Abstract
Leguminous plants and bacteria from the family Rhizobiaceae form a symbiotic relationship, which culminates in novel plant structures called root nodules. The indeterminate symbiosis that forms between Sinorhizobium meliloti and alfalfa requires biosynthesis of Nod factor, a β-1,4-linked lipochitooligosaccharide that contains an essential 6-O-sulfate modification. S. meliloti also produces sulfated cell surface polysaccharides, such as lipopolysaccharide (LPS). The physiological function of sulfated cell surface polysaccharides is unclear, although mutants of S. meliloti with reduced LPS sulfation exhibit symbiotic abnormalities. Using a bioinformatic approach, we identified a homolog of the S. meliloti carbohydrate sulfotransferase, LpsS, in Mesorhizobium loti. M. loti participates in a determinate symbiosis with the legume Lotus japonicus. We showed that M. loti produces sulfated forms of LPS and capsular polysaccharide (KPS). To investigate the physiological function of sulfated polysaccharides in M. loti, we identified and disabled an M. loti homolog of the sulfate-activating genes, nodPQ, which resulted in undetectable amounts of sulfated cell surface polysaccharides and a cysteine auxotrophy. We concomitantly disabled an M. loti cysH homolog, which disrupted cysteine biosynthesis without reducing cell surface polysaccharide sulfation. Our experiments demonstrated that the nodPQ mutant, but not the cysH mutant, showed an altered KPS structure and a diminished ability to elicit nodules on its host legume, Lotus japonicus. Interestingly, the nodPQ mutant also exhibited a more rapid growth rate and appeared to outcompete wild-type M. loti for nodule colonization. These results suggest that sulfated cell surface polysaccharides are required for optimum nodule formation but limit growth rate and nodule colonization in M. loti.
Most soils are limited in reduced forms of nitrogen. Thus, plants undergo symbioses with microorganisms that provide the plants with reduced nitrogen. The best studied of these symbioses occur between leguminous plants and a family of gram-negative bacteria known as rhizobia. The symbiosis culminates in the formation of novel structures on the plant root called nodules. To enter and colonize the nodules, the bacteria elicit a series of morphological changes in specialized epidermal cells called root hairs. The bacteria induce curling of the root hairs, trapping bacterial microcolonies in the center of the structure. The bacteria then elicit the formation of a plant-derived tubular structure that emanates from the center of the curl, extending through the root hair and ultimately penetrating through the epidermis into the cortical layers of the root. This structure, known as an infection thread, is occupied by the bacteria and allows their entry into the interior of the root. The bacteria are released from the infection thread into the cytoplasm of plant cells within the nodule, where they differentiate into intracellular forms called bacteroids, which then reduce molecular dinitrogen for use by the plant (7, 8, 24, 52, 68).
The symbiosis between rhizobia and legumes is highly specific. Typically, a single Rhizobium species can form a nitrogen-fixing symbiosis with only a small subset of host legume species. This specificity is maintained by the exchange of chemical signals between the symbiotic partners. For example, rhizobia produce lipochitooligosaccharides called Nod factors in response to plant-derived inducer molecules. Nod factor is a β-1,4-linked lipochitooligosaccharide that is required for the initiation of plant developmental pathways leading to nodule formation (15-17, 23, 67). Every species of Rhizobium produces a Nod factor backbone consisting of three to five N-acetylglucosamine residues, which are adorned with host-specific modifications (37, 64). For example, Sinorhizobium meliloti produces a Nod factor covalently modified by a 6-O-sulfate on the reducing end of the N-acetylglucosamine backbone (34). The presence of this sulfate modification is essential for the formation of nodules on the root of its symbiotic partner, Medicago sativa (alfalfa) (61).
S. meliloti produces not only sulfated Nod factor, but also a sulfate-modified form of lipopolysaccharides (LPS) (10). The production of sulfated cell surface polysaccharides is prevalent in eukaryotic cells but appears to be relatively rare in prokaryotes. To date, only three bacterial genera, Mycobacterium, Sinorhizobium, and Pseudoalteromonas, have been reported to contain sulfated polysaccharides (10, 42, 53, 56). While the function of sulfated Nod factor is relatively well understood, the function of sulfated cell surface polysaccharides is poorly characterized.
Examination of the physiological role played by sulfated polysaccharides in S. meliloti has proven challenging. Due to a functional redundancy in carbohydrate sulfotransferases, constructing mutants that lack sulfation of cell surface polysaccharides requires the identification and inactivation of multiple sulfotransferase genes (14). An alternative method involves the disruption of the synthesis of biochemical precursors required for polysaccharide sulfation. For example, the covalent modification of polysaccharides by sulfate requires synthesis of 3′-phosphoadenosine-5′-phosphosulfate (PAPS), which is produced by the nodP and nodQ gene products in S. meliloti (59-61). However, disabling PAPS production in S. meliloti prevents Nod factor sulfation, which is required for its biological activity (61). Since exogenous Nod factor will not rescue symbiosis of the nodPQ mutant, this approach is not suitable for studying the symbiotic function of sulfated cell surface polysaccharides in S. meliloti.
A recent study reported the identification of an open reading frame (ORF) in Mesorhizobium loti whose sequence is similar to that of the LPS sulfotransferase, LpsS, of S. meliloti (14). Thus, we were interested in determining if M. loti produces sulfated cell surface polysaccharides and, if so, examining their physiological function. M. loti is the N2-fixing symbiont of Lotus species, in which it elicits the formation of determinate nodules. Interestingly, some strains of M. loti can also elicit the formation of indeterminate nodules on Leucaena species (46). Determinate nodules lack a persistent meristem and contain bacteroids that morphologically resemble free-living cells and can be cultured following recovery from nodules (41). Conversely, indeterminate nodules maintain an active meristem, which allows the symbiotic bacteria to constantly infect new cells within a single nodule. Indeterminate nodules contain polyploid bacteroids that have hyper-permeable membranes and cannot be cultured following recovery from nodules (41). While previous studies with S. meliloti have shown that mutants with reduced LPS sulfation exhibit alterations in symbiosis, there has been no study to examine the role of bacterium-derived sulfated polysaccharides with symbiotic legumes that form determinate nodules.
M. loti strains produce Nod factor structures that contain an N-acetylglucosamine backbone adorned with a 6′-O-fucosyl residue on the reducing end and an N-methyl modification on the nonreducing end (38). However, unlike S. meliloti, M. loti produces a Nod factor that is not decorated with a covalent sulfate modification. Thus, inactivation of genes involved in the biosynthesis of sulfate precursors such as PAPS would not be expected to affect the biological activity of Nod factor. This would allow a means to disrupt the synthesis of sulfated polysaccharides and examine their physiological function during free-living growth and symbiosis. Here, we show that M. loti produces two distinct sulfated polysaccharides and that inactivation of the nodPQ gene disrupts PAPS biosynthesis and results in alterations in polysaccharide structure and sulfation. We further show that the nodPQ mutant of M. loti elicits nodules on Lotus japonicus at a decreased rate compared to the wild type.
MATERIALS AND METHODS
Bacterial strains and media.
All M. loti strains used are derivatives of NZP-2235 (26) and are described in Table 1. All strains were cultured in tryptone-yeast extract (TY) (3) or rhizobium defined medium (RDM) (55). Selective media contained antibiotic concentrations as follows: gentamicin, 10 μg/ml; neomycin, 10 μg/ml; spectinomycin, 10 μg/ml; streptomycin, 50 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype | Reference(s) or source |
|---|---|---|
| M. loti strains | ||
| NZP-2235 | Wild-type M. loti | 26 |
| GTO015 | NZP-2235/pMS03 | This study |
| GTO016 | NZP-2235/pMS03::nodPQ | This study |
| GTO100 | NZP-2235 nodPQ::Nmr | This study |
| GTO101 | NZP-2235 nodPQ::Nmr/pMS03::nodPQ | This study |
| GTO108 | NZP-2235 nodPQ::Nmr/pMS03 | This study |
| GTO110 | NZP-2235 cysH::Nmr | This study |
| GTO112 | NZP-2235 nodS::Nmr | This study |
| S. meliloti Rm41 | AK684 Strr | 48 |
| E. coli strains | ||
| MG1655 | Wild-type E. coli | 5 |
| DM63 | cysN96::kan proC leu thi ara gal lac hsd Strr | 36 |
| JM96 | thr-1 leuB(Am) fhuA2 lacY1 glnV44(AS) gal-6 λ−trp-1 hisG1(Fs) rfbC1 cysH56 galP63 Δ(gltB-gltF)500 malT1(λr) xyl-7 mtlA2 ΔargH1 rplL9 thi-1 | 27, 28 |
| CAG12182 | λ−cysI3152::Tn10kan rph-1 | 63 |
| AT2427 | λ− e14− cysJ43 relA1 spoT1 thi-1 creC510 | 65 |
| RL165 | thr-1 leuB(Am) fhuA2 lacY1 glnV44(AS) gal-6 λ−trp-1 hisG1(Fs) cysK511 malT1(λr) xyl-7 mtlA2 ΔargH1 rplL9 thi-1 | 19 |
| GET130 | cysN96::kan proC leu thi ara gal lac hsd Strr/pMS03 | This study |
| GET131 | cysN96::kan proC leu thi ara gal lac hsd Strr/pMS03::nodPQ | This study |
| Top10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 deoR araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| DH5α | F− φ80lacZΔM15 Δ(lacZYA-argF)U169 deoR recA endA1 hsdR17(rK− mK+) phoA supE44 thi-1 gyrA96 relA1 | 58 |
| MT616 | E. coli pRK600 (conjugation helper strain) | 33 |
| Plasmids | ||
| pGTO100 | pJQ200::nodPQ::Nmr (including 500 bp immediately 5′ and 3′ of nodPQ) | This study |
| pGTO101 | pMS03::nodPQ | This study |
| pGTO102 | pDW33::nodP (internal fragment) | This study |
| pGTO110 | pJQ200::cysH::Nmr (including 500 bp immediately 5′ and 3′ of cysH) | This study |
| pGTO112 | pJQ200::nodS::Nmr (including 500 bp immediately 5′ and 3′ of nodS) | This study |
| pGET069 | pJQ200::Nmr | This study |
| pMS03 | Broad-host-range plasmid containing trp promoter | 2 |
| pDW33 | Insertional inactivation plasmid | 14 |
| pCR2.1 | Topo cloning vector | Invitrogen |
Strain construction.
Plasmids were introduced into M. loti via triparental mating using strain MT616 (pRK600) as the conjugation helper (33). Strain GTO100 was constructed by the introduction of plasmid pGTO100, a derivative of pJQ200 (49) harboring an Nmr-marked deletion of mll7575 and mll7576, into NZP-2235 and selection for neomycin-resistant colonies. Plasmid pGTO100 cannot replicate within M. loti, and therefore, neomycin-resistant colonies arise from recombination events between pGTO100 and regions immediately 5′ of mll7575 or 3′ of mll7576. Neomycin-resistant colonies were then cultured on TY containing neomycin and 5% sucrose, which selects against cells carrying the sacB gene carried on pGTO100. The neomycin-resistant colonies that survived sucrose challenge were then screened for gentamicin resistance, which is also encoded by the vector. Colonies that were gentamicin sensitive and neomycin resistant were presumed to have undergone allelic replacement, which was then verified by PCR. Strains GTO110 and GTO112 were constructed in a similar manner, except the integrated plasmids pGTO110 and pGTO112 contained a neomycin resistance cassette flanked by 5′ and 3′ regions of mll3228 and mll6161, respectively.
