Abstract
The reason for genetic exchange remains a crucial question in evolutionary biology. Acinetobacter baylyi strain ADP1 is a highly competent and recombinogenic bacterium. We compared the parallel evolution of wild-type and engineered noncompetent lineages of A. baylyi in the laboratory. If transformability were to result in an evolutionary benefit, it was expected that competent lineages would adapt more rapidly than noncompetent lineages. Instead, regardless of competency, lineages adapted to the same extent under several laboratory conditions. Furthermore, competent lineages repeatedly evolved a much lower level of transformability. The loss of competency may be due to a selective advantage or the irreversible transfer of loss-of-function alleles of genes required for transformation within the competent population.
Bacteria may achieve genetic exchange by several means (38). Conjugation is typically mediated by extrachromosomal elements that direct the transfer of their own genetic material directly from one organism to another. This also results in the occasional transfer of host genetic material. Transduction involves the accidental packaging and genetic transfer of host DNA in phage capsids. Finally, transformation involves the uptake of free DNA from the environment followed by recombination into the genome. A number of bacterial species, such as Bacillus subtilis, Neisseria gonorrhoeae, and Helicobacter pylori, are known to be naturally competent for transformation (17, 21, 26). Conjugation and transduction are caused by extrachromosomal genetic elements acting in a “selfish” manner and are mechanistically independent of the recipient's genotype. However, the evolutionary derivation of active, recipient-driven competence remains obscure.
The widespread distribution of these mechanisms that result in genetic exchange, as well as eukaryotic sexual recombination, suggests that such genetic recombination provides some benefit (4, 32, 38). However, sex involves the pairing of gametes from individuals that will then undergo meiosis, a fundamentally different process from the mechanisms of genetic transfer listed above. Therefore, a different set of benefits and costs may exist when the mechanism of genetic exchange is transformation rather than sex (38). In addition to recombining beneficial mutations (as in sex), possible benefits of transformation include the uptake of DNA as a nutrient source, the repair of DNA damage, the generation of variation within a population, and the reduction of mutational load.
Sources of DNA for transformation include the genomes and extrachromosomal elements of dead cells of the same species or of unrelated organisms and living cells of species that actively release DNA (see reference 42 and references therein). With regard to reducing the mutational load of a population, simulations have suggested that transformation is actually more likely to decrease fitness when the source of the DNA is from closely related cells that died, due to overrepresentation of low-fitness mutants among dead cells compared to their living counterparts (39). It was suggested by simulations that, in mixed populations of competent and noncompetent bacteria, the potential benefit of transformation with regard to reducing the mutation load was exceeded by the risk of transformation to a noncompetent genetic background (40). This would suggest that a noncompetent phenotype should dominate the population via unidirectional genetic transfer of defective competence alleles from noncompetent mutants to competent recipients. In this case, the possibility of reversion to competence is obviated by the nature of the allele being transferred. In addition, transformation may carry a fitness cost simply due to the metabolic demand of synthesizing the proteins involved in the active uptake of DNA from the environment and recombination of DNA fragments into the genome (Table 1), favoring loss of the trait through mutation and subsequent selection.
TABLE 1.
Identified genes related to transformability
| Gene category | Gene name | bp |
|---|---|---|
| Competencya | comFc | 435 |
| comE | 510 | |
| comC | 4,353 | |
| comB | 969 | |
| smf | 1,152 | |
| comM | 1,488 | |
| pilD | 861 | |
| pilC | 1,227 | |
| pilB | 1,740 | |
| pilU | 1,152 | |
| pilT | 1,035 | |
| comL | 1,056 | |
| comFd | 642 | |
| comP | 444 | |
| comA | 2,379 | |
| Recombinationb | recA | 1,050 |
| recB | 3,705 | |
| recC | 3,666 | |
| recD | 1,779 | |
| recG | 2,061 | |
| recJ | 1,707 | |
| recN | 1,662 | |
| recO | 711 | |
| ruvA | 615 | |
| ruvB | 1,005 | |
| ruvC | 576 | |
| pepA | 1,449 | |
| xerD | 918 | |
| xerC | 918 | |
| himD | 306 | |
| himA | 732 | |
| ssb | 579 | |
| polA | 2,763 | |
| ligA | 2,031 | |
| gyrB | 2,469 | |
| gyrA | 2,730 |
Recombination-associated genes are those present in the A. baylyi genome of those listed previously (38) and genes listed as recombination associated (3).
comF (ACIAD3314) is part of the comFECB operon (7) that was disrupted in this study.
comF (ACIAD3236) is a homologue of Haemophilus influenzae comF (3).
We sought to address the issue of evolutionary costs and benefits of transformation by using Acinetobacter baylyi strain ADP1 (previously Acinetobacter calcoaceticus BD413 and Acinetobacter sp. strain ADP1). A. baylyi is a gram-negative soil bacterium that displays a broad metabolic versatility (3, 45); indeed, one-quarter of its genome contains a majority of genes dedicated to the breakdown of a large variety of compounds, a feature so unique that this region has been termed an “archipelago of catabolic diversity” (3). Importantly, A. baylyi is characteristically highly competent and recombinogenic (27, 43, 45). Furthermore, A. baylyi does not have any sequence requirements for DNA uptake (though genomic homology greatly increases the recombination rate), and the species is maximally competent at the onset of exponential growth phase (35). These features make A. baylyi an attractive target for genetic manipulation (33). Indeed, a number of operons and genes whose functions are associated with competency have been identified by targeted mutagenesis (7, 20, 25, 31, 36) and by genomic sequence analysis (3). Of these genes, knockouts of comB and comC result in noncompetent phenotypes, while knockouts of comE and comF result in 10- and 1,000-fold-diminished transformability, respectively. By comparing rates of evolution in competent and engineered noncompetent A. baylyi strains, we sought to investigate the effect of genetic exchange on laboratory evolution of bacteria and the reciprocal effect of laboratory evolution on genetic exchange mechanisms.
MATERIALS AND METHODS
Strain construction.
Wild-type Acinetobacter baylyi strain ADP1 (PS8004) was streaked out, and five randomly picked clones were used to initiate five cultures which were stored at −80°C and later used to initiate five wild-type lineages of A. baylyi. These were designated PS8135 through PS8139. (Strains are summarized in Table 2.)
TABLE 2.
