Abstract
A novel mutant of the marine oil-degrading bacterium Alcanivorax borkumensis SK2, containing a mini-Tn5 transposon disrupting a “tesB-like” acyl-coenzyme A (CoA) thioesterase gene, was found to hyperproduce polyhydroxyalkanoates (PHA), resulting in the extracellular deposition of this biotechnologically important polymer when grown on alkanes. The tesB-like gene encodes a distinct novel enzyme activity, which acts exclusively on hydroxylated acyl-CoAs and thus represents a hydroxyacyl-CoA-specific thioesterase. Inactivation of this enzyme results in the rechanneling of CoA-activated hydroxylated fatty acids, the cellular intermediates of alkane degradation, towards PHA production. These findings may open up new avenues for the development of simplified biotechnological processes for the production of PHA as a raw material for the production of bioplastics.
Alcanivorax borkumensis strain SK2 is a cosmopolitan marine bacterium with a specialized metabolism adapted to the degradation of petroleum oil hydrocarbons, enabling it to degrade a wide range of hydrocarbons (26). A. borkumensis is usually the most abundant member of microbial communities that develop following an oil spill at sea and is assumed to be globally one of the most important microbes involved in removing oil from marine environments (9). The genome of SK2 was recently sequenced and annotated (21). It is the best studied—and the paradigm—of the so-called hydrocarbonoclastic bacteria, a recently discovered group of oligotrophic marine microbes belonging to the Gammaproteobacteria that utilize hydrocarbons but not most other common bacterial sources of carbon and energy.
In this study, we identify and describe a new “tesB-like” gene of A. borkumensis that encodes a novel hydroxyacyl-coenzyme A (CoA)-specific thioesterase. Acyl-CoA thioesterases that hydrolyze acyl-CoA molecules have thus far been studied mainly in Escherichia coli, which possesses two of such enzymes: (i) thioesterase I, encoded by the tesA gene, cleaves C12 to C18 acyl-CoA molecules (4), and (ii) thioesterase II, encoded by the tesB gene, acts on C6-C18 acyl-CoA thioesters as well as on C12-C18 3-hydroxyacyl-CoA thioesters (3). Little is known about the exact physiological role of the TesB protein in bacterial metabolism except that it releases free fatty acids and also, at least in one case, hydroxylated fatty acids from the corresponding CoA-activated forms, thus producing free 3-hydroxyalkanoic acids (3-HAA) (27). CoA-activated hydroxylated fatty acids in turn are cellular precursor intermediates for the synthesis of polyhydroxyalkanoates (PHA), well-known bacterial storage compounds, which usually are produced as insoluble intracellular granules by many microorganisms during times of carbon surfeit (24), and they have long been explored as a renewable resource for biodegradable thermoplastics and biopolymers (2, 18, 24). We describe here that a disruption of the tesB-like gene of A. borkumensis by a mini-Tn5 transposon causes hyperproduction and extracellular deposition of medium-chain-length PHA when grown on alkanes. Since commercial exploitation of the biological production of PHA has thus far been hampered by the need for the costly recovery of intracellularly stored granules from whole cells (13), the present mutant allows this costly recovery step to be circumvented, as large amounts of PHA can easily be obtained from culture medium.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A. borkumensis strain SK2 (DSM 11573) is the wild-type parental strain in all experiments. A mini-Tn5 mutant named C9 was generated by standard procedures using the mini-Tn5 Str/Sp element (7). Wild-type SK2 and C9 mutant strains were grown at 30°C in modified ONR7a medium (26) containing 0.27 g/liter of NH4Cl and either 1.5% (wt/vol) octadecane or 2% (wt/vol) pyruvate as carbon sources. E. coli strains DH5α (Invitrogen, Carlsbad, CA) and RosettaBlue DE3 (Novagen, Madison, Wisconsin), which were used for cloning and expression studies, were grown at 37°C in Luria-Bertani medium supplemented with kanamycin (50 μg/ml) or streptomycin (50 μg/ml) and/or chloramphenicol (34 μg/ml) where appropriate.
Construction of an A. borkumensis SK2 mini-Tn5 transposon library.
Transposon mutagenesis was based on the mini-Tn5 Str/Sp element constructed as described previously by de Lorenzo et al. (7). A. borkumensis SK2 was grown at 30°C on ONR7a medium until the stationary phase of growth, and cells were centrifuged at 3,200 × g at 4°C. Donor strain E. coli (CC118 λpir) cultures and helper (HB101 λpir) cultures of E. coli were grown overnight at 37°C on LB medium with either streptomycin or chloramphenicol, respectively, washed with fresh LB, and centrifuged at 3,200 × g at 4°C. The pellets of A. borkumensis and E. coli donor and helper strains were mixed in proportion (4:1:1 [by volume]) and placed onto a membrane filter on a plate with LB agar and salts (0.45 g/liter Na2HPO4 · 2H2O, 2.5 g/liter NaNO3, 11.5 g/liter NaCl, 0.38 g/liter KCl, 0.7 g/liter CaCl2 · 2H2O) and 2% (wt/vol) pyruvate as a carbon and energy source. The plate was incubated for 24 h at 30°C. The cells were then washed with 10 mM MgSO4, and transconjugants were selected on ONR7a medium with 0.5% (wt/vol) pyruvate and 0.5% (wt/vol) acetate as carbon sources and nalidixic acid (10 μg/ml) and streptomycin as antibiotics as required.
Inverse PCR.