Plasmid construction.
The plasmid pGTO100 was constructed by amplifying an extragenic region immediately 5′ of the mll7575 open reading frame from NZP-2235 chromosomal DNA using the following primers: 5′-TGCCGGGTACCGATCTGCTAGAACG-3′ and 5′-GCGGATCCCAGCCCAAAGTGCGGAA-3′. The extragenic region immediately 3′ of the mll7576 ORF was also amplified from NZP-2235 chromosomal DNA using the following primers: 5′-CTGCAGCGCCGAACGGCGCATGGCG-3′ and 5′-GTCTCGAGAAGAAGCGGCCGGTGCA-3′. The resulting 5′ and 3′ PCR fragments were cloned into plasmid pCR2.1 (Invitrogen), generating plasmids pGET011 and pGET012, respectively. The presence of each fragment was verified by PCR and restriction digestion. The 5′ upstream fragment of mll7575 was isolated from pGET011 by restriction enzyme digestion with SpeI and BamHI and subsequently ligated into pGET069 (pJQ200 containing a neomycin resistance cassette cloned into the SmaI restriction site) to generate pGTO099. The 3′ downstream fragment from mll7576 was isolated from pGET012 by restriction enzyme digestion with PstI and XhoI and cloned into pGTO099 to generate pGTO100.
The plasmid pGTO110 was constructed by amplifying an extragenic region immediately 5′ of the mll3228 open reading frame from NZP-2235 chromosomal DNA using the following primers: 5′-CGTCTAGAAATACGCCTTCGACGAG-3′ and 5′-TGGATCCGCCAGCATAGAGCGCCTC-3′. The extragenic region immediately 3′ of the mll3228 ORF was also amplified from NZP-2235 chromosomal DNA using the following primers: 5′-TCTGCAGCCGGCCTGCGAACGGCGA-3′ and 5′-GGGCCCGAACGGCACCCTCGAAGTG-3′. Both the 5′ and 3′ fragments were cloned into plasmid pCR2.1 to generate pGET021 and pGET022, respectively, and verified by PCR and restriction digestion. The 5′ fragment upstream of mll3228 was isolated from pGET021 by restriction digestion with XbaI and BamHI and ligated into pGET069 to form pGTO109. The 3′ downstream fragment was isolated from pGET022 by restriction digestion with ApaI and XhoI and cloned into pGET109 to generate pGTO110.
The plasmid pGTO112 was constructed by amplifying an extragenic region immediately 5′ of the mll6161 ORF from NZP-2235 chromosomal DNA with the primers 5′-GGGCCCCGCTCTACCATGCTACGAA-3′ and 5′-TCTCGAGTCAGCAGTTCATGATCGA-3′. The extragenic region immediately 3′ of the mll6161 ORF was also amplified from NZP-2235 chromosomal DNA with the primers 5′-TGGATCCGGTTGATGGTAACAACCG-3′ and 5′-GTCTGGAACACACAGCTTGAGCACC-3′. The resulting 5′ and 3′ PCR fragments were cloned into plasmid pCR2.1 (Invitrogen), generating plasmids pGET013 and pGET014, respectively. The presence of each fragment was verified by PCR and restriction digestion. The 5′ upstream fragment of mll6161 was isolated from pGET013 by restriction enzyme digestion with ApaI and XhoI and subsequently ligated into pGET069 (pJQ200 containing a neomycin resistance cassette cloned into the SmaI restriction site) to generate pGTO111. The 3′ downstream fragment from mll6161 was isolated from pGET012 by restriction enzyme digestion with PstI and XhoI and cloned into pGTO013 to generate pGTO112.
The plasmid pGTO101 was constructed by amplifying both mll7575 and mll7576 from NZP-2235 chromosomal DNA using the primers 5′-GCCGGGTACCGATCTGCTAGAACGC-3′ and 5′-ACGGTACCCCTCGGTCACCGGCGAG-3′ and cloning the resulting fragment into pCR2.1 to yield pGET106. The presence of mll7575 and mll7576 was verified by PCR and restriction digestion. The pair of ORFs was isolated from pGET106 following restriction digestion with KpnI and ligated into pMS03 (2) to generate pGTO101. The presence of mll7575 and mll7576 was verified by PCR and DNA sequencing.
Preparation of polysaccharide extracts.
Cell surface extracts were prepared using a hot phenol-water procedure as described previously (50) with the following modifications: extracts were prepared from 2 ml of cells cultured in minimal medium (RDM) and grown to stationary phase (optical density at 600 nm [OD600] equal to 2.0). Cultures were centrifuged at 8,000 × g and washed in 1 ml of sterile water. The cells were once again centrifuged at 8,000 × g, and the pellet was resuspended in 150 μl of solution A (0.05 M Na2HPO4, 0.005 M EDTA, pH 7). A 150-μl volume of 90% phenol was added to the cell suspension, and the mixture was vortexed vigorously and subsequently incubated at 65°C for 15 min. The mixture was subsequently incubated on ice for an additional 10 min and then centrifuged at 8,000 × g. The aqueous phase was removed and subjected to digestion with 25 μg/ml RNase and 1 μg/ml DNase, followed by digestion with 25 μg/ml pronase E. The resulting material was fractionated through a Microspin Sephadex G-25 column (Amersham). The flowthrough was collected and lyophilized. The sample was dissolved in 20 μl sodium dodecyl sulfate (SDS) sample buffer, and 10 μl was fractionated by Tris-Tricine-polyacrylamide gel electrophoresis (PAGE) using 10% acrylamide gels (44).
In vivo labeling of polysaccharides.
Both wild-type and mutant strains of NZP-2235 were cultured in TY to saturation. Cells were then diluted to an OD600 of 0.1 in either TY or RDM in a final volume of 2 ml. Either 5 μCi of Na235SO4 or 1 μCi of [U-14C]glucose (MP Biomedicals) was added to these 2-ml cultures, and cultures were grown to saturation (OD600 of approximately 2.0). Polysaccharides were extracted as described above and resuspended in 20 μl SDS sample buffer. Ten microliters of each sample was fractionated by Tris-Tricine-SDS-PAGE as described above (44). The incorporation of 35SO4 or 14C was visualized by autoradiography and quantified by phosphorimaging.
Nodulation assay.
The ability of wild-type and mutant strains of M. loti to undergo symbiosis with Lotus japonicus (B-129-S9 Gifu) was examined by counting the number of nodules formed when the plant and bacteria were cocultured. L. japonicus seeds were scarified in concentrated sulfuric acid for 20 min and subsequently sterilized in 70% ethanol and 3% H2O2 for 10 min. The sterilized seeds were permitted to imbibe overnight and germinated in an inverted petri dish in the dark. The sterile L. japonicus seedlings were transferred onto Broughton and Dilworth (B&D) (9) agar slants containing 40 nM aminoethoxyvinylglycine (AVG) and allowed to grow for 48 h. The plants were then inoculated with bacterial strains cultured to log phase (an OD600 equivalent to 0.5) in TY and diluted 1:200 in 10 mM MgSO4. Ten milliliters of the diluted M. loti culture was poured onto the sterile L. japonicus plants and then removed. At various times postinoculation, the plants were observed and the numbers of nodules were counted. Between 10 and 20 plants were assayed under each condition.
Symbiotic competition assay.
Wild-type, nodPQ, and cysH M. loti cells were cultured separately overnight to saturation and diluted to an OD600 equivalent to 0.5. Cultures of the wild type were mixed with either the nodPQ or cysH mutant in 1:1 or 1:10 ratios and then diluted to 1:200 in 10 ml of 10 mM MgSO4. Each diluted mixture of cells was then poured onto 10 replicate sterile L. japonicus plants that were grown for 48 h on Broughton and Dilworth agar slants (9). After 30 days following inoculation, nodules from each condition were excised from the root of each plant and pooled. The nodules were subjected to treatment with 20% sodium hypochlorite for 2 min and subsequently washed five times with sterile water. One milliliter of TY with 0.3 M sucrose was added to the surface-sterilized nodules, and the mixture was homogenized. The resulting suspension was serially diluted, plated on TY agar, and incubated at 30°C for 4 days. The colonies that formed were patched onto TY agar containing neomycin (both nodPQ and cysH deletions contain a neomycin resistance cassette) and allowed to incubate at 30°C for 3 days.
PAPS analysis by TLC.
The wild type and nodPQ and cysH mutants were cultured in 1 ml of RDM with Na235SO4 to saturation. The cells were centrifuged at 8,000 × g, resuspended in 1 ml of water, and centrifuged again at 8,000 × g. The resulting cell mass was resuspended in 0.2 ml of water, and 20 μl of 11 N formic acid was added. The mixture was homogenized by vigorous vortexing, incubated on ice for 30 min, and centrifuged at 8,000 × g for 10 min. The supernatant was transferred to a new tube, and then 50 μl of each supernatant was spotted on a polyethyleneimine (PEI)-cellulose thin-layer chromatography (TLC) plate (Baker) in 10-μl increments (60). The plate was bathed in methanol and allowed to dry before being placed in a TLC chamber containing 100 ml of 0.9 M LiCl2. After the solvent front reached the top of the TLC plate, the plate was again bathed in methanol for 2 min and allowed to dry. The TLC plate was developed, bathed in methanol, and dried a second time. The 35SO4 incorporation was visualized by autoradiography and quantified by phosphorimaging.
Polysaccharide isolation and fractionation.
Cells from 1-liter cultures were extracted using the modified hot phenol-water procedure described above. The extracts were treated sequentially with RNase, DNase, and pronase as described above and then dialyzed and lyophilized, yielding 12 to 17 mg of crude polysaccharide preparation from each bacterial strain. The lyophilized residues were subjected to size-exclusion chromatography under dissociative conditions (0.25% sodium deoxycholate, 0.2 M NaCl, 1.0 mM EDTA, 10 mM Tris, pH 9.2) on a column of Sephadex G-150 (1.1 by 100 cm; 10 to 40 μM; superfine). This procedure is capable of separating rough LPS (lacking O-polysaccharide) from smooth LPS (containing O-polysaccharide) and also effectively separates many capsular PS (KPS; K antigens) from LPS (21). The eluant was monitored by the refractive index using a RID-10A detector (Shimadzu Corp., Kyoto, Japan) and by PAGE.
Deoxycholate SDS-PAGE analysis.
Deoxycholate SDS-PAGE analysis was performed as previously described (12, 51).
Polysaccharide composition analysis.
Fractions obtained by size-exclusion chromatography were dialyzed to remove detergent as described elsewhere (50) and then lyophilized. Aliquots were subjected to glycosyl composition analysis by preparing the trimethylsilyl methyl glycoside derivatives. Gas chromatography-mass spectrometry analysis was performed using a 30-m DB-5 capillary column (J&W Scientific, Folsom, CA) on a 5890A gas chromatograph equipped with a mass selective detector (Agilent Technologies, Palo Alto, CA). Inositol was used as an internal standard, and retention times were compared to authentic sugars.
