Abstract
Inorganic carbon (IC), such as bicarbonate or carbon dioxide, stimulates the growth of Lactobacillus plantarum. At low IC levels, one-third of natural isolated L. plantarum strains are nutritionally dependent on exogenous arginine and pyrimidine, a phenotype previously defined as high-CO2-requiring (HCR) prototrophy. IC enrichment significantly decreased the amounts of the enzymes in the pyrimidine biosynthetic pathway encoded by the pyrR1BCAa1Ab1DFE operon, as demonstrated by proteomic analysis. Northern blot and reverse transcription-PCR experiments demonstrated that IC levels regulated pyr genes mainly at the level of transcription or RNA stability. Two putative PyrR regulators with 62% amino acid identity are present in the L. plantarum genome. PyrR1 is an RNA-binding protein that regulates the pyr genes in response to pyrimidine availability by a mechanism of transcriptional attenuation. In this work, the role of PyrR2 was investigated by allelic gene replacement. Unlike the pyrR1 mutant, the ΔpyrR2 strain acquired a demand for both pyrimidines and arginine unless bicarbonate or CO2 was present at high concentrations, which is known as an HCR phenotype. Analysis of the IC- and pyrimidine-mediated regulation in pyrR1 and pyrR2 mutants suggested that only PyrR2 positively regulates the expression levels of the pyr genes in response to IC levels but had no effect on pyrimidine-mediated repression. A model is proposed for the respective roles of PyrR1 and PyrR2 in the pyr regulon expression.
Lactobacillus plantarum is an aerotolerant, homofermentative lactic acid bacterium (11). Its growth is stimulated by increasing supplies of inorganic carbon (IC), such as carbon dioxide or its predominant soluble form in cells, bicarbonate. An optimal growth rate was obtained when 4% CO2 was present in the gas phase or when 2 g/liter KHCO3 was directly added to the culture media (1). L. plantarum strains can be naturally present along with CO2-producing cells, such as heterofermentative lactic acid bacteria, yeast in sourdough or beer, or eukaryotic cells in the gastrointestinal tract. L. plantarum is a versatile bacterium with metabolic abilities that favor its adaptation to a large range of biotopes (17). The nutritional richness of its biotopes has allowed L. plantarum, like other lactic acid bacteria, to evolve toward auxotrophy for several amino acids, vitamins, and nucleobases. When the arginine and pyrimidine requirements of 150 L. plantarum strains isolated from various origins were analyzed, nutritional needs were found to be strain specific and modulated by the addition of bicarbonate or CO2. While most strains were prototrophs, one-third of the strains were defined as high-CO2-requiring (HCR) prototrophs for both arginine and pyrimidine, i.e., these strains were prototrophs only when CO2 partial pressure was increased or bicarbonate was added to the medium (5). These observations suggested that IC would modulate L. plantarum arginine and pyrimidine metabolism either by changing the activities of enzymes with CO2-bicarbonate-related substrates or products or by regulating the expression levels of the enzyme-encoding genes.
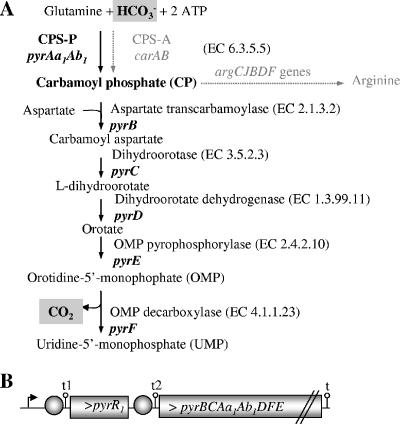
Bicarbonate is the substrate for carbamoyl phosphate synthesis, an enzymatic step common in the biosynthesis pathways of arginine and pyrimidine. This step is catalyzed by the heterodimeric carbamoyl phosphate synthases (CPS) (Fig. 1A). Two CPS are functional in L. plantarum: CPS-P, encoded by pyrAa1Ab1, and CPS-A, encoded by carAB (20). The expression of CPS-P is regulated by pyrimidine availability, whereas CPS-A expression is subject to regulation by arginine, both at the transcriptional level. In the arginine pathway, carbamoyl phosphate is a substrate for citrulline synthesis catalyzed by the ornithine transcarbamoylase ArgF. In UMP synthesis, carbamoyl phosphate is fused to aspartate, yielding carbamoyl aspartate catalyzed by the aspartate transcarbamoylase PyrB. The genes involved in arginine and pyrimidine biosynthesis have been genetically characterized in the prototrophic strain CCM 1904 and mutant derivatives (4, 18-20). Bicarbonate concentrations affected the activities of CPS-A and CPS-P differently, as demonstrated with ΔpyrAa1Ab1 and ΔcarAB mutants. At low CO2 concentrations, such as those in ordinary air, CPS-A activity was too low to provide carbamoyl phosphate for arginine and pyrimidine biosynthesis, as the ΔpyrAa1Ab1 mutant FB331 did not grow. On the other hand, CPS-P could satisfy the carbamoyl phosphate requirement for both arginine and pyrimidine biosynthesis pathways since the ΔcarAB mutant FB335 was a prototroph. When CPS-P expression was repressed by uracil, the wild-type prototroph strain CCM 1904 was unable to grow in the absence of arginine. The resulting phenotype, referred to as the uracil-sensitive phenotype, is due to cellular arginine depletion (20). In addition, the growth rate of the L. plantarum prototroph strain was reduced in ordinary air compared to that in bicarbonate-enriched air (1). All these observations revealed that the limiting reactant in carbamoyl phosphate synthesis in ordinary air may result from limiting amounts of either the substrate bicarbonate or the enzyme CPS-P.
