. Nucleotide sequencing confirmed that the IUI is present as a single, contiguous locus in B. holmesii but interrupted by an IS481 element at the same position as the Tohama I breakpoint (Fig. 2). PCR screening of 45 B. pertussis strains identified one strain, 18323, that carried the same contiguous but IS481-disrupted allele of BP0787-BP2450 as B. holmesii (GenBank accession numbers DQ420073 to DQ420074).
Fourteen PCR primer pairs that amplify overlapping fragments from the B. pertussis Tohama I IUI sequence were used to map the IUIs in 12 B. holmesii strains by PCR amplification (see Table S2 in the supplemental material). As shown by agarose gel electrophoresis, most B. holmesii PCR amplicons were identical in size to those from B. pertussis, suggesting conserved genomic organization, but small insertions or deletions or point mutations that could result in pseudogene formation would not have been detected by electrophoresis of PCR amplicons or CGH. Two primer pairs yielded PCR products from B. holmesii that were approximately 1 kb shorter than the corresponding Tohama I products; sequencing of these products indicated that the IS481 elements BP2485 and BP2492 were missing in B. holmesii. Like B. holmesii, B. pertussis 18323 lacked these two IS481 elements. A third IS481 element, BP2477, was variably present among the B. holmesii strains.
PCR amplification of one genomic region in the IUI, downstream of the alc operon, revealed a 4.8-kb insertion in B. holmesii which is not present in the classical bordetellae (Fig. 2). Nucleotide sequencing identified the left terminus of this insert 237 nucleotides downstream of the BP2464 stop codon and its right terminus in BP2465, which has been partially deleted (GenBank accession number DQ402419).
BLASTX comparison of the 4.8-kb B. holmesii-specific sequence against GenBank identified five putative genes, designated bhoABCDE, with highly significant hits (E value < 1 × 10−17) to known proteins (Table 2). bhoA is predicted to encode an IS3 family transposase with two out-of-frame overlapping ORFs (orfA and orfB), as previously described for IS3 transposases (39). However, a frameshift mutation detected in the 3′ ORF may have inactivated the transposase. Southern blot hybridization with a bhoA-specific probe identified 40 to 50 copies in the genome of B. holmesii but no copies in the genome of B. pertussis (data not shown). bhoB and bhoC are homologous to adjacent genes from the Ralstonia metallidurans CH34 genome. These encode a probable extracytoplasmic solute receptor and a d-isomer-specific 2-hydroxyacid dehydrogenase. bhoD encodes a putative extracytoplasmic function sigma factor, and bhoE encodes a homolog of the FecR proteins (Table 2). The GC content of the insert is 62%, similar to the average GC content of B. holmesii genes (61.5 to 62.3%) (48) but lower than the shared component of the IUI (66.0%).
TABLE 2.
Annotation of the B. holmesii-specific insertion in IUI
| Gene | Predicted product | BLASTX top hit
|
Pfam significant hit(s)
|
||||
|---|---|---|---|---|---|---|---|
| Entrez locus | Species | E value | Accession no. | Domain | E value | ||
| bhoA | IS3 family transposase (pseudogene) | YP_107572 | Burkholderia pseudomallei | 2E−74 | PF00665 | Integrase core domain | 9E−16 |
| bhoB | Extracytoplasmic solute receptor | EAN49994 | Ralstonia metallidurans | 7E−79 | PF03401 | Bordetella uptake gene (bug) product | 4E−55 |
| bhoC | d-Isomer-specific 2-hydroxyacid dehydrogenase | EAN49993 | R. metallidurans | 3E−76 | PF00389 | d-Isomer-specific 2-hydroxyacid dehydrogenase, catalytic domain | 0.27 |
| bhoD | ECF sigma factor | CAD84465 | Nitrosomonas europaea | 2E−21 | PF04542 | Sigma-70, region 2 | 2E−10 |
| PF04545 | Sigma-70, region 4 | 5E−08 | |||||
| bhoE | FecR protein | YP_349526 | Pseudomonas fluorescens | 6E−18 | PF04773 | FecR protein | 4E−33 |
The CGH data suggested a sharp chromosomal boundary between the IUI and the flanking B. holmesii genomic sequence, prompting a search for the discrete breakpoints that define the ends of the IUI. The left breakpoint in B. holmesii was mapped by PCR and sequencing (GenBank accession numbers DQ420096 to DQ420098) to approximately codon 31 of BP0794. The right breakpoint was mapped by chromosomal walking and sequencing (GenBank accession number DQ402420) to approximately codon 276 of BP2499. In both cases, the breakpoints were approximated as the point at which sequence identity, reading out from the IUI, dropped from greater than 99% to below 90%. The ORFs at both breakpoints are intact in B. holmesii. No evidence was found at either end of the IUI for the presence of phage, plasmid, or IS sequences.
The IUI sequence was also compared to the B. avium 197N genome sequence (38). Using the BLAST programs, a syntenic region with homology extending beyond the ends of the IUI was identified (Fig. 2). The average nucleotide sequence identity of the B. avium region to the B. holmesii IUI was 73%, indicating that these sequences are not highly conserved across all members of the genus, but sequences near the ends of the IUI were more highly conserved (at least 88%) than sequences in the middle. Although gene order is otherwise largely conserved, the B. avium genome shows no detectable homology to the alcABCDERS, the fauA, the bhoABCDE, and the IS481 loci (Fig. 2; see also Table S1 in the supplemental material).