Cationic peptide assays.
For all cationic peptide assays, M. loti strains were grown to an OD600 of 0.5 in TY medium. A 100-μl aliquot of cells was transferred to a microcentrifuge tube, and either polymyxin B (20 μg/ml) or poly-l-lysine (50 μg/ml) was added. The tubes were incubated at room temperature for 1 h. The number of viable bacteria in the culture was then determined by plating a series of dilutions on TY plates.
RESULTS
Mesorhizobium loti produces sulfated polysaccharides.
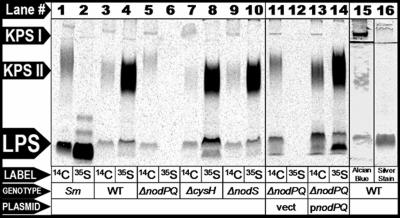
A recent report identified an open reading frame in the genome of Mesorhizobium loti strain MAFF303099 that showed similarity to the Sinorhizobium meliloti lipopolysaccharide sulfotransferase, LpsS (14). To determine if M. loti produces sulfated polysaccharides, we cultured wild-type M. loti (strain NZP-2235) in the minimal medium RDM, in the presence of either Na235SO4 or 14C-uniformly labeled glucose. We purified the polysaccharides from each labeled culture and then fractionated them by Tris-Tricine-SDS-PAGE. We subsequently visualized the incorporated 35S or 14C by autoradiography (Fig. 1). We identified three 14C-labeled species in these extracts, which we categorized as one distinct LPS and two distinct non-LPS species that we refer to as KPS. The highest-molecular-weight 14C-labeled species comigrated with a band that could be detected with Alcian blue but not silver staining (which is consistent with acidic capsular polysaccharide), which we refer to as KPS I (Fig. 1, lanes 3, 15, and 16). We also detected a diffuse lower-molecular-weight 14C-labeled species that comigrated with a band that weakly stained positive with Alcian blue and stained negatively with silver stain and which we refer to as KPS II (Fig. 1, lanes 3, 15, and 16). Furthermore, we detected a low-molecular-weight radiolabeled species which comigrated with a band that could be detected with both Alcian blue and silver staining (which is consistent with lipopolysaccharide) that we refer to as LPS (Fig. 1, lanes 3, 15, and 16). M. loti cells cultured in the presence of Na35SO4 exhibited radiolabeled material that comigrated with both KPS II and LPS but not KPS I (Fig. 1, lane 4). To determine if these molecules are secreted, we isolated total supernatant precipitant collected from M. loti cell cultures grown in the presence of uniformly labeled [14C]glucose and fractioned them by SDS-PAGE. We subsequently visualized 14C incorporation by autoradiography. We failed to detect any of the three distinct 14C-labeled species in culture supernatant precipitants (66). Thus, M. loti produces at least three distinct cell-associated polysaccharide species as judged by PAGE and staining with Alcian blue, of which two are modified by the covalent addition of sulfate.
FIG. 1.
Analysis of sulfated polysaccharides produced by wild-type S. meliloti and M. loti and M. loti mutants lacking either nodPQ, cysH, or nodS. Lanes 1 and 2, wild-type S. meliloti (strain Rm41); lanes 3 and 4, wild-type M. loti (strain NZP-2235); lanes 5 and 6, M. loti harboring chromosomal deletions of nodPQ (GTO100); lanes 7 and 8, cysH (GTO110); lanes 9 and 10, nodS (GTO100); lanes 11 and 12, nodPQ mutant harboring the multicopy empty vector pMS03 (GTO108); lanes 13 and 14, nodPQ mutant harboring a multicopy plasmid encoding nodPQ (GTO101). Cells were cultured in minimal medium in the presence of either uniformly labeled [14C]glucose (odd-numbered lanes) or Na235SO4 (even-numbered lanes) for 72 h. Polysaccharide extracts were prepared from each radiolabeled culture and fractionated by Tris-Tricine-SDS-PAGE as described in Materials and Methods. 14C and 35SO4 incorporation was visualized using autoradiography. Lane 15 contains extracts from wild-type M. loti visualized by Alcian blue staining. Lane 16 contains extracts prepared from wild-type M. loti visualized by silver staining. The lines between KPS I and KPS II in lanes 15 and 16 are the nonspecific accumulation of Alcian blue and silver stain at the interface between the stacker and separating gels.
Identification of M. loti genes responsible for PAPS biosynthesis.
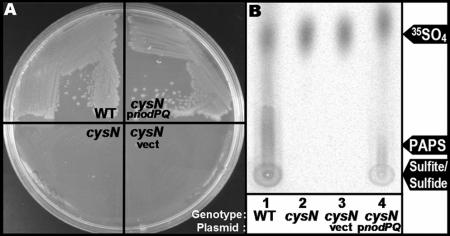
The covalent attachment of sulfate to polysaccharides in all organisms requires biosynthesis of the activated sulfate donor PAPS. We searched the published M. loti genomic sequence (strain MAFF303099) (30) for ORFs with significant sequence identity to the enzymes required for PAPS biosynthesis (ATP sulfurylase and APS kinase) which are encoded by the nodPQ gene products in S. meliloti (59-61). We identified a single genomic locus containing two tandem ORFs, mll7575 and mll7576. ORF mll7575 exhibited 71% amino acid identity to nodP, while mll7576 shared 67% amino acid sequence identity with nodQ. To determine if these ORFs were sufficient to produce PAPS, we cloned the pair into the broad-host-range plasmid pMS03 and introduced them into DM63, an Escherichia coli cysN mutant that is incapable of producing PAPS (36, 60). In E. coli, PAPS is a biosynthetic precursor of cysteine and methionine. Thus, strain DM63 cannot grow in the absence of exogenous cysteine and methionine (32, 36). We cultured DM63 harboring either pGTO101 (which encodes both mll7575 and mll7576) or empty vector on solid M9 minimal medium (39), which lacks exogenous cysteine or methionine. Strain DM63 was capable of forming colonies on M9 minimal medium only when it harbored pGTO101 (Fig. 2A). Thus, ORFs mll7575 and mll7576 encode enzymes capable of restoring cysteine and methionine prototrophy to DM63.
FIG. 2.
Complementation of PAPS-deficient E. coli with mll7575 and mll7576. (A) Colony formation on minimal medium. The cysN mutant (strain DM63, lower left), DM63 harboring vector (lower right), DM63 harboring pGTO101 (which encodes ORFs mll7575 and mll7576) (upper right), or wild-type E. coli (upper left) was grown on solid M9 minimal medium for 48 h at 37°C. (B) PAPS biosynthesis. Wild-type E. coli (lane 1), the cysN mutant (lane 2), or the cysN mutant harboring either empty vector (lane 3) or pGTO101 (lane 4) was cultured in minimal medium in the presence of Na235SO4. PAPS was extracted and fractionated by thin-layer chromatography, and incorporation of 35SO4 was visualized by autoradiography as described in Materials and Methods.
To determine if these two ORFs produce PAPS, we measured PAPS production in DM63 harboring either pGTO101 or empty vector. We cultured both strains on minimal medium lacking cysteine and methionine but supplemented with 35SO4. We subsequently extracted PAPS and its biosynthetic intermediates by formic acid extraction. The formic acid-extractable material was then fractionated by PEI-cellulose TLC, and 35SO4 incorporation was visualized by autoradiography (Fig. 2B). The autoradiograph revealed that both DM63 harboring pGTO101 and wild-type E. coli produced radiolabeled material that was not observed in DM63 harboring vector alone. Comparison of the mobilities of this radiolabeled material to mutants blocked at various stages of the cysteine biosynthetic pathway (see Fig. 4B, below) suggests that the material is sulfite and/or sulfide. Since the biosynthesis of sulfite and sulfide is PAPS dependent, we conclude that ORFs mll7575 and mll7576 encode gene products capable of producing PAPS when expressed in E. coli.
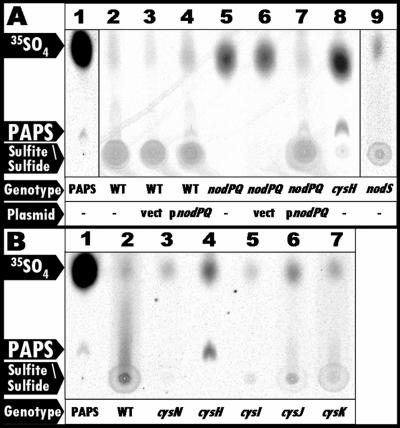
FIG. 4.
PAPS biosynthesis in nodPQ, cysH, and nodS mutants. Formic acid-extractable material was fractionated by thin-layer chromatography, and the incorporation of 35SO4 was visualized by autoradiography as described in Materials and Methods. (A) In vitro-synthesized 35S-labeled PAPS was added to 40 μl of an unlabeled M. loti formic acid extract and spotted onto a TLC plate (lane 1). Wild-type M. loti (lane 2), wild-type M. loti harboring either empty vector (lane 3) or pGTO101 (plasmid carrying nodPQ) (lane 4), the nodPQ mutant (lane 5), the nodPQ mutant harboring either empty vector (lane 6) or pGTO101 (lane 7), the cysH mutant (lane 8), and the nodS mutant (lane 9) were cultured in minimal medium in the presence of Na235SO4 for 48 h. (B) E. coli mutants were cultured in minimal medium in the presence of Na235SO4 for 12 h, and formic acid extracts were examined as for panel A. Lane 1, PAPS; lane 2, wild type (MG1655); lane 3, cysN (DM63); lane 4, cysH (JM96); lane 5, cysI (CAG12182); lane 6, cysJ (AT2427); lane 7, cysK (RL165).
Construction of M. loti mutants lacking PAPS synthase and PAPS reductase.
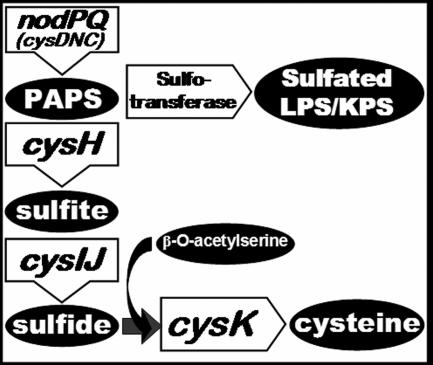
The M. loti genome encodes a single locus resembling nodPQ from S. meliloti; thus, we expected that deletion of mll7575 and mll7576 from the M. loti chromosome would eliminate PAPS biosynthesis. To determine if mll7575 and mll7576 are necessary for PAPS biosynthesis in M. loti, we constructed a chromosomal deletion of mll7575 and mll7576 that was replaced with a neomycin resistance cassette. PAPS is necessary not only for polysaccharide sulfation but also for the biosynthesis of cysteine, methionine, and S-adenosyl methionine (SAMe) (32). Thus, we expected that the M. loti mutant lacking mll7575 and mll7576 would exhibit a cysteine and methionine auxotrophy and a defect in SAMe production. SAMe is a required substrate for the N methylation of M. loti Nod factor(s) (20). Because the cysteine/methionine auxotrophy, SAMe deficiency, and lack of sulfated polysaccharides could all alter symbiosis with Lotus japonicus, we sought to construct additional M. loti mutants that retained the ability to produce PAPS but would be unable to produce cysteine and methionine or be unable to utilize SAMe for Nod factor N methylation. CysH is necessary for the reduction of PAPS to sulfite, which is an essential intermediate for cysteine biosynthesis (Fig. 3) (27, 29). We identified a single ORF within the M. loti genome that exhibited 35% amino acid identity to CysH in S. meliloti, which is annotated as mll3228. We constructed an M. loti mutant that harbored a deletion of mll3228 that was replaced with a neomycin resistance cassette. Additionally, we identified a single ORF within the M. loti genome, annotated as mll6161, that exhibited 71.5% amino acid identity to NodS in Rhizobium tropici. NodS utilizes SAMe for Nod factor N methylation in several Rhizobium species (20, 25, 35, 69). Eliminating PAPS production could affect intracellular SAMe levels, resulting in decreased Nod factor N methylation. Thus, we constructed an additional mutant that harbored a chromosomal deletion of the mll6161 ORF, which also was replaced with a neomycin resistance cassette.