FIG. 1.
UMP and arginine biosynthetic pathways in L. plantarum. (A) The pyrBCAa1Ab1DFE operon codes for the enzymes involved in UMP synthesis. For each step, the corresponding enzyme and gene are indicated. CPS-P stands for carbamoyl phosphate synthase pyrimidine-regulated protein. The carAB operon codes for CPS-A, the carbamoyl phosphate synthase arginine-regulated protein (20). (B) Genetic organization of pyr genes. Flag, transcriptional start; t, transcriptional terminator; circle, transcriptional attenuators as defined previously (19).
The CPS-P-encoding pyrAa1Ab1 is cotranscribed with the pyrR1BCAa1Ab1DFE operon, encoding the enzymes catalyzing UMP synthesis (Fig. 1B) (19). The first gene in the operon encodes the transcriptional repressor PyrR1. This protein was demonstrated to regulate the transcription of the pyr operon in response to pyrimidine availability in L. plantarum, most probably by transcriptional attenuation (19), a mechanism first elucidated in Bacillus subtilis (23). A second copy of the pyrR gene was found in the L. plantarum genome (12). PyrR2 shares 62% identity at the amino acid level with PyrR1 (19), suggesting that PyrR2 might also be a pyr operon regulator. In B. subtilis, a single copy of PyrR is present, and when activated by UMP, PyrR is able to stabilize the formation of an RNA loop (the anti-antiterminator loop) formed upstream of the coding sequence. This binding favors the formation of a terminator loop leading to transcriptional termination. When UMP pools are low, PyrR binding is less efficient and the formation of the antiterminator is favored, preventing termination. In L. plantarum, two such attenuators have been identified upstream of pyrR1 and between pyrR1 and pyrB. A similar attenuator was also found upstream of pyrP, a monocistronic, pyrimidine-repressed gene located elsewhere in the genome that codes for a uracil permease (2). Low pyrimidine-independent expression of the pyr operon was obtained by antiterminator site-directed mutagenesis. The resulting strain, AE1023, had reduced UTP and CTP pools and could grow in the absence of pyrimidines and arginine only if the bicarbonate concentration was high, which corresponds to the HCR phenotype (19). Therefore, repression of the pyr operon prevents the ability of L. plantarum to grow at low carbon dioxide levels (19). Since the HCR phenotype was linked to lower pyr expression, it became evident that it was necessary to investigate whether CO2-bicarbonate concentration affects pyr gene expression in L. plantarum.
In this work, pyr gene expression in response to CO2-bicarbonate concentrations was tested using a proteomic approach as well as by quantification of pyr transcripts in the wild type and mutants of each of the two PyrR regulator-like orthologs. PyrR2 was found to regulate pyr genes in response to IC independently of pyrimidine availability.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
L. plantarum subsp. plantarum CCM 1904 and NC8 are wild-type arginine and uracil prototrophs; they and their derivatives (Table 1) were grown on MRS agar plates (Difco Laboratories) at 30°C. Liquid cultivations were performed in agitated DLA chemically defined medium (4). When required, uracil and arginine were added at 50 μg/liter. IC-enriched medium was obtained either by adding KHCO3 at a final concentration of 2 g/liter, with pH adjusted to 6.5 by using 2 M HCl, or by increasing the CO2 tension in the gas phase to 4%. To prevent CO2 loss during incubation when CO2 or KHCO3 was added, the Erlenmeyer flasks were closed with gas-tight corks. To obtain 4% CO2 in the gas phase, 10.8 ml of pure CO2 gas was injected with a sterile syringe through the gas-tight cork of an Erlenmeyer flask (internal volume, 320 ml; volume of gas phase, 270 ml; liquid volume, 50 ml). For RNA and protein studies, cells were first cultivated in DLA without uracil in normal-air-agitated precultures. These precultures were used as an inoculum for 50-ml cultures in DLA: under “growing” conditions, the inoculum was added to obtain an optical density at 600 nm (OD600) of 0.05 and the culture was stopped when the OD600 reached 0.5 (approximately 4 h and two generations in the exponential growth phase); under “nongrowing” conditions (with the presence of uracil and no inorganic carbon enrichment), cells were inoculated to obtain an OD600 of 0.4 and incubated for 4 h.
TABLE 1.
List of studied strains
| Strain | Genotype | Comment(s)a | Source or reference |
|---|---|---|---|
| AE1026 | pyrR1 | PyrR1 D104Y mutation in CCM 1904 | 19 |
| CCM 1904 | Wild type | Prototroph | 5 |
| FB335 | ΔcarAB | Site mutagenesis in CCM 1904 | 20 |
| U17 | ΔcarABΔpyrR1 | Derivative of FB335 | 19 |
| U32 | ΔcarAB pyrR1 | PyrR1 truncated from Q17-E180 in FB335 | 19 |
| NC8 | Wild type | Prototroph | 5 |
| FB421 | pyrR1 | PyrR1 D104Y mutated in NC8 | This work |
| FB422 | ΔpyrR2 | Entire pyrR2 coding sequence deletion in NC8 | This work |
Sequence data refer to EMBL accession number Z54240.