Strains used in this study
| Strain(s) | Genotype or description | Reference |
|---|---|---|
| PS8004 | Wild-type A. baylyi | This laboratory |
| PS8135 to PS8139 | Five clones of PS8004 | This study |
| PS8130 to PS8134 | Five clones of PS8004 ΔcomFECB::Kanr-sacB | This study |
| PS8455 | PS8004 ΔmutS::Kanr-sacB | This study |
| PS8471 to PS8475 | Five clones of PS8455 ΔmutS | This study |
| PS8025 | PS8004 ΔilvC::Specr-sacB | = PS6324 (33) |
| PS8041 | PS8004 ΔtrpGDC::Kanr-tdk | = PS6372 (33) |
Strains were constructed essentially as described previously (33). In order to eliminate the competence of A. baylyi, we made a DNA construct in which regions external to comFECB flanked a cassette that consisted of the kanamycin resistance (Kanr, which allows positive selection) gene and the saccharase B gene (sacB, which allows negative selection due to induced sensitivity to sucrose). This construct was then used to transform A. baylyi and select for Kanr clones, which should eliminate the comFECB operon and result in a noncompetent strain. The regions flanking comFECB were amplified from genomic DNA of PS8004 by using the primers comOpACF (5′-GCACGTCCGCTGATTCCATAAGCAGTGAT) and CO-ACR-KSB (5′-ggttgtaacactggcagagcATGCAAATTCAAAACTGTGGATAAGCCAA) for the 5′ region and CO-BNF-KSB (5′-gagacacaacgtggctttccTTAGTACGCCTCCAGAAACAAACACGTTGTA) and comOpBNR3 (5′-TTAAACAAGTGATTCAGCGTTTACAGGACTGGGGTGCAGAAGC GCC) to amplify the 3′ flank; CO-ACR-KSB and CO-BNF-KSB add regions of homology to the Kanr-sacB cassette (lowercase). The cassette was amplified using the primers SacBKanF (5′-GGAAAGCCACGTTGTGTCTC) and SacBKanR (5′-GCTCTGCCAGTGTTACAACC) with genomic DNA of strain PS6308 (A. baylyi ΔilvC::Kanr-sacB) (33). Conditions for amplification were identical among the three reactions: HiFi Platinum Taq (Invitrogen, Carlsbad, CA) with 0.2 μM of each primer, 200 μM deoxynucleoside triphosphates, and buffer and MgSO4 as supplied by the manufacturer. Reactions were cycled 35 times at 94°C for 30 s, 58°C for 30 s, and 68°C for 2 min and then at 68°C for 5 min. For PCR of flanking regions, outer primers were used at 0.8 μM. Reaction products were mixed at 3.3 μl for each reaction in a new 100-μl reaction mixture and cycled again, without additional primers, as described above. The entire reaction mixture was added to a growing, 0.5-ml culture of PS8004. This culture was grown for an additional 4 h, and then 200 μl was plated on Luria-Bertani medium (LB) with kanamycin (15 μg/ml) (LB + Kan). This resulted in hundreds of kanamycin-resistant colonies, only ∼1% of which were also sucrose sensitive. Of these, two were subjected to PCR with the primers comOpACF and comOp-BNR3, and both yielded amplified products of a size that was predicted given the cassette replacing comFECB. Strain PS8032 (A. baylyi ΔcomFECB::Kanr-sacB) was streaked out on LB + Kan, and five colonies were stored at −80°C and used to initiate noncompetent lineages (PS8130 to PS8134).
A similar strategy was employed to eliminate the mutS gene. The flanks of mutS were amplified with the primers mutSNF (5′GAGCTGGCAATTGGTGATCAAA) and mutS-NR-KSB (5′-gagacacaacgtggctttccGGTCAGCCATTGTTTCTGTGCTAT) for the 5′ flank and mutS-CF-KSB (5′-ggttgtaacactggcagagcCTAATTACGCTCAAACAGTC) and mutSCR (5′-GGTACGAACAATTCCTTTTA) for the 3′ flank. Overlaps to the Kanr-sacB cassette were included (lowercase). Three-way assembly PCR was carried out as described above and used to transform PS8004, generating strain PS8455 (A. baylyi ΔmutS::Kanr-sacB). The 5′ flank was again amplified, replacing mutS-NR-KSB with the primer RCCFmutSNR (5′-gactgtttgagcgtaattagGGTCAGCCATTGTTTCTGTGCTAT), which includes the reverse complement of primer mutSCF on the 5′ end (lowercase). Similarly, the 3′ flank was reamplified, replacing mutS-CF-KSB with mutSCF (5′-CTAATTACGCTCAAACAGTC). These products were then spliced by two-way assembly PCR with external primers mutSNF and mutSCR. This product was used to transform PS8455 and was plated on LB plus 6% sucrose (without NaCl) (LB + Suc) to take advantage of negative selection against sacB. Surviving colonies were tested by PCR with mutSNF and mutSCR and were found to have the products that would indicate the clean deletion of the mutS gene. Five such clones were isolated and used to generate lineages for selection (PS8471 to PS8475).
Serial dilution experiments.
Strains PS8130 to PS8139, representing five cultures each of Δcom and wild-type A. baylyi, were inoculated from frozen culture in 1 ml of LB and grown to stationary phase at 30°C and 250 rpm. Cultures were diluted 1:10,000 to a bottleneck size of ∼2 × 105, and this process was repeated. Each culture represents ∼13 generations (g), with an effective population size of 2.3 × 106 (Ne = N0 × g) (30). Samples were stored at −80°C every five cultures, representing ∼66 generations. Because of the suspicion that combining the separate lineages might enhance the rate of evolution if recombination was a factor (potentially due to increasing the population diversity), after 400 generations, each lineage was severely bottlenecked (∼200 CFU) and mixed to form one lineage per wild-type and com genotype. These new, mixed lineages were propagated for five serial cultures. These cultures were diluted to initiate five new lineages per genotype, which were each serially diluted 10 times. The process of bottlenecking, serial dilution, and restoration of lineages was repeated, with the restored lineages (five per genotype) being propagated for five transfers when the experiment was halted. This resulted in a total of ∼730 generations of adaptation to benign laboratory conditions.
Later experiments were conducted without the severe bottlenecking steps and subsequent mixing of lineages. To compare the evolution of wild-type and mutator strains of A. baylyi, PS8135 to PS8139 and PS8471 to PS8475 were serially diluted for ∼730 generations in 1 ml LB at 30°C and 250 rpm. To compare the adaptation of Δcom and wild-type A. baylyi strains in harsh conditions, PS8130 to PS8139 were similarly propagated for ∼400 generations in 1 ml LB plus 300 mM NaCl at 40°C and 250 rpm.
Growth rate determination.
Growth rates were determined essentially as described previously (2). In brief, overnight cultures were diluted 1:1,000 into 250-μl microplate wells. Growth was monitored by absorbance at 595 nm in a PowerWave 200 microplate reader (Bio-Tek, Winooski, VT), with incubation at the appropriate temperature and shaking between readings. Growth curves were fit by nonlinear regression to the logistic growth equation
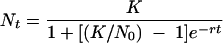 |
(1) |
in which K is the carrying capacity, N is the population size, t is time, and r is the specific growth rate. Rates shown are means and standard deviations (SDs) for five replicate lineages, measured at least twice.
Fitness assays.