FIG. 3.
Molecular destinations of PAPS.
nodPQ is required for PAPS biosynthesis in M. loti.
To determine if mll7575 and mll7576 are required for PAPS biosynthesis in M. loti, we radiolabeled cultures of wild-type M. loti and GTO100 (which harbors the disrupted form of nodPQ) with Na235SO4 for 48 h and subsequently subjected them to extraction with formic acid. To demonstrate the position of spots corresponding to PAPS, APS, sulfite, and sulfide, we radiolabeled E. coli mutants harboring mutations at various cysteine biosynthetic loci (Fig. 3). We incubated the wild type (MG1655) and cysN (the E. coli equivalent to nodP; DM63), cysH (JM96), cysI (CAG12182), cysJ (AT2427), and cysK (RL165) mutants in the presence of Na235SO4. The formic acid-soluble material was then fractionated by PEI-cellulose TLC, and 35SO4 incorporation was visualized by autoradiography (Fig. 4). We were unable to detect PAPS in either wild-type M. loti or GTO100, which harbors a disruption of mll7575 and mll7576 (Fig. 4A, lanes 2 and 5). However, we observed radiolabeled material that migrated as a smear in wild-type M. loti extracts (Fig. 4A, lane 2), which is consistent with the material observed in extracts prepared from E. coli cysI, cysJ, and cysK mutants (Fig. 4B, lanes 4 to 6) (19, 62, 63, 65). While we were unable to detect PAPS, sulfite, or sulfide in extracts prepared from the mll7575-mll7676 mutant, the introduction of a multicopy plasmid carrying mll7575 and mll7676 (pGTO101) into this mutant resulted in the detection of PAPS, sulfite, and sulfide (Fig. 4A, lane 7). Consistent with extracts prepared from the cysH E. coli mutant (Fig. 4B, lane 3), we were able to detect only PAPS but not sulfite or sulfide in the mll3228 mutant (Fig. 4A, lane 8). Since the production of sulfite requires the biosynthesis and reduction of PAPS or APS (PAPS lacking the 3′-phosphate) in all systems studied to date, these results suggest that mll7575 and mll7576 encode the enzymes necessary for PAPS biosynthesis and mll3228 encodes an enzyme necessary for PAPS reduction. Therefore, we propose renaming mll7575 and mll7576 as nodP and nodQ, respectively, and mll3228 as cysH.
The nodPQ mutant fails to produce sulfated polysaccharides.
The M. loti nodPQ mutant lacks the ability to synthesize PAPS and thus would be expected to be unable to produce sulfated polysaccharides. We cultured wild-type M. loti, GTO100 (nodPQ), GTO110 (cysH), and GTO112 (nodS) in the presence of either Na235SO4 or 14C-uniformly labeled glucose, fractionated total cellular polysaccharide extracts by Tris-Tricine-SDS-PAGE, and subsequently visualized 14C or 35S incorporation by autoradiography (Fig. 1). When we radiolabeled with [14C]glucose, we detected KPS I, KPS II, and LPS in all M. loti strains. However, when we labeled with 35SO4, we were unable to detect LPS or KPS II in extracts prepared from M. loti lacking nodPQ (Fig. 1, lane 6). Additionally, the migration of KPS II prepared from the M. loti nodPQ mutant was retarded compared to the wild type or the cysH mutant when fractionated by Tris-Tricine-SDS-PAGE (Fig. 1, lane 5). Sulfation of cell surface polysaccharides was restored in GTO101, which contains a chromosomal deletion of nodPQ and harbors pGTO101, a multicopy plasmid containing the M. loti nodPQ genes (Fig. 1, lane 14). GTO101 produced KPS II that migrated similarly to the wild type. Consistent with their ability to produce PAPS, we observed radiolabeled LPS and capsular polysaccharides in extracts prepared from both the cysH and nodS M. loti mutants (Fig. 1, lanes 8 and 10).
Compositional analysis of M. loti polysaccharides.
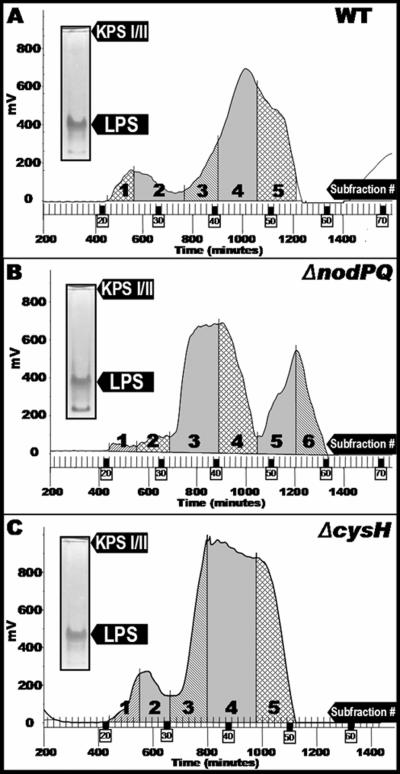
To characterize the polysaccharides produced by wild-type and nodPQ M. loti, we prepared hot phenol-water extracts from 1-liter cultures of wild-type, nodPQ, and cysH strains of M. loti. Each extract was subjected to size-exclusion chromatography under dissociative conditions, which affords a general class separation of the low-molecular-weight, monomeric LPS from the higher-molecular-weight KPS when present (Fig. 5). Similar conditions were used to effectively separate rough LPS from smooth LPS and KPS from Sinorhizobium sp. strain NGR234 (21). The polysaccharide profiles of extracts prepared from both wild-type (Fig. 5A) and cysH (Fig. 5C) M. loti strains showed two distinct peaks, a high-molecular-weight peak migrating near the column void volume and a lower-molecular-weight peak that was partially retained by the column. In contrast, the polysaccharide profile for extracts prepared from the M. loti nodPQ mutant showed an additional, late-moving peak (centered around 1,200 min), and the high-molecular-weight peak was virtually absent in this mutant (Fig. 5B). The profiles of both the wild type and the cysH mutant were subdivided into five fractions, while that of the nodPQ mutant was subdivided into six fractions. Each of the subfractions was subjected to compositional analysis (Table 2). Fraction 1 consists primarily of three sugars, xylose, mannose, and glucose, while fraction 2 contains abundant levels of fucose and is also enriched in 3-O-methyl-6-deoxy hexose and other neutral sugars. The large difference in composition between fraction 1 and fraction 2 suggests that they represent separate polysaccharide components, both having very high molecular weights. The acidic nature of these polysaccharides could arise from noncarbohydrate substituents, such as pyruvate, succinate, or sulfate, which were not examined during the glycosyl composition analysis. For all three bacterial strains, subfractions 3, 4, and 5 were enriched in sugars that are characteristic of LPS, including heptose, 2-keto-3-deoxy-octulosonic acid (Kdo) (a typical LPS core region component), and 2,3-diamino-2,3-dideoxy-glucose (DAG), characteristic of the Mesorhizobium sp. lipid A moiety (11, 57). These subfractions also contained hydroxy-fatty acids, indicating the presence of LPS/lipid A (detected during gas chromatography-mass spectrometry analysis as the trimethylsilyl methyl-esters) (66).
FIG. 5.
Polysaccharide profiles produced by the wild type and nodPQ- and cysH-deficient strains of M. loti during size-exclusion chromatography. Water layer extracts were prepared as described in Materials and Methods and then chromatographed on Sephadex G-150 in detergent (dissociative conditions). The eluants were monitored by refractive index detection from M. loti NZP-2235 (wild type) (A), the M. loti nodPQ mutant (B), and the M. loti cysH mutant (C). (Inset) Prior to chromatography, polysaccharide extracts were analyzed by deoxycholate-PAGE and stained sequentially with Alcian blue, periodate, and silver reagent. KPS I and KPS II comigrate on this gel system.
TABLE 2.
Compositional analysis of polysaccharides purified from wild-type, nodPQ, and cysH M. loti strains
| Residue | G-150 fraction (mol%)
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild type
|
ΔnodPQ mutant
|
cysH mutant
|
||||||||||||||
| 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | 6 | 1 | 2 | 3 | 4 | 5 | |
| 3Me6dHexa | 3.4 | 1.4 | 0.3 | 1.3 | 4.1 | 1.1 | 0.6 | 0.6 | 0.3 | 4.1 | 1.3 | 0.2 | ||||
| Arabinose | 3.8 | 2.0 | 1.6 | 1.5 | 2.1 | 1.7 | 3.2 | 2.3 | 2.5 | 1.7 | 2.0 | 4.6 | 3.0 | 1.9 | 1.0 | |
| Xylose | 21.0 | 7.0 | 2.9 | 1.1 | 0.9 | 21.1 | 4.0 | 1.4 | 1.1 | 1.1 | 1.3 | 18.5 | 5.3 | 1.6 | 0.8 | 0.4 |
| Fucose | 55.8 | 24.0 | 6.0 | 0.2 | 5.7 | 67.7 | 23.7 | 14.5 | 17.3 | 5.7 | 1.1 | 42.0 | 25.3 | 6.7 | 1.0 | |
| Mannose | 32.5 | 12.9 | 6.2 | 2.0 | 1.6 | 23.4 | 6.2 | 2.6 | 2.0 | 1.6 | 2.2 | 22.8 | 12.2 | 2.8 | 1.4 | 0.9 |
| Galactose | 0.6 | 13.9 | 19.3 | 13.5 | 15.1 | 10.3 | 11.4 | 11.4 | 2.0 | 15.1 | 16.9 | 8.2 | ||||
| Glucose | 46.5 | 15.6 | 18.1 | 29.2 | 55.5 | 46.4 | 10.9 | 20.9 | 47.4 | 40.3 | 54.7 | 55.7 | 26.9 | 19.8 | 34.9 | 70.7 |
| gluco-Heptose | 5.7 | 7.2 | 5.6 | 1.8 | 7.2 | 5.1 | 5.3 | 6.3 | 1.1 | 7.2 | 6.1 | 2.8 | ||||
| QuiNAcb | 0.8 | 0.2 | 0.7 | 0.8 | 0.1 | 0.3 | 0.3 | 0.7 | 0.4 | |||||||
| Kdo | 16.4 | 23.4 | 15.1 | 2.8 | 18.8 | 13.2 | 14.7 | 13.7 | 1.5 | 19.1 | 21.2 | 7.1 | ||||
| DAG | 8.6 | 9.6 | 5.1 | 5.3 | 3.3 | 5.0 | 2.7 | 4.1 | 9.4 | 7.9 | ||||||
3Me6dHex, 3-O-methyl-6-deoxyhexose.
