Abstract
Mycoplasma genitalium is a human pathogen that mediates cell adhesion by a complex structure known as the attachment organelle. This structure is composed of cytadhesins and cytadherence-associated proteins, but few data are available about the specific role of these proteins in M. genitalium cytadherence. We have deleted by homologous recombination the mg191 and mg192 genes from the MgPa operon encoding the P140 and P110 cytadhesins. Molecular characterization of these mutants has revealed a reciprocal posttranslational stabilization between the two proteins. Loss of either P140 or P110 yields a hemadsorption-negative phenotype and correlates with decreased or increased levels of cytoskeleton-related proteins MG386 and DnaK, respectively. Scanning electron microscopy analysis reveals the absolute requirement of P140 and P110 for the proper development of the attachment organelle. The phenotype described for these mutants resembles that of the spontaneous class I and class II cytadherence-negative mutants [G. R. Mernaugh, S. F. Dallo, S. C. Holt, and J. B. Baseman, Clin. Infect. Dis. 17(Suppl. 1):S69-S78, 1993], whose genetic basis remained undetermined until now. Complementation assays and sequencing analysis demonstrate that class I and class II mutants are the consequence of large deletions affecting the mg192 and mg191-mg192 genes, respectively. These deletions originated from single-recombination events involving sequences of the MgPa operon and the MgPa island located immediately downstream. We also demonstrate the translocation of MgPa sequences to a particular MgPa island by double-crossover events. Based on these observations, we propose that in addition to being a source of antigenic variation, MgPa islands could be also involved in a general phase variation mechanism switching on and off, in a reversible or irreversible way, the adhesion properties of M. genitalium.
Mycoplasmas are microorganisms derived from gram-positive bacteria with low G+C content, and they are characterized by streamlined genomes and the absence of a cell wall. Their reduced metabolic abilities (32) lead them to a parasitic lifestyle, and many of them are pathogens for humans and animals (5). In particular, Mycoplasma genitalium is the leading cause of chlamydia-negative, nongonococcal urethritis (13), and it has been also associated with several extragenitourinary diseases (43). With just 517 genes (9), M. genitalium is considered one of the smallest self-replicating cells, and it is currently used as a model for the studies of the minimal gene complement (11, 14). Despite its apparent simplicity, M. genitalium adheres to host cells via its complex tip structure, known as the attachment or terminal organelle. This polar membrane extension confers a flask-shaped appearance to the M. genitalium cells, and it is characterized by an electron-dense core that is a part of the mycoplasma cytoskeleton. The terminal organelle is present in several Mycoplasma species, and it has been extensively studied in Mycoplasma pneumoniae. In the latter case, characterization of cytadherence-negative mutants (3, 17, 35) allowed the identification of specific membrane cytadhesins (P1 and P30) as well as cytadherence-associated proteins (HMW1 to -3, P65, B, and C). These cytadherence-associated proteins are the key components of the mycoplasma cytoskeleton (33). They are implicated in both the maintenance and integrity of the terminal organelle and are also involved in the clustering and anchoring of cytadhesins at the tip structure (16). In addition, the intimate and dynamic interactions observed between cytadherence-associated proteins and cytadhesins suggest a spatial and hierarchical development of the attachment organelle (16).
M. genitalium and M. pneumoniae are morphologically and serologically related pathogens, sharing both chromosomal organization and sequence identity among their cytadhesin-related genes and proteins (6, 36). In fact, both mycoplasmas have been coisolated occasionally from nasopharyngeal throat swabs of patients with acute respiratory disease (4) and from synovial fluids from patients with arthritis (44, 45). It is thought that M. genitalium adheres to host cells through the P140 adhesin (23), which shows an extensive immunological cross-reactivity with the major M. pneumoniae cytadhesin P1 (25, 26). M. genitalium mutants possessing small amounts of P140 (designated class I mutants) or lacking the P140 protein (designated class II mutants) exhibit a cytadherence-negative phenotype, reinforcing the importance of P140 as a major adhesin (23). Interestingly, these cytadherence-negative mutants appear spontaneously at unexpectedly high rates, but the genetic basis underlying this phenomenon remains to be defined. Concerning gene organization, mg190 or mgpA (encoding a predicted 29-kDa protein), mg191 or mgpB (encoding the P140 protein), and mg192 or mgpC (encoding the P110 protein) are transcriptionally linked and comprise the MgPa operon (27). Some repetitive DNA elements (MgPa islands) containing sequences from the MgPa operon are distributed along the M. genitalium genome (9). Although these MgPa islands do not appear to directly express protein-coding sequences, it has been proposed that recombination between these repetitive DNA elements and the MgPa operon contributes to the antigenic variation of the MgPa proteins (15, 29).
Finally, as stated above, most of the information on cytadherence proteins is derived from studies based on unstable spontaneous mutants, leading to difficulties in their genetic characterization. Although transposons have been revealed as useful tools to isolate and study cytadherence-negative mutants, more precise and accurate data can be obtained from the analysis of mutants obtained by gene replacement. In this study, we have taken advantage of the possibility of deleting target genes by homologous recombination in M. genitalium (8) to obtain mg191 and mg192 deletion mutants. Since all nonadhering M. genitalium mutants isolated lacked both the P140 and P110 proteins (8, 23), we aimed to unravel the striking relationship between these two polypeptides and determine their roles in M. genitalium cytadherence. Here we show that deletion of either the mg191 or mg192 gene yields a noncytadherent phenotype with concomitant loss of the terminal organelle. In addition, detailed analysis of these mutants also reveals a reciprocal posttranslational stabilization between P140 and P110 and downstream events associated with several proteins that comprise the mycoplasma cytoskeleton. Moreover, mg192 and mg191 deletion mutants mimic the protein profiles of spontaneous class I and class II mutants, respectively. Supporting this observation, we have found that the nonadherent phenotype in class I and class II mutants is the consequence of large deletions affecting the mg192 gene and the mg191 and mg192 genes, respectively. These large deletions seem to be the product of recombination events involving DNA repetitive elements and the MgPa operon, and they could be the basis of a general phase variation mechanism switching the adhesion properties of M. genitalium on and off.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Escherichia coli strain XL-1Blue, Mycoplasma genitalium wild-type (WT) strain G37, and nonadhering class I and class II mutants (23) were grown as previously described (31).
DNA manipulations. (i) Molecular cloning.
General DNA manipulations were performed as described previously (39). Genomic DNA of the WT strain, isolated as previously described (31), was used as a template for PCRs. Primers used in this work are summarized in Table 1. A 1-kb PCR fragment encompassing the upstream region and the first ≈100 bp of the mg191 gene was obtained by using primers 5′BEKOP140 and 3′BEKOP140. These primers incorporate at their ends Acc65I and EcoRI restriction sites, respectively. Another 1-kb PCR fragment containing the last ≈100 bp of the mg191 gene and a downstream region was obtained by using primers 5′BDKOP140 and 3′BDKOP140. These primers contained at their ends BamHI and XbaI restriction sites, respectively. Both PCR fragments were cloned into EcoRV-digested pBE (31), excised with the corresponding restriction enzyme (Roche), and ligated together with a 2-kb fragment containing the tetM438 selectable marker (31) and an Acc65I/XbaI-digested pBSKII(+) (Invitrogen). The tetM438 selectable marker was released from plasmid pMTnTetM438 by digestion with EcoRI and BamHI, and this ligation mixture resulted in the creation of pΔmg191. Alternatively, a 1-kb PCR fragment containing the upstream region and the first ≈100 bp of the mg192 gene was obtained by using primers 5′BEKOP110 and 3′BEKOP110. These primers incorporate at their ends the Acc65I and SalI sites, respectively. Otherwise, primer 5′BDKOP110, which includes a BamHI site, and primer 3′BDKOP110, which includes an XbaI site, were used to amplify a 1-kb PCR fragment that comprised the last ≈100 bp of the mg192 gene, followed by a downstream region. Finally, both PCR fragments digested as described above were ligated with the tetM438 marker released with SalI and BamHI and the Acc65I/XbaI-digested pBSKII(+), creating plasmid pΔmg192.