Fitness was assayed by direct competition at high and low densities. Overnight cultures of lineages to be competed were diluted and mixed into the same culture at either 1:100,000 for low-density competitions (∼103 CFU/ml) or 1:10 for high-density competitions (∼108 CFU/ml). Low-density competitions lasted for ∼15 generations, but the linear relation between number of generations and time did not change at the densities examined (r2 was at least 0.98 for all experiments), indicating that culture conditions had not dramatically changed. High-density cultures reached stationary phase quickly and then stayed at high density (∼109 CFU/ml) for the remainder of the competition. At various time points (on the scale of hours for low-density competition and ∼12 h apart for high-density competitions), samples from the competition were serially diluted, and titers were determined in duplicate on LB + Kan (specifying Δcom lineages) or LB + Suc (specifying wild-type lineages due to the sensitivity of the Δcom lineage to sucrose). While the Sucs phenotype is lost at a frequency of ∼10−7, competitions were carried out such that the difference between types was much greater than this frequency and therefore was insensitive to reversion. The change in the ratio of the two types being competed was fit to the equation
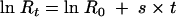 |
(2) |
in which R is the ratio of the two types, t is time, and s is the selection coefficient, i.e., the difference in fitness between the two types (29). Fitness values are reported in terms of hours because in high-density fitness competitions, there is no change in population size. Even though there is turnover within the population, the generation time cannot be measured. Although the actual change in total number of bacteria is known for the low-density cultures, t is reported in terms of hours as well so that the two measures can be directly compared. Calculating the low-density fitness value in terms of generations does not change the conclusions reached here.
Transformability assay.
Strains to be tested for transformability were diluted from an overnight culture 1:10 into 250-μl microplate wells. After 2 h of shaking and incubation at 30°C, 1 μg of genomic DNA was added to each well. DNA was prepared as described previously (1) from either PS8025 (= PS6315; A. baylyi ΔilvC::Specr-sacB) or PS8041 (= PS6372; A. baylyi ΔtrpGDC::Kanr-tdk) (33). Microplates were shaken and incubated for an additional 4 h. Cultures were serially diluted, and titers were determined on selective (LB + Kan or LB plus spectinomycin [200 μg/ml] [LB + Spec], as appropriate) and nonselective (LB) media to determine the fraction transformed. It should be noted that this procedure was optimized for throughput rather than absolute transformability.
In order to test the distribution of transformability, the ancestral and evolved lineages were tested as described above, with 1 μg of genomic DNA from each of PS8025 and PS8041. The fractions of single and double transformants were determined by measuring titers on LB, LB + Kan, LB + Spec, and LB + Kan + Spec. The expected number of double transformants was the product of the Kanr and Specr frequencies multiplied by the total titer of the culture.
DNA release assay.
Strains were tested for the amount of free DNA produced in culture at stationary phase. These strains were grown for 16 h at 30°C and 250 rpm. Cultures were spun down at 10,000 × g for 5 min. The supernatant was then filtered through 0.22-μm polyvinylidene difluoride syringe filters. The filtrate was phenol-chloroform extracted, chloroform extracted, and ethanol precipitated. The pellet was resuspended in 10 mM Tris, pH 8.0. A portion of the purified DNA was used in a transformation assay in which PS8025 was transformed with DNA from all tested strains. Transformability was assayed by determining the fraction of the culture that had reverted to ilvC+ by comparing the number of colonies formed on minimal medium plus glucose (MSglc) (41) to the number formed on LB. Because of the linearity of transformability over the DNA ranges found here (data not shown), the amount of transformable DNA released in cultures could be estimated by generating a standard curve and relating the fraction of PS8025 transformed to known concentrations of transforming DNA.
Mutation fixation calculations.
A slight fitness differential at high density was observed in parallel fitness assays between the noncompetent and competent variants of A. baylyi. It can be asked whether the measured fitness differential can account for the observed loss of transformability. The number of generations required for a spontaneous loss-of-function mutation with this selective advantage to go to fixation can be calculated by several methods, provided that several assumptions are made (see below). If a difference exists between the calculated number of generations required for the loss of competency to sweep to high frequency and the observed number of generations required to lose competency in the actual experiment (no more than 730 generations), then we can conclude that competitive growth rate differences alone cannot account for the rate at which we observe the evolution of diminished competency in A. baylyi.
All of the assumptions required for the calculations favor a faster sweep of the loss of competency and therefore minimize the number of generations required for selective fixation of com alleles in the following estimates. The result is that these estimates of the number of generations required for the loss of transformability should be taken as the minimal number of generations required, and almost certainly the true number is greater than what is calculated. The assumptions used include that (i) the selection coefficient is converted to per generation, rather than per hour, which effectively results in estimating the doubling time as 1 hour at high density (from the growth rate of the ancestral wild-type clones, the doubling time in logarithmic growth is 49.6 min), and (ii) the high-density fitness benefit is in effect at all growth phases (lag, exponential, and stationary). These assumptions almost certainly cause an overestimate of the total fitness difference between the wild-type and Δcom lineages; any calculated difference is therefore an estimate of the minimal difference between the two genotypes.
In addition, assume that (iii) the upper estimate for mutation frequency (see below) is the mutation rate after adjusting to a per-generation basis and (iv) a point mutation in any gene responsible for either competency or recombination will result in the loss of transformability that was observed. Finally, for simplicity it is assumed that (v) a single mutation was required for the observed loss of transformability and that this mutation swept to fixation in the culture during the last generation of the experiment (at 730 generations).
One method to estimate the time in generations, t, for a mutation to sweep to high frequency can be given as (5)
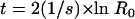 |
(3) |
The initial ratio, R0, can be calculated in two ways. The first of these requires an estimate of the mutation rate. This can be estimated as 7.5 × 10−11 generation−1 base pair−1 (from the upper measured limit of 1.5 × 10−9 spontaneous mutants generated over 20 generations [data not shown]). The number of base pairs is taken from the genes predicted to be involved in competency and recombination in the A. baylyi genome (Table 1). Together, they total 53,166 base pairs. The mutation rate is calculated as μ = 7.5 × 10−11 generation−1 base pair−1 × 53,166 base pairs = 3.99 × 10−6 generation−1. Estimates of R0 may now be made as μ/s, the fraction of mutations that remain after selection, which assumes that the mutation was present in the initial population but was initially deleterious: R0 = μ/s = 3.99 × 10−6 generation−1/0.012 generation−1 = 3.3 × 10−4. Alternatively, assume that the mutation was immediately beneficial as soon as it appeared in the first generation of the experiment. In this case, the initial frequency was 1/Ne, the effective population size was 2.3 × 106, and therefore R0 = 3.7 × 10−7.
Another approach would be to estimate the time for the mutant fraction to become 95% of the population. In this case, we can use the equation (13)
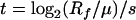 |
(4) |
in which the final ratio Rf = 0.95/0.05 and μ is estimated as described above. Because the selection coefficient is much greater than the mutation rate (s ≫ μ), selection will account for driving the mutations to fixation (13).