Mutagenesis by allelic exchange in NC8.
Mutagenesis by allelic replacement of pyr genes has been successfully used for L. plantarum with pGID023-derived plasmids such as plasmid pAE1020, which harbors the pyrR1 mutation (19). Another derivative of pGID023, plasmid pΔpyrR2, was designed to delete pyrR2 from its start codon until its stop codon. This plasmid was constructed as follows. Two amplifications, using L. plantarum CCM 1904 genomic DNA template and primer set Lp1781f1/ΔpyrR2r or primer set ΔpyrR2f/Lp1783r1, were performed. Since primers ΔpyrR2r and ΔpyrR2f have an overlapping 25-nucleotide-long sequence (Table 2), the two PCR products were merged by a second PCR with primers Lp1781f1/Lp1783r1. The resulting 1.15-kb PCR product was PstI-EcoRI restricted and cloned into the linearized PstI-EcoRI pGID023 vector. For allelic gene replacement, the ΔpyrR2 allele present in plasmid pΔpyrR2 was electroporated into the two wild-type prototroph L. plantarum strains, NC8 and CCM1904. However, no transformants were obtained with CCM 1904, which has reduced electro-competence. The selection of the recombinants harboring the mutated allele was performed as previously described (20). The constructs were confirmed by sequencing the corresponding PCR-amplified chromosome locus. The NC8-derivative strains FB421 and FB422 contained the pyrR1 and pyrR2 mutations, respectively.
TABLE 2.
Primer list
| Genetic locusa | PCR product parameter
|
Primer names and sequences (5′→3′)b | |
|---|---|---|---|
| Use | Size (bp) | ||
| pyrP (AJ012720) | P probe and RT-PCR | 298 | pyrP-g4, AATCATTCACAAAAAGGGGTGCG (F); pyrP-d2, TCATCAGAGACTGCATCACAACG (R) |
| pyrR2 (AJ617795) | pyrR2 mutant | 596 | lp1781f1, ACTTACTGCAGTACGTCGCACTCGTTCACGGAG (F); ΔpyrR2r, TGACTTTGTCGTAAATTGCCTCCCAATTTGTGG (R) |
| pyrR2 mutant | 579 | ΔpyrR2f, TGGGAGGCAATTTACGACAAAGTCAAACAGATG (F); lp1783r1, GCCAAAGAATTCGATGACCACGACATTGAGC (R) | |
| pyrR1 (Z54240) | R1 probe | 240 | 2072, CTGACAAACTAATGGCACG (F); 2068, ACCGCATGATGATCATCCCG (R) |
| RT-PCR | 379 | pyrR631, CAATTAAACAAGCACCTTTAAC (F); 2068, ACCGCATGATGATCATCCCG (R) | |
| pyrAb1 (Z54240) | RT-PCR or Ab1#1 probe | 364 | N58, CAGCTTCTGGTATTGCGC (F); 2005, ATCTTCAAAGCAGCATCCCG (R) |
| RT-PCR or Ab1#2 probe | 291 | N49′, TCAAAGTTGAATCGTGTGG (F); 2019, AATCATGCTCGCCACTATCG (R) | |
| pyrB (Z54240) | B probe and RT-PCR | 260 | N1, TGAGAATAGCACCCGCACAC (F); 2042, TGTGACGGATGTTGCCCAC (R) |
| pyrD (Z54240) | D probe | 313 | N35, TGATGAATACGTGGCAGTCG (F); N41, GATAAACCGCCCGTACCATG (R) |
| rrn (AL935263) | RT-PCR or rrn probe | 400 | 530f, GTGCCAGCAGCCGCGG (F); 907r, CCGTCAATTCCTTTGAGTTT (R) |
The EMBL accession numbers are shown in parentheses.
F, Forward primer; R, reverse primer. Underlined sequences show the PstI and EcoRI restriction sites.
Protein extractions and separations.
Cells were grown in 50 ml DLA medium (see above) and harvested by centrifugation at an OD600 of 0.5. The cell pellet was washed once in 10 mM Tris, 1 mM EDTA, pH 7.6, and resuspended in 600 μl of the same buffer. Cells were disrupted by grinding them with glass beads (0.3 mm in diameter; 1 g per 0.6-ml extract) at the maximal speed in an MM2 mixer mill (Retsh Haan, Germany).
For first-dimension (1D) gel electrophoresis, the cells were disrupted by grinding them for 6 min, followed by centrifugation. Proteins (40 μg) of the supernatant were diluted in Laemmli buffer (50 mM Tris-HCl, pH 6.8, 2 mM EDTA, 1% sodium dodecyl sulfate [SDS], 1% β-mercaptoethanol, 10% glycerol, 0.01% bromophenol blue) (1:1), separated by electrophoresis in 10% acrylamide gel, and stained using Coomassie blue (Amersham Pharmacia Biotech, General Electric, Germany).