TABLE 1.
Primers
| Primer | Sequence (5′ to 3′)a |
|---|---|
| 5′BEKOP140 | GGTACCTCTTTCACCATGTGCGCCC |
| 3′BEKOP140 | GAATTCGGTTAAAGAGAAAATAACCACC |
| 5′BDKOP140 | GGATCCTTATCCTTAGTGTTACTTTGGG |
| 3′BDKOP140 | TCTAGACTCCAGAAACACTGGGCGC |
| 5′BEKOP110 | GGTACCAAGCGAAATTGGGGAAGGG |
| 3′BEKOP110 | GTCGACATCCTCTTTGACAAGGAAGG |
| 5′BDKOP110 | GGATCCTGCATTGAAAGCTGCTAATCC |
| 3′BDKOP110 | TCTAGAATTGATGTTAGAAAAAGTGGGG |
| 5′2PrP140 | GTCGACCTTTTATAATCTTTGTTATGAAAA |
| 3′P140 | GTCGACTTATTGTTTTACTGGAGGTTTT |
| 5′P110Pr | GTCGACTAGTATTTAGAATTAATAAAGTATGAAAACAATGAGAAAACAG |
| 3′P110 | GTCGACCTAACTTTTGGTTTCTTCTG |
| 3′P140XbaI | TCTAGATAACTGCCTTTGAAAC |
| 5′P140XbaI | TCTAGACAGTGATGGTACCC |
| 3′P110ApaI | GGGCCCCTAACTTTTGGTTTCTTCTGAC |
| 5′SP140 | GATGAAGGACAAGCTAAGGC |
| 3′SP140 | CATTGTCCTGGATATCAGGG |
| 5′SP140C-t | CTCAGTAGAAAACCAAACCTTAGG |
| 5′SP110 | GCAGATGGATTTAAGCGTCC |
| 3′SP110 | CTGAATTCCTCACTAAGTGGG |
| 5′mg192N-t | AGAATGTTACTGCTTACACCC |
| 3′mg194N-t | GTAATTAACTTGTTTTGATCAATC |
| 3′down mg192 | AATTCGACCACATAGGAGGG |
| 5′mg260C-t | GATCCAAGTGACTTTAGGAAC |
Underlining indicates the restriction site introduced at the 5′ end of the primer. Boldface indicates the 22 nucleotides from the putative mg438 gene promoter region (31).
The constructions used for the complementation assay were designed as follows. The WT mg191 allele was amplified by using primers 5′ 2PrP140 and 3′P140, both incorporating SalI sites at their ends. In order to maintain the transcriptional context, this fragment also contained the mg190 gene and the two transcriptional start sites previously described for the mg191 gene, PrmMG190 and PrmMG191 (27). Otherwise, the WT mg192 coding region was obtained using 5′P110Pr and 3′P110 primers. Both primers incorporate SalI sites at their ends, while the 5′P110Pr primer additionally included the 22-bp promoter sequence of the previously described mg438 gene (31). Both PCR fragments were cloned into the SalI-digested pMTnTetM438 gene, creating pTnTetMG191 and pTnTetMG192, respectively. In addition, the entire MgPa operon was amplified by PCR in two steps, taking advantage of the XbaI site in the mg191 gene. In this way, a first fragment of 4.3 kb was amplified using primers 5′ 2PrP140 and 3′P140XbaI, and a second fragment of 4.2 kb was amplified using primers 5′P140XbaI and 3′P110ApaI, which included an ApaI site. Finally, both PCR fragments were ligated together with SalI/ApaI-digested pMTnTetM438, creating pTnTetMG191-192.
(ii) Southern blotting.
Genomic DNAs of pΔmg191 or pΔmg192 transformants were digested with HindIII and BglII, respectively, while genomic DNAs of class I and class II mutants were digested with several enzymes (HindIII, BglII, EcoRI, ScaI, Acc65I, and AvaI) and probed as previously described (31) with fragments S1, S2, and S3 (Fig. 1), which were amplified by PCR using primers 5′SP140 and 3′SP140 (S1), 5′SP140C-t and 3′P140 (S2), and 5′SP110 and 3′SP110 (S3).
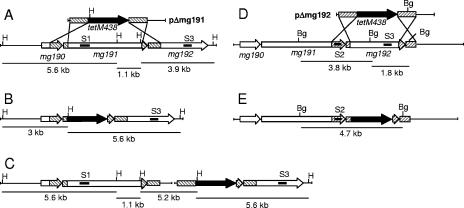
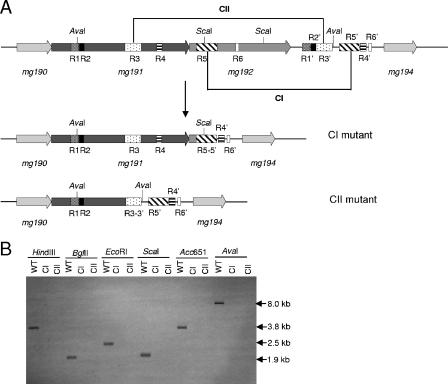
FIG. 1.
Homologous recombination at the MgPa operon in M. genitalium. (A and D) Schematic representation of the two possible crossover events of the pΔmg191 and pΔmg192 deletion constructs with the MgPa operon. (B and E) mg191 and mg192 genes, respectively, disrupted by double-crossover recombination. (C) mg191 gene disrupted by single-crossover recombination through the downstream flanking region. Hatched boxes represent flanking regions of the mg191 (lines from right to left) and mg192 (lines from left to right) genes in suicide plasmids pΔmg191 and pΔmg192, respectively. Black, tetM438 selectable marker; white, mg191 and mg192 genes; lines inside genes, probes S1, S2, and S3. The sizes of fragments resulting from HindIII (H) and BglII (Bg) digestions of the WT and each recombinant mutant DNA are also indicated.
(iii) DNA sequencing.
Deletion breakpoints were amplified using primers 5′mg192N-t and 3′mg194N-t for class I mutant and primers 5′ SP140 and 3′downmg192 for class II mutants. Sequencing of these PCR products was performed with primers 5′mg192N-t and 3′downmg192, respectively, as previously described (31).
Electroporation and transformation.
Transformation of M. genitalium strain WT with suicide plasmid pΔmg191 or pΔmg192 was performed by electroporation as previously described (31), using 30 μg of DNA. For transformation of class I and class II mutants, cells were grown to mid-log phase, centrifuged, and passed five times through a 25-gauge needle. Cells were then electroporated with 5 μg of plasmid pTnTetMG191, pTnTetMG192, or pTnTetMG191-192.
RNA manipulations.
RNA was extracted from 20 ml of mid-log phase cultures by using the RNAaqueous kit (Ambion). For reverse transcription-PCR assay, mycoplasma RNAs treated with DNase I (Invitrogen) were reverse transcribed by using the SuperScript first-strand synthesis system (Invitrogen) kit and the 3′BDKOP140 primer. The resulting cDNAs were amplified using primers 5′mg192N-t and 3′BDKOP140, producing a final product that comprises 668 bp of the 5′ end of the mg192 gene.