RESULTS AND DISCUSSION
The comFECB operon of A. baylyi strain ADP1 was replaced with a Kanr-sacB cassette, which resulted in a 10-million-fold diminished level of transformability and conferred kanamycin resistance and sucrose sensitivity (strains are listed in Table 2). The growth rates of the isogenic wild-type and ΔcomFECB::Kanr-sacB (Δcom) strains were measured, and a nested analysis of variance (ANOVA) revealed a significant difference in growth rate between the strains (Table 3) (F[1,8] = 16.2; P < 0.01). At low density the Δcom lineage had a nonsignificant apparent fitness advantage in direct competition with the wild type (s = 0.02 ± 0.06 h−1 [mean ± SD]; P = 0.22 by two-tailed t test; n = 10), while at high density, the Δcom lineage appeared to be significantly more fit (s = 0.012 ± 0.0127 h−1 [mean ± SD]; P = 0.016 by two-tailed t test; n = 10) (see Materials and Methods for a discussion of units used for fitness assays). An average of 131 colonies were counted at each of four time points for each strain, and competing strains were distinguished by resistance to either kanamycin or sucrose.
TABLE 3.
Nested ANOVA testing for differences in growth rates between ancestral and evolved Δcom and wild-type clonesa
| Clone type | Source of variation | df | SSb | MSc | Fs |
|---|---|---|---|---|---|
| Ancestral | Among genotypes | 1 | 0.009001 | 0.009001 | 16.24152 |
| Among lineages | 8 | 0.004434 | 0.000554 | 0.512541 | |
| Within lineages | 20 | 0.021625 | 0.001081 | ||
| Total | 29 | ||||
| Evolved | Among genotypes | 1 | 0.029142 | 0.029142 | 2.603207 |
| Among lineages | 8 | 0.089556 | 0.011195 | 0.050514 | |
| Within lineages | 20 | 4.432271 | 0.221614 | ||
| Total | 29 |
F0.05[1,8] = 5.318, F0.01[1,8] = 11.259, and F0.05[8,20] = 3.564.
SS, sum of squares.
MS, mean of squares.
We recognized that one-way transfer of the Δcom::Kanr-sacB marker to competent wild-type cells (rendering them identical to Δcom cells) would reveal itself in a similar way as would competitive changes in frequency through differential growth rates, and this test cannot directly discriminate between these two possible mechanisms. However, one-way genetic transfer should operate only transiently. Since A. baylyi is most competent at the transition to log-phase growth, genetic transfer is mistimed for operation in the high-density regimen.
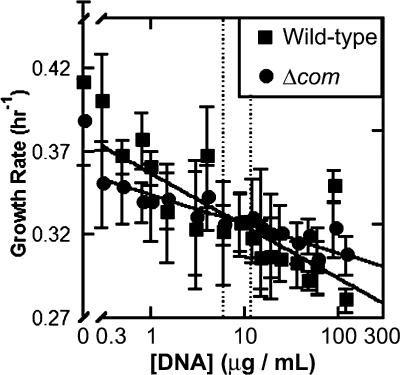
It has been argued that competency in bacteria may exist for the sake of the organism-level advantage it may confer, particularly the import of nucleotides for salvage (19, 38). Growth rate analyses of the ancestral strains were carried out in MSglc in the presence of a broad range of concentrations of genomic DNA from A. baylyi ΔilvC::Specr-sacB. Importantly, free DNA conferred no growth benefit to the competent cells; conversely, the growth rates of all strains declined in response to free DNA, with growth rates of the wild-type strains possibly declining at an increased rate relative to those of the Δcom strains (Fig. 1). This supports the previous conclusion that competence is not maintained in A. baylyi for the sake of nutrient acquisition (34).
FIG. 1.
Free DNA is detrimental to the growth of A. baylyi. Competent and noncompetent lineages were grown in minimal medium in the presence of genomic DNA from A. baylyi ΔilvC::Specr-sacB. Dashed lines indicate the range of concentrations of genomic DNA found in the medium after 16 h of growth of ancestral, competent A. baylyi. Curve fits have r2 values of 0.85 and 0.67 for Δcom and wild-type lineages, respectively; shown are means and standard deviations for five lineages.
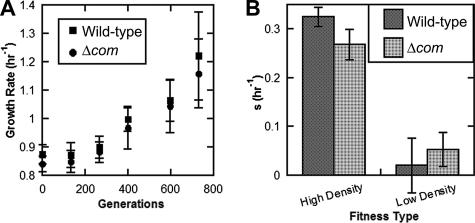
We then sought to test whether competency could be of evolutionary benefit by allowing a higher rate of adaptation. Wild-type and Δcom strains were adapted in parallel in LB at 30°C in a laboratory environment. Five clones of each genotype were selected randomly to initiate lineages. These were thereafter diluted 10,000-fold every 12 h. This allowed the cultures to reach stationary phase, resulting in a maximum population density of ∼2 × 109 CFU in 1 ml. Because the effective population (Ne) is the bottleneck size (∼2 × 105 CFU) multiplied by the number of generations that the culture is permitted to grow, serial dilution resulted in an Ne of ∼2.7 × 106. Every five serial transfers (corresponding to ∼66 generations), aliquots were stored at −80°C for later analysis (see Materials and Methods). After 55 serial transfers (∼730 generations), the growth rate had improved in all evolved populations (Fig. 2A). However, no consistent or significant differences in evolved growth rate were noted between the evolved competent and noncompetent lineages, as confirmed by a nested ANOVA (Table 3). In order to assess the fitness differences between the evolved competent and noncompetent lineages, a fitness assay was conducted in which evolved populations were competed against the ancestral clone of the opposite genotype. Fitness was determined at low density (representing the stage of fresh dilutions in the serial transfer experiment) and at high density (representing the stationary-phase culture that was inevitably achieved) (see Materials and Methods) (Fig. 2B). The fitness increase at high density was considerable and was significantly greater in the competent wild-type lineages (the difference between means is s = 0.057 ± 0.012 h−1 [mean and standard error of the mean]; P = 0.00012 by two-tailed t test; n = 10; two measurements for each of five lineages). However, the fitness increase at low density was not significantly different for the competent and noncompetent lineages (the difference between the means is s = 0.033 ± 0.021 h−1 [mean and standard error of the mean]; P = 0.13 by two-tailed t test; n = 10; two measurements for each of five lineages). As can be observed by the similar growth rates over the course of the evolution experiment (Fig. 2A), it seems likely that the population dynamics experienced by each lineage over the course of evolution were similar. In other words, even though serial transfers occurred approximately every 12 hours for the entire experiment (all 10 lineages, 5 wild-type and 5 Δcom, were diluted at the same time), lineages would have spent more time in log phase in the earlier stages of the experiment. As the growth rate increased, more time would have been spent at stationary phase awaiting the next transfer. Nevertheless, it remains unclear what about these culture conditions might have favored the wild-type lineages at high density, since A. baylyi becomes nontransformable in stationary phase. These data, along with the similarly ambiguous and mixed findings related to growth rate (Fig. 2A), suggest that neither strain has a consistent evolutionary advantage under laboratory conditions.
FIG. 2.