Alcaligin expression and detection.
To determine whether the IUI alcaligin biosynthesis, export, and uptake locus is expressed in B. holmesii, the transcript levels of alcA and fauA were measured by quantitative reverse transcription-PCR of B0437 grown under iron-depleted and iron-replete conditions. The transcript abundances of alcA and fauA were 3.3- and 4.8-fold higher, respectively, in the iron-depleted sample than in the nondepleted sample, indicating that the locus is expressed and iron regulated in vitro.
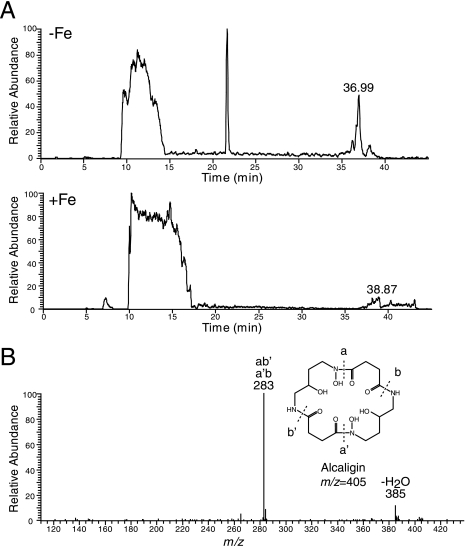
LC-MS of culture supernatant from iron-starved B. holmesii detected a compound in the iron-depleted supernatant but not in the iron-replete supernatant with m/z 405 (M + H)+ (Fig. 3), corresponding to the mass of desferri-alcaligin (27). MS fragmentation analysis detected two fragment ions with m/z 283 and m/z 387, corresponding to previously described alcaligin fragment ions (27).
FIG. 3.
LC-MS spectra of B. holmesii supernatants from iron-depleted and iron-replete cultures. (A) LC-MS spectra of iron-depleted (−Fe) and iron-replete (+Fe) B. holmesii supernatants, depicting ions with m/z 405 (corresponding to alcaligin) and with retention times between 0 and 45 min. (B) Fragment ions detected after collisional activation (35% energy) of the peak from (A), with a retention time of 36.99 min. After measurement of the reference standard, the calibration deviated by approximately 1.5 mass units, explaining the difference in m/z values compared to those of the fragment ions as reported in reference 22. The nomenclature of fragment ions is according to that in reference 27.
Evolutionary relationship of B. holmesii to other bordetellae.
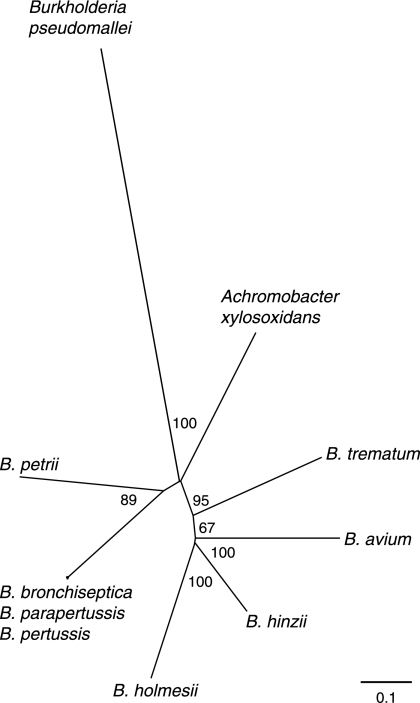
Contrary to 16S rRNA-based phylogenies, our CGH data suggested substantial sequence divergence between B. pertussis and B. holmesii. To determine the phylogenetic position of B. holmesii within the Bordetella genus, conserved regions of the atpD, rpoB, tuf, and rnpB genes, which have been used for phylogenetic inference in other groups of closely related bacterial species (18, 26, 29), were sequenced from seven B. holmesii strains and from representative isolates of B. hinzii, B. trematum, B. petrii, and the closely related β-proteobacterial species Achromobacter xylosoxidans (GenBank accession numbers DQ420062 to DQ420072 and DQ420099 to DQ420164). Orthologous sequences from the classical Bordetella species (GenBank accession numbers NC_002927, NC_002928, and NC_002929), B. avium, and another β-proteobacterial species, Burkholderia pseudomallei (GenBank accession numbers NC_006350 and NC_006351), were obtained from available genome sequence data. A neighbor-joining tree, based on 3,559 fully informative characters in the alignment of concatenated nucleotide sequences of all these genes, indicated that B. holmesii is more closely related to B. avium and B. hinzii than to the mammalian bordetellae (Fig. 4). The exclusion of B. holmesii from the mammalian Bordetella clade was supported by all individual gene trees. Comparison of sequences from seven independent B. holmesii strains revealed only two single-nucleotide polymorphisms (one synonymous and one nonsynonymous) among 3,666 aligned bases, both of which were variant only in Bho29. Because of the near identity of these seven strains, no further B. holmesii isolates were sequenced.