The mini-Tn5 insertion sites of the selected mini-Tn5 mutants were determined by inverse PCR as described previously (15). Briefly, total DNA of the mutant was isolated and digested with ClaI, which does not cut within the mini-Tn5 element. The resulting DNA fragments were circularized with DNA T4 ligase, and the flanking regions of the inserted mini-Tn5 were amplified with two primers corresponding to the O and I ends of the Tn5 transposon (GGC CGC ACT TGT GTA TAA GAG TCA G and GCG GCC AGA TCT GAT CAA GAG ACA G, respectively). The conditions for the PCR were as follows: 94°C for 1.5 min, 48°C for 1 min, and 70°C for 4 min for 30 cycles. The PCR products were gel purified and used for automatic DNA sequencing with BigDye terminators on an ABI Prism 377 sequencer (AP Biosystems). To determine the precise site of transposon insertion, additional primers have been designed to read the flanking regions of the disrupted gene, i.e., primers 1086 (TTA CTG GCT TCG CAG GAA TGG) and intSM160 (CTT GGC ACC CAG CAT GCG CGA GCA GG).
RT-PCR.
To determine whether the two genes ABO_1111 and ABO_1112 constitute an operon, reverse transcription (RT)-PCR was performed on DNase I-treated total RNA extracted with a Fast Blue RNA isolation kit (Qbiogene, Heidelberg, Germany) from a 10-ml culture of SK2 grown to early stationary phase (optical density at 600 nm of 1.0) on either 2% (wt/vol) pyruvate or 1.5% (wt/vol) octadecane. Primers used were Oligo I (TAT GGT CAA AGT CAG GCG GTG) and Oligo II (CAC ATC CAA GCG CAA AGA CTG), which are specific for a 311-bp region spanning the 3′ end of ABO_1111 and the 5′ end of ABO_1112 (21). The same primers were also used for RT-PCR with RNA isolated from the C9 tesB-like::Tn5 mutant in order to determine whether the mini-Tn5 mutation had a polar effect on the transcription of the downstream gene(s). RT-PCR was performed using the SuperScript first-strand synthesis system for RT-PCR (Invitrogen) according to the instructions of the supplier. Briefly, the reaction mixture, consisting of 2 μl of template RNA, 1 μl of a 10 mM deoxynucleoside triphosphate mix, 1 μl of 2 μM primer Oligo II, and 6 μl of diethylpyrocarbonate-treated water, was incubated at 65°C for 5 min; placed on ice; mixed with a solution consisting of 2 μl 10× RT buffer, 4 μl of 25 mM MgCl2, 2 μl of 0.1 M dithiothreitol, and 1 μl of RNaseOUT recombinant RNase inhibitor; and incubated further at 42°C for 2 min, and 1 μl (50 units) of SuperScript II RT was then added to each tube (except the RT control tubes), and incubation continued at 42°C for 50 min. The RT reaction was then stopped by raising the temperature to 70°C for 15 min. One μl of RNase H was then added, and the mixture was incubated for 20 min at 37°C. Subsequent PCR amplification was performed under standard conditions, and the RT-PCR products were separated by electrophoresis on a 1.8% (wt/vol) agarose gel.
PHA isolation and analysis of its monomer composition.
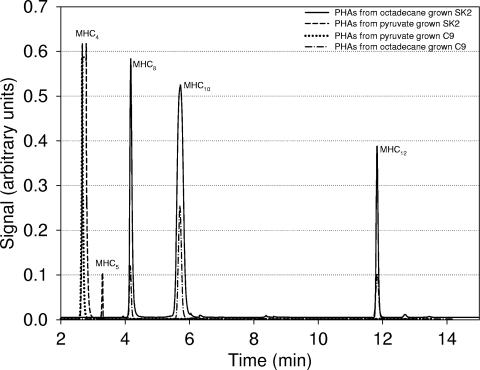
Bacteria were cultured in ONR7a medium with either 2% (wt/vol) pyruvate or 1.5% (wt/vol) octadecane as a carbon source on a rotary shaker (100 rpm) at 30°C until the late stationary phase of growth. Cell pellets and supernatant fluids of the wild type were separated by centrifugation (60 min at 12,000 × g). As C9 mutant cells could not be entirely separated from the culture media by centrifugation, total cell cultures of both the wild type and the C9 mutant were also included in the chemical analysis. The cell pellets, supernatants, or total cell cultures were lyophilized, rinsed with ice-cold water, dried again overnight at 80°C, and stored at room temperature until use. To quantify PHA, the polyesters were extracted from accurately weighed freeze-dried samples by Soxhlet extraction with hot chloroform (95°C) as described previously by Cromwick et al. (6). Briefly, chloroform extracts were filtered through Whatman paper to remove cell debris and then concentrated and added to cold methanol to precipitate PHA. The precipitated PHA was washed with methanol, dried, and subjected to gel filtration to select only those cell polymers with molecular masses higher than 100,000 Da, thus eliminating any contamination of the samples by glucolipids or free 3-HAA, which are also potentially produced under these conditions. To determine the polyester content and composition, 2 mg of purified PHA was incubated with a mixture of chloroform-methanol-sulfuric acid (1:0.85:0.15 [by volume]) for 2 h at 100°C to degrade PHA to its constituent, 3-hydroxycarboxylic acid methyl esters (FAMEs), by methanolysis. Distilled water (0.5 ml) was then added, tubes were shaken for 1 min, and the phases were then allowed to separate. The organic phase was transferred into a vial, and the FAMEs were analyzed using a gas chromatograph-mass spectrometer (gas chromatograph model Varian 3400CX [Varian Chromatography Systems, Sugar Land, TX] and a VG Autospec spectrometer) equipped with a 30-m by 0.25-mm HP-5 (5% diphenyl and 95% dimethylpolysiloxane)-fused silica capillary column with a flow rate of 1 ml/min (with helium as the carrier gas), a sample input temperature to 230°C at a rate of 8°C/min, an interface temperature of 250°C, an ion source temperature of 175°C, the electron impact mode at 70 eV, and scanning from 45 to 450 atomic mass units at 0.5 s/scan. The degree of purity of the PHA samples used for analysis was about 99.5%, as no additional peaks on gas chromatograms were observed (Fig. 1). Molecular weights were determined by gel permeation chromatography using a Spectra Physics gel permeation chromatograph under the following conditions: 50°C column temperature; isocratic gradient; tetrahydrofuran mobile phase, 1.0-ml/min flow rate, and light-scattering detector.