Competent and noncompetent Acinetobacter baylyi strain ADP1 lineages adapt equivalently to laboratory conditions. (A) Growth rates of competent and noncompetent lineages are improved to comparable extents. Shown are means and standard deviations of three measurements each for five lineages of each genotype. (B) The fitness of each evolved lineage was determined by competition with the ancestor of the opposite type at high and low densities. The high-density fitness of the evolved competent lineages had increased by ∼0.06 h−1 greater than the increase of the noncompetent lineages. Shown are the means and standard deviations for five lineages, with two replicates each.
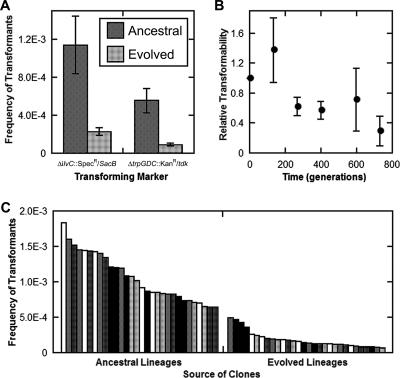
If the extent of transformability itself were to evolve, this could help reveal the role of that characteristic in evolution (6). In all five evolved competent lineages, the fraction of the population transformed in a standardized assay had decreased by 80 to 85% (Fig. 3A). Furthermore, the level of transformability decreased over the course of the selection (Fig. 3B). In order to disentangle the contributions of lineage and evolution, we performed an ANOVA to determine whether the variation observed was significant (F = 3.3; P < 0.05) (Table 4). The variation was found to be significant, which suggests that some lineages may have evolved in significantly different ways in terms of transformability. In particular, the variation at ∼130 and ∼600 generations (Fig. 3B) may indicate that some lineages were acquiring an evolutionary benefit from transformability. As they adapted to the culture conditions, the evolved populations acquired one or more mutations that resulted in the observed changes in transformability levels. These mutations may have spread either by selection against competence or by the process of unidirectional transformation from competence to noncompetence described above.
FIG. 3.
A. baylyi evolves a diminished transformability in response to adaptation to laboratory conditions. (A) Transformability in the evolved and ancestral competent lineages was assayed. Transformation was tested with two markers in order to ensure that the transforming marker was not having an effect. Regardless of marker, evolved lineages have transformability that is ∼15 to 20% of that of the ancestral lineages. Shown are means and standard deviations for five lineages. (B) Transformability of lineages over the course of the evolution experiment. Transformability was normalized to the initial fraction transformed within each lineage; means and standard deviations are shown for five lineages. (C) Six clones from each of five lineages, evolved and ancestral, were assayed for transformability. Rank-ordered clones are shown, with similar fill patterns of bars indicating clones from the same lineage.
TABLE 4.
ANOVA testing response of transformability to evolutiona
| Source of variation | df | SSb | MSc | Fs |
|---|---|---|---|---|
| Among lineages | 4 | 3.226852 | 0.806713 | 3.285856 |
| Within lineages | 20 | 4.910215 | 0.245511 | |
| Total | 24 |
F0.05[4,20] = 2.866; F0.01[4,20] = 4.431.
SS, sum of squares.
MS, mean of squares.
The diminished average transformability may have been due to complete fixation of a partial loss-of-function com mutation or to the presence of a more complete loss-of-function com mutation in a subpopulation. The majority of clones from the evolved populations have diminished transformability comparable to that measured on the population level (Fig. 3C). A more sensitive assay measured the distribution of transformability within a culture. A mixture of genomic DNAs from two donors, each with a distinct marker, was used to transform each of the evolved and ancestral competent lineages. The observed frequency of double transformants was greater than the product of the frequencies of single transformants in the ancestral lineages, suggesting that fewer than 100% of com+ cells are simultaneously competent. In cultures of the ancestral populations, 86% ± 8% (mean ± SD) of A. baylyi cells became competent under the conditions of the transformability assay. The fraction of transformable cells in the evolved competent lineages was 70% ± 30% (mean ± SD), showing no significant change in transformable fraction over the course of evolution. The variance in the level of competence within the ancestral and evolved populations, on the other hand, differed to a significant extent (F[1,8] = 21.4231; P = 0.0017). While this altered distribution of competence may have contributed to the evolution of some lineages and not others, it does not sufficiently account for the fraction that retains significant competency, which is at most 20% (Fig. 3A). Overall, then, it was observed that over the short term (e.g., ∼130 generations), some lineages may increase in transformability (Fig. 3B and Table 4). However, it appears that partial loss-of-function mutations in competence genes appear to inevitably become fixed in the population over the full course of these experiments.
Diminished transformability may have been selected because transformability was failing to provide any of a number of benefits. As demonstrated, DNA taken up by A. baylyi was not used as a nutrient (Fig. 1). Since these populations are essentially clonal, both the reduction of the mutational load and the repair of genetic damage may be considered straightforward benefits that can be derived from transformability. However, the loss of competence suggests that this was not the case, or at least that such putative benefits were not sufficiently advantageous to overcome any mechanisms that resulted in the loss of transformability.
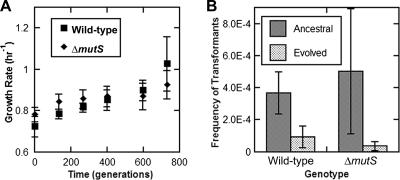
Similarly, one potential benefit of competence—acquisition of novel, immediately advantageous alleles such as, for example, antibiotic resistance in nosocomial environments—is obviated by the nearly clonal nature of the lineages. It has been shown that the benefit from sex in Chlamydomonas reinhardtii increases in larger populations due to the increased availability of mutations (8). It was thought that increasing the mutation rate might make recombination more beneficial by increasing the total number of mutations available for recombination in the population. To this end, the mutS gene of A. baylyi was disrupted with the Kanr-sacB cassette, which was in turn replaced with a clean deletion of mutS. The ΔmutS strain demonstrated ∼100-fold increased mutation frequency, as measured by the fraction of the population that spontaneously acquired rifampin resistance in 20 generations (data not shown). This is a mutation frequency comparable to what has previously been measured for disrupted mutS genes in A. baylyi (44). The growth rate of the ancestral ΔmutS lineage did not differ significantly from that of the wild-type ancestor. Five ΔmutS lineages were adapted in parallel with the five lineages of the wild-type strain under conditions similar to those in the initial serial evolution experiment. After ∼730 generations of adaptation, the growth rate had improved to comparable extents in wild-type and ΔmutS lineages (Fig. 4A). Mutation frequencies were generally unchanged over the course of evolution, although the mutation frequency of one mutS+ lineage did appear to increase mildly (∼6-fold [data not shown]). This may be indicative of a role for transformability as a source of moderate genetic variation. In principle, this could result from the uptake of single-stranded DNA, causing the induction of the SOS response and the error-prone DNA polymerases IV and V; however, A. baylyi apparently lacks lexA (3), and the expression of both recA and umuD is unusual (24, 37). This indicates that even with a dramatically increased frequency of mutation, there remains no evolutionary benefit of competence and transformation in pure laboratory cultures of A. baylyi. Finally, a decrease in transformability was again observed across all strains (Fig. 4B) (wild type, P = 0.003 by two-tailed t test [n = 5]; ΔmutS, P = 0.03 by two-tailed t test [n = 5]).