QuiNAC, N-acetyl quinovosamine.
For all three bacterial strains, glycosyl composition data (Table 2) indicate that the highest-molecular-weight material (subfraction 1) is composed of xylose, mannose, and glucose. The proportion of this material is greatly diminished in nodPQ, although it is still detectable. These glycosyl components could reflect a KPS that consists of xylose, mannose, and glucose in approximately a 1:1:2 ratio. Alternatively, secondary polysaccharides, such as separate xylomannans and glucans, could also account for these glycosyl components. In all three bacterial strains, fucose is the prominent component of the high-molecular-weight subfraction 2, along with a variety of other neutral sugars. The variety of carbohydrates present in subfraction 2 suggests that multiple high-molecular-weight polysaccharide components are also present within this subfraction. The relative absence of LPS marker components (e.g., Kdo, DAG, heptose) from this fraction further indicates that KPS or other surface polysaccharides are present in this subfraction 2 while intact LPS are absent. Degradation of KPS prevented analysis of the polysaccharides after G-150 size-exclusion chromatography. Thus, we were unable to determine whether a particular subfraction was enriched in KPS I or KPS II and therefore could not assign a composition to a particular capsular polysaccharide. However, we observed that those fractions expected to contain LPS based on the compositional analysis (subfractions 3, 4, and 5 in wild-type M. loti) were enriched in LPS (66). Additional chromatography steps would be required to obtain homogeneous polysaccharide preparations, allowing the assignment of fine structural details.
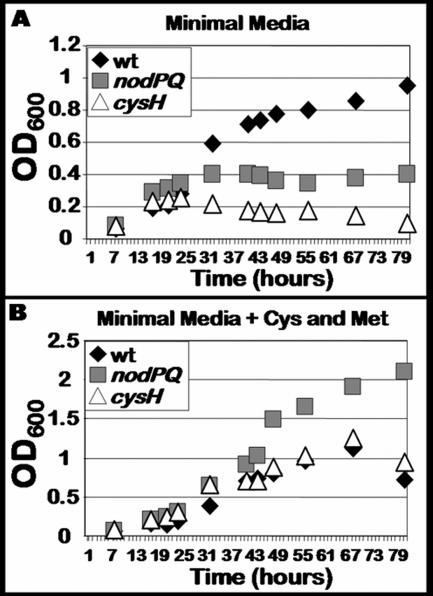
The nodPQ and cysH mutants are cysteine and methionine auxotrophs.
Since the M. loti genome encodes a single copy of the genes necessary for PAPS biosynthesis and the M. loti nodPQ mutant lacks detectable amounts of PAPS and its reduced form, sulfite, we expected this mutant to exhibit a cysteine auxotrophy. Thus, we measured the ability of both the nodPQ and cysH mutants to grow in minimal medium in the presence or absence of exogenous cysteine and methionine (Fig. 6). Both the nodPQ and cysH mutants were unable to grow in liquid minimal medium lacking cysteine and methionine (Fig. 6A). However, the nodPQ mutant was able to grow approximately twice as rapidly as either wild-type or cysH-deficient M. loti when cultured in minimal medium supplemented with 10 μM cysteine and methionine (Fig. 6B).
FIG. 6.
Growth kinetics of wild-type M. loti and nodPQ and cysH mutants. (A) Wild-type M. loti (wt) and the nodPQ and cysH mutants were cultured in minimal medium in the absence of exogenous cysteine and methionine. (B) Wild-type M. loti and the nodPQ and cysH mutants were cultured in minimal medium in the presence of 10 μM cysteine and methionine. Aliquots were collected from each culture approximately every 8 h, and their optical densities were measured using spectrophotometry.
The nodPQ mutants show a reduced ability to elicit nodules on L. japonicus.
Studies with S. meliloti have reported symbiotic phenotypes among mutants with reduced LPS sulfation (13, 14, 31). Thus, we asked whether M. loti mutants unable to produce sulfated polysaccharides exhibit an altered symbiosis with Lotus japonicus. The nodPQ mutant of M. loti lacks the ability to produce PAPS, which results in an inability to produce sulfated polysaccharides and a cysteine auxotrophy in liquid medium. Conversely, the M. loti cysH mutant retained the ability to produce sulfated polysaccharides but also exhibited a cysteine auxotrophy. The nodS mutant would be expected to lack the ability to modify its Nod factor by N methylation while retaining cysteine prototrophy and the ability to produce sulfated polysaccharides. Thus, we reasoned that comparing the symbiotic phenotypes of the three mutants to that of the wild type would allow us to determine if the ability to synthesize PAPS-dependent molecules is required for symbiosis.
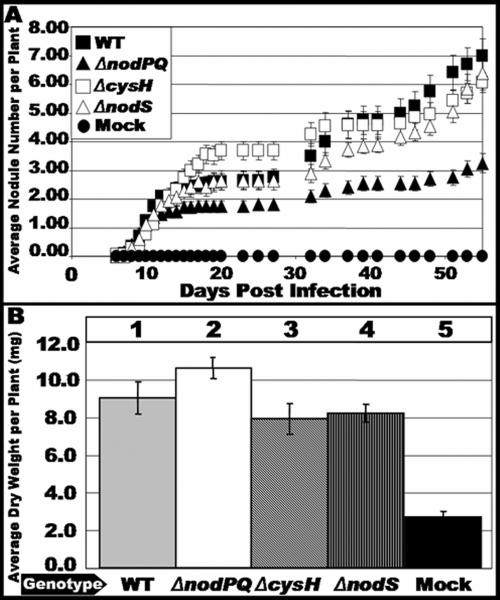
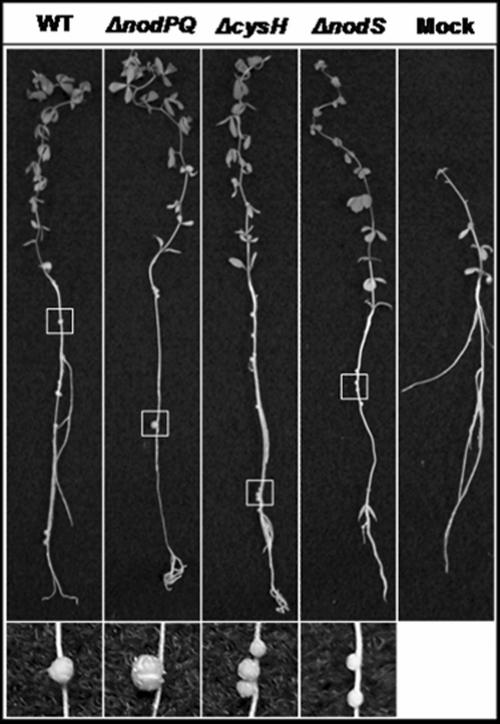
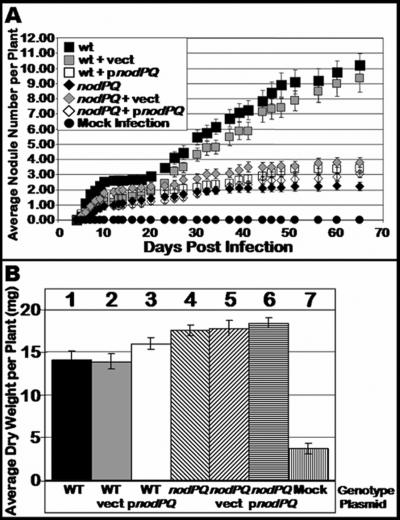
We inoculated sterile Lotus japonicus seedlings with either the wild type or nodPQ, cysH, or nodS mutant strains of M. loti. We subsequently counted the number of nodules that formed on each plant over a 45-day period (Fig. 7A). The rate of nodule formation exhibited by the nodPQ mutant was decreased by approximately 50% compared to the wild type. However, both the cysH and nodS mutants exhibited a rate of nodule formation similar to that of the wild type. This suggests that the cysteine auxotrophy associated with the nodPQ mutation is not the cause of the reduced rate of nodule formation. Accordingly, the loss of nodS did not alter the kinetics of nodule formation. The resulting nodules elicited by either the nodPQ, cysH, or nodS mutant resembled those elicited by wild-type M. loti (Fig. 8).
FIG. 7.
Kinetics of nodule formation exhibited by the wild type and nodPQ, cysH, and nodS mutants. Sterile L. japonicus seedlings were cultured on B&D agar slants supplemented with 40 nM AVG. Plants were inoculated 48 h after transfer to B&D agar slants with either wild-type M. loti, the nodPQ, cysH, or nodS mutant, or sterile 10 mM MgSO4. (A) At various intervals following inoculation, the number of nodules was counted as described in Materials and Methods for wild-type M. loti (wt), the nodPQ, cysH, or nodS mutant, or sterile 10 mM MgSO4. (B) Plants were harvested at 120 days postinoculation, and their dry weight was measured. Lane 1, wild-type M. loti; lane 2, nodPQ; lane 3, cysH; lane 4, nodS; lane 5, sterile 10 mM MgSO4.
FIG. 8.
L. japonicus plants inoculated with either wild-type, nodPQ, cysH, or nodS M. loti strains. L. japonicus plants inoculated with either the wild type, the nodPQ, cysH, or nodS mutant, or 10 mM MgSO4 were collected following 65 days of incubation and photographed. An area of each plant root (highlighted by the white box) that contained a nodule(s) was selected and enlarged.
Interestingly, complementation of the nodPQ mutant with a plasmid-borne copy of nodPQ (which restored PAPS production and sulfated polysaccharide production) (Fig 1, lane 14, and Fig. 4, lane 6) did not restore normal nodule formation kinetics to the nodPQ mutant (Fig. 9A). We do not currently understand the inability of this plasmid construct to complement the symbiotic defect exhibited by the nodPQ mutant. However, the wild-type strain harboring the same plasmid-borne copy of nodPQ also exhibited reduced rates of nodule formation (Fig. 9A), suggesting that the presence of the plasmid-borne nodPQ has a negative effect on symbiosis.
FIG. 9.
Nodule formation kinetics exhibited by the nodPQ mutant harboring plasmid-borne nodPQ. Sterile L. japonicus seedlings were cultured on B&D agar slants containing 40 nM AVG. Plants were inoculated 48 h after transfer to B&D agar slants with either wild-type M. loti, wild-type M. loti harboring either empty vector (GTO015) or pGTO101 (GTO016), the nodPQ mutant or the nodPQ mutant harboring either empty vector (GTO108) or pGTO101 (GTO101), or sterile 10 mM MgSO4. (A) At various intervals following inoculation the plants were analyzed and the number of nodules was counted as described in Materials and Methods for wild-type M. loti (wt), GTO015, GTO016, the nodPQ mutant, GTO108, GTO101, or sterile 10 mM MgSO4. (B) Plants were harvested at 120 days postinoculation, and their dry weight was measured. Lane 1, wild-type M. loti; lane 2, GTO015; lane 3, GTO016; lane 4, nodPQ; lane 5, GTO108; lane 6, GTO101; lane 7, sterile 10 mM MgSO4.