FIG. 4.
Neighbor-joining phylogeny of Bordetella spp. and related β-proteobacteria based on four genes outside the rRNA operon. The unrooted tree is based on 3,559 fully informative nucleotides from an alignment of concatenated sequences of atpD, rpoB, tuf, and rnpB from seven B. holmesii strains, B. pertussis Tohama I, B. parapertussis 12822, B. bronchiseptica RB50, B. avium 197N, B. hinzii BC304, B. trematum DSM11334, and B. petrii SE1111R, and from A. xylosoxidans ATCC 15173 and B. pseudomallei K96243. Bootstrapping values greater than 50% (in 1,000 resamplings) are indicated at branches.
Molecular characterization of B. holmesii rRNA loci.
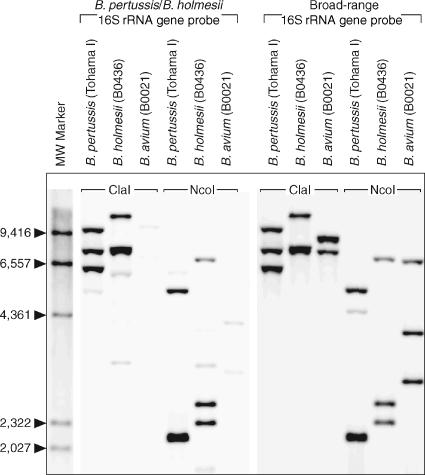
In light of the phylogeny determined above, the near identity of the B. pertussis and B. holmesii 16S rRNA genes (99.7%) is anomalous. One possible explanation for this discrepancy is lateral transfer of the 16S rRNA gene from B. pertussis to B. holmesii. To determine the number of B. pertussis-like 16S rRNA gene copies and search for possibly divergent 16S rRNA genes, Southern blot hybridization was performed. A B. pertussis-specific 16S rDNA probe that did not hybridize to the B. avium genome detected three copies in the B. holmesii genome (Fig. 5). A broad-range 16S rRNA gene probe (37) that hybridized equally well to B. pertussis and B. avium DNA did not detect any additional bands in the B. holmesii genome, suggesting that the only 16S rRNA loci are the three B. pertussis-like copies (Fig. 5). 16S rRNA broad-range PCR was also employed to identify possible variant 16S rRNA sequences. Twelve cloned PCR products from each of three B. holmesii strains were sequenced. All were at least 99.5% identical to the B. pertussis 16S rRNA, further suggesting that all copies of the 16S rRNA in B. holmesii are essentially identical to the B. pertussis gene (GenBank accession numbers DQ420056 to DQ420058).
FIG. 5.
Southern blot hybridizations of B. pertussis, B. holmesii, and B. avium with 16S rRNA gene probes. Genomic DNA from each of the three strains was restriction digested with ClaI or NcoI as indicated and hybridized to a B. pertussis-specific probe (left panel) and a broad-range 16S rRNA gene probe (B-16S8F [right panel]) (15). Biotinylated DNA markers are in the first lane, and numbers at the left are fragment sizes (in base pairs).
No copies of the 16S rRNA gene were identified within the boundaries of the IUI as defined above. To test further whether a 16S rRNA gene was located near either end of the IUI in B. holmesii, PCR amplification was attempted using primers in the 16S rRNA gene and in the putative 5′ and 3′ termini of the IUI. PCR amplification was unsuccessful, suggesting that no 16S rRNA genes are in close proximity to the IUI.
Among the sequenced Bordetella genomes, the intergenic transcribed spacer region (ITS) between the 16S and 23S rRNA genes is more varied than the sequences of the structural rRNA genes (Table 3). However, ITS sequences from seven B. holmesii strains, all of which were identical to each other, were 99.6% identical to that from B. pertussis (GenBank accession numbers DQ420075 to DQ420095) and more similar than even the ITS sequence from B. bronchiseptica. A partial sequence of the 23S rRNA gene (B. pertussis bases 1043 to 1919) was also determined following broad-range PCR of three B. holmesii strains. Nine sequences (three from each strain; GenBank accession numbers DQ420059 to DQ420061) were identical to each other but only 97.2% identical to B. pertussis and more divergent than the B. avium sequence (Table 3). For comparison, B. pertussis and B. bronchiseptica vary at only a single nucleotide in this region. Therefore, the region of unexpectedly high identity between the B. pertussis and B. holmesii rRNA gene operons includes the 16S rRNA gene and the ITS but does not include the 3′ end of the 23S rRNA gene. The boundary between nearly identical (>99.5%) and divergent sequences lies in the first kilobase of the 23S gene, which was not sequenced in this study.
TABLE 3.