For two-dimensional (2D) gel electrophoresis studies, prior to being ground (six cycles, each with 1 min shaking and a 1-min pause at 4°C), cells were incubated for 30 min at 4°C in the presence of 1 mM phenylmethylsulfonyl fluoride and benzonase (348 units; Roche) to prevent protease degradation and nucleic acid contamination. The lyzate was decanted overnight at 4°C, and the clear lysate was centrifuged at 14,000 × g for 1 h at 4°C. Protein concentration in the supernatant was quantified using the method of Bradford (3), standardized with known concentrations of bovine serum albumin, and subsequently adjusted to 3 mg/ml using Tris-EDTA (TE) buffer. Two hundred fifty microliters of rehydratation buffer (7 M urea, 2 M thiourea) purified by mixing for 1 hour with 10 g/liter Amberlite IRN-150L (Amersham Pharmacia Biotech, Germany), 2% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 0.5% immobilized pH gradient (IPG) buffer (pH range, 4 to 7), 3.4 mg/ml dithiothreitol (DTT), and a few bromophenol blue grains was added to the protein extracts (100 μl). Proteins were first separated according to pI. Proteins (300 μg) were loaded on 18-cm IPG strips (linear gradient pH, 4 to 7) with the IPGphor isoelectric focusing system as recommended by the manufacturer (Amersham Pharmacia Biotech, Germany). Rehydratation (6 h at 0 V and 6 h at 30 V) was followed by four 2-hour increments in which the voltage was increased stepwise to 150 V, 500 V, 1,000 V, and 3,000 V. Finally, separation was obtained using 8,000 V until a minimum of 45,000 V·h was reached. Strips were frozen at −80°C for at least 1 day, thawed at room temperature, and placed in the equilibration solution (50 mM Tris-HCl, pH 8.8, 6 M urea, 30% glycerol, 2% SDS, and traces of bromophenol blue) two times for 15 min, first in the presence of 10 mg/ml DTT and then in the presence of 25 mg/ml iodoacetamide. The second-dimension gel experiment was performed with an 11.5% SDS-polyacrylamide gel electrophoresis (PAGE) gel. Proteins were stained with brilliant blue G-colloidal (SIGMA). Computer analysis was performed using the ImageMaster 2D platinum program (Amersham Pharmacia Biotech); 16 gels were grouped in two classes according to the two tested conditions (ordinary air or a 4% CO2-enriched atmosphere), and each class contained 8 independent gels. After spot detection (around 300 spots per gel), gels were matched with one reference gel (extracts from cells cultivated in ordinary air). For each spot, the relative volume corresponded to the normalized volume of the spot compared to the normalized volume of the entire gel coloration. The statistical analysis was performed by calculating for each spot the Student t value and the “ratio” (the average for 8 relative volumes obtained at low CO2 concentrations divided by the average for 8 relative volumes obtained at high CO2 concentrations). Only spots sharing Student t values higher than 1.4 (corresponding to a P value of <0.1) and ratios higher than 2 were analyzed by mass spectrometry (MS). The selected spots were cut from 1D or 2D gel and stored at −20°C.
Protein digestion and mass spectrometry.
For in-gel digestion, picked spots were washed with 100 μl of 25 mM NH4HCO3 buffer and dehydrated with 100 μl of acetonitril (ACN), and the two operations were repeated twice. The samples were vacuum dried for 10 min, reduced (100 μl of 10 mM 1,4-DTT-25 mM NH4HCO3 buffer at 56°C for 1 h), and alkylated (100 μl of 25 mM iodoacetamide-25 mM NH4HCO3 buffer at room temperature in the dark for 1 h). After three washes for 5 min each in 25 mM NH4HCO3 and ACN alternately, samples were vacuum dried and rehydrated overnight at room temperature in the presence of trypsin (3 volumes of 12.5 ng trypsin [V5111; Promega]/μl in 25 mM NH4HCO3 buffer freshly diluted). Tryptic peptides were extracted from the gels by sonication for 30 min in 5 μl of 35% H2O-60% ACN-5% HCOOH. Mass measurements were performed with a BIFLEX III matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) MS (Bruker Daltonics, Bremen, Germany) equipped with SCOUT high-resolution optics with an X-Y multisample probe and griddle reflectors. This instrument was used in positive-ion-reflector mode with a maximum accelerating potential of 19 kV. A saturated solution of α-cyano-4-hydroxycinnamic acid (Sigma, Saint Louis, MO) in acetone was used as a matrix. Spreading and fast evaporation of 0.5 μl of matrix solution yielded a fine layer of crystals, on which a droplet of 0.5 μl of 5% aqueous HCOOH solution was mixed first with 0.5 μl of peptide-containing digest medium and then with 0.3 μl saturated matrix solution (in 50% H2O-50% ACN). The preparation was vacuum dried and washed once with 0.7 μl of 5% aqueous HCOOH. Mass spectra were internally calibrated with trypsin autolysis peaks (m/z = 842.510 and m/z = 2,211.105). Monoisotopic peptide masses were assigned, and the peak list was transferred to the MS BioTools program (Bruker Daltonics, Bremen, Germany) and then compared to data in the NCBI protein database by using MASCOT software (Matrix Science, London, United Kingdom). Tryptic mass searches retained only data with up to one missed tryptic cleavage and optional methionine oxidation, with mass accuracy limited to 50 ppm.
Transcription analysis.