FIG. 1.
Gas chromatograms of polyesters produced by Alcanivorax borkumensis SK2 and mutant strain C9 under different culture conditions. MHC4, monohydroxybutyric acid; MHC5, monohydroxypentanoic acid; MHC8, monohydroxyoctanoic acid; MHC10, monohydroxydecanoic acid; MHC12, monohydroxydodecanoic acid.
Cloning of the tesB-like gene in E. coli and preparation of cell extracts for thioesterase assay.
The ABO_1111 gene, coding for TesB-like acyl-CoA thioesterase, was amplified using primers 1086F (5′-TTA CTG GCT TCG CAG GAA TGG-3′) and 1086R (5′-CTT GCT TAC CTA AAG TCC GCG-3′), and the resulting PCR product of 896 bp was cloned into the pCR2.1 TOPO cloning vector (Invitrogen). The cloned gene was then excised from the recombinant plasmid as an EcoRI fragment and inserted into the EcoRI site of the pCDFDuet-1 expression vector (Novagen), and the resulting construct was then transformed into competent E. coli DH5α cells (Invitrogen), selecting for transformants on LB medium containing streptomycin (50 μg/ml). The clones obtained were checked for the correct orientation of the cloned gene, positive plasmid constructs were then transformed into RosettaBlue DE3 competent cell, and transformants were selected on streptomycin (50 μg/ml) and chloramphenicol (34 μg/ml). For expression and purification of the enzyme, cultures of E. coli RosettaBlue DE3, harboring the tesB-like gene in the expression vector, were diluted 1:10 and grown overnight at 37°C in LB liquid medium containing appropriate antibiotics until an absorbance at 600 nm of 1.0 was reached. At that point, overexpression was induced by the addition of 1.0 mM isopropyl thio-β-d-galactoside (IPTG), and after 6 h of growth, cells were harvested, washed with buffer A (50 mM potassium phosphate buffer, pH 8.0, 10 mM EDTA), and stored at 4°C until use. Approximately 0.5 g (wet weight) of E. coli cells expressing or not expressing the tesB-like gene was suspended in 1 ml of buffer A supplemented with 200 μg of phenylmethylsulfonyl fluoride, 5 μg DNase I grade II, and 1 μg lysozyme per ml, and cells were then disrupted by sonification for a total of 4 min (30-s pulses and 1-min pauses) at 4°C in a W250 sonifier (Branson Schallkraft GmbH, Germany). Soluble cell fractions were obtained as supernatants after 30 min of centrifugation at 15,000 × g at 4°C. The resulting supernatants were tested for thioesterase activity (see below). The total protein concentration was determined by the Bradford method using bovine serum albumin as a standard (5).
Thioesterase assay.
The hydrolysis of acyl-CoAs and hydroxyacyl-CoAs by E. coli cell extracts containing or not containing the TesB-like enzyme was determined using a 5,5-dithio-bis(2-nitrobenzoic acid) (DTNB)-based assay, as described elsewhere previously (28). Briefly, reactions for DTNB-based test were carried out in buffer A, and 5-thio-2-nitrobenzoate, produced from DTNB reacting with CoA released by hydrolysis from the acyl-CoA substrate, was monitored through its absorbance at 412 nm (molar extinction coefficient of 13,600 M−1). A 1-ml reaction mixture contained 4 μM acyl-CoAs or hydroxyacyl-CoA (chain length in each case ranging from acetyl to decanoyl), 1 mM DTNB, and 100 μl of crude cell extract containing 25 μg of total protein (obtained as described above) in a quartz cuvette with a 1-cm light path length. One unit of enzymatic activity was defined as the amount of protein releasing 1 μmol of CoA per min. (R,S)-3-Hydroxyacyl-CoAs (from 3-hydroxyhexadecyl-CoA to 3-hydroxydecanoyl-CoA) were synthesized as described previously by Rehm et al. (19). (R,S)-3-Acyl-CoAs were obtained from Sigma Chemical Co. (St. Louis, Missouri). Quantification of the hydrolysis of hydroxyacyl-CoAs by extracts from A. borkumensis wild-type strain SK2 and the C9 mutant was performed essentially as described above by using protein extracts of A. borkumensis cells cultured in ONR7a medium with either 2% (wt/vol) pyruvate or 1.5% (wt/vol) octadecane as a carbon source on a rotary shaker (100 rpm) at 30°C until the late stationary phase of growth (conditions for disruption were the same as those for E. coli cells).
Electron microscopy.
For scanning electron microscopy, cells were grown on Permanox slides (Nalge Nunc) in ONR7a medium containing 1.5% (wt/vol) octadecane (slides covered with octadecane), or 2% (wt/vol) pyruvate, and 0.27 g/liter NH4Cl in liquid culture, and cells were harvested in their stationary phase of growth. Scanning electron microscopy was prepared and carried out as described previously (14).