Percentages of nucleotide identity in the rRNA operon between B. holmesii and selected Bordetella species
| Species | % Nucleotide identity
|
||
|---|---|---|---|
| 16S rRNA | ITS | 23S rRNA | |
| B. pertussis | 99.7 | 99.6 | 97.2 |
| B. bronchiseptica | 99.4 | 96.4 | 96.6 |
| B. avium | 97.2 | 72.5 | 98.1 |
DISCUSSION
Although 16S rRNA phylogeny and clinical criteria had placed B. holmesii close to B. pertussis, other evidence, including gene sequencing and cellular fatty acid analysis, had in fact suggested a more distant evolutionary relationship (15, 16, 48). The multilocus sequence analysis presented here provides further evidence that B. holmesii is not, in fact, a member of the classical Bordetella lineage. Rather, phylogenetic analysis based on the nucleotide sequences of four different housekeeping genes strongly suggested that B. holmesii is a member of the phylogenetically diverse nonclassical Bordetella group (Fig. 4). Furthermore, at these loci, B. holmesii and B. avium were similarly distant from the classical bordetellae (89.1% and 90.0%, respectively). By demonstrating that the tree based on the 16S rRNA gene differs significantly from the tree based on four independent housekeeping genes, these results underscore the importance of using multiple loci when reconstructing evolutionary histories. An explanation for the anomalously high sequence identity of the B. holmesii and B. pertussis 16S rRNA genes is proposed below.
Only 2 of 3,666 nucleotide positions in the housekeeping genes were polymorphic among seven geographically distinct B. holmesii strains. This level of sequence conservation is comparable to the three polymorphisms of 3,666 nucleotides observed between B. bronchiseptica and B. pertussis, which diverged and expanded clonally from the B. bronchiseptica lineage 2 to 5 million years ago (12, 30). Likewise, B. holmesii may have diverged and expanded within a similar time frame.
Comparative genomic hybridization identified a 66-kb DNA region, designated the IUI, which, unlike most of the B. holmesii genome, was highly conserved in B. holmesii and the classical bordetellae. No conserved chromosomal regions were identified when B. avium genomic DNA was hybridized to the classical Bordetella microarray (data not shown). Detailed molecular characterization verified that the shared sequences are more than 99% identical, much higher even than the expectation based on strongly conserved phylogenetic marker genes (under the assumption that the nucleotide substitution rates are similar across the genome). Cloning of the IUI ends also demonstrated sharp boundaries between the conserved IUI sequence and the weakly conserved flanking sequences. These data are consistent with the recent transfer of the IUI from a classical Bordetella genome to the B. holmesii genome. Transfer from a B. bronchiseptica or B. parapertussis genome cannot be ruled out, but the presence in the IUI of IS481 elements, which have been found in all B. pertussis and B. holmesii strains analyzed but in only two B. bronchiseptica strains (12, 45), argues strongly that a B. pertussis strain was the donor of this DNA. Although the IUI region is rearranged in B. pertussis Tohama I relative to this region in B. holmesii and the other mammalian bordetellae, another B. pertussis strain, 18323, had a genomic architecture and an IS481 element distribution very similar to those observed in the B. holmesii IUI, suggesting that the B. holmesii IUI may have been transferred from an 18323-like B. pertussis strain. B. pertussis 18323, isolated from a pertussis patient in 1947, is an atypical B. pertussis strain by many criteria, including pulsed-field gel electrophoresis (17), multilocus sequence typing (12), multilocus enzyme electrophoresis (45), and CGH (11) results, but a genetically related strain, CZ, was isolated from a pertussis case in 1993 (4, 17), suggesting that this is a rare, but circulating, variant.
The data presented here do not rule out the possibility that the IUI was transferred in the other direction, from B. holmesii to the classical bordetellae. However, our interpretation is supported by two lines of evidence. First, the GC content of the IUI is similar to that of the classical Bordetella chromosomes but significantly higher than that of B. holmesii protein-coding genes outside this region. Because atypical nucleotide composition is a hallmark of recently acquired DNA (reviewed in reference 13), this suggests that the IUI was recently transferred into B. holmesii. Second, the presence of multiple IS481 elements in the B. holmesii and B. pertussis IUIs, but not in the other classical Bordetella genomes, is more easily explained by our model, which does not require transfer of an IS481-containing DNA fragment to the ancestor of the classical Bordetella lineage, followed by precise excision of those transposons in two of three derived species. B. pertussis and B. holmesii have both been isolated from the respiratory tracts of humans, making the human airway a likely environment in which the transfer of the IUI could have occurred. At the resolution of CGH and PCR analysis, IUIs were identical in all examined B. holmesii isolates, with the exception of a variably present IS481 element, suggesting that the IUI was acquired recently or is under selective pressure to maintain its genomic organization.
A region of synteny between B. avium, B. holmesii, and the classical bordetellae extended through the IUI and into the flanking sequences on either side, indicating that the gene order of this chromosomal region is conserved across distantly related Bordetella species (Fig. 2). This finding, together with the failure to identify signatures of mobile elements (e.g., phage or conjugative transposon genes) at the ends of the IUI, suggests that the most likely mechanism for the proposed integration of the IUI into the B. holmesii genome is homologous recombination between the laterally transferred IUI and the ancestral B. holmesii genome, which may have diverged from the B. pertussis sequence to an extent similar to the extent of the divergence of the B. avium sequence. The degree of nucleotide sequence identity between the B. holmesii IUI and the orthologous sequences in B. avium is highest in the vicinities of the left and right insertion breakpoints, indicating that these genes may be more highly conserved across the Bordetella genus than the average gene. These sequences are proposed to have served as the substrates for the putative double-recombination event that replaced the B. holmesii sequence with a transferred B. pertussis fragment.