SDS-PAGE, immunoblotting, and matrix-assisted laser desorption ionization-time-of-flight MS analyses.
Total mycoplasma cell proteins were prepared for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electrophoresed on 8% polyacrylamide gels by standard procedures. Gels were stained with Coomassie blue or transferred electrophoretically to nitrocellulose membranes and probed with monoclonal anti-P140 antibody (1:1,000) or polyclonal rabbit antiserum against P110 (1:1,000) as previously described (39). The Coomassie blue-stained protein bands of interest were excised from the acrylamide gel, destained, and trypsin digested, and the eluted peptides were analyzed by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MS) as previously described (30).
HA assays.
Qualitative hemadsorption activity (HA) assays for mg191− and mg192− mutants and the different class I and class II transformants were performed as previously described (30).
Scanning electron microscopy.
Cells were grown to mid-log phase, pelleted by centrifugation, and suspended in SP-4 medium. Cell suspensions were passed three times through a 25-gauge needle to break aggregates and passed through a 0.45-μm low-protein-binding filter before seeding individual wells of 24-well cell culture dishes containing poly-l-lysine-coated glass coverslips (Becton Dickinson). After 2 h of incubation at 37°C under 5% CO2, the medium was removed and the coverslips were washed three times with phosphate-buffered saline and fixed with 1% glutaraldehyde for 1 h. Samples were washed three times with phosphate-buffered saline and then dehydrated sequentially with 30, 50, 70, 90, and 100% ethanol for 10 min each. Immediately, samples were critical point dried (K850 critical point drier; Emitech Ashfort, United Kingdom) and sputter coated with 20 nm of gold. Samples were observed using a Hitachi S-570 (Tokyo, Japan) scanning electron microscope. Proportions of rounded and pleomorphic forms were estimated from digital image analysis of 700 individual cells for each mutant.
RESULTS
Construction of pΔmg191 and pΔmg192 and transformation assay.
The M. genitalium mg191 and mg192 genes were deleted by homologous recombination. For this purpose, suicide plasmids pΔmg191 and pΔmg192 were engineered to contain the tetM438 selectable marker (31) enclosed by the flanking regions of the mg191 or mg192 gene, respectively (Fig. 1A and D). A double-crossover recombination event between plasmid pΔmg191 and the mycoplasma genome resulted in the deletion from bases 105 to 4093 of the mg191 gene (92% of the coding sequence). Similarly, plasmid pΔmg192 promotes the deletion of base 98 to 3029 of the mg192 gene (92.8% of the coding sequence). Deletions were designed to preserve approximately 100 bp from both ends of the target genes in order to minimize undesired effects in the transcriptional and translational status of the remaining flanking genes. Deleted sequences were replaced by the tetM438 selectable marker, which confers tetracycline resistance to transformed cells. When the M. genitalium WT strain was electroporated with plasmid pΔmg191 or pΔmg192, transformation efficiencies were 5.5 × 10−7 and 2.4 × 10−7 transformants per viable cell, respectively. These values represent a 20-fold increase with respect to those previously reported in studies aimed to knock out target genes from M. genitalium by homologous recombination (7, 8). This increment could be related to the use of the tetM438 selectable marker as we previously described (31) or could reflect an increased recombination frequency in the MgPa locus.
Genomic analyses of pΔmg191 and pΔmg191 transformants.
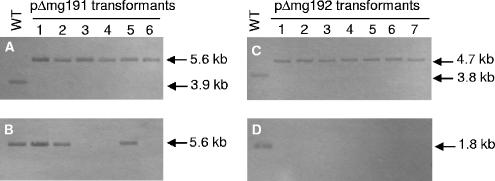
Six tetracycline-resistant colonies arising from transformation with pΔmg191 and seven from transformation with pΔmg192 were picked and propagated in SP-4 medium containing tetracycline. All pΔmg192 transformants isolated and three pΔmg191 transformants (clones 3, 4, and 6) did not adhere to the cell culture plastic surface and grew aggregated in cell suspension. In contrast, the remaining pΔmg191 transformants (clones 1, 2, and 5) grew attached to cell culture flasks. To confirm the absence of the deleted sequences, genomic DNAs from the WT strain and from pΔmg191 and pΔmg192 transformants were isolated and subjected to Southern blot analyses. Because of the presence in the chromosome of DNA repetitive elements of the MgPa operon, specific probes containing unique sequences were designed to avoid cross-hybridization to multiple parts of the M. genitalium genome. Genomic DNAs from the WT strain and pΔmg191 transformants were digested with HindIII and probed with a specific internal fragment of the mg192 gene (probe S3 [Fig. 1]). A single 3.9-kb band was detected for WT DNA, while a 5.6-kb band was detected for all pΔmg191 transformants (Fig. 2A). The 5.6-kb band is compatible with either a double-crossover event or a single-crossover event through the 3′ mg191 flanking region (Fig. 1B and C). Clones originating from a double-crossover event were detected by reprobing digested DNAs with a specific internal fragment of the mg191 gene (probe S1 [Fig. 1]). A predicted band of 5.6 kb was detected for the WT and clones 1, 2, and 5, indicating a nondisruptive single-crossover event. Otherwise, clones 3, 4, and 6 (designated mg191− mutants) failed to hybridize with probe S1, indicating the presence of the expected deletion in the mg191 gene (Fig. 2B).
FIG. 2.
Southern hybridization profiles of genomic DNAs from the M. genitalium WT strain and pΔmg191 and pΔmg192 transformants. A and B) HindIII digestions of genomic DNAs from the WT strain and pΔmg191 transformants hybridized with probes S3 and S1, respectively. (C and D) BglII digestions of DNAs from the WT strain and pΔmg192 transformants hybridized with probes S2 and S3, respectively. Probes S1, S2, and S3 are described in Fig. 1.
Genomic DNAs from WT and pΔmg192 transformants were digested with BglII and probed with a fragment from the 3′end of the mg191 gene (probe S2 [Fig. 1]). A single 3.8-kb band was detected for the WT, while a 4.7-kb band was detected for all pΔmg192 transformants (Fig. 2C). The 4.7-kb band is compatible with either a double-crossover event (Fig. 1E) or a single-crossover event through the 5′ mg192 flanking region. Finally, clones originating from a double-crossover event were detected by reprobing digested DNAs with a specific internal fragment of the mg192 gene (probe S3). A predicted 1.8-kb band was detected for the WT, while no hybridization was detected for pΔmg192 transformants (designated mg192− mutants), indicating that all seven mutants had the expected deletion in the mg192 gene (Fig. 2D).
Transcriptional analysis of the mg192 gene in mg191− mutants.
Reverse transcription-PCR analysis of two individual mg191− mutants (clones 3 and 4) was performed to rule out transcriptional defects in the mg192 gene as a consequence of the deletion of the mg191 gene. An expected 668-bp product corresponding to an internal fragment of the mg192 gene was amplified, demonstrating the transcriptional integrity of the mg192 gene in mg191− mutants (data not shown).
Analyses of mg191− mutants and mg192− mutants.