RESULTS AND DISCUSSION
A. borkumensis produces polyesters.
For marine oil-degrading bacteria including A. borkumensis, oil pollution constitutes a temporary condition of carbon excess coupled with limiting nitrogen, i.e., a high carbon/nitrogen (C/N) ratio, which is precisely the condition that would prompt bacteria to embark on the formation of storage compounds such as PHA or other cellular storage substances (24). Such storage compounds permit oligotrophic bacteria like A. borkumensis to survive during less nutritious periods interceding those periods when carbon sources are abundant, e.g., during events of oil pollution. Since PHA is known to be a common polymeric storage compound, and since A. borkumensis was reported previously not only to express one of two putative phaC synthase genes (ABO_1418) under conditions of alkane excess (20) but also to be able to synthesize the monomeric PHA precursor compound 3-HAA (1), we inspected this organism's ability to produce polyhydroxyalkanoates.
Chemical analysis of cell contents from wild-type A. borkumensis SK2 grown under conditions of C excess by gas chromatography-mass spectrometry revealed that A. borkumensis produces either medium-chain-length PHA, consisting of 3-hydroxyacyl monomers of 6 to 12 carbon units, or polyhydroxybutyrate (PHB) (four carbon units), depending on the carbon source used for growth (Fig. 1 and Table 1). The composition of the polymer produced on alkanes corresponds well to the respective PHA monomer composition typical for Pseudomonas species (25). The mean molecular mass of the polymer as determined by gel permeation chromatography was found to be around 280 kDa under both growth conditions tested (Table 1). However, although the amount of PHA produced from alkane (i.e., under conditions of a high C/N ratio) was three times higher (18 mg/liter) than the amount of PHB produced during growth on pyruvate (6.5 mg/liter), such rather low concentrations in absolute terms are far below the amounts typically found for intracellular storage polymers in PHA- or PHB-storing bacteria (up to 1.6 g/liter). The chemical analysis was also confirmed by nuclear magnetic resonance (data not shown). We conclude that while A. borkumensis clearly has the genetic equipment to synthesize PHA and PHB polyesters, the wild type does so by producing only small amounts, suggesting that neither PHB nor PHA is likely to be a major storage compound in this bacterium, which instead probably employs other types of storage compounds to serve as carbon/energy source storage during periods of carbon/energy limitation.
TABLE 1.
PHA production in SK2 wild-type and C9 mutant strains grown on different carbon sources
| Strain | Substrate | Mean ± SD
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| PHA yield (mg/liter)a | Monomer composition of hydroxyalkanoates (mol%)b
|
Molecular mass (kDa)c | |||||||
| C4 | C5 | C6 | C8 | C10 | C12 | ||||
| SK2 wild type | Pyruvate | 6.5 ± 1.2 | 100 | —d | — | — | — | — | 279 ± 23 |
| SK2 wild type | Octadecane | 18.0 ± 3.8 | — | — | 2 ± 0.3 | 20 ± 2.5 | 48 ± 3.7 | 30 ± 2.1 | 277 ± 38 |
| C9 mutant | Pyruvate | 112 ± 16.8 | 98 ± 0.4 | 2 ± 0.1 | — | — | — | — | 352 ± 16 |
| C9 mutant | Octadecane | 2,560 ± 165.1 | — | — | 4 ± 0.2 | 18 ± 2.0 | 37 ± 2.5 | 39 ± 1.8 | 350 ± 42 |
To quantify PHA, the polyesters were extracted with chloroform as described in Materials and Method section and accurately weighed after gel permeation and gas chromatography analysis.
The molar fraction of polyester was calculated by gas chromatography according to the area of the peak of FAMEs obtained after methanolysis. Pure FAMEs were used for calibration. In all cases, data are means ± standard deviations (two independent cultures subjected to three independent analyses).
Molecular mass of polymer; means were determined by a gel permeation chromatograph calibrated with polystyrene.
—, not detected.
Isolation of a mutant showing hyperproduction and extracellular deposition of PHA.
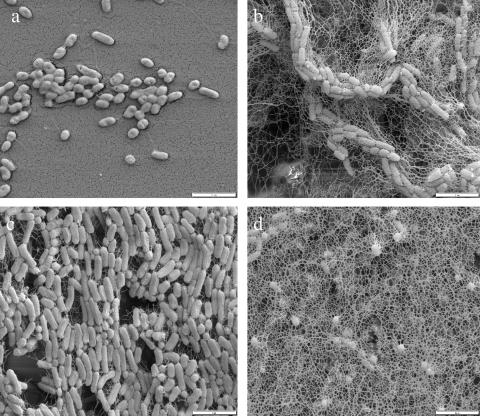
In the course of the screening of a mini-Tn5 transposon library based on the mini-Tn5 Str/Sp element (7), applying Kolter's assay (17) to look for biofilm-deficient mutants, a number of mutants that failed to form a biofilm in 96-well microtiter plates were isolated (J. S. Sabirova et al., unpublished data). The Kolter assay is routinely used for the detection of biofilm-deficient mutants: it involves growing the cells in 96-well microtiter polyvinyl chloride plates, staining biofilm-forming cells with crystal violet, and finally washing out and measuring the retained dye with ethanol to quantify the degree of biofilm formation. Cells that are able to form a biofilm normally produce a violet-stained circle at the air-water interface. In one of the biofilm-deficient mutants, designated the C9 mutant, the formation of biofilm was prevented by the excessive production of secreted polymeric material, later identified as the bacterial storage compound PHA (see below). Scanning electron micrographs of SK2 wild-type and C9 mutant (Fig. 2) cells grown on Permanox hydrophobic slides covered with octadecane in ONR7a medium showed that the mutant cells are embedded in a dense extracellular network of material, whereas the wild-type cells are not. As the cells were grown in excess of the alkane source, i.e., under conditions favoring the production of PHA storage material, we suspected that the extracellularly deposited polymer was PHA. To test this assumption, both wild-type strain SK2 and mutant strain C9 were grown on ONR7a medium with 1.5% (wt/vol) octadecane as the carbon and energy source, and the polymer was extracted from each of the total cell cultures and was analyzed as described in Materials and Methods. As no PHA was detected in the culture supernatant of alkane-grown SK2 wild-type cells, we conclude that essentially all of the PHA produced by wild-type strain SK2 on alkane was intracellular. As C9 mutant cells could not be entirely separated from the culture media by centrifugation (most likely due to the extracellular PHA tightly attached to them), we determined PHA yields in total cell culture. We found that in the C9 mutant grown on octadecane, the amount of PHA was almost 2.6 g/liter, which is about 140 times that produced by the SK2 parental strain grown on octadecane (0.018 g/liter) (Table 1). PHA production was thus significantly higher in the C9 mutant strain than in the wild type under conditions where PHA precursor intermediates are made from alkane substrates.