Given the evidence presented above for lateral transfer of the IUI from B. pertussis to B. holmesii, lateral transfer is also a plausible explanation for the unexpectedly high sequence similarity of their 16S rRNA genes. The failure to identify a 16S rRNA gene within, or in close proximity to, the IUI suggests that it was independently transferred from B. pertussis or that a subsequent rearrangement separated the 16S rRNA gene from the IUI. The proposed exchange includes the 16S rRNA gene and the ITS and may include a 5′ fragment of the 23S rRNA gene.
Immediately following the proposed 16S rRNA gene transfer event, the recipient B. holmesii chromosome would have carried a single copy of the B. pertussis 16S rRNA gene and, unless there was only a single rRNA locus that was replaced by homologous recombination with the foreign DNA, one or more copies of its ancestral 16S rRNA gene. Other microbial genomes have been found to harbor one or more foreign copies of the small-subunit rRNA locus in addition to their presumed native copies (1, 37, 49). In order to detect putative ancestral B. holmesii 16S rRNA loci, independent hybridization- and PCR-based methods were used to search for copies of the 16S rRNA gene that diverged from the B. pertussis-like sequence. However, only B. pertussis-like 16S rRNA genes were found, suggesting that all copies of the ancestral recipient strain's 16S rRNA gene have been either lost or converted by recombination since acquisition of the B. pertussis-like 16S rRNA locus. The putative replacement of the ancestral B. holmesii 16S rRNA, which, by analogy to B. avium, might have been more than 98% identical to the B. pertussis sequence, may not have had serious functional consequences for the organism, because heterologous rRNA genes have been shown to function in some cases. For example, the Salmonella enterica serovar Typhimurium rRNA operon, although only 97% identical to that of Escherichia coli, can functionally replace the native E. coli loci (2).
The complement of genes carried by the IUI is of particular interest because it may include virulence factors or host specificity determinants that have contributed to the emergence of B. holmesii as a human pathogen. Of the 57 putative and confirmed genes (excluding transposons) carried on the IUI, the alcaligin locus, consisting of 8 genes, is the most recognizable candidate for a role in virulence. Alcaligin is an important iron-scavenging siderophore in B. pertussis and B. bronchiseptica (3, 6, 7, 19, 31) and is required for the maximal virulence of B. bronchiseptica in a piglet infection model (33). The demonstration of iron-regulated alcA and fauA transcription and the detection of alcaligin in the supernatants of iron-limited B. holmesii cultures indicated that the alcaligin biosynthesis and export locus was functional in B. holmesii. Although B. avium is capable of acquiring iron through other mechanisms (e.g., heme uptake [28]), alcaligin production has not been detected in B. avium, and its genome does not appear to encode an alcaligin biosynthesis locus, indicating that not all Bordetella species possess the ability to produce this siderophore. Therefore, prior to the IUI acquisition, the hypothetical progenitor of B. holmesii may not have been competent to produce alcaligin. Wholesale acquisition of this function by lateral transfer from B. pertussis could have provided B. holmesii with a new, highly efficient iron uptake system, potentially leading to an enhancement of its ability to colonize the human host, in which free iron is sequestered. Additionally, BhoD and BhoE, encoded by ORFs in the B. holmesii-specific IUI fragment, are homologous to E. coli FecI and FecR, which regulate ferric citrate transport (5, 14, 46, 47), and to related Bordetella heme uptake regulators (20, 32, 43, 44), raising the possibility that these may regulate iron uptake systems in B. holmesii.
Other genes in the IUI may also contribute to B. holmesii host specificity or virulence. For example, mar, adjacent to the alcaligin locus, encodes a member of the MarC family of multiple antibiotic resistance efflux pumps. Multidrug resistance transporters have been shown to confer resistance to specific host antimicrobial peptides in diverse bacterial pathogens (e.g., Neisseria meningitidis [40] and Staphylococcus aureus [23]). Perhaps the mar gene product enhances colonization of the host airway by B. holmesii by increasing resistance to host defensins and other endogenous antimicrobial peptides. Some ORFs in the IUI encode putative cell surface or exported proteins, including a putative lipoprotein (BP0793) and a putative exported protein (BP2486), that could influence pathogenesis by modifying interactions with the host epithelium and immune system. The presence of the B. pertussis Bvg-repressed gene vrg6 in the IUI is interesting, but this gene is not required for virulence in B. bronchiseptica (24), making its role in B. holmesii virulence unlikely. The numerous IUI genes encoding so-called housekeeping functions (e.g., metabolic enzymes, cytochromes, potassium transporters, cell division proteins, and molecular chaperones) might contribute to pathogenicity by enhancing fitness and so cannot be ruled out as virulence factors without further functional characterization. The potential roles of the hypothetical and conserved hypothetical ORFs, which account for about 20% of the genes in IUIs, remain to be explored.