RNA extraction and semiquantitative reverse transcription (RT)-PCR protocols have been previously described (18). RT-PCR was performed with primers listed in Table 2, using optimized amounts of total RNA to target different transcripts (20 ng for pyrAb1 and rrn, 30 ng for pyrP, and 150 ng for pyrR1 and pyrB). For Northern hybridizations, DNA probes were PCR amplified (95°C for 1 min, followed by 35 three-step cycles at 94°C for 40s, 50°C for 40s, and 72°C for 2 min; the program was completed with a postelongation step at 72°C for 10 min), using primers listed in Table 2. The PCR products were digoxigenin labeled using a digoxigenin-labeling kit (Roche). For each probe, the optimal amount of total RNA to be used was tested (0.1 μg for rrn, 5 μg for pyrD, 2 μg for pyrAb1 probe Ab1#1 [see Fig. 4A] or Ab1#2, and 10 μg for pyrR1 and pyrB). Slot blot hybridizations were performed and quantified as described previously (2), with Quantity One software (Bio-Rad). At least three independent hybridizations were performed for each tested condition. The background level was subtracted, and the relative signal was calculated as follows: the measured signal for each probe was divided by the signal obtained with the rrn probe to obtain the relative amount for each pyr gene. Under the tested conditions (uracil and IC supplementation or not), the constitutive expression levels of the rrn genes were checked using the upp gene, whose transcription is not regulated by IC and pyrimidine availability (2; data not shown).
FIG. 4.
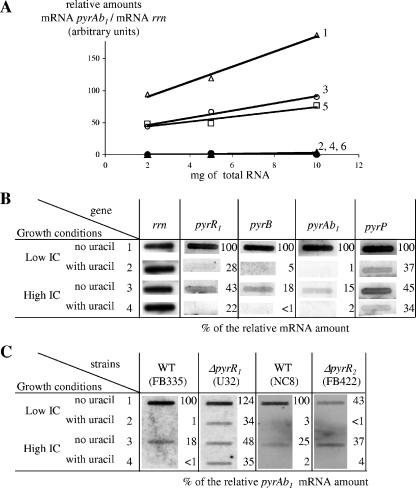
Transcription of pyr genes in response to IC and uracil. Slot blot hybridizations detecting pyr mRNA were performed with probes specific to pyr genes (described in Table 2) and standardized with probe rrn, specific to 16S RNA encoding genes. The relative amounts (arbitrary units) were calculated by dividing the signal for each pyr probe by the signal obtained with probe rrn. RNAs were extracted from cells cultivated in DLA under different conditions, in normal air (1 and 2), in a 4% CO2-enriched atmosphere (3 and 4), with KHCO3 added at 2 g/liter (5 and 6), without uracil (1, 3, and 5), or with uracil (2, 3, and 6). (A) Quantification test with probe Ab1#1, with different amounts of total RNA prepared from strain CCM1904. (B) Transcription of different pyr genes in the wild-type strain CCM 1904. (C) Gene pyrAb1 transcription efficiencies in strains harboring wild-type or mutated pyrR1 or pyrR2 genes. Relative amounts calculated for different growth conditions were compared to relative amounts obtained for the corresponding wild-type isogenic strain grown at low IC levels without uracil added (condition marked 1). Each “% of the relative mRNA amount” is therefore expressed as a percentage of the amount obtained for condition 1, defined as 100%. Names of strains are indicated in parentheses.
Nucleotide sequence accession number.
The pyrR1BCAa1Ab1DFE operon and pyrR2 genes were sequenced in strain NC8. These sequences have been submitted to the EMBL database and assigned the accession numbers AM228716 and AM228717, respectively.
RESULTS
The expression of the pyr operon is repressed at high bicarbonate concentrations.
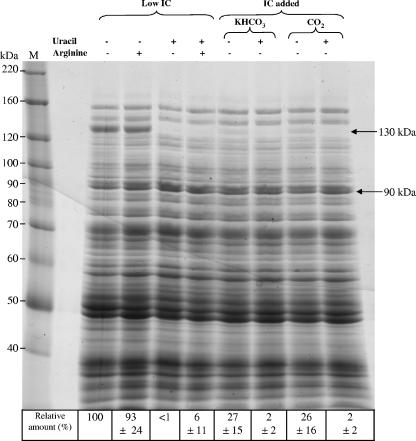
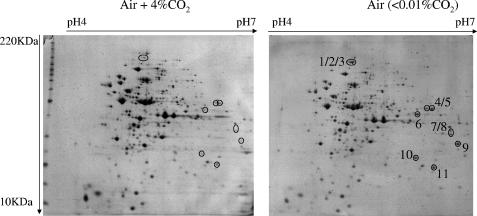
The effect of IC supplementation on gene expression was first analyzed by 1D gel electrophoresis (Fig. 2). Proteins were extracted from L. plantarum CCM 1904 cultivated in DLA medium with either CO2 or KHCO3 in combination with uracil and arginine. The amount of one protein with an apparent mass of 130 kDa decreased fourfold upon addition of CO2 or KHCO3. Arginine addition had no effect, whereas exogenous uracil addition led to a 10- to 100-fold decrease in the concentration of the 130-kDa protein. The 130-kDa protein band obtained under three different growth conditions was cut from the gel for mass spectrometry protein identification (Table 3) . In all three cases, the best scores matched those of the L. plantarum large-chain carbamoyl phosphate synthase PyrAb1 (EMBL database accession no. P77886). In a second set of experiments, proteins were separated using 2D gel electrophoresis (Fig. 3). Two growth conditions for the wild-type strain CCM 1904 were compared: a 4% CO2-enriched atmosphere and a low-IC condition found in ordinary air. Spots with the most-significant differential expression levels were excised from the gel. The corresponding proteins were identified using MALDI-TOF mass spectrometry. Among those, the PyrAb1 protein previously identified in the 1D gel experiment was found (Table 3, spots 1, 2, and 3). Moreover, the expression levels of all the other proteins encoded by the pyrR1BCAa1Ab1DFE operon, except PyrF, were decreased in response to IC (Table 3, spots 4 to 11). With a pI of 7.7, PyrF was excluded from the gels that targeted proteins with pI values between 4 and 7.