FIG. 2.
Scanning electron micrograph images of A. borkumensis SK2 wild-type and C9 mutant cells. Shown are SK2 wild-type (a and c) and C9 mutant (b and d) cells grown on Permanox slides covered with octadecane in ONR7a medium containing 1.5% (wt/vol) octadecane and 0.27 g/liter of NH4Cl featuring either the external, medium-exposed side of the biofilm (a and b) or the internal side of it, adjacent to the slide (c and d).
The gene mutated in mutant strain C9 codes for a TesB-like acyl-CoA thioesterase.
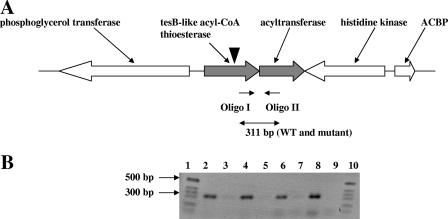
The site of insertion of the mini-Tn5 element in the C9 mutant was determined by inverse PCR as described elsewhere previously (15) and was found to be located between nucleotides 557 and 558 of the coding sequence (CDS) of the gene ABO_1111, which is annotated as coding for a putative TesB-like acyl-CoA thioesterase II (21). Analysis of the A. borkumensis genome sequence revealed that A. borkumensis possesses three different acyl-CoA thioesterase-encoding genes, namely, tesA, tesB, and the above-mentioned tesB-like gene, with the tesB and tesB-like genes being the closest homologues. The site of the mini-Tn5 insertion in the tesB-like gene predicts a disruption of the gene's function. Since the inverse PCR produced only one amplicon, we conclude that the phenotype of C9 results from the single transposition event identified. Inspection of the 3′ downstream region of ABO_1111 revealed the presence of a second CDS, ABO_1112, of 645 bp in length, which overlaps the last codon of ABO_1111 (Fig. 3A). The predicted encoded protein exhibits similarity to acylglycerolacyl transferase PlsC proteins of other bacterial species (amino acid identity and similarity of 41% and 55%, respectively, to PlsC from Pseudomonas aeruginosa PAO-1). The close proximity of ABO_1111 and ABO_1112 suggests that these two CDSs may form an operon. To determine whether the mutating transposon might exert a potential polar effect on the expression of the downstream ABO_1112 gene by preventing the expression of a potential operon-spanning single transcript, RT-PCR was employed (using primers Oligo I and Oligo II) to specifically amplify the 311-bp region spanning the ABO_1111 and ABO_1112 junction (Fig. 3A). In both the mutant and the wild type, we obtained the expected PCR product of approximately 311 bp. This confirms that ABO_1112 is also well expressed in the C9 mutant either as a part of an operon with ABO_1111 with no polar effect of the insertion or with ABO_1112 being transcribed from its own promoter. In any case, the amplified transcript indicative of ABO_1112 expression appears to be of comparable intensities in both the wild type and the C9 mutant (Fig. 3B).
FIG. 3.
RT-PCR analysis of DNase I-treated RNA extracted from A. borkumensis SK2 and a tesB-like acyl-CoA thioesterase mutant from a 10-ml culture of SK2 grown to stationary phase (optical density at 600 nm of 1.0) on either 2% (wt/vol) pyruvate or 1.5% (wt/vol) octadecane. Primers used were Oligo I and Oligo II, specific for a 311-bp region spanning the 3′ end of ABO_1111 and the 5′ end of ABO_1112. (A) Organization of the operon and adjacent genes, location of the primers used, and predicted size of the RT-PCR product. WT, wild type. (B) RT-PCR products were obtained from total RNA extracted from SK2 and mutant C9 using primers Oligo I and Oligo II. Lanes: 1, 1-kb marker, 2, SK2 on pyruvate; 4, SK2 on octadecane; 6, C9 on pyruvate; 8, C9 on octadecane. Lanes 3, 5, 7, 9 are corresponding negative controls (without reverse transcriptase).
Expression of the TesB-like protein.