Although the nodPQ mutant produces nodules on L. japonicus, we wanted to test whether the bacteria were capable of colonizing the nodules and fixing nitrogen. Since the plants are cultured on nitrogen-free medium, they can grow only if reduced nitrogen is supplied by bacteroids within the nodule. Thus, the dry weight of the infected plant can be used as an indirect measure of nitrogen fixation (18, 40). We measured the dry weight of plants inoculated with either wild-type, nodPQ, or cysH M. loti cells. In plants harvested at 120 days postinoculation, there was no significant difference in the dry weight of plants inoculated with either wild-type or nodPQ-, cysH-, or nodS-deficient M. loti, suggesting that although nodules are formed at a reduced rate when plants are inoculated with the nodPQ mutant, the nodules are capable of reducing nitrogen (Fig. 7B). Additionally, despite the reduced rates of nodule formation, both wild-type and nodPQ-deficient M. loti harboring pGTO101 (a multicopy plasmid carrying nodPQ) did not exhibit a difference in dry weight relative to the wild type (Fig. 9B). These results indicate that the reduced rates of nodule formation do not affect nitrogen fixation.
The nodPQ mutant outcompetes the wild type in mixed infections.
The nodPQ mutant elicited nodules at rates lower than that of wild-type M. loti, whereas the cysH mutant shared a rate of nodule formation similar to that observed with the wild type. We then asked whether the reduced rate of nodule formation observed with the nodPQ mutant results in a differential ability to compete with the wild type in mixed-infection experiments. We inoculated sterile L. japonicus seedlings with mixtures of the wild type and either the nodPQ or cysH mutant. Approximately 30 days following inoculation, we recovered the bacteria from within the nodule and determined the relative number of wild-type and mutant M. loti cells by plating on medium containing neomycin (the nodPQ and cysH mutants are neomycin resistant) (Table 3). When a 1:1 ratio of wild-type and nodPQ M. loti was inoculated onto plants, 88% of colonies recovered from the nodules were neomycin resis-tant. In contrast, 59% of colonies recovered from nodules collected from plants inoculated with a 1:1 mixture of the wild type and the cysH mutant were neomycin resistant. Thus, we recovered more nodPQ than wild-type bacteria from nodules elicited by a 1:1 mixture of both strains under these conditions.
TABLE 3.
Quantification of bacteria recovered from nodules following coinoculation with wild-type and either nodPQ::Nmr or cysH::Nmr M. loti
| Inoculated strain(s) | No. of wild-type bacteria | No. of mutant bacteria | Mutants/total recovered bacteria (%) |
|---|---|---|---|
| Wild type | 400 | 0 | NAd |
| nodPQ::Nmr | 0 | 400 | NA |
| cysH::Nmr | 0 | 200 | NA |
| Wild type and nodPQ::Nmr (1:1)a | 49 | 351 | 87.8 |
| Wild type and nodPQ::Nmr (1:10)b | 41 | 359 | 89.8 |
| Wild type and nodPQ::Nmr (10:1)c | 72 | 328 | 82.0 |
| Wild type and cysH::Nmr (1:1)a | 82 | 118 | 59.0 |
| Wild type and cysH::Nmr (1:10)b | 28 | 204 | 87.9 |
| Wild type and cysH::Nmr (10:1)c | 280 | 20 | 6.7 |
The 1:1 designation refers to an inoculum containing an equal ratio of wild-type to mutant bacteria.
The 1:10 designation refers to an inoculum containing 10% wild-type and 90% mutant bacteria.
The 10:1 designation refers to an inoculum containing 90% mutant and 10% wild-type bacteria.
NA, not applicable.
DISCUSSION
The covalent modification of polysaccharides by sulfate is common in eukaryotic cells, and mutants with reduced polysaccharide sulfation exhibit a variety of defects (6). In bacteria, sulfation appears to be a rare type of polysaccharide modification whose physiological function is poorly characterized. Utilizing a bioinformatic approach, we identified an ORF in the genome of Mesorhizobium loti, mll3788, that exhibited sequence similarity to the S. meliloti sulfotransferase, LpsS. We subsequently demonstrated that M. loti produces sulfated polysaccharides (although ORF mll3788 was not required for their sulfate modification) (66). We then constructed a mutant disrupted for the sulfate-activating genes, nodPQ, and showed that it produced undetectable amounts of sulfated polysaccharides, a change in KPS structure or degree of polymerization, and an altered symbiosis, eliciting nodules on L. japonicus roots at a decreased rate compared to the wild type. A mutant with a mutation in cysH which was auxotrophic for cysteine and methionine but produced normal amounts of sulfated polysaccharides and a mutant with a mutation in nodS, which is responsible for the N methylation of Nod factor, elicited nodules on L. japonicus at rates similar to that of the wild type. Because the only known metabolic destinations of PAPS are cysteine and methionine biosynthesis (which appears to be dispensable for symbiosis), the N-methyl group on Nod factor, and sulfated polysaccharides, our data suggest that sulfated polysaccharides are important during interactions with Lotus japonicus.
Sulfated polysaccharide production has been reported in only four bacterial genera to date, Sinorhizobium, Mycobacterium, Pseudoalteromonas, and now Mesorhizobium (10, 42, 53, 56). Several Sinorhizobium species produce sulfated polysaccharides (10, 22). For example, Sinorhizobium meliloti produces a sulfated form of Nod factor as well as LPS (10, 22, 34). The addition of sulfate to the nonreducing sugar of the Nod factor backbone affects both the activity of Nod factor and its ability to be secreted by S. meliloti (54, 70). Mutants with reduced LPS sulfation show altered symbiosis, which can be reflected by changes in nodule numbers. For example, the lpsS mutant of S. meliloti exhibits an increased number of nodules during symbiosis with alfalfa (14). Conversely, the lps212 mutant of S. meliloti exhibits two phenotypes: a decreased number of nodules with respect to the wild type and an inability to colonize the nodules that do form (31). Thus, although the mechanism is not understood, sulfated cell surface polysaccharides appear to be required for optimal symbiosis with alfalfa. Mycobacterium species have also been shown to produce sulfated polysaccharides in the form of glycolipids (42, 53). While some studies suggested that sulfated trehalose glycolipids may be involved in M. tuberculosis pathogenesis during disease progression in the lung, another group failed to observe this requirement (42, 53). Thus, the function of sulfated polysaccharides in Mycobacterium species is currently unclear. Finally, a bacterium isolated from a deep-sea hydrothermal vent, Pseudoalteromonas sp. strain HYD721, has also been reported to produce an exopolysaccharide on which a covalent sulfate modification adorns the 3′-hydroxyl group of mannose phosphate (56). The function of this sulfated exopolysaccharide has not been reported. Similar to S. meliloti and M. tuberculosis, M. loti produces sulfated cell surface polysaccharides, and mutants blocked in PAPS biosynthesis elicit reduced numbers of nodules during symbiosis with L. japonicus, although the nodules are capable of fixing nitrogen. Thus, these data suggest that sulfated polysaccharides produced by M. loti, like those of S. meliloti and perhaps M. tuberculosis, are required for optimal interaction with eukaryotic hosts.
The activation of sulfate to PAPS is a required step in the biosynthesis of cysteine and methionine in many organisms. However, other organisms utilize APS (PAPS lacking the 3′ phosphate group) as a source of activated sulfate for cysteine biosynthesis (1, 4). Our analysis of the M. loti genome identified only two genes, mll7575 and mll7576, that shared significant amino acid sequence similarity to the sulfate-activating genes nodPQ. Additionally, we identified a single M. loti gene (mll3228) that shared significant amino acid sequence identity with the gene responsible for PAPS reduction, cysH. Therefore, we expected both the M. loti nodPQ and cysH mutants to exhibit a cysteine and methionine auxotrophic phenotype. Surprisingly, while both the nodPQ and cysH mutants exhibited a cysteine auxotrophy when grown in liquid medium, neither mutant exhibited an auxotrophic phenotype when grown on solid minimal medium (66). The nodPQ mutant of M. loti was capable of growth on a range of solid media that lacked cysteine and methionine, including those containing highly purified agar and agarose. Thus, either M. loti is able to utilize a contaminant of agar that is present even in highly enriched agar preparations or the bacterium is able to utilize reduced forms of sulfate within the agar itself to generate cysteine biosynthetic intermediates. The mechanism of this sulfur assimilation during M. loti growth on solid medium is currently under investigation.
Since M. loti Nod factor is not normally sulfated, the inability to produce PAPS would not be expected to alter its sulfation. However, the nodPQ mutation might be expected to influence Nod factor structure through its effect on the cysteine and methionine biosynthetic pathway. The Nod factors produced by some rhizobia contain an acylated derivative of N-acetylglucosamine that is N methylated (25, 38). These modifications are required for Rhizobium tropici and Sinorhizobium sp. strain NGR234 to elicit nodules on common bean plants (Phaseolus vulgaris) (35, 69). The N-methyl modification is dependent on the nodS gene product, which has been shown to covalently attach an N-methyl residue to the nonreducing end of Nod factor, using SAMe as a donor (20, 25). SAMe is produced from homocysteine, an intermediate in the conversion of cysteine to methionine; thus, mutants blocked in cysteine biosynthesis would be expected to produce Nod factor lacking this modification (32). We generated a nodS mutant of M. loti and showed that this mutant produces nodules at rates similar to that of the wild type. Thus, nodS is not essential for M. loti symbiosis with L. japonicus.
Since the nodPQ mutant exhibited a reduced number of nodules with respect to wild-type M. loti, we anticipated that complementing the nodPQ mutant with nodPQ on a multicopy plasmid in trans (pGTO101) would restore the rate of nodule formation to that of the wild type. Although pGTO101 restored sulfation of cell surface polysaccharides, pGTO101 did not restore normal rates of nodule formation to the nodPQ mutant. Furthermore, the introduction of pGTO101 into wild-type M. loti also resulted in decreased rates of nodule formation, suggesting that nodPQ in high copy can negatively affect nodule formation. Our measurements of internal PAPS concentrations in M. loti suggest that there is usually only a very small pool of intracellular PAPS in wild-type cells, whereas wild-type M. loti harboring a multicopy version of nodPQ exhibits a ca. 400% increase in its pool of intracellular PAPS (66). Previous reports have suggested that the accumulation of PAPS can exert toxic effects on cells, although the mechanism of this toxicity is controversial (43). Therefore, the decreased number of nodules elicited by the M. loti strain encoding multicopy nodPQ may result from toxicity of intracellular PAPS. Despite the negative effect of overproducing nodPQ on the kinetics of nodule formation, L. japonicus seedlings inoculated with these strains exhibited an otherwise normal symbiosis as evidenced by the final average dry weight, which was approximately equivalent to that of the wild type. Additionally, the bacteria recovered from within the nodules retained pGTO101 (66), suggesting that the bacteria that did colonize the nodules were overproducing PAPS. Interestingly, the cysH mutant of M. loti also accumulates approximately equivalent intracellular PAPS pools compared to what is observed in the strain harboring a multicopy nodPQ, yet the cysH mutant does not exhibit a symbiotic phenotype. Perhaps the biosynthesis of PAPS is downregulated during symbiosis and the strain harboring plasmid pGTO101 constitutively produces PAPS, resulting in a decreased symbiotic efficiency.