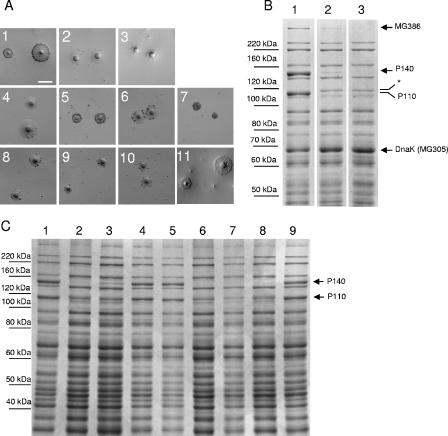
Adherence of mg191− and mg192− mutants was qualitatively assessed by their ability to bind erythrocytes. As expected, both mutants exhibited an HA− phenotype (Fig. 3A, panels 2 and 3). Transformants were also screened by SDS-PAGE for the absence of the P140 and P110 proteins, and, in accordance with their adhering phenotype on plastic, clones 1, 2, and 5 from pΔmg191 transformants showed a WT protein profile (data not shown). Otherwise, the protein profiles from mg191− mutants were clearly different from the WT protein profile (Fig. 3B). As expected, the 140-kDa protein corresponding to the mg191 gene product was completely absent. Surprisingly, we found that the P110 protein was also absent but a new band of approximately 115 kDa was evident. The new band was excised from the gel and digested with trypsin, but MS analysis could not unequivocally assign this band to any M. genitalium protein. Further investigation will be needed to determine the exact nature of the 115-kDa band. The protein profile analysis of mg191− clones revealed other changes that could be clearly assigned. A high-molecular-mass band recently identified by MS as the product of the mg386 gene (30) was markedly reduced. Finally, although some variations in band intensities were observed in other proteins, a protein with an apparent molecular mass of 68 kDa was found to be consistently increased in all mg191− mutants examined (Fig. 3B). That protein was identified by MS as DnaK (MG305). When protein profiles of mg192− mutants were analyzed, the 110-kDa protein corresponding to the mg192 gene product was completely absent, as expected (Fig. 3B). Interestingly, although the 140-kDa protein band was present, its intensity was dramatically reduced. Finally, variations in the levels of the MG386 and DnaK proteins previously observed in the mg191− mutants were also found in the mg192− mutants.
FIG. 3.
(A) Qualitative HA assessment of the different mutants and the WT strain. Panels 1 to 3, WT strain, mg192− mutant (clone 1), and mg191− mutant (clone 3), respectively. Panels 4 to 7, class I mutant, class I mutant plus pTnTetMG192, class I mutant plus pTnTetMG191, and class I mutant plus pTnTetMG191-192, respectively. Panels 8 to 11, class II mutant, class II mutant plus pTnTetMG192, class II mutant plus pTnTetMG191, and class II mutant plus pTnTetMG191-192, respectively. Bar, 100 μm. (B) SDS-8% PAGE protein profiles of the WT strain (lane 1) and the mg192− (clone 1) and mg191− (clone 3) mutants (lanes 2 and 3, respectively). Arrows show the differences observed between WT and mutant strains. *, new 115-kDa band. (C) SDS-8% PAGE protein profiles of the WT strain (lane 1), class I mutant (lane 2), class I mutant plus pTnTetMG191 (lane 3), class I mutant plus pTnTetMG192 (lane 4), class I mutant plus pTnTetMG191-192 (lane 5), class II mutant (lane 6), class II mutant plus pTnTetMG191 (lane 7), class II mutant plus pTnTetMG192 (lane 8), and class II mutant plus pTnTetMG191-192 (lane 9).
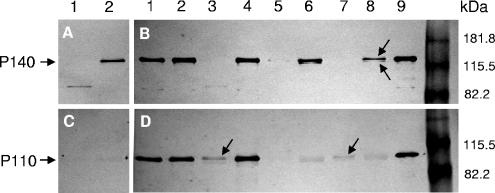
Western blot analyses using an anti-P140 monoclonal antibody confirmed the absence of P140 in mg191− mutants (Fig. 4B) and revealed the presence of a double band corresponding to the P140 protein in the mg192− mutants (Fig. 4B). Protein profiles of mg191− and mg192− mutants were comparable to those previously observed for class II and class I spontaneous mutants of M. genitalium, respectively (Fig. 4A). However, in class II mutants, a faint band of 85 kDa not detected earlier (23) was detected by anti-P140 antibodies (Fig. 4A, lane 1). As expected, when anti-P110 polyclonal antibodies were used, no reactivity was detected in mg192− mutants (Fig. 4D, lane 8). Otherwise, in mg191− mutants a very faint band was detected in the 110-kDa region (Fig. 4D, lane 7). These results suggest a mutual posttranslational stabilization effect between the P140 and P110 proteins.
FIG. 4.
Western blot analyses of the different mutants and the WT strain. Blots were probed with an anti-P140 monoclonal antibody (A and B) or anti-P110 polyclonal antibody (C and D). In panels A and C, lanes 1 and 2 correspond to class II and class I mutants, respectively. In panels B and D, lanes 1 and 2 correspond to class II and class I mutants transformed with pTnTetMG191-192, respectively. Lanes 3 and 4, class II and class I mutants transformed with pTnTetMG192, respectively; lanes 5 and 6, are class II and class I mutants transformed with pTnTetMG191, respectively; lanes 7 and 8, mg191− mutant clone 4 and mg192− mutant clone 5, respectively; lane 9, WT strain. The arrows in panel B indicate the P140 doublet, while those in panel D indicate the presence of low quantities of P110.
Cell morphology of mg191− and mg192− mutants.
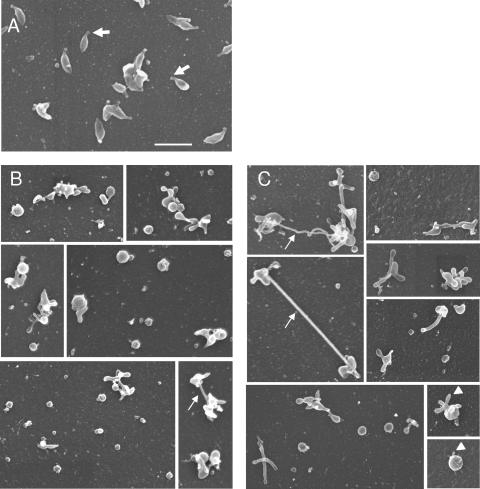
To assess the effect of the loss of the P140 and P110 proteins on cell morphology, mg191− and mg192− mutants were analyzed by scanning electron microscopy. Because of the nonadhering property of these mutants, coverslips coated with poly-l-lysine were used to promote cell attachment. However, some background of cell debris was observed in the preparations after the poly-l-lysine treatment. WT cells showed a flask-shaped appearance with a well defined tip structure characterized by a thickened end (Fig. 5A), as previously described (21). However, a small proportion of WT cells showed atypical morphologies (data not shown), indicating a variability of the population as previously described for other mycoplasmas (38). In contrast, cells from mg191− and mg192− mutants showed dramatic morphological changes compared to the WT strain, with the most remarkable feature being the loss of the attachment organelle. In particular, we found that 76.9% of mg191− mutant cells displayed a round appearance, while the remaining 23.1% of cells were multilobed or pleomorphic (Fig. 5B). As seen in Fig. 5, a certain degree of variability in the size of rounded forms from both mutants was observed. Both rounded and multilobed cells (74.8% and 25.2%, respectively) were also found in preparations from the mg192− mutant (Fig. 5C). Noteworthy was the presence of long filaments or membrane extensions connecting cells or protruding from a single cell (Fig. 5C). Such structures were reminiscent of those previously found in Mycoplasma penetrans strain GTU-54 preparations (10). In addition, some cells exhibited small cell membrane extensions or buds that seemed to be a preliminary step in the development of the long filaments (Fig. 5, lower right panels).