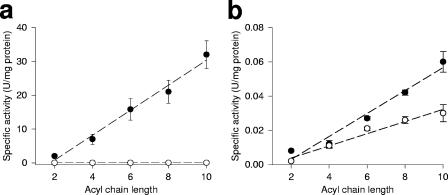
To determine the substrate specificity of the predicted TesB-like protein of A. borkumensis, we cloned the tesB-like gene (ABO_1111) into the expression vector pCDFDuet-1 (Novagen) and expressed the gene in E. coli RosettaBlue DE3 competent cells (Novagen). E. coli crude extracts containing the expressed tesB-like gene product were then tested for enzymatic activity of the TesB-like protein. Acyl-CoA and (R,S)-3-hydroxyacyl-CoA were provided as substrates, and the reaction products were analyzed by a DTNB assay as described in Materials and Methods, with E. coli harboring only vector pCDF as a negative control. The data shown in Fig. 4a clearly demonstrate that the TesB-like enzyme efficiently hydrolyzes 3-hydroxyacyl-CoAs ranging from 3-hydroxyhexanoyl-CoA to 3-hydroxydecanoyl-CoA, with a clear preference for long-chain derivatives. By contrast, when the corresponding nonhydroxylated acyl-CoAs (ranging from hexanoyl to decanoyl) were provided as substrates, the TesB-like protein exhibited little ability to hydrolyze these nonhydroxylated acyl-CoA substrates (Fig. 4b). Since crude extracts containing the cloned A. borkumensis tesB-like gene display a very high ratio of hydroxyacyl-CoA- to acyl-CoA-specific activity (approximately 500:1 for C10 derivatives), we conclude that the tesB-like gene encodes a product that specifically acts on hydroxylated acyl-CoAs and can therefore be named a hydroxyacyl-CoA-specific thioesterase. For future reference, we suggest that the tesB-like gene encoding the hydroxyacyl-CoA-specific thioesterase now be designated tesB2, as opposed to the previously described tesB gene known to hydrolyze acyl-CoAs (Fig. 4). Comparative measurement of 3-hydroxyacyl-CoA thioesterase activity in A. borkumensis wild-type and C9 mutant strains revealed that the mutation in the tesB-like gene resulted in the complete disruption of this enzymatic activity in the C9 mutant (Fig. 5), thus confirming that this novel enzymatic activity is indeed encoded by the tesB-like gene.
FIG. 4.
Enzymatical hydrolysis of (R,S)-3-hydroxyacyl-CoAs (a) and acyl-CoAs (b) by crude extracts of E. coli harboring only vector pCDF as the control (○) or the recombinant insertion construct pCDF::tesB-like (•). Data given are means with standard deviations of three independent culture samples and three independent assays. The specific activity of the crude extract of E. coli harboring only vector pCDF was lower than 0.025 U/mg for all substrates tested, which is in the range of previously published data.
FIG. 5.
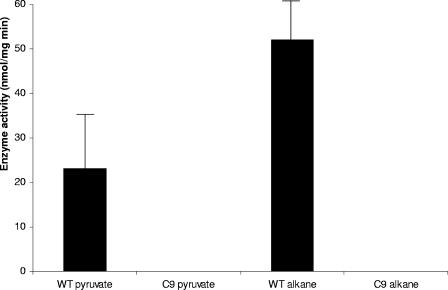
Enzyme hydrolysis of (R,S)-3-hydroxyacyl-CoAs by wild-type A. borkumensis (WT) and its C9 mutant. 3-Hydroxy-dodecanoyl-CoA was the substrate for enzyme determinations. The assay was performed as described in Materials and Methods, using 100 μl of crude cell extracts containing 25 μg of total protein. Activity is shown as the amount of protein releasing 1 nmol of CoA per min. Values represent the averages of three determinations ± standard deviations.
Inactivation of the tesB-like gene channels 3-hydroxyacyl-CoA intermediates towards polyhydroxyalkanoate production.
An amino acid similarity search of the mutated gene ABO_1111 against the entire A. borkumensis genome identified an acyl-CoA thioesterase II protein, encoded by the tesB gene, as its closest homologue. Two acyl-CoA thioesterases have been studied in E. coli: (i) acyl-CoA thioesterase I (encoded by the tesA gene), which is specific for C12-C18 acyl-CoA esters (4), and (ii) acyl-CoA thioesterase II (encoded by the tesB gene), which cleaves C6-C18 acyl-CoA esters as well as C12-C18 3-hydroxyacyl-CoA esters (3). Whereas TesA has been implicated in the hydrolysis of the thioester bond that links nascent fatty acids to the biosynthetic acyl carrier protein-containing biosynthetic multienzyme complex, thus generating free fatty acids (12), little is known about the exact physiological role of TesB in bacterial metabolism, except that it releases free fatty acids and, at least in one case, hydroxylated fatty acids from their respective CoA-activated forms (27). Since 3-hydroxyacyl-CoAs are the substrates of PHA synthase in the formation of PHA, acyl-CoA thioesterases of the tesB-encoded type releasing free 3-HAAs and PHA synthase compete for the same intermediates as their substrates, namely, 3-hydroxyacyl-CoAs. Hence, inactivation of a tesB gene encoding such an acyl-CoA thioesterase II that is also able to act on hydroxylated acyl-CoAs, or inactivation of a tesB-like gene encoding such an enzyme acting specifically on hydroxylated acyl-CoAs only, as observed here in the case of A. borkumensis, would in principle channel all 3-hydroxyacyl-CoA into PHA synthesis (Fig. 6), which can explain the hyperproduction of PHA observed in the A. borkumensis C9 mutant that is deficient in the tesB-like gene. Thus, it appears that in A. borkumensis, the existence of two genes, tesB and the tesB-like gene (tesB2), reflects distinct functions of the TesB and TesB-like proteins, with the former acting specifically on nonhydroxylated acyl-CoAs and the latter acting on hydroxylated acyl-CoAs exclusively. Indeed, a tesB-encoded acyl-CoA thioesterase II that is unable to act on 3-hydroxyacyl-CoA has also been described previously for Rhodobacter sphaeroides (22). A plausible explanation for the phenotype of mutant strain C9 therefore seems to be that the mutation inactivates the tesB-like gene, whose protein product acts specifically on hydroxylated acyl-CoAs and thus abolishes the release of free 3-HAA from 3-HAA-CoA, which leads to an increase in the pool of the PHA precursor molecule 3-hydroxyacyl-CoA and consequently to enhanced PHA formation. The potential metabolic pathways relevant to this scenario in A. borkumensis are depicted in Fig. 6.