FIG. 2.
SDS-PAGE protein patterns in response to inorganic carbon, arginine, and uracil. Proteins were extracted from L. plantarum wild-type strain CCM 1904 under different growth conditions: in ordinary air (Low IC), with KHCO3 added at 2 g/liter, in a 4% CO2-enriched atmosphere, or with arginine and uracil (+). M, protein size marker (BenchMark protein ladder; Invitrogen). The 90-kDa constitutive band of an unknown protein served as the internal standard for quantification of the signal corresponding to PyrAb1 (band shown at 130 kDa). Numbers indicate the relative amounts of the PyrAb1 polypeptide expressed as percentages of the value obtained for cells cultivated at low IC levels, in the absence of arginine and uracil. The values shown are averages for at least four independent experiments.
TABLE 3.
Mass spectrometry analysis of IC-regulated proteins observed either by 1D or 2D gel electrophoresis
| Proteina | Genea | Spotb | Gel type | Protein characteristicc
|
Mass spectrometry valued
|
Air/IC ratio (P)f | |||
|---|---|---|---|---|---|---|---|---|---|
| pI | MM (Da) | Error (ppm) | Coverage (%) | Score | |||||
| Carbamoyl phosphate synthase (large subunit) | pyrAb1 | 1 | 1D | 4.93 | 115,757 | 20 | 46 | 484 | - |
| 2 | 1D | 4.93 | 115,757 | 22 | 56 | 429 | - | ||
| 5 | 1D | 4.93 | 115,757 | 25 | 47 | 305 | - | ||
| 1 | 2D | 4.93 | 115,757 | 31 | 42 | 334 | 4.7 (1.5) | ||
| 2 | 2D | 4.93 | 115,757 | 42 | 54 | 371 | 4.7 (1.5) | ||
| 3 | 2D | 4.93 | 115,757 | 42 | 56 | 458 | 4.7 (1.5) | ||
| Dihydroorotase | pyrC | 4 | 2D | 5.76 | 45,677 | 40 | 52 | 123 | 4.3 (1.9) |
| 5 | 2D | 5.76 | 45,677 | 44 | 64 | 130 | 4.3 (1.9) | ||
| Carbamoyl phosphate synthase (small subunit) | pyrAa1 | 6 | 2D | 5.67 | 40,143 | 49 | 48 | 212 | 2.5 (1.8) |
| Aspartate carbamoyltransferase | pyrB | 7 | 2D | 6.06 | 34,689 | 50 | 63 | 189 | 5.4 (2.2) |
| 8 | 2D | 6.06 | 34,689 | 38 | 62 | 205 | 5.4 (2.2) | ||
| Dihydroorotate dehydrogenase | pyrD | 9 | 2D | 6.14 | 31,318 | 51 | 53 | 139 | 5.5 (2.6) |
| Orotate phosphoribosyltransferase | pyrE | 10 | 2D | 5.64 | 22,691 | 50 | 72 | 147 | 3.3 (3.4) |
| Transcriptional repressor | pyrR1 | 11 | 2D | 5.64 | 19,812 | NDe | 71 | 127 | 2.0 (1.6) |
EMBL accession no. Z54240.
For 1D gels, numbers refer to growth conditions (1, no IC added; 2, with arginine; 5, addition of 2g KHCO3/liter), and for 2D gels, numbers refer to spots shown in Fig. 3.
pI, Isoelectric point; MM, molecular mass.
Score, probability-based Mowse score calculated using MASCOT software (Matrix Science, London, United Kingdom); error, mass accuracy; coverage, percentage of the protein sequence covered by the matched peptide.
Internal calibration not determined.
P values were determined by Student's t test. -, results are shown in Fig. 2.
FIG. 3.
Comparison of proteomic profiles at low and high inorganic carbon levels. L. plantarum wild-type strain CCM 1904 was cultivated in defined DLA medium (in the absence of arginine or uracil), with agitation in CO2-enriched or ordinary air. Spots corresponding to the proteins encoded by the pyr genes are circled and identified by numbers referenced in Table 3. Sizes of protein markers (BenchMark protein ladder; Invitrogen) are shown in 10-kDa increments from 10 to 220 kDa.
These results clearly suggest that the expression levels of the genes in the pyr operon are subject to regulation by bicarbonate-CO2 availability in either a direct or an indirect manner.
IC-dependent transcription of the pyr genes.