Only recently has B. holmesii been identified as a human pathogen and recognized to cause pertussis-like disease. However, in spite of its similarities in host range and clinical course to those of B. pertussis, B. holmesii is not as closely related to B. pertussis as are the other mammalian bordetellae. The results presented here suggest that lateral transfer of a genomic island from B. pertussis may have contributed to the emergence of B. holmesii and its adaptation to the human host. These results represent a significant advance in the characterization of B. holmesii and in our understanding of the evolution of virulence in the important group of pathogens comprising the genus Bordetella.
Supplementary Material
Acknowledgments
We are grateful to J. ten Hove and A. de Jong (NVI, Unit Research and Development) for assistance with LC-MS experiments and analysis.
This work was supported by a travel grant from The Netherlands Organization for Scientific Research (NWO). C.A.C. was supported by an American Lung Association research training fellowship. D.A.R. received grant support from the NIH (grants AI054970 and AI057188).
Footnotes
Published ahead of print on 13 October 2006.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Acinas, S. G., L. A. Marcelino, V. Klepac-Ceraj, and M. F. Polz. 2004. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J. Bacteriol. 186:2629-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asai, T., D. Zaporojets, C. Squires, and C. L. Squires. 1999. An Escherichia coli strain with all chromosomal rRNA operons inactivated: complete exchange of rRNA genes between bacteria. Proc. Natl. Acad. Sci. USA 96:1971-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaumont, F. C., H. Y. Kang, T. J. Brickman, and S. K. Armstrong. 1998. Identification and characterization of alcR, a gene encoding an AraC-like regulator of alcaligin siderophore biosynthesis and transport in Bordetella pertussis and Bordetella bronchiseptica. J. Bacteriol. 180:862-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boursaux-Eude, C., S. Thiberge, G. Carletti, and N. Guiso. 1999. Intranasal murine model of Bordetella pertussis infection: II. Sequence variation and protection induced by a tricomponent acellular vaccine. Vaccine 17:2651-2660. [DOI] [PubMed] [Google Scholar]
- 5.Braun, V., S. Mahren, and M. Ogierman. 2003. Regulation of the FecI-type ECF sigma factor by transmembrane signalling. Curr. Opin. Microbiol. 6:173-180. [DOI] [PubMed] [Google Scholar]
- 6.Brickman, T. J., and S. K. Armstrong. 2005. Bordetella AlcS transporter functions in alcaligin siderophore export and is central to inducer sensing in positive regulation of alcaligin system gene expression. J. Bacteriol. 187:3650-3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brickman, T. J., and S. K. Armstrong. 1999. Essential role of the iron-regulated outer membrane receptor FauA in alcaligin siderophore-mediated iron uptake in Bordetella species. J. Bacteriol. 181:5958-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Celentano, L. P., M. Massari, D. Paramatti, S. Salmaso, and A. E. Tozzi. 2005. Resurgence of pertussis in Europe. Pediatr. Infect. Dis. J. 24:761-765. [DOI] [PubMed] [Google Scholar]
- 9.Connell, T. D., A. Dickenson, A. J. Martone, K. T. Militello, M. J. Filiatraut, M. L. Hayman, and J. Pitula. 1998. Iron starvation of Bordetella avium stimulates expression of five outer membrane proteins and regulates a gene involved in acquiring iron from serum. Infect. Immun. 66:3597-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cummings, C. A., H. J. Bootsma, D. A. Relman, and J. F. Miller. 2006. Species- and strain-specific control of a complex, flexible regulon by Bordetella BvgAS. J. Bacteriol. 188:1775-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cummings, C. A., M. M. Brinig, P. W. Lepp, S. van de Pas, and D. A. Relman. 2004. Bordetella species are distinguished by patterns of substantial gene loss and host adaptation. J. Bacteriol. 186:1484-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diavatopoulos, D. A., C. A. Cummings, L. M. Schouls, M. M. Brinig, D. A. Relman, and F. R. Mooi. 2005. Bordetella pertussis, the causative agent of whooping cough, evolved from a distinct, human-associated lineage of B. bronchiseptica. PLoS Pathogens 1:e45. [Online.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobrindt, U., B. Hochhut, U. Hentschel, and J. Hacker. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2:414-424. [DOI] [PubMed] [Google Scholar]
- 14.Enz, S., S. Mahren, U. H. Stroeher, and V. Braun. 2000. Surface signaling in ferric citrate transport gene induction: interaction of the FecA, FecR, and FecI regulatory proteins. J. Bacteriol. 182:637-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerlach, G., S. Janzen, D. Beier, and R. Gross. 2004. Functional characterization of the BvgAS two-component system of Bordetella holmesii. Microbiology 150:3715-3729. [DOI] [PubMed] [Google Scholar]