FIG. 4.
A. baylyi evolves a diminished transformability regardless of mutation frequency. (A) Over the course of adaptation to LB, wild-type and ΔmutS lineages adapted to similar extents. (B) After ∼730 generations of parallel adaptation, wild-type and ΔmutS lineages achieved similar reductions in transformability. Shown are means and standard deviations for five lineages.
An alternative reason that competency might not be of benefit could be due to the relatively benign environment (i.e., rich medium and appropriate temperature) of the selection conditions. If the population is less fit in a given environment, then more mutations are required for adaptation and it is likely that a population may harbor a greater fraction of beneficial mutations. This principle was recently demonstrated using the model organism Saccharomyces cerevisiae (22). SPO11/SPO13 mutants of yeast, which are deficient in recombination and meisosis, were adapted in parallel with wild-type yeast under conditions that included punctuated episodes of sex; all lineages were adapted to both benign and harsh laboratory conditions. Under benign conditions, no change in fitness was observed in sexual or asexual S. cerevisiae lineages. However, harsh conditions yielded both positive adaptation and an evolutionary advantage of sex.
A similar experiment was carried out with A. baylyi. The addition of 300 mM salt to LB medium resulted in a ∼35% decrease in growth rate. In addition, the temperature was increased to 40°C, resulting in an additional 15 to 25% decrease in growth rate. The temperature change affected the noncompetent lineage to a slightly greater extent, possibly due to a disruption of the surface of the bacteria that lack the competency-related pilins. Competent and noncompetent lineages of A. baylyi were adapted to these “harsh” conditions (LB plus 300 mM NaCl, 40°C) over the course of 30 serial 1:10,000 transfers, corresponding to ∼400 generations. The competent and noncompetent lineages adapted to harsh conditions in very similar manners (Fig. 5A). The growth rates in benign conditions of bacteria adapted to harsh conditions remained unchanged over the course of the experiment (Fig. 5B). The competent lineage appeared to have again evolved a diminished level of competence, in approximately half as many generations as in previous experiments (Fig. 5C) (P = 0.043 by paired t test comparing within lineages; n = 5). The lack of advantage to the competent lineages and the repeated loss of competence under all circumstances suggest that competence provides no evolutionary benefit under the specific harsh conditions examined here.
FIG. 5.
Adaptation to harsh conditions results in the evolution of diminished transformability. (A) Competent and noncompetent lineages were adapted to LB plus 300 mM NaCl at 40°C for ∼400 generations. Growth rates changed at a greater rate in the Δcom lineages than in the wild-type lineages. (B) Growth rates of competent and noncompetent lineages were measured in benign conditions over the course of adaptation to harsh conditions. Adaptation to harsh conditions elicited neither correlated benefits nor trade-offs with regard to growth rates when measured in benign conditions. (C) Similar to the case for experiments in benign conditions, adaptation to harsh conditions resulted in a diminished transformability. Shown are means and standard deviations for five lineages.
The process of sexual recombination in eukaryotes allows evolution to proceed at a rate that exceeds the mutation rate in individual lineages (8, 14). The results presented here suggest that such a rate increase is less (and possibly nonexistent) for bacterial transformation. This may be related to the difference in recombination rates between meiosis (0.25) and transformation (∼10−3/μg available DNA, as determined here [Fig. 2]). The amount of DNA free in a stationary-phase culture was determined using a transformation assay and was found to be 6 to 12 μg/ml (data not shown); the transformation rate can therefore be considered to be no more than 1.2 × 10−2, and it would be dramatically lower for much of the growth cycle. In addition to the relative proportion of mutations in a population, factors that contribute to the probability of successful transformation combining beneficial mutations into one genome include the fraction of the genome that is incorporated in any one transformation event, the relative frequencies of clones carrying beneficial mutations, and the fraction of the population that is transformable. This may explain different outcomes with A. baylyi compared with Chlamydomonas (8, 9, 28) and Saccharomyces (22, 23, 47).
Computer simulations have been used to model the characteristics of transformable cells in culture. Such models have been used to ask whether one evolutionary benefit of transformability might be the active reversion of deleterious mutations via transformation and recombination with wild-type alleles that constitute the majority of the DNA pool (39). However, simulations of a mixed culture of transformable and nontransformable bacteria suggested that any potential benefit of tranformability would be overwhelmed by the rate of acquisition of com alleles that result in a loss of transformability in previously com+ lineages (40). While DNA from Com− cells can transform Com+ cells, making them Com−, the opposite process is impossible by definition (Com− cells cannot be transformed). Therefore, mixed populations of Com+ and Com− cells (whether derived by spontaneous mutation or by deliberate introduction) should tend to become homogeneously Com−, all else being equal. This biased transfer effect is similar in many ways to the effects attributed to alleles that alter their own segregation dynamics in diploid species (“molecular drive”) (15, 16). This process should theoretically operate in the absence of selection against Com+ and possibly even overwhelm selective pressure that favors Com+. We sought to test whether the data presented here fulfill this prediction.
One way to carry this out would be to estimate the number of generations required for the observed loss of competency mutations to sweep the population. With the observed fitness difference of 0.012 h−1 and provided that several assumptions are made (see Materials and Methods), two equations can be used to estimate the number of generations required for these mutations to sweep the population (5, 11). By the use of equation 3, we can calculate the number of generations required for a sweep of the loss of competency by using two estimates of the initial fraction, R0, of the population that has lost competency (see Materials and Methods). If R0 is a fraction of mutants in the initial population, then t = 1,335, nearly twice the number of generations than were found for the loss of transformability. If the mutation that conferred the loss of competence was present once in the initial population, then t = 2,468, more than three times the number of generations that it took in the described evolution experiment for the loss of transformability to go to high frequency. Similarly, by the use of equation 4, we can calculate that the number of generations required for 95% of the population to carry the mutation is t = 1,861, more than twice the number of actual generations. Predicting the number of generations required for the loss of transformability to go to high frequency demonstrates that a nonselective mechanism is acting to favor com alleles over wild-type com+ alleles. This mechanism seems likely to be the unidirectional transfer of com alleles as predicted by modeling efforts.