The pyrR1BCAa1Ab1DFE and pyrP genes are part of the pyr regulon, whose transcription is regulated in response to exogenous pyrimidine (2, 19). Transcription regulation of the pyr regulon in response to IC concentration was assessed using slot blot hybridization and RT-PCR amplifications. For this study, five pyr genes were chosen because of their genetic positions: four genes within the pyr operon, pyrR1 located upstream of the first attenuator, pyrB located just after the second attenuator, and pyrAb1 and pyrD located in the middle and at the end of the operon, respectively. The last gene was pyrP, a gene not found in the pyr operon (Fig. 1B). For each probe, the intensity of the signal was proportional to the amount of spotted RNA (Fig. 4A for pyrAb1, data not shown for other pyr genes). For each pyr gene, the detected mRNA amount decreased significantly when RNA originated from cells incubated under IC enrichment conditions, either by increasing the gas phase CO2 partial pressure or by adding KHCO3 to the medium (Fig. 4A for pyrAb1; also Fig. 4B, compare condition 1 and 3 or 5). After quantification, the “IC regulation factor” (the relative amount obtained at low IC levels divided by the relative amount obtained at high IC levels) was in the order of five- to sixfold for the pyrBCAa1Ab1DFE operon and of twofold for both the pyrR1 and the pyrP genes (Fig. 4B). Arginine addition had no detectable effect on the transcription levels of the tested pyr genes (data not shown). As expected from characterized uracil-repressed genes, in the presence of exogenous uracil, transcription of pyrAb1, pyrB (Fig. 4, compare conditions 1 and 2) and pyrD (data not shown) was repressed in the order of 20- to 100-fold, whereas pyrR1 and pyrP transcription was reduced 3-fold. Similar IC-mediated repression levels were obtained using semiquantitative RT-PCR experiments (data not shown), with specific primer sets detecting pyrAb1, pyrB, pyrP, and pyrR1 transcripts. These results are in agreement with the observations made by proteomics that the expression of the pyr regulon responds to changes in bicarbonate and CO2 concentration. This finding adds to the previously known regulation that occurs in response to pyrimidine availability.
IC-responding regulation of the pyr genes did not require a functional PyrR1.
Both pyrimidine and IC concentrations regulate the transcription of the pyr genes. Since PyrR1 is known as the repressor of the pyr operon in response to exogenous pyrimidines (19), the role of PyrR1 in IC-responding regulation was first analyzed by comparing the phenotypes of the wild-type and pyrR1 strains in the genetic context of two prototroph strains, CCM 1904 and NC8 (Table 1). PyrR1 displays 99.4% amino acid identity in these two strains. Like wild-type strains, pyrR1 mutants were prototrophs and had similar nutritional IC requirements (19). The expression levels of the pyr genes in response to IC levels were estimated by quantification of the amounts of PyrAb1 after SDS-PAGE. Several pyrR1 mutants were tested in the genetic context of CCM 1904 and its ΔcarAB-derivative strain FB335; the CCM 1904-derivative strain AE1026 harbors the D104Y mutation in PyrR1, and strains U32 and U17 are ΔpyrR1 ΔcarAB mutants (19). The same patterns of regulation were obtained for strains CCM 1904 and FB335, suggesting that deletion of the carAB genes had no detectable effect on the tested IC- and uracil-responding regulation. In all three pyrR1 mutants, the regulations in response to uracil or IC levels were similar (data not shown). In addition, similar results were also obtained when wild-type strains NC8 or CCM1904 or their pyrR1 derivatives (AE1026 and FB421, respectively) were compared (data not shown). Thus, PyrR1 plays the same role in the L. plantarum wild-type prototroph CCM 1904 and NC8 strains.
We used strain U32 (ΔpyrR1) to quantify the amounts of pyrBCAa1Ab1DFE transcript by using slot blot hybridization with probes specific to pyrAb1 and pyrB (Fig. 4C and data not shown). The IC regulation factors were similar in the reference strain FB335 (factors 5 and 6) and in the pyrR1 U32 mutant (factors 3 to 5) both at the protein and at the mRNA levels. Nevertheless, in the absence of uracil and in normal air, we observed that pyr expression in the pyrR1 mutant was increased compared to that in the wild type, indicating that the pyr operon is not fully induced in the absence of pyrimidines. As expected from previous work, uracil-dependent repression was found to be PyrR1 dependent: a 100-fold repression was observed with the wild-type strain (Fig. 4C, compare lanes 1 and 2), but only a 3- to 4-fold repression was observed with mutated pyrR1. Finally, the amounts of detected pyrAb1 mRNA in pyrR1 mutants in the presence of uracil were similar whether the cells grew at low or high IC concentrations (Fig. 4C, compare lanes 2 and 4), as observed in wild-type strains. These observations suggested that PyrR1 has no effect on pyr operon repression in response to IC concentration and, moreover, is not the sole sensor of pyrimidine availability.
pyrR2 deletion confers higher CO2 needs for growth in the absence of arginine and uracil.
A pyrR2 mutant was constructed to test the effect of PyrR2 on the ability of L. plantarum to grow at low IC concentrations and its role in IC-dependent regulation. The function of pyrR2 was analyzed in strain FB422, a Δ pyrR2 derivative of NC8. The effects of the addition of CO2, arginine, and uracil on the growth of CCM1904 and NC8 were analyzed. The ΔpyrR2 mutant was an HCR prototroph (5), since it was unable to grow in the absence of uracil and arginine in ordinary air but grew in 4% CO2-enriched air. Thus, pyrR2 deletion confers the HCR phenotype.
IC-mediated regulation of pyr genes depends on a functional PyrR2.