FIG. 5.
Scanning electron microscopy analyses of the WT strain (A) and mg191− (B) and mg192− (C) mutants. Thick arrows indicate the terminal organelle, thin arrows show long filaments, and arrowheads show buds. All pictures are shown at the same magnification. Bar, 1 μm.
Complementation of class I and class II mutants with mg191 and mg192 genes.
As we stated above, the phenotypes exhibited for mg191− and mg192− mutants, summarized in Table 2, agree with those observed for the spontaneous class II and class I cytadherence-negative mutants, respectively, possibly linking these mutants through common genetic lesions. This fact was demonstrated by the reintroduction of mg191 and mg192 WT copies (separately or simultaneously) in class I and class II mutants by transposition using pTnTetMG191, pTnTetMG192, and pTnTetMG191-192 constructions. Transformation of a class I mutant with either the pTnTetMG192 or pTnTetMG191-192 construct restored both HA+ activity (Fig. 3A, panels 5 and 7) and the WT protein profile as demonstrated by SDS-PAGE (Fig. 3C) and Western blotting (Fig. 4B and D). However, neither HA+ activity nor the WT protein profile was restored after transformation with pTnTetMG191 (Fig. 3A [panel 6] and C and 4B and D). Noticeable was a slight increase in the intensity of the doublet in the 140-kDa region detected by SDS-PAGE and Western blotting, which was consistent with the higher dosage of P140. These results suggest that spontaneous class I mutants arise as a consequence of mutations in the mg192 gene.
TABLE 2.
Summary of phenotypes of the mg191− and mg192− mutants
| Straina | Protein levelb
|
HA activity | Cell morphology | |||
|---|---|---|---|---|---|---|
| P140 | P110 | MG386 | DnaK | |||
| G37 | +++ | +++ | +++ | +++ | Positive | Flask shape |
| mg191− mutant | − | ± | + | ++++ | Negative | Rounded, pleomorphic |
| mg192− mutant | + | − | + | ++++ | Negative | Rounded, pleomorphic |
Class I and class II mutants have the phenotypes found in mg192− and mg191− mutants, respectively.
Relative amounts of protein: −, none; ±, very reduced (detectable only by Western blotting); +, significantly reduced; +++, WT levels; ++++, overexpressed.
Complementation of a class II mutant was achieved only after transformation with the pTnTetMG191-192 construction (Fig. 3A [panels 8 to 11] and C and 4B and D). This suggests that mutations in both the mg191 and mg192 genes are the origin of class II mutants. Accordingly, when this mutant was transformed with pTnTetMG191 or pTnTetMG192 constructions, only very faint bands corresponding to P140 or P110, respectively, could be detected by Western blotting (Fig. 4B and D). These results provide additional support for the mutual stabilization between the P140 and P110 proteins.
Genetic basis of class I and class II mutants.
We were unable to amplify by PCR either the mg192 gene from the class I mutant or the mg191 and mg192 genes from the class II mutant, even using alternative primers, suggesting the presence of deletions in these mutants. To map these possible deletions in both mutants, Southern blot analyses were performed using several restriction enzymes and probes S1 and S2 (Fig. 1). The Southern blots (data not shown) revealed the presence of a ∼4-kb deletion close to the AvaI site of the mg191 gene in the class I mutant and a ∼6-kb deletion close to the ScaI site at the 5′ end of the mg192 gene in the class II mutant (Fig. 6A). On the basis of the restriction fragment analysis, specific PCR primers were designed to selectively amplify the deletion breakpoint for each mutant. Sequence analyses of the PCR fragments from the class I mutant revealed a deletion within the mg192 gene from base 226524 to 230986 of M. genitalium genome. In a similar way, a deletion from base 224176 to 230169 of M. genitalium genome was found, affecting both the mg191 and mg192 genes in the class II mutant. The breakpoint identified in the class II mutant introduces a premature stop codon in the P140-coding region, which is in agreement with the presence of the 85-kDa truncated polypeptide of P140 detected by Western blotting (Fig. 4A, lane 1). Detailed analysis of the regions around the sequenced breakpoints suggests that these deletions have been generated by a single-recombination event between sequences within the MgPa operon and MgPa island V, which is located immediately downstream of this operon. In particular, in the class I mutant the sequences involved in the rearrangement are boxes R5 and R5′, while those in the class II mutant are boxes R3 and R3′ (Fig. 6A). The permanent loss of the sequences between the two boxes implicated in each particular single-crossover event was further confirmed by the absence of signal hybridization in Southern blots of class I and class II mutant DNAs with probe S3 (Fig. 6B).
FIG. 6.
(A) Schematic representation of the DNA repetitive elements of the MgPa operon and MgPa island V located immediately downstream (bases 220000 to 234000 of the M. genitalium genome). A single-recombination event between the R5 and R5′ boxes and the R3 and R3′ boxes may generate class I and class II mutants, respectively. Base coordinates refer to accession number NC_000908 from the NCBI database. Boxes R1 to R6 refer to the DNA repetitive sequences of the MgPa operon, which are also found in the MgPa island (R1′ to R6′) located immediately downstream. CI, class I mutant; CII, class II mutant. (B) Southern hybridization profiles using probe S3 and genomic DNAs from the WT strain, class I mutant, and class II mutant.
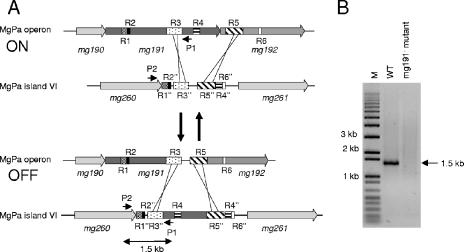
As we have shown, irreversible deletions in the MgPa operon are the result of single-crossover events involving MgPa island V. This result prompted us to find out whether reversible deletions could arise from a double-recombination event between sequences from the MgPa operon and any MgPa island interspersed in the M. genitalium genome (Fig. 7A). Specific primers were designed to test by PCR the presence of such reversible mutants within the WT population. The first primer (3′P140XbaI) is located between the R3 and R4 boxes of the MgPa operon (Fig. 7A), and the second primer (5′mg260C-t) is derived from unique sequences at the 3′ end of the mg260 gene, located immediately upstream of MgPa island VI. Because the two primers are 90 kb apart in the M. genitalium genome, PCR amplification is expected only when a double-recombination event translocates the sequence corresponding to 3′P140XbaI primer from the MgPa operon to the MgPa island VI. This can be accomplished by several sorts of double-recombination events between boxes from the MgPa operon and the corresponding ones present in MgPa island VI (Fig. 7A). To ensure the specificity of the PCR amplification, genomic DNA from one mg191− mutant was used as a negative control. No PCR amplification is expected in the negative control, because the deletion present in the mg191− mutant comprises the sequence used to design primer 3′P140XbaI. As shown in Fig. 7B, a 1.5-kb fragment is clearly amplified from WT genomic DNA. This PCR product is compatible with a double-recombination event involving R3 and downstream boxes R4, R5, or R6 from the MgPa operon and the corresponding ones present in MgPa island VI. The recombinative origin of this PCR fragment was further confirmed by sequencing (data not shown) and demonstrates the translocation of long stretches of MgPa operon sequence to a particular MgPa island.
FIG. 7.