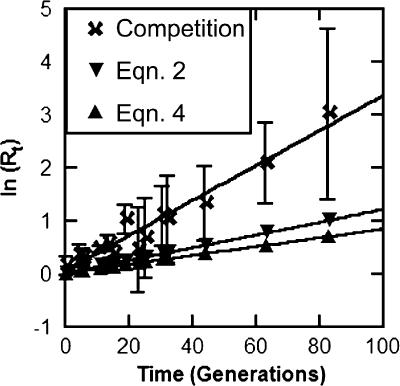
The ancestral wild-type and Δcom strains were allowed to compete in serial transfer (incorporating high- and low-density regimens) for >80 generations (Fig. 6). The apparent competitive fitness of the Δcom strains exceeds that predicted based on equation 2 (see Materials and Methods), in which R, the ratio of noncompetent to competent bacteria, was determined based on the selection coefficient. Equation 4 was also used to predict the ratio of noncompetent to competent bacteria in culture, based on the mutation rate and the selection coefficient (Fig. 6). These data show that the ratio of competent and noncompetent variants changes at approximately three times the rate expected based on two methods to calculate the ratio using the high-density fitness differential that was observed. An important difference between this repeated serial transfer competition experiment and those in which the high-density fitness value was determined is the cycling between phases of growth (corresponding to the regimens in which high fitness and low fitness were measured). The competence of A. baylyi is known to be dependent on growth phase (35); if there is a significant transfer of Δcom alleles, this experiment will enhance the ability to observe that effect relative to the strictly high-density fitness experiment. These results suggest that any potential evolutionary benefits derived from transformation are exceeded by the risk of loss of transformability in a mixed culture of wild-type and com strains (40).
FIG. 6.
Noncompetent lineages of A. baylyi overtake competent lineages more rapidly than predicted in direct competition. The rate at which the noncompetent lineages overtake the population is higher than predicted by two equations: (i) using equation 2, which was used to determine selection coefficients in the initial competitions, assuming that s = 0.012 generation−1 (see Materials and Methods) (29), and (ii) using equation 4 (see Materials and Methods) (13). Shown are the means and standard deviations for five competitions, as well as predicted values based on the two equations discussed.
It is also possible that the loss of transformability was partly due to a corresponding increase in fitness. It is unusual for populations to lose metabolic functions with such high reproducibility, unless such a loss results in a fitness benefit. For example, long-term experimental evolution of Escherichia coli found changes in niche breadth such that 75% of informative substrates did not show parallel decay in the first 10,000 generations of adaptation (12). However, mutations in d-ribose catabolism and spoT were both shown to be important in this adaptation, resulting in selection coefficients of 0.014 and 0.094, respectively (10, 13). These sorts of genes show parallel decay in serial transfer experiments. Similarly, mutations in rpoS have been shown to be critical to growth advantage in stationary phase (18, 46). The repeated loss of transformability by the competent lineages under three selection regimens may suggest that transformability is costly. We cannot strictly conclude that either unidirectional genetic transfer or adaptive evolution was the sole mechanism by which transformability so quickly diminished in these evolutionary experiments. Comparisons of independent growth rates suggest that selection alone is not responsible for loss of competence in these serial cultures. However, independent measurement of growth rate does not always correlate perfectly with competitive fitness, and hence this method still leaves significant room for doubt. On the other hand, in mixed populations of competent wild-type cells and mutants with diminished competency (which will certainly arise spontaneously), com alleles should inexorably spread through horizontal transfer unless strongly counterselected. Whatever the reason for the loss of transformability, it must be noted that these results may for the moment be interpreted only in the context of the laboratory, with limited environmental and genetic diversity. The evolutionary benefit conferred on an organism by competence remains to be discovered, and A. baylyi is an ideal organism to carry out such investigations.
Acknowledgments
We thank Rosie Redfield, Graham Bell, Cliff Zeyl, and Jim Bull for helpful comments and discussion, as well as Christian Hansen for discussions concerning statistical analyses. We thank Paul Schimmel for guidance and support.
This work was funded by NSF grant MCB-0128901 to V.D.C.-L. We also acknowledge support to Paul Schimmel from NIH grant GM23562 and the National Foundation for Cancer Research.
Footnotes
Published ahead of print on 6 October 2006.
REFERENCES
- 1.Ausubel, F. M. 1997. Short protocols in molecular biology: a compendium of methods from Current Protocols in Molecular Biology, 3rd ed. Wiley, New York, N.Y.
- 2.Bacher, J. M., V. de Crecy-Lagard, and P. R. Schimmel. 2005. Inhibited cell growth and protein functional changes from an editing-defective tRNA synthetase. Proc. Natl. Acad. Sci. USA 102:1697-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbe, V., D. Vallenet, N. Fonknechten, A. Kreimeyer, S. Oztas, L. Labarre, S. Cruveiller, C. Robert, S. Duprat, P. Wincker, L. N. Ornston, J. Weissenbach, P. Marliere, G. N. Cohen, and C. Medigue. 2004. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 32:5766-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell, G. 1982. The masterpiece of nature: the evolution and genetics of sexuality. University of California Press, Berkeley.
- 5.Bell, G. 1997. Selection: the mechanism of evolution. Chapman & Hall, New York, N.Y.
- 6.Burt, A. 2000. Perspective: sex, recombination, and the efficacy of selection—was Weismann right? Evol. Int. J. Org. Evol. 54:337-351. [DOI] [PubMed] [Google Scholar]
- 7.Busch, S., C. Rosenplanter, and B. Averhoff. 1999. Identification and characterization of ComE and ComF, two novel pilin-like competence factors involved in natural transformation of Acinetobacter sp. strain BD413. Appl. Environ. Microbiol. 65:4568-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colegrave, N. 2002. Sex releases the speed limit on evolution. Nature 420:664-666. [DOI] [PubMed] [Google Scholar]
- 9.Colegrave, N., O. Kaltz, and G. Bell. 2002. The ecology and genetics of fitness in Chlamydomonas. VIII. The dynamics of adaptation to novel environments after a single episode of sex. Evol. Int. J. Org. Evol. 56:14-21. [DOI] [PubMed] [Google Scholar]
- 10.Cooper, T. F., D. E. Rozen, and R. E. Lenski. 2003. Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc. Natl. Acad. Sci. USA 100:1072-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper, V. S., A. F. Bennett, and R. E. Lenski. 2001. Evolution of thermal dependence of growth rate of Escherichia coli populations during 20,000 generations in a constant environment. Evol. Int. J. Org. Evol. 55:889-896. [DOI] [PubMed] [Google Scholar]
- 12.Cooper, V. S., and R. E. Lenski. 2000. The population genetics of ecological specialization in evolving Escherichia coli populations. Nature 407:736-739. [DOI] [PubMed] [Google Scholar]