We observed a noticeable difference in migration on Tris-Tricine-SDS-PAGE between the KPS II produced by the wild-type and nodPQ strains of M. loti. We predicted that this shift in mobility could be attributed to an alteration in polysaccharide structure or degree of polymerization resulting from the loss of sulfation. Thus, we ascertained the composition of polysaccharides produced from the wild-type, nodPQ, and cysH strains of M. loti. We observed similar polysaccharide profiles in extracts prepared from wild-type and cysH-deficient M. loti with three compositionally distinct subfractions, supporting the observation of three distinct polysaccharide species observed by SDS-PAGE. In contrast, the polysaccharide profile obtained from extracts prepared from the nodPQ mutant differed from those of the wild type and the cysH mutant. The compositions of fractions 1, 2, and 3 were altered in the nodPQ mutant compared to the wild type, which is consistent with the data showing nodPQ KPS II having an altered mobility when fractionated by SDS-PAGE (Fig. 1, lane 5). For example, fraction 1 derived from the wild type contains glucose, mannose, and xylose, whereas the same fraction from the nodPQ mutant contains not only glucose, mannose and xylose but also arabinose, xylose, glucose, mannose, fucose, and 3-O-methyl-6-deoxyhexose. One potential explanation for these results is that KPS II derived from the nodPQ mutant exhibits a greater degree of polymerization than that of the wild type, resulting in comigration of KPS I and II during G-150 gel filtration. However, since we were unable to conclusively assign a specific composition to either KPS I or KPS II, we cannot conclude that this shift in composition reflects a shift in KPS I or II abundance or polymerization. The altered composition and Mr we observed in the nodPQ mutant strongly suggest that sulfation is an integral part of the biosynthesis of KPS II.
There are several explanations for the symbiotic phenotype exhibited by the nodPQ mutant. For example, nodPQ could be required for the synthesis or modification of an unknown molecule that is required for optimum nodule formation. However, we are unaware of any additional roles for PAPS other than cysteine and methionine biosynthesis and polysaccharide sulfation. If polysaccharide sulfation is required for optimum symbiosis, there are several possible mechanisms. First, the integrity of the cell surface is dependent upon the molecules that line the barrier between the cell wall and the external environment. The sulfate modifications on LPS and KPS generate additional negative charges that could potentially be involved in the maintenance of cell wall integrity. Thus, elimination of nodPQ could subsequently disrupt cell wall stability and potentially increase the permeability of the cell wall to antimicrobial compounds and reactive oxygen species that are likely encountered during symbiosis. The nodPQ mutant did not, however, display an increased sensitivity to poly-l-lysine or polymyxin B compared to wild-type M. loti (Table 4). Second, the altered symbiosis exhibited by the nodPQ mutant could be attributed to its altered KPS and LPS structures. Alternatively, sulfated LPS and KPS could act as a signal that is required for plant recognition of the symbiotic bacteria.
TABLE 4.
Results of cationic peptide killing assaysa
| Cationic agent (concn [μg/ml]) | % Survival
|
Survival ratio (WT/ΔnodPQ mutant) | % Survival of ΔcysH mutant | Survival ratio (WT/ΔcysH mutant) | |
|---|---|---|---|---|---|
| Wild type | ΔnodPQ mutant | ||||
| Polymyxin B (20) | 99.6 | 112.7 | 88.4 | 100.3 | 99.3 |
| Poly-l-lysine (50) | 73.6 | 103.2 | 71.3 | 96.5 | 76.3 |
Cationic peptide killing assays were performed with polymyxin B and poly-l-lysine. Results were recorded as percent survival after a 1-h exposure to the cationic peptide at the concentration indicated. The results shown are the average of three experiments. WT, wild type.
Despite the reduced nodule number observed in symbiotic assays, the nodPQ mutant exhibited a surprising ability to outcompete the wild type in competition experiments. Our data show that the nodPQ mutant grows more rapidly than the wild type on minimal medium in the presence of exogenous cysteine and methionine. Therefore, the enhancement in nodule colonization could simply be attributed to the ability of nodPQ to grow faster than the wild type in planta. The increased growth rate could also affect the recovery of bacteria from nodules, complicating the interpretation of the competition experiments. Further experiments using fluorescently and chromatically tagged M. loti strains are required to rule out this hypothesis. Alternatively, accumulation of by-products produced from the polysaccharide sulfotransferase reaction could potentially exert toxic effects on the bacterial cell, thereby inhibiting the efficiency of nitrogen fixation by the bacteroids within the nodule. Thus, eliminating PAPS could enhance the efficiency of nodule colonization and nitrogen fixation of the nodPQ mutant. Previous studies have shown that plants regulate the number of nodules produced and the number of infection threads that penetrate into the nodule (45, 47). Therefore, if the nodPQ mutant were capable of colonizing the nodule and/or supplying the plant with reduced nitrogen more efficiently than wild-type M. loti, this could explain the reduction in the nodule number and enhancement in competition. We are currently investigating each of these possibilities.
Acknowledgments
We thank Maike Müller, David Thach, and Brittany Mortensen for helpful comments on the manuscript, Jeanne Harris for the gift of Lotus japonicus seeds, and Makoto Hayashi for technical expertise regarding culturing L. japonicus seedlings.
This study was funded by award 2005-35319-15304 from the U.S. Department of Agriculture. The Complex Carbohydrate Research Center was supported in part by Department of Energy grant DE-FG02-93ER20097.
Footnotes
Published ahead of print on 6 October 2006.
REFERENCES
- 1.Abola, A. P., M. G. Willits, R. C. Wang, and S. R. Long. 1999. Reduction of adenosine-5′-phosphosulfate instead of 3′-phosphoadenosine-5′-phosphosulfate in cysteine biosynthesis by Rhizobium meliloti and other members of the family Rhizobiaceae. J. Bacteriol. 181:5280-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnett, M. J., V. Oke, and S. R. Long. 2000. New genetic tools for use in the Rhizobiaceae and other bacteria. BioTechniques 29:240-242. [DOI] [PubMed] [Google Scholar]
- 3.Beringer, J. E. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84:188-198. [DOI] [PubMed] [Google Scholar]
- 4.Bick, J. A., J. J. Dennis, G. J. Zylstra, J. Nowack, and T. Leustek. 2000. Identification of a new class of 5′-adenylylsulfate (APS) reductases from sulfate-assimilating bacteria. J. Bacteriol. 182:135-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Bowman, K. G., and C. R. Bertozzi. 1999. Carbohydrate sulfotransferases: mediators of extracellular communication. Chem. Biol. 6:R9-R22. [DOI] [PubMed] [Google Scholar]
- 7.Brewin, N. J. 1991. Development of the legume root nodule. Annu. Rev. Cell Biol. 7:191-226. [DOI] [PubMed] [Google Scholar]
- 8.Brewin, N. J. 1992. Nodule formation in legumes. Encycl. Microbiol. 3:229-248. [Google Scholar]
- 9.Broughton, W. J., and M. J. Dilworth. 1971. Control of leghaemoglobin synthesis in snake beans. Biochem. J. 125:1075-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cedergren, R. A., J. Lee, K. L. Ross, and R. I. Hollingsworth. 1995. Common links in the structure and cellular localization of Rhizobium chitolipooligosaccharides and general Rhizobium membrane phospholipid and glycolipid components. Biochemistry 34:4467-4477. [DOI] [PubMed] [Google Scholar]
- 11.Choma, A., and P. Sowinski. 2004. Characterization of Mesorhizobium huakuii lipid A containing both D-galacturonic acid and phosphate residues. Eur. J. Biochem. 271:1310-1322. [DOI] [PubMed] [Google Scholar]
- 12.Corzo, J., R. Perez-Galdona, M. Leon-Barrios, and A. M. Gutierrez-Navarro. 1991. Alcian blue fixation allows silver staining of the isolated polysaccharide component of bacterial lipopolysaccharides in polyacrylamide gels. Electrophoresis 12:439-441. [DOI] [PubMed] [Google Scholar]
- 13.Cronan, G. E., G. R. O. Campbell, G. C. Walker, and D. H. Keating. Unpublished results.