Translocation of sequences from the MgPa operon to MgPa islands in the M. genitalium genome. (A) Schematic representation exemplifying how a double-recombination event between the R3-R3′ and R5-R5′ boxes translocates, in a reversible way, sequences from the MgPa operon (bases 220000 to 230000) to MgPa island VI (bases 310000 to 319000). Boxes R1 to R6 refer to the DNA repetitive sequences of the MgPa operon, which are also found in the MgPa island VI (R1′ to R6′) located 90 kb downstream. P1 and P2 indicate the position of the 3′P140XbaI and 5′mg260C-t primers, respectively. Base coordinates refer to accession number NC_000908 from the NCBI database. A number of additional double-recombination events are also possible but are not included in the drawing for clarity. (B) PCR amplification from M. genitalium WT and mg191− mutant DNAs, using primers P1 and P2. The 1.5-kb product results from a double-recombination event between R3 and downstream box R4, R5, or R6 of the MgPa operon and the corresponding boxes in the MgPa island VI. M, 1-kb plus DNA ladder marker.
DISCUSSION
Adhesion of mycoplasmas to host target cells is considered a prior step for infection and tissue colonization. Cell adhesion of many mycoplasmas is mediated by the attachment organelle, which also plays a role in cell division and gliding motility (16). This complex structure contains membrane cytadhesins and cytadherence accessory proteins required for successful surface parasitism (1). Such proteins are characterized by their novel nature, showing no homology with any other known protein. P140 and its operon-related protein P110 are currently considered the major M. genitalium cytadhesins (23), but no data are currently available about the role of these cytadhesins in the architecture and organization of the attachment organelle. Analysis of mg191− or mg192− M. genitalium null mutants has revealed the requirement of the P140 and P110 proteins for the proper assembly and development of the terminal organelle. In addition, these mutants exhibited an HA− phenotype, reinforcing the role of P140 and P110 in M. genitalium cytadherence.
Protein profile analysis of these mutants reveal that P110 levels are drastically reduced in mg191− mutants, while low levels of P140 and a truncated form of this cytadhesin are detected in mg192− mutant strains. Similar results have been previously reported for M. pneumoniae, where stability of the B (P90) and C (P40) proteins, the counterparts of P110, depends on the presence of P1, the counterpart of P140 (47). However, the converse is not seen, because the B and C proteins seem to be required for the maintenance of P1 in the cytoskeletal fraction (18) but not for P1 stability (47). These functional differences between the B/C and P110 proteins could be a consequence of the different nature of the functional products of the orf6 (M. pneumoniae) and mg192 (M. genitalium) genes. While in M. pneumoniae the 130-kDa polypeptide encoded by the orf6 gene is posttranslationally cleaved to yield two independent proteins, B and C (19), M. genitalium does not undergo such posttranslational modification of its mg192 gene product. In addition, it seems that some but not all synthesis of B or C is dependent on coupling to the translation of P1 (47). In this sense, the dramatically reduced levels of P110 observed in mg191− mutants could be explained by a failure of P110 translation in these mutants. However, despite the likelihood of translational coupling of P110 to P140, our results clearly show that stability of P110 depends almost exclusively on the presence of P140. This is supported by the observation that reintroduction of pTnTetMG192 in the class II mutant does not restore P110 at WT levels. Taken together, our results indicate that the P140 and P110 proteins are reciprocally stabilized in M. genitalium, and they provide an explanation for the previous findings showing the instability of P140 in class I mutants (23).
In addition to the effects on the P140 and P110 proteins, an interesting finding of this work is the observation of increased or decreased levels of additional proteins in mg191− and mg192− mutants. Protein profiles of these mutants show decreased levels of the MG386 protein. MG386 is homologous to P200, a protein found in the Triton X-100 shell of M. pneumoniae (37) that shares common features with other cytadherence accessory proteins (34). Interestingly, we have recently described the involvement of the MG386 protein in M. genitalium gliding motility (30). These observations reinforce the close connection between gliding motility and adherence machineries in motile mycoplasmas (40).
Our results also show increased levels of MG305 (DnaK) protein when P140 or P110 is absent or much reduced. Interestingly, DnaK is a component of the mycoplasma cytoskeleton (37), and it has been described as a protein complexed to the P1 adhesin in M. pneumoniae (20). Up-regulation of DnaK has been also reported after heat shock (48), opening the possibility that loss of P140/P110 proteins and/or the lack of the attachment organelle could initiate a stress signal response in M. genitalium similar to that found after a heat shock. However, since DnaK is associated with the cytoskeleton and is a member of the actin superfamily proteins with potential polymerization activity, additional cytoskeleton-related roles for DnaK have been suggested (30, 37). Thus, we cannot exclude the possibility that DnaK is up-regulated as a consequence of possible cytoskeletal rearrangements after the loss of the terminal organelle. At the same time, the loss of the attachment organelle observed in the rounded and pleomorphic cells from the mg191− and mg192− mutant cells suggests the absolute requirement of both the P140 and P110 proteins for the proper development of such a structure. Studies performed with an M. pneumoniae mutant deficient in the B/C proteins reached similar conclusions (18), despite the fact that neither cell pleomorphism nor long filaments were described. Since duplication of the attachment organelle seems to precede cell division (24), it is reasonable to presume that the absence of the terminal organelle hampers cell division, generating these pleomorphic forms with multiple filaments (38).
In the present study we also aimed to determine the genetic basis of the spontaneous cytadherence-negative class II and class I mutants (23), since their phenotype and proteins profiles resemble those exhibited for the mg191− and mg192− mutants, respectively. From the similarities observed we hypothesized that the mg191 and mg192 genes could be affected in some way in these mutants. Nucleotide sequence analyses of the MgPa operons from the two individual spontaneous mutants revealed the presence of large deletions affecting the mg192 gene in class I mutants and affecting both the mg191 and mg192 genes in class II mutants. In addition, we succeeded in restoring the phenotypes and protein profiles of both mutants after reintroduction of WT copies of the mg192 or mg191-mg192 genes, suggesting the absence of additional changes in other cytadhesin-related genes. The sequence analyses also seem to indicate that these deletions originated from a single-crossover event between the mg191 or mg192 gene and sequences from the MgPa island located immediately downstream of the MgPa operon. The recombinative origin of these deletions provides a plausible explanation for the unexpectedly high rates of appearance of these spontaneous cytadherence-negative mutants. Similar mutation rates are also observed among the spontaneous cytadherence-negative mutants isolated in M. pneumoniae (17, 41) despite the fact that frameshifts but not large deletions have been identified. In addition, by PCR analysis we have also detected the presence of cells harboring these deletions in cultures of the WT strain (data not shown), evidencing the heterogeneity that can be found in a dynamic mycoplasma population.