- 13.Cooper, V. S., D. Schneider, M. Blot, and R. E. Lenski. 2001. Mechanisms causing rapid and parallel losses of ribose catabolism in evolving populations of Escherichia coli B. J. Bacteriol. 183:2834-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Visser, J. A. G. M., C. W. Zeyl, P. J. Gerrish, J. L. Blanchard, and R. E. Lenski. 1999. Diminishing returns from mutation supply rate in asexual populations. Science 283:404-406. [DOI] [PubMed] [Google Scholar]
- 15.Dover, G. 2002. Molecular drive. Trends Genet. 18:587-589. [DOI] [PubMed] [Google Scholar]
- 16.Dover, G. 1982. Molecular drive: a cohesive mode of species evolution. Nature 299:111-117. [DOI] [PubMed] [Google Scholar]
- 17.Dubnau, D. 1991. Genetic competence in Bacillus subtilis. Microbiol. Rev. 55:395-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farrell, M. J., and S. E. Finkel. 2003. The growth advantage in stationary-phase phenotype conferred by rpoS mutations is dependent on the pH and nutrient environment. J. Bacteriol. 185:7044-7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finkel, S. E., and R. Kolter. 2001. DNA as a nutrient: novel role for bacterial competence gene homologs. J. Bacteriol. 183:6288-6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedrich, A., T. Hartsch, and B. Averhoff. 2001. Natural transformation in mesophilic and thermophilic bacteria: identification and characterization of novel, closely related competence genes in Acinetobacter sp. strain BD413 and Thermus thermophilus HB27. Appl. Environ. Microbiol. 67:3140-3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fussenegger, M., T. Rudel, R. Barten, R. Ryll, and T. F. Meyer. 1997. Transformation competence and type-4 pilus biogenesis in Neisseria gonorrhoeae—a review. Gene 192:125-134. [DOI] [PubMed] [Google Scholar]
- 22.Goddard, M. R., H. C. Godfray, and A. Burt. 2005. Sex increases the efficacy of natural selection in experimental yeast populations. Nature 434:636-640. [DOI] [PubMed] [Google Scholar]
- 23.Greig, D., R. H. Borts, and E. J. Louis. 1998. The effect of sex on adaptation to high temperature in heterozygous and homozygous yeast. Proc. Biol. Sci. 265:1017-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hare, J. M., S. N. Perkins, and L. A. Gregg-Jolly. 2006. A constitutively expressed, truncated umuDC operon regulates the recA-dependent DNA damage induction of a gene in Acinetobacter baylyi strain ADP1. Appl. Environ. Microbiol. 72:4036-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herzberg, C., A. Friedrich, and B. Averhoff. 2000. comB, a novel competence gene required for natural transformation of Acinetobacter sp. BD413: identification, characterization, and analysis of growth-phase-dependent regulation. Arch. Microbiol. 173:220-228. [DOI] [PubMed] [Google Scholar]
- 26.Hofreuter, D., S. Odenbreit, J. Puls, D. Schwan, and R. Haas. 2000. Genetic competence in Helicobacter pylori: mechanisms and biological implications. Res. Microbiol. 151:487-491. [DOI] [PubMed] [Google Scholar]
- 27.Juni, E., and A. Janik. 1969. Transformation of Acinetobacter calcoaceticus (Bacterium anitratum). J. Bacteriol. 98:281-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaltz, O., and G. Bell. 2002. The ecology and genetics of fitness in Chlamydomonas. XII. Repeated sexual episodes increase rates of adaptation to novel environments. Evol. Int. J. Org. Evol. 56:1743-1753. [DOI] [PubMed] [Google Scholar]
- 29.Lenski, R. E. 1991. Quantifying fitness and gene stability in microorganisms. Biotechnology 15:173-192. [DOI] [PubMed] [Google Scholar]
- 30.Lenski, R. E., M. R. Rose, S. C. Simpson, and S. C. Tadler. 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Nat. 138:1315-1341. [Google Scholar]
- 31.Link, C., S. Eickernjager, D. Porstendorfer, and B. Averhoff. 1998. Identification and characterization of a novel competence gene, comC, required for DNA binding and uptake in Acinetobacter sp. strain BD413. J. Bacteriol. 180:1592-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maynard Smith, J. 1978. The evolution of sex. Cambridge University Press, Cambridge, United Kingdom.
- 33.Metzgar, D., J. M. Bacher, V. Pezo, J. Reader, V. Doring, P. Schimmel, P. Marliere, and V. de Crecy-Lagard. 2004. Acinetobacter sp. ADP1: an ideal model organism for genetic analysis and genome engineering. Nucleic Acids Res. 32:5780-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palmen, R., P. Buijsman, and K. J. Hellingwerf. 1994. Physiological regulation of competence induction for natural transformation in Acinetobacter calcoaceticus. Arch. Microbiol. 162:344-351. [Google Scholar]
- 35.Palmen, R., B. Vosman, P. Buijsman, C. K. Breek, and K. J. Hellingwerf. 1993. Physiological characterization of natural transformation in Acinetobacter calcoaceticus. J. Gen. Microbiol. 139:295-305. [DOI] [PubMed] [Google Scholar]
- 36.Porstendorfer, D., U. Drotschmann, and B. Averhoff. 1997. A novel competence gene, comP, is essential for natural transformation of Acinetobacter sp. strain BD413. Appl. Environ. Microbiol. 63:4150-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rauch, P. J., R. Palmen, A. A. Burds, L. A. Gregg-Jolly, J. R. van der Zee, and K. J. Hellingwerf. 1996. The expression of the Acinetobacter calcoaceticus recA gene increases in response to DNA damage independently of RecA and of development of competence for natural transformation. Microbiology 142:1025-1032. [DOI] [PubMed] [Google Scholar]
- 38.Redfield, R. J. 2001. Do bacteria have sex? Nat. Rev. Genet. 2:634-639. [DOI] [PubMed] [Google Scholar]
- 39.Redfield, R. J. 1988. Evolution of bacterial transformation: is sex with dead cells ever better than no sex at all? Genetics 119:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Redfield, R. J., M. R. Schrag, and A. M. Dean. 1997. The evolution of bacterial transformation: sex with poor relations. Genetics 146:27-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richaud, C., D. Mengin-Lecreulx, S. Pochet, E. J. Johnson, G. N. Cohen, and P. Marliere. 1993. Directed evolution of biosynthetic pathways. Recruitment of cysteine thioethers for constructing the cell wall of Escherichia coli. J. Biol. Chem. 268:26827-26835. [PubMed] [Google Scholar]
- 42.Thomas, C. M., and K. M. Nielsen. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3:711-721. [DOI] [PubMed] [Google Scholar]
- 43.Vaneechoutte, M., D. M. Young, L. N. Ornston, T. De Baere, A. Nemec, T. Van Der Reijden, E. Carr, I. Tjernberg, and L. Dijkshoorn. 2006. Naturally transformable Acinetobacter sp. strain ADP1 belongs to the newly described species Acinetobacter baylyi. Appl. Environ. Microbiol. 72:932-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Young, D. M., and L. N. Ornston. 2001. Functions of the mismatch repair gene mutS from Acinetobacter sp. strain ADP1. J. Bacteriol. 183:6822-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Young, D. M., D. Parke, and L. N. Ornston. 2005. Opportunities for genetic investigation afforded by Acinetobacter baylyi, a nutritionally versatile bacterial species that is highly competent for natural transformation. Annu. Rev. Microbiol. 59:519-551. [DOI] [PubMed] [Google Scholar]
- 46.Zambrano, M. M., D. A. Siegele, M. Almiron, A. Tormo, and R. Kolter. 1993. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science 259:1757-1760. [DOI] [PubMed] [Google Scholar]
- 47.Zeyl, C., and G. Bell. 1997. The advantage of sex in evolving yeast populations. Nature 388:465-468. [DOI] [PubMed] [Google Scholar]