- 14.Cronan, G. E., and D. H. Keating. 2004. Sinorhizobium meliloti sulfotransferase that modifies lipopolysaccharide. J. Bacteriol. 186:4168-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Bruijn, F. 1991. Biochemical and molecular studies: symbiotic nitrogen fixation. Curr. Opin. Biotechnol. 2:184-192. [DOI] [PubMed] [Google Scholar]
- 16.Denarie, J., and J. Cullimore. 1993. Lipo-oligosaccharide nodulation factors: a new class of signaling molecules mediating recognition and morphogenesis. Cell 74:951-954. [DOI] [PubMed] [Google Scholar]
- 17.Downie, J. A. 1994. Signalling strategies for nodulation of legumes by rhizobia. Trends Microbiol. 2:318-324. [DOI] [PubMed] [Google Scholar]
- 18.Dymov, S. I., D. J. Meek, B. Steven, and B. T. Driscoll. 2004. Insertion of transposon Tn5tac1 in the Sinorhizobium meliloti malate dehydrogenase (mdh) gene results in conditional polar effects on downstream TCA cycle genes. Mol. Plant-Microbe Interact. 17:1318-1327. [DOI] [PubMed] [Google Scholar]
- 19.Fimmel, A. L., and R. E. Loughlin. 1977. Isolation and characterization of cysK mutants of Escherichia coli K12. J. Gen. Microbiol. 103:37-43. [DOI] [PubMed] [Google Scholar]
- 20.Geelen, D., B. Leyman, P. Mergaert, K. Klarskov, M. Van Montagu, R. Geremia, and M. Holsters. 1995. NodS is an S-adenosyl-L-methionine-dependent methyltransferase that methylates chitooligosaccharides deacetylated at the non-reducing end. Mol. Microbiol. 17:387-397. [DOI] [PubMed] [Google Scholar]
- 21.Gudlavalleti, S. K., and L. S. Forsberg. 2003. Structural characterization of the lipid A component of Sinorhizobium sp. NGR234 rough and smooth form lipopolysaccharide. Demonstration that the distal amide-linked acyloxyacyl residue containing the long chain fatty acid is conserved in rhizobium and Sinorhizobium sp. J. Biol. Chem. 278:3957-3968. [DOI] [PubMed] [Google Scholar]
- 22.Hanin, M., S. Jabbouri, D. Quesada-Vincens, C. Freiberg, X. Perret, J. C. Prome, W. J. Broughton, and R. Fellay. 1997. Sulphation of Rhizobium sp. NGR234 Nod factors is dependent on noeE, a new host-specificity gene. Mol. Microbiol. 24:1119-1129. [DOI] [PubMed] [Google Scholar]
- 23.Higashi, S. 1993. (Brady)Rhizobium-plant communications involved in infection and nodulation. J. Plant Res. 106:206-211. [Google Scholar]
- 24.Hirsch, A. M. 1992. Developmental biology of legume nodulation. New Phytol. 122:211-237. [DOI] [PubMed] [Google Scholar]
- 25.Jabbouri, S., R. Fellay, F. Talmont, P. Kamalaprija, U. Burger, B. Relic, J. C. Prome, and W. J. Broughton. 1995. Involvement of nodS in N-methylation and nodU in 6-O-carbamoylation of Rhizobium sp. NGR234 nod factors. J. Biol. Chem. 270:22968-22973. [DOI] [PubMed] [Google Scholar]
- 26.Jarvis, B. D. W., C. E. Pankhurst, and J. J. Patel. 1982. Rhizobium loti, a new species of legume root nodule bacteria. Int. J. Syst. Bacteriol. 32:378-380. [Google Scholar]
- 27.Jones-Mortimer, M. C. 1973. Mapping of structural genes for the enzymes of cysteine biosynthesis in Escherichia coli K12 and Salmonella typhimurium LT2. Heredity 31:213-221. [DOI] [PubMed] [Google Scholar]
- 28.Jones-Mortimer, M. C. 1968. Positive control of sulphate reduction in Escherichia coli. Isolation, characterization and mapping of cysteineless mutants of E. coli K12. Biochem. J. 110:589-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones-Mortimer, M. C., J. F. Wheldrake, and C. A. Pasternak. 1968. The control of sulphate reduction in Escherichia coli by O-acetyl-L-serine. Biochem. J. 107:51-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesawa, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7:331-338. [DOI] [PubMed] [Google Scholar]
- 31.Keating, D. H., M. G. Willits, and S. R. Long. 2002. A Sinorhizobium meliloti lipopolysaccharide mutant altered in cell surface sulfation. J. Bacteriol. 184:6681-6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kredich, N. M. 1987. Biosynthesis of cysteine, p. 419-427. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol. 1. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 33.Leigh, J. A., E. R. Signer, and G. C. Walker. 1985. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc. Natl. Acad. Sci. USA 82:6231-6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lerouge, P., P. Roche, C. Faucher, F. Maillet, G. Truchet, J. C. Prome, and J. Denarie. 1990. Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344:781-784. [DOI] [PubMed] [Google Scholar]
- 35.Lewin, A., E. Cervantes, W. Chee-Hoong, and W. J. Broughton. 1990. nodSU, two new nod genes of the broad host range Rhizobium strain NGR234 encode host-specific nodulation of the tropical tree Leucaena leucocephala. Mol. Plant-Microbe Interact. 3:317-326. [DOI] [PubMed] [Google Scholar]
- 36.Leyh, T. S., J. C. Taylor, and G. D. Markham. 1988. The sulfate activation locus of Escherichia coli K12: cloning, genetic, and enzymatic characterization. J. Biol. Chem. 263:2409-2416. [PubMed] [Google Scholar]
- 37.Long, S. R. 1996. Rhizobium symbiosis: nod factors in perspective. Plant Cell 8:1885-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopez-Lara, I. M., J. D. van den Berg, J. E. Thomas-Oates, J. Glushka, B. J. Lugtenberg, and H. P. Spaink. 1995. Structural identification of the lipo-chitin oligosaccharide nodulation signals of Rhizobium loti. Mol. Microbiol. 15:627-638. [DOI] [PubMed] [Google Scholar]
- 39.Maniatis, T. E., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 40.Marroqui, S., A. Zorreguieta, C. Santamaria, F. Temprano, M. Soberon, M. Megias, and J. A. Downie. 2001. Enhanced symbiotic performance by Rhizobium tropici glycogen synthase mutants. J. Bacteriol. 183:854-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mergaert, P., T. Uchiumi, B. Alunni, G. Evanno, A. Cheron, O. Catrice, A. E. Mausset, F. Barloy-Hubler, F. Galibert, A. Kondorosi, and E. Kondorosi. 2006. Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium-legume symbiosis. Proc. Natl. Acad. Sci. USA 103:5230-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mougous, J. D., R. E. Green, S. J. Williams, S. E. Brenner, and C. R. Bertozzi. 2002. Sulfotransferases and sulfatases in mycobacteria. Chem. Biol. 9:767-776. [DOI] [PubMed] [Google Scholar]
- 43.Neuwald, A. F., B. R. Krishnan, I. Brikun, S. Kulakauskas, K. Suziedelis, T. Tomcsanyi, T. S. Leyh, and D. E. Berg. 1992. cysQ, a gene needed for cysteine synthesis in Escherichia coli K-12 only during aerobic growth. J. Bacteriol. 174:415-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niehaus, K., A. Largares, and A. Puhler. 1998. A Sinorhizobium meliloti lipopolysaccharide mutant induces effective nodules on the host plant Medicago sativa (alfalfa) but fails to establish a symbiosis with Medicago truncatula. Mol. Plant-Microbe Interact. 11:906-914. [Google Scholar]
- 45.Oldroyd, G. E., E. M. Engstrom, and S. R. Long. 2001. Ethylene inhibits the Nod factor signal transduction pathway of Medicago truncatula. Plant Cell 13:1835-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pankhurst, C. E., D. H. Hopcroft, and W. T. Jones. 1987. Comparative morphology and flavolan content of Rhizobium loti induced effective and ineffective root nodules on Lotus species, Leuceana leucocephala, Carmichaelia flagelliformis, Ornithopus sativus, and Clianthus puniceus. Can. J. Bot. 65:2676-2685. [Google Scholar]
- 47.Penmetsa, R. V., and D. R. Cook. 1997. A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 275:527-530. [DOI] [PubMed] [Google Scholar]
- 48.Putnoky, P., E. Grosskopf, D. T. Ha, G. B. Kiss, and A. Kondorosi. 1988. Rhizobium fix genes mediate at least two communication steps in symbiotic nodule development. J. Cell Biol. 106:597-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 50.Reuhs, B. L., R. W. Carlson, and J. S. Kim. 1993. Rhizobium fredii and Rhizobium meliloti produce 3-deoxy-d-manno-2-octulosonic acid-containing polysaccharides that are structurally analogous to group II K antigens (capsular polysaccharides) found in Escherichia coli. J. Bacteriol. 175:3570-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reuhs, B. L., D. P. Geller, J. S. Kim, J. E. Fox, V. S. Kolli, and S. G. Pueppke. 1998. Sinorhizobium fredii and Sinorhizobium meliloti produce structurally conserved lipopolysaccharides and strain-specific K antigens. Appl. Environ. Microbiol. 64:4930-4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ridge, R. 1992. A model for legume root hair growth and Rhizobium infection. Symbiosis 14:359-373. [Google Scholar]
- 53.Rivera-Marrero, C. A., J. D. Ritzenthaler, S. A. Newburn, J. Roman, and R. D. Cummings. 2002. Molecular cloning and expression of a novel glycolipid sulfotransferase in Mycobacterium tuberculosis. Microbiology 148:783-792. [DOI] [PubMed] [Google Scholar]
- 54.Roche, P., F. Debelle, F. Maillet, P. Lerouge, C. Faucher, G. Truchet, J. Denarie, and J. C. Prome. 1991. Molecular basis of symbiotic host specificity in Rhizobium meliloti: nodH and nodPQ genes encode the sulfation of lipo-oligosaccharide signals. Cell 67:1131-1143. [DOI] [PubMed] [Google Scholar]
- 55.Ronson, C. W., B. T. Nixon, L. M. Albright, and F. M. Ausubel. 1987. Rhizobium meliloti ntrA (rpoN) gene is required for diverse metabolic functions. J. Bacteriol. 169:2424-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rougeaux, H., J. Guezennec, R. W. Carlson, N. Kervarec, R. Pichon, and P. Talaga. 1999. Structural determination of the exopolysaccharide of Pseudoalteromonas strain HYD 721 isolated from a deep-sea hydrothermal vent. Carbohydr. Res. 315:273-285. [DOI] [PubMed] [Google Scholar]
- 57.Russa, R., T. Urbanik-Sypniewska, K. Lindstrom, and H. Mayer. 1995. Chemical characterization of two lipopolysaccharide species isolated from Rhizobium loti NZP2213. Arch. Microbiol. 163:345-351. [DOI] [PubMed] [Google Scholar]
- 58.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 59.Schwedock, J., and S. R. Long. 1990. ATP sulphurylase activity of the nodP and nodQ gene products of Rhizobium meliloti. Nature 348:644-647. [DOI] [PubMed] [Google Scholar]
- 60.Schwedock, J. S., C. Liu, T. S. Leyh, and S. R. Long. 1994. Rhizobium meliloti NodP and NodQ form a multifunctional sulfate-activating complex requiring GTP for activity. J. Bacteriol. 176:7055-7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schwedock, J. S., and S. R. Long. 1992. Rhizobium meliloti genes involved in sulfate activation: the two copies of nodPQ and a new locus, saa. Genetics 132:899-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sekowska, A., H. F. Kung, and A. Danchin. 2000. Sulfur metabolism in Escherichia coli and related bacteria: facts and fiction. J. Mol. Microbiol. Biotechnol. 2:145-177. [PubMed] [Google Scholar]
- 63.Singer, M., T. A. Baker, G. Schnitzler, S. M. Deischel, M. Goel, W. Dove, K. J. Jaacks, A. D. Grossman, J. W. Erickson, and C. A. Gross. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53:1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spaink, H. P., D. M. Sheeley, A. A. van Brussel, J. Glushka, W. S. York, T. Tak, O. Geiger, E. P. Kennedy, V. N. Reinhold, and B. J. Lugtenberg. 1991. A novel highly unsaturated fatty acid moiety of lipo-oligosaccharide signals determines host specificity of Rhizobium. Nature 354:125-130. [DOI] [PubMed] [Google Scholar]
- 65.Taylor, A. L., and C. D. Trotter. 1967. Revised linkage map of Escherichia coli. Bacteriol. Rev. 31:332-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Townsend, G. E., II, L. S. Forsberg, and D. H. Keating. Unpublished results.
- 67.van Rhijn, P., and J. Vanderleyden. 1995. The Rhizobium-plant symbiosis. Microbiol. Rev. 59:124-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vijn, I., L. das Nevas, A. van Kammen, H. Franssen, and T. Bisseling. 1993. Nod factors and nodulation in plants. Science 260:1764-1765. [DOI] [PubMed] [Google Scholar]
- 69.Waelkens, F., T. Voets, K. Vlassak, J. Vanderleyden, and P. van Rhijn. 1995. The nodS gene of Rhizobium tropici strain CIAT899 is necessary for nodulation on Phaseolus vulgaris and on Leucaena leucocephala. Mol. Plant-Microbe Interact. 8:147-154. [DOI] [PubMed] [Google Scholar]
- 70.Wais, R. J., D. H. Keating, and S. R. Long. 2002. Structure-function analysis of Nod factor-induced root hair calcium spiking in Rhizobium-legume symbiosis. Plant Physiol. 129:211-224. [DOI] [PMC free article] [PubMed] [Google Scholar]