FIG. 6.
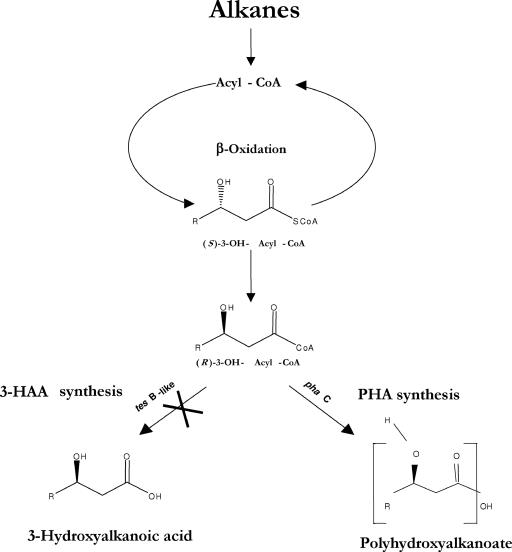
Suggested pathway of PHA biosynthesis in A. borkumensis SK2 and mutant strain C9 grown on hydrocarbons. Hydrocarbons are degraded via terminal oxidation to produce free fatty acids, which are then activated by an acyl-CoA synthase and subjected to β-oxidation. The (S)-3-OH-acyl-CoAs produced by β-oxidation are isomerized into (R)-3-OH-acyl-CoAs by the action of an isomerase. (R)-3-OH-Acyl-CoAs produced during β-oxidation are converted to either 3-HAA through the action of TesB-like acyl-CoA thioesterase or PHA through the action of PhaC synthase. The mutation in the TesB-like acyl-CoA thioesterase abolishes the production of free 3-HAA and channels (R)-3-OH-acyl-CoAs exclusively into PHA synthesis.
The existence of a hydroxyacyl-CoA-specific TesB-like thioesterase in A. borkumensis may be strongly linked to the alkane metabolism of this oil-degrading bacterium such that this TesB-like thioesterase together with PHA synthase represent two “valve” enzymes allowing either the storage of the metabolic precursors in the form of PHAs or the ability to hydrolyze and possibly excrete them in the form of 3-HAAs. The latter have been shown either to possess biosurfactant properties themselves (8) or to be constituents of biosurfactants (23). In fact, A. borkumensis has been shown to produce biosurfactants of a glycolipid nature in excessive amounts, with some of them containing 3-hydroxyalkanoic acid moieties (1), which should be highly advantageous during growth on alkane-containing oil spills.
To conclude, we report here a new enzyme found in the marine oil-degrading bacterium A. borkumensis that specifically hydrolyzes hydroxylated acyl-CoA and that a mini-Tn5 mutation abolishing this 3-hydroxyacyl-CoA-specific thioesterase activity leads to the hyperproduction of extracellularly deposited PHA. This mutant's ability to deposit overproduced PHA extracellularly provides an interesting starting point for studying the biological mechanisms by which PHA is translocated into the culture medium, particularly with regard to contrasting reports on mutants of other organisms that store overproduced PHA exclusively intracellularly (16). Apart from gaining insights into biological mechanisms, our findings present a novel system to potentially generate high yields of biotechnologically important PHA, which can easily be recovered from the culture medium and thus which circumvents the need for costly procedures for the extraction of PHA granules from producer cells.
Acknowledgments
This research was supported by grants from the German Ministry for Education and Research (BMBF) and European Community project MERG-CT-2004-505242, “BIOMELI.” M.F. thanks the Spanish Ministerio de Ciencia y Tecnología (Ramón y Cajal contract). K.N.T. thanks Fonds der Chemischen Industrie for generous support.
J.S.S. is grateful to S. Lang (Technical University of Braunschweig), who was the first to give us a hint that the extracellular polymer might be PHA. J.S.S. also thanks Angelika Arnscheidt for technical assistance.
Footnotes
Published ahead of print on 22 September 2006.
REFERENCES
- 1.Abraham, W.-R., H. Meyer, and M. Yakimov. 1998. Novel glycine containing glucolipids from the alkane using bacterium Alcanivorax borkumensis. Biochim. Biophys. Acta 1393:57-62. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, A. J., and E. A. Dawes. 1990. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 54:450-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes, E. M., A. C. Swindell, and S. J. Wakil. 1970. Purification and properties of a palmityl thioesterase II from E. coli. J. Biol. Chem. 245:3122-3128. [PubMed] [Google Scholar]
- 4.Bonner, W. M., and K. Bloch. 1972. Purification and properties of fatty acyl thioesterase I from E. coli. J. Biol. Chem. 247:3123-3133. [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Cromwick, A.-M., T. Foglia, and R. W. Lenz. 1996. The microbial production of poly(hydroxyalkanoates) from tallow. Appl. Microbiol. Biotechnol. 46:464-469. [Google Scholar]
- 7.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Déziel, E., F. Lépine, S. Milot, and R. Villemur. 2003. rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 149:2005-2013. [DOI] [PubMed] [Google Scholar]
- 9.Golyshin, P. N., V. A. Martins Dos Santos, O. Kaiser, M. Ferrer, Y. S. Sabirova, H. Lunsdorf, T. N. Chernikova, O. V. Golyshina, A. Pühler, and K. T. Timmis. 2003. Genome sequence completed of Alcanivorax borkumensis, a hydrocarbon-degrading bacterium that plays a global role in oil removal from marine systems. J. Biotechnol. 106:215-220. [DOI] [PubMed] [Google Scholar]
- 10.Reference deleted.