MgPa islands are found interspersed along the M. genitalium genome, and they seem to contribute to antigenic variation of the major cytadhesin P140 and its operon-related protein P110 via recombination (29). Supporting this hypothesis, recent studies have shown that sequence variation in the mg191 gene occurs in vitro as well in vivo within a single M. genitalium strain (15). Restriction fragment length polymorphism analyses of clinical M. genitalium strains have also revealed DNA sequence divergences at the proximal region of the mg192 gene (28). However, in this study we demonstrate that, in addition to antigenic variation, the repetitive MgPa element located immediately downstream of the MgPa operon could generate irreversible deletions by a looping-out configuration mechanism when a single-crossover recombination event occurs between this repetitive MgPa element and homologous sequences of the mg191 or mg192 gene (Fig. 6). It seems unlikely for a microorganism with an optimized streamlined genome to keep genomic redundancies in a location that can be potentially adverse for the integrity of the MgPa operon. Thus, the recombination system described here, by which P140 and/or P110 is lost, could be assumed to be an irreversible mechanism of phase variation that could provide important advantages to the whole mycoplasma population (12, 46). For example, since the P140 and P110 proteins are considered the most immunological proteins of M. genitalium (2, 42), the lost of these cytadhesins could contribute both to evasion of the host defense mechanisms and to the infection perseverance of M. genitalium. Such permanent losses of genetic material are not uncommon in mycoplasmas. It is known, for example, that one of the consequences of the formation of chimeric vsp genes in Mycoplasma bovis is the irreversible lost of coding sequences (22). Alternatively, reversible phase variation in the Mycoplasma gallisepticum gapA gene (homologous to mg191) has been described, providing variable adhesive properties to this avian pathogen and promoting the consecutive colonization of either several hosts or various niches within a single host (49). In this study, we show translocation of MgPa sequences via recombination to a particular MgPa island. Based on these observations, we propose the existence of a reversible mechanism to switch on and off the expression of the M. genitalium P140 and P110 cytadhesins, based on a double-recombination event involving sequences from two different boxes of a given MgPa island and the corresponding homologues from the MgPa operon (Fig. 7A). Although it remains to be determined whether these phenomena take place in the course of natural infection, these rearrangements as seen from the population point of view could provide important advantages in tissue colonization and perseverance to this human pathogen.
Collectively, the results presented in this study support previous data and provide new insights into the relationships between the cytoskeleton and the P140 and P110 proteins, as well their roles in the M. genitalium cytadherence. Finally, we show experimental data demonstrating the high genome dynamism of this minimal microorganism.
Acknowledgments
This work was supported by grant BFU2004-06377-C02-01 to E.Q. R.B. acknowledges an FPU predoctoral fellowship from Ministerio de Educación y Ciencia, and O.Q.P. acknowledge a predoctoral fellowship from CeRBa (Centre de Referència en Biotecnologia).
We thank the staff of Servei de Microscòpia (UAB) for processing scanning electron microscopy samples.
Footnotes
Published ahead of print on 6 October 2006.
REFERENCES
- 1.Baseman, J. B. 1993. The cytadhesins of Mycoplasma pneumoniae and M. genitalium. Subcell. Biochem. 20:243-259. [DOI] [PubMed] [Google Scholar]
- 2.Baseman, J. B., M. Cagle, J. E. Korte, C. Herrera, W. G. Rasmussen, J. G. Baseman, R. Shain, and J. M. Piper. 2004. Diagnostic assessment of Mycoplasma genitalium in culture-positive women. J. Clin. Microbiol. 42:203-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baseman, J. B., R. M. Cole, D. C. Krause, and D. K. Leith. 1982. Molecular basis for cytadsorption of Mycoplasma pneumoniae. J. Bacteriol. 151:1514-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baseman, J. B., S. F. Dallo, J. G. Tully, and D. L. Rose. 1988. Isolation and characterization of Mycoplasma genitalium strains from the human respiratory tract. J. Clin. Microbiol. 26:2266-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baseman, J. B., and J. G. Tully. 1997. Mycoplasmas: sophisticated, reemerging, and burdened by their notoriety. Emerg. Infect. Dis. 3:21-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dallo, S. F., A. Chavoya, C. J. Su, and J. B. Baseman. 1989. DNA and protein sequence homologies between the adhesins of Mycoplasma genitalium and Mycoplasma pneumoniae. Infect. Immun. 57:1059-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhandayuthapani, S., M. W. Blaylock, C. M. Bebear, W. G. Rasmussen, and J. B. Baseman. 2001. Peptide methionine sulfoxide reductase (MsrA) is a virulence determinant in Mycoplasma genitalium. J. Bacteriol. 183:5645-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhandayuthapani, S., W. G. Rasmussen, and J. B. Baseman. 1999. Disruption of gene mg218 of Mycoplasma genitalium through homologous recombination leads to an adherence-deficient phenotype. Proc. Natl. Acad. Sci. USA 96:5227-5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraser, C. M., J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, J. M. Kelley, R. D. Fritchman, J. F. Weidman, K. V. Small, M. Sandusky, J. Fuhrmann, D. Nguyen, T. R. Utterback, D. M. Saudek, C. A. Phillips, J. M. Merrick, J. F. Tomb, B. A. Dougherty, K. F. Bott, P. C. Hu, T. S. Lucier, S. N. Peterson, H. O. Smith, C. A. Hutchison III, and J. C. Venter. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397-403. [DOI] [PubMed] [Google Scholar]
- 10.Giron, J. A., M. Lange, and J. B. Baseman. 1996. Adherence, fibronectin binding, and induction of cytoskeleton reorganization in cultured human cells by Mycoplasma penetrans. Infect. Immun. 64:197-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glass, J. I., N. Assad-Garcia, N. Alperovich, S. Yooseph, M. R. Lewis, M. Maruf, C. A. Hutchison III, H. O. Smith, and J. C. Venter. 2006. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. USA 103:425-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henderson, I. R., P. Owen, and J. P. Nataro. 1999. Molecular switches—the ON and OFF of bacterial phase variation. Mol. Microbiol. 33:919-932. [DOI] [PubMed] [Google Scholar]
- 13.Horner, P. J., C. B. Gilroy, B. J. Thomas, R. O. Naidoo, and D. Taylor-Robinson. 1993. Association of Mycoplasma genitalium with acute non-gonococcal urethritis. Lancet 342:582-585. [DOI] [PubMed] [Google Scholar]
- 14.Hutchison, C. A., S. N. Peterson, S. R. Gill, R. T. Cline, O. White, C. M. Fraser, H. O. Smith, and J. C. Venter. 1999. Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286:2165-2169. [DOI] [PubMed] [Google Scholar]
- 15.Iverson-Cabral, S. L., S. G. Astete, C. R. Cohen, E. P. Rocha, and P. A. Totten. 2006. Intrastrain heterogeneity of the mgpB gene in Mycoplasma genitalium is extensive in vitro and in vivo and suggests that variation is generated via recombination with repetitive chromosomal sequences. Infect. Immun. 74:3715-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krause, D. C., and M. F. Balish. 2004. Cellular engineering in a minimal microbe: structure and assembly of the terminal organelle of Mycoplasma pneumoniae. Mol. Microbiol. 51:917-924. [DOI] [PubMed] [Google Scholar]