- 16.Gerlach, G., F. von Wintzingerode, B. Middendorf, and R. Gross. 2001. Evolutionary trends in the genus Bordetella. Microbes Infect. 3:61-72. [DOI] [PubMed] [Google Scholar]
- 17.Guiso, N. 1997. Isolation, identification and characterization of Bordetella pertussis. Dev. Biol. Stand. 89:233-238. [PubMed] [Google Scholar]
- 18.Herrmann, B., B. Pettersson, K. D. Everett, N. E. Mikkelsen, and L. A. Kirsebom. 2000. Characterization of the rnpB gene and RNase P RNA in the order Chlamydiales. Int. J. Syst. Evol. Microbiol. 50:149-158. [DOI] [PubMed] [Google Scholar]
- 19.Kang, H. Y., T. J. Brickman, F. C. Beaumont, and S. K. Armstrong. 1996. Identification and characterization of iron-regulated Bordetella pertussis alcaligin siderophore biosynthesis genes. J. Bacteriol. 178:4877-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirby, A. E., D. J. Metzger, E. R. Murphy, and T. D. Connell. 2001. Heme utilization in Bordetella avium is regulated by RhuI, a heme-responsive extracytoplasmic function sigma factor. Infect. Immun. 69:6951-6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ko, K. S., K. R. Peck, W. S. Oh, N. Y. Lee, J. H. Lee, and J.-H. Song. 2005. New species of Bordetella, Bordetella ansorpii sp. nov., isolated from the purulent exudate of an epidermal cyst. J. Clin. Microbiol. 43:2516-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotilainen, P., J. Jalava, O. Meurman, O.-P. Lehtonen, E. Rintala, O.-P. Seppälä, E. Eerola, and S. Nikkari. 1998. Diagnosis of meningococcal meningitis by broad-range bacterial PCR with cerebrospinal fluid. J. Clin. Microbiol. 36:2205-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kupferwasser, L. I., R. A. Skurray, M. H. Brown, N. Firth, M. R. Yeaman, and A. S. Bayer. 1999. Plasmid-mediated resistance to thrombin-induced platelet microbicidal protein in staphylococci: role of the qacA locus. Antimicrob. Agents Chemother. 43:2395-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez de Tejada, G., P. A. Cotter, U. Heininger, A. Camilli, B. J. Akerley, J. J. Mekalanos, and J. F. Miller. 1998. Neither the Bvg− phase nor the vrg6 locus of Bordetella pertussis is required for respiratory infection in mice. Infect. Immun. 66:2762-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meiring, H. D., E. van der Heeft, G. J. ten Hove, and A. P. J. M. de Jong. 2002. Nanoscale LC-MS(n): technical design and applications to peptide and protein analysis. J. Sep. Sci. 25:557-568. [Google Scholar]
- 26.Mollet, C., M. Drancourt, and D. Raoult. 1997. rpoB sequence analysis as a novel basis for bacterial identification. Mol. Microbiol. 26:1005-1011. [DOI] [PubMed] [Google Scholar]
- 27.Moore, C. H., L.-A. Foster, D. G. Gerbig, Jr., D. W. Dyer, and B. W. Gibson. 1995. Identification of alcaligin as the siderophore produced by Bordetella pertussis and B. bronchiseptica. J. Bacteriol. 177:1116-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy, E. R., R. E. Sacco, A. Dickenson, D. J. Metzger, Y. Hu, P. E. Orndorff, and T. D. Connell. 2002. BhuR, a virulence-associated outer membrane protein of Bordetella avium, is required for the acquisition of iron from heme and hemoproteins. Infect. Immun. 70:5390-5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paradis, S., M. Boissinot, N. Paquette, S. D. Belanger, E. A. Martel, D. K. Boudreau, F. J. Picard, M. Ouellette, P. H. Roy, and M. G. Bergeron. 2005. Phylogeny of the Enterobacteriaceae based on genes encoding elongation factor Tu and F-ATPase β-subunit. Int. J. Syst. Evol. Microbiol. 55:2013-2025. [DOI] [PubMed] [Google Scholar]
- 30.Parkhill, J., M. Sebaihia, A. Preston, L. D. Murphy, N. Thomson, D. E. Harris, M. T. Holden, C. M. Churcher, S. D. Bentley, K. L. Mungall, A. M. Cerdeno-Tarraga, L. Temple, K. James, B. Harris, M. A. Quail, M. Achtman, R. Atkin, S. Baker, D. Basham, N. Bason, I. Cherevach, T. Chillingworth, M. Collins, A. Cronin, P. Davis, J. Doggett, T. Feltwell, A. Goble, N. Hamlin, H. Hauser, S. Holroyd, K. Jagels, S. Leather, S. Moule, H. Norberczak, S. O'Neil, D. Ormond, C. Price, E. Rabbinowitsch, S. Rutter, M. Sanders, D. Saunders, K. Seeger, S. Sharp, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, L. Unwin, S. Whitehead, B. G. Barrell, and D. J. Maskell. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 35:32-40. [DOI] [PubMed] [Google Scholar]