- 11.Reference deleted.
- 12.Klinke, S., Q. Ren, B. Witholt, and B. Kessler. 1999. Production of medium-chain-length poly(3-hydroxyalkanoates) from gluconate by recombinant Escherichia coli. Appl. Environ. Microbiol. 65:540-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, S. Y. 1996. Bacterial polyhydroxyalkanoates. Biotechnol. Bioeng. 49:1-14. [DOI] [PubMed] [Google Scholar]
- 14.Lünsdorf, H., C. Strompl, A. M. Osborn, A. Bennasar, E. R. Moore, W. R. Abraham, and K. N. Timmis. 2001. Approach to analyse interactions of microorganisms, hydrophobic substrates and soil colloids leading to formation of composite biofilms, and to study initial events in microbiogeological processes. Methods Enzymol. 336:317-331. [DOI] [PubMed] [Google Scholar]
- 15.Ochman, H., A. S. Gerber, and D. L. Hartl. 1998. Genetic applications of an inverse polymerase chain reaction. Genetics 120:621-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olivera, E. R., D. Carnicero, R. Jondra, B. Minambres, B. Garcia, G. A. Abraham, A. Gallardo, J. S. Roman, J. L. Garcia, G. Naharro, and J. M. Luengo. 2001. Genetically engineered Pseudomonas: a factory of new bioplastics with broad applications. Environ. Microbiol. 3:612-618. [DOI] [PubMed] [Google Scholar]
- 17.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 18.Pandey, J. K., A. P. Kumar, M. Misra, A. K. Mohanty, L. T. Drzal, and R. P. Singh. 2005. Recent advances in biodegradable nanocomposites. J. Nanosci. Nanotechnol. 5:497-526. [DOI] [PubMed] [Google Scholar]
- 19.Rehm, B. H. A., N. Krüger, and A. Steinbüchel. 1998. A new metabolic link between fatty acid de novo synthesis and polyhydroxyalkanoic acid synthesis. The phaG gene from Pseudomonas putida KT2440 encodes a 3-hydroxyacyl-acyl carrier protein-coenzyme A transferase. J. Biol. Chem. 273:24044-24051. [DOI] [PubMed] [Google Scholar]
- 20.Sabirova, J. S., M. Ferrer, D. Regenhardt, K. N. Timmis, and P. N. Golyshin. 2006. Proteomic insights into metabolic adaptations in Alcanivorax borkumensis induced by alkane utilization. J. Bacteriol. 188:3763-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneiker, S., V. A. Dos Santos, D. Bartels, T. Bekel, M. Brecht, J. Buhrmester, T. N. Chernikova, R. Denaro, M. Ferrer, G. Gertler, A. Goesmann, O. V. Golyshina, F. Kaminski, A. N. Khanane, S. Lang, B. Linke, A. C. McHardy, F. Meyer, T. Nechitaylo, A. Puhler, D. Regenhardt, O. Rupp, J. S. Sabirova, W. Selbitschka, M. M. Yakimov, K. N. Timmis, F. J. Vorholter, S. Weidner, O. Kaiser, and P. N. Golyshin. 2006. Genome sequence of the ubiquitous hydrocarbon-degrading marine bacterium Alcanivorax borkumensis. Nat. Biotechnol. 24:997-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seay, T., and D. R. Lueking. 1986. Purification and properties of acyl coenzyme A thioesterase II from Rhodopseudomonas sphaeroides. Biochemistry 25:2480-2485. [DOI] [PubMed] [Google Scholar]
- 23.Soberon-Chavez, G., F. Lepine, and E. Deziel. 2005. Production of rhamnolipids by Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 68:718-725. [DOI] [PubMed] [Google Scholar]
- 24.Steinbüchel, A. 1991. Polyhydroxyalkanoic acids, p. 123. In D. Byrom (ed.), Biomaterials. MacMillan Publishers, Basingstoke, United Kingdom.
- 25.Timm, A., and A. Steinbüchel. 1990. Formation of polyesters consisting of medium-chain-length 3-hydroxyalkanoic acids from gluconate by Pseudomonas aeruginosa and other fluorescent pseudomonads. Appl. Environ. Microbiol. 56:3360-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yakimov, M. M., P. N. Golyshin, S. Lang, E. R. Moore, W. R. Abraham, H. Lunsdorf, and K. N. Timmis. 1998. Alcanivorax borkumensis gen. nov., sp. nov., a new, hydrocarbon-degrading and surfactant-producing marine bacterium. Int. J. Syst. Bacteriol. 48:339-348. [DOI] [PubMed] [Google Scholar]
- 27.Zheng, Z., Q. Gong, T. Liu, Y. Deng, J. C. Chen, and Q. Q. Chen. 2004. Thioesterase II of Escherichia coli plays an important role in 3-hydroxydecanoic acid production. Appl. Environ. Microbiol. 70:3807-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhuang, Z., F. Song, B. M. Martin, and D. Dunaway-Mariano. 2002. The YbgC protein encoded by the ybgC 27 gene of the tol-pal gene cluster of Haemophilus influenzae catalyzes acyl-coenzyme A thioester hydrolysis. FEBS Lett. 10:161-173. [DOI] [PubMed] [Google Scholar]