- 17.Krause, D. C., D. K. Leith, R. M. Wilson, and J. B. Baseman. 1982. Identification of Mycoplasma pneumoniae proteins associated with hemadsorption and virulence. Infect. Immun. 35:809-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Layh-Schmitt, G., and M. Harkenthal. 1999. The 40- and 90-kDa membrane proteins (ORF6 gene product) of Mycoplasma pneumoniae are responsible for the tip structure formation and P1 (adhesin) association with the Triton shell. FEMS Microbiol. Lett. 174:143-149. [DOI] [PubMed] [Google Scholar]
- 19.Layh-Schmitt, G., and R. Herrmann. 1992. Localization and biochemical characterization of the ORF6 gene product of the Mycoplasma pneumoniae P1 operon. Infect. Immun. 60:2906-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Layh-Schmitt, G., A. Podtelejnikov, and M. Mann. 2000. Proteins complexed to the P1 adhesin of Mycoplasma pneumoniae. Microbiology 146:741-747. [DOI] [PubMed] [Google Scholar]
- 21.Luo, D., W. Xu, G. Liang, S. Wang, Z. Wang, Z. Bi, and W. Zhu. 1999. Isolation and identification of Mycoplasma genitalium from high risk populations of sexually transmitted diseases in China. Chin. Med. J. 112:489-492. [PubMed] [Google Scholar]
- 22.Lysnyansky, I., Y. Ron, K. Sachse, and D. Yogev. 2001. Intrachromosomal recombination within the vsp locus of Mycoplasma bovis generates a chimeric variable surface lipoprotein antigen. Infect. Immun. 69:3703-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mernaugh, G. R., S. F. Dallo, S. C. Holt, and J. B. Baseman. 1993. Properties of adhering and nonadhering populations of Mycoplasma genitalium. Clin. Infect. Dis. 17(Suppl. 1):S69-S78. [DOI] [PubMed] [Google Scholar]
- 24.Miyata, M., and S. Seto. 1999. Cell reproduction cycle of mycoplasma. Biochimie 81:873-878. [DOI] [PubMed] [Google Scholar]
- 25.Morrison-Plummer, J., D. H. Jones, K. Daly, J. G. Tully, D. Taylor-Robinson, and J. B. Baseman. 1987. Molecular characterization of Mycoplasma genitalium species-specific and cross-reactive determinants: identification of an immunodominant protein of M. genitalium. Isr. J. Med. Sci. 23:453-457. [PubMed] [Google Scholar]
- 26.Morrison-Plummer, J., A. Lazzell, and J. B. Baseman. 1987. Shared epitopes between Mycoplasma pneumoniae major adhesin protein P1 and a 140-kilodalton protein of Mycoplasma genitalium. Infect. Immun. 55:49-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Musatovova, O., S. Dhandayuthapani, and J. B. Baseman. 2003. Transcriptional starts for cytadherence-related operons of Mycoplasma genitalium. FEMS Microbiol. Lett. 229:73-81. [DOI] [PubMed] [Google Scholar]
- 28.Musatovova, O., C. Herrera, and J. B. Baseman. 2006. Proximal region of the gene encoding cytadherence-related protein permits molecular typing of Mycoplasma genitalium clinical strains by PCR-restriction fragment length polymorphism. J. Clin. Microbiol. 44:598-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peterson, S. N., C. C. Bailey, J. S. Jensen, M. B. Borre, E. S. King, K. F. Bott, and C. A. Hutchison III. 1995. Characterization of repetitive DNA in the Mycoplasma genitalium genome: possible role in the generation of antigenic variation. Proc. Natl. Acad. Sci. USA 92:11829-11833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pich, O. Q., R. Burgos, M. Ferrer-Navarro, E. Querol, and J. Pinol. 2006. Mycoplasma genitalium mg200 and mg386 genes are involved in gliding motility but not in cytadherence. Mol. Microbiol. 60:1509-1519. [DOI] [PubMed] [Google Scholar]
- 31.Pich, O. Q., R. Burgos, R. Planell, E. Querol, and J. Pinol. 2006. Comparative analysis of antibiotic resistance gene markers in Mycoplasma genitalium: application to studies of the minimal gene complement. Microbiology 152:519-527. [DOI] [PubMed] [Google Scholar]
- 32.Pollack, J. D., M. V. Williams, and R. N. McElhaney. 1997. The comparative metabolism of the mollicutes (mycoplasmas): the utility for taxonomic classification and the relationship of putative gene annotation and phylogeny to enzymatic function in the smallest free-living cells. Crit. Rev. Microbiol. 23:269-354. [DOI] [PubMed] [Google Scholar]
- 33.Proft, T., and R. Herrmann. 1994. Identification and characterization of hitherto unknown Mycoplasma pneumoniae proteins. Mol. Microbiol. 13:337-348. [DOI] [PubMed] [Google Scholar]
- 34.Proft, T., H. Hilbert, H. Plagens, and R. Herrmann. 1996. The P200 protein of Mycoplasma pneumoniae shows common features with the cytadherence-associated proteins HMW1 and HMW3. Gene 171:79-82. [DOI] [PubMed] [Google Scholar]
- 35.Reddy, S. P., W. G. Rasmussen, and J. B. Baseman. 1996. Isolation and characterization of transposon Tn4001-generated, cytadherence-deficient transformants of Mycoplasma pneumoniae and Mycoplasma genitalium. FEMS Immunol. Med. Microbiol. 15:199-211. [DOI] [PubMed] [Google Scholar]
- 36.Reddy, S. P., W. G. Rasmussen, and J. B. Baseman. 1995. Molecular cloning and characterization of an adherence-related operon of Mycoplasma genitalium. J. Bacteriol. 177:5943-5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Regula, J. T., G. Boguth, A. Gorg, J. Hegermann, F. Mayer, R. Frank, and R. Herrmann. 2001. Defining the mycoplasma ‘cytoskeleton’: the protein composition of the Triton X-100 insoluble fraction of the bacterium Mycoplasma pneumoniae determined by 2-D gel electrophoresis and mass spectrometry. Microbiology 147:1045-1057. [DOI] [PubMed] [Google Scholar]
- 38.Romero-Arroyo, C. E., J. Jordan, S. J. Peacock, M. J. Willby, M. A. Farmer, and D. C. Krause. 1999. Mycoplasma pneumoniae protein P30 is required for cytadherence and associated with proper cell development. J. Bacteriol. 181:1079-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 40.Seto, S., T. Kenri, T. Tomiyama, and M. Miyata. 2005. Involvement of P1 adhesin in gliding motility of Mycoplasma pneumoniae as revealed by the inhibitory effects of antibody under optimized gliding conditions. J. Bacteriol. 187:1875-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su, C. J., A. Chavoya, and J. B. Baseman. 1989. Spontaneous mutation results in loss of the cytadhesin (P1) of Mycoplasma pneumoniae. Infect. Immun. 57:3237-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Svenstrup, H. F., J. S. Jensen, K. Gevaert, S. Birkelund, and G. Christiansen. 2006. Identification and characterization of immunogenic proteins of mycoplasma genitalium. Clin. Vaccine Immunol. 13:913-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor-Robinson, D. 2002. Mycoplasma genitalium—an up-date. Int. J. STD AIDS 13:145-151. [DOI] [PubMed] [Google Scholar]
- 44.Taylor-Robinson, D., C. B. Gilroy, S. Horowitz, and J. Horowitz. 1994. Mycoplasma genitalium in the joints of two patients with arthritis. Eur. J. Clin. Microbiol. Infect. Dis. 13:1066-1069. [DOI] [PubMed] [Google Scholar]
- 45.Tully, J. G., D. L. Rose, J. B. Baseman, S. F. Dallo, A. L. Lazzell, and C. P. Davis. 1995. Mycoplasma pneumoniae and Mycoplasma genitalium mixture in synovial fluid isolate. J. Clin. Microbiol. 33:1851-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Woude, M. W., and A. J. Baumler. 2004. Phase and antigenic variation in bacteria. Clin. Microbiol. Rev. 17:581-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waldo, R. H., III, and D. C. Krause. 2006. Synthesis, stability, and function of cytadhesin P1 and accessory protein B/C complex of Mycoplasma pneumoniae. J. Bacteriol. 188:569-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiner, J., III, C. U. Zimmerman, H. W. Gohlmann, and R. Herrmann. 2003. Transcription profiles of the bacterium Mycoplasma pneumoniae grown at different temperatures. Nucleic Acids Res. 31:6306-6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winner, F., I. Markova, P. Much, A. Lugmair, K. Siebert-Gulle, G. Vogl, R. Rosengarten, and C. Citti. 2003. Phenotypic switching in Mycoplasma gallisepticum hemadsorption is governed by a high-frequency, reversible point mutation. Infect. Immun. 71:1265-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]