- 31.Pradel, E., N. Guiso, and C. Locht. 1998. Identification of AlcR, an AraC-type regulator of alcaligin siderophore synthesis in Bordetella bronchiseptica and Bordetella pertussis. J. Bacteriol. 180:871-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pradel, E., and C. Locht. 2001. Expression of the putative siderophore receptor gene bfrZ is controlled by the extracytoplasmic-function sigma factor BupI in Bordetella bronchiseptica. J. Bacteriol. 183:2910-2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Register, K. B., T. F. Ducey, S. L. Brockmeier, and D. W. Dyer. 2001. Reduced virulence of a Bordetella bronchiseptica siderophore mutant in neonatal swine. Infect. Immun. 69:2137-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reischl, U., N. Lehn, G. N. Sanden, and M. J. Loeffelholz. 2001. Real-time PCR assay targeting IS481 of Bordetella pertussis and molecular basis for detecting Bordetella holmesii. J. Clin. Microbiol. 39:1963-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Relman, D. A., T. M. Schmidt, R. P. MacDermott, and S. Falkow. 1992. Identification of the uncultured bacillus of Whipple's disease. N. Engl. J. Med. 327:293-301. [DOI] [PubMed] [Google Scholar]
- 36.Russell, F. M., J. M. Davis, M. J. Whipp, P. H. Janssen, P. B. Ward, J. R. Vyas, M. Starr, S. M. Sawyer, and N. Curtis. 2001. Severe Bordetella holmesii infection in a previously healthy adolescent confirmed by gene sequence analysis. Clin. Infect. Dis. 33:129-130. [DOI] [PubMed] [Google Scholar]
- 37.Schouls, L. M., C. S. Schot, and J. A. Jacobs. 2003. Horizontal transfer of segments of the 16S rRNA genes between species of the Streptococcus anginosus group. J. Bacteriol. 185:7241-7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sebaihia, M., A. Preston, D. J. Maskell, H. Kuzmiak, T. D. Connell, N. D. King, P. E. Orndorff, D. M. Miyamoto, N. R. Thomson, D. Harris, A. Goble, A. Lord, L. Murphy, M. A. Quail, S. Rutter, R. Squares, S. Squares, J. Woodward, J. Parkhill, and L. M. Temple. 2006. Comparison of the genome sequence of the poultry pathogen Bordetella avium with those of B. bronchiseptica, B. pertussis, and B. parapertussis reveals extensive diversity in surface structures associated with host interaction. J. Bacteriol. 188:6002-6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sekine, Y., N. Eisaki, and E. Ohtsubo. 1994. Translational control in production of transposase and in transposition of insertion sequence IS3. J. Mol. Biol. 235:1406-1420. [DOI] [PubMed] [Google Scholar]
- 40.Shafer, W. M., X. Qu, A. J. Waring, and R. I. Lehrer. 1998. Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc. Natl. Acad. Sci. USA 95:1829-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shepard, C. W., M. I. Daneshvar, R. M. Kaiser, D. A. Ashford, D. Lonsway, J. B. Patel, R. E. Morey, J. G. Jordan, R. S. Weyant, and M. Fischer. 2004. Bordetella holmesii bacteremia: a newly recognized clinical entity among asplenic patients. Clin. Infect. Dis. 38:799-804. [DOI] [PubMed] [Google Scholar]
- 42.Tanaka, M., C. R. Vitek, F. B. Pascual, K. M. Bisgard, J. E. Tate, and T. V. Murphy. 2003. Trends in pertussis among infants in the United States, 1980-1999. JAMA 290:2968-2975. [DOI] [PubMed] [Google Scholar]
- 43.Vanderpool, C. K., and S. K. Armstrong. 2001. The Bordetella bhu locus is required for heme iron utilization. J. Bacteriol. 183:4278-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vanderpool, C. K., and S. K. Armstrong. 2004. Integration of environmental signals controls expression of Bordetella heme utilization genes. J. Bacteriol. 186:938-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Zee, A., F. Mooi, J. van Embden, and J. Musser. 1997. Molecular evolution and host adaptation of Bordetella spp.: phylogenetic analysis using multilocus enzyme electrophoresis and typing with three insertion sequences. J. Bacteriol. 179:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Hove, B., H. Staudenmaier, and V. Braun. 1990. Novel two-component transmembrane transcription control: regulation of iron dicitrate transport in Escherichia coli K-12. J. Bacteriol. 172:6749-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Visca, P., L. Leoni, M. J. Wilson, and I. L. Lamont. 2002. Iron transport and regulation, cell signalling and genomics: lessons from Escherichia coli and Pseudomonas. Mol. Microbiol. 45:1177-1190. [DOI] [PubMed] [Google Scholar]
- 48.Weyant, R. S., D. G. Hollis, R. E. Weaver, M. F. M. Amin, A. G. Steigerwalt, S. P. O'Connor, A. M. Whitney, M. I. Daneshvar, C. W. Moss, and D. J. Brenner. 1995. Bordetella holmesii sp. nov., a new gram-negative species associated with septicemia. J. Clin. Microbiol. 33:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yap, W. H., Z. Zhang, and Y. Wang. 1999. Distinct types of rRNA operons exist in the genome of the actinomycete Thermomonospora chromogena and evidence for horizontal transfer of an entire rRNA operon. J. Bacteriol. 181:5201-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yih, W. K., E. A. Silva, J. Ida, N. Harrington, S. M. Lett, and H. George. 1999. Bordetella holmesii-like organisms isolated from Massachusetts patients with pertussis-like symptoms. Emerg. Infect. Dis. 5:441-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.