The impact of pyrR2 deletion on pyrBCAa1Ab1DFE operon expression was assessed both at the protein level by comparing the PyrAb1 polypeptide expression pattern (data not shown) and at the transcriptional level by quantifying transcripts with probes specific for pyrAb1 (Fig. 4C) or pyrB (data not shown). In the mutant strain in ordinary air, the expression levels of these genes, and by assumption that of the entire pyr operon, were reduced compared to that of the wild type (Fig. 4C, condition 1). Moreover, whereas the expression of the pyr genes was two- to sixfold repressed by IC in the wild-type strain, their level of expression in the pyrR2 mutant was unaffected by changes in IC availability (Fig. 4C, strain FB422 in conditions 1 and 3). Deletion of the pyrR2 gene resulted in reduced pyr operon transcription under low-IC conditions, with loss of the observed IC-dependent regulation. This suggested that PyrR2 is required for maximal pyr gene expression when IC concentration is low.
A functional PyrR2 is not involved in the pyrimidine-dependent repression of the pyr operon.
The role of PyrR2 in pyrimidine dependent repression of the pyr operon was analyzed by quantification of the protein PyrAb1 (SDS-PAGE data not shown) and of the transcripts of genes pyrAb1 (Fig. 4C) and pyrB (data not shown). In the presence of uracil, the same level of repression was obtained for the wild type and for the ΔpyrR2 strains (Fig. 4C). These results showed that pyrR2 is not involved in pyrimidine-dependent repression of the pyr operon.
DISCUSSION
The transcription-dependent regulation of the pyr operon in response to exogenous pyrimidines has been extensively studied (23). Using proteomic and transcriptional quantitative analysis, this work gives evidence of transcription regulation of the pyr operon in response to inorganic carbon but not to arginine.
Inorganic carbon levels modulate the transcription of pyr genes.
Similar levels of regulation were obtained either by adding CO2 to the gas phase or by adding KHCO3 to the medium. The values for IC-mediated repression calculated at the levels of protein and RNA were similar (four- to fivefold for the pyrBCAa1Ab1DFE operon, twofold for pyrR1 and pyrP). Thus, inorganic carbon concentrations are sensed by the cell to regulate pyr gene expression mainly at the level of transcription or RNA stability for at least two noncontiguous loci (the pyr operon and the pyrP gene). Regulation of gene expression in response to IC has been observed in phototrophs and chemolithotrophs (Rhodobacter sphaeroides, Ralstonia eutropha, and cyanobacteria) in genes involved in inorganic carbon assimilation (14) but also in chemoorganotrophs (Bacillus anthracis, Escherichia coli, group A streptococci, Pseudomonas spp., and Staphylococcus aureus) (10, 13, 22), in general stress or in purine metabolism in cyanobacteria (21, 26), and in amino acid decarboxylation in E. coli (24). To our knowledge, this report constitutes the first description that proteins involved in pyrimidine biosynthesis are regulated in response to IC. Differential proteomic experiments with increased detection thresholds (two-dimensional differential gel electrophoresis) are currently in progress to further characterize IC-responding genes in L. plantarum.
Only PyrR2 is involved in the IC-mediated regulation of pyr gene expression.
Analysis of knockout mutants in each gene of the two L. plantarum PyrR homologs shows that PyrR1 is involved in the uracil-mediated repression but not in the IC response (model proposed in Fig. 5). As a pyrR1 mutant does not lead to total loss of pyrimidine-dependent regulation (19) (Fig. 4C), another unknown mechanism controls pyr mRNA levels in response to intracellular pyrimidine nucleotide pools. Neither of the two pyrimidine-responding regulations involved PyrR2. It is not uncommon to find two different regulatory mechanisms that control CPS expression in response to pyrimidine availability. For instance, in Escherichia coli, two independent, pyrimidine-specific regulatory systems controlling carAB have been identified: UTP-sensitive reiterative transcription, which accounts for 30% of the repression (9), and a multiprotein-dependent regulation at the level of transcription initiation (7).
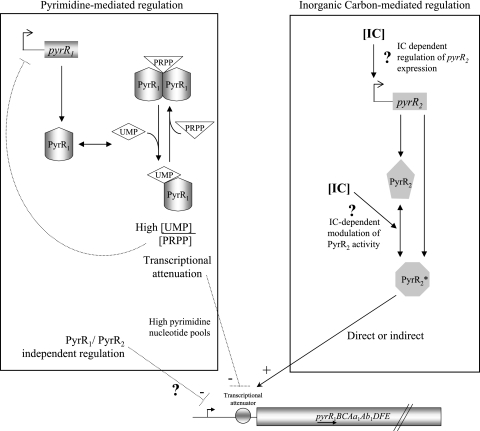
FIG. 5.
Model for PyrR1 and PyrR2 regulation of L. plantarum pyr regulon in response to pyrimidine availability and inorganic carbon. The regulated pyr genes studied include the pyrR1BCAa1Ab1DFE operon and pyrP. The gray circle schematizes pyr gene cis transcription regulatory elements that are involved in response to pyrimidine availability and IC at the DNA or the mRNA level. The activity of the RNA-binding PyrR1 protein is regulated by binding to antagonist effectors such as 5-phospho-d-ribosyl-1-pyrophosphate (PRPP) and UMP. The UMP-PyrR1 complex binds to an attenuation site of the pyr mRNA, leading to terminated transcription (19). Another unknown mechanism independent of PyrR1 and PyrR2 activity operates under conditions of elevated intracellular pyrimidine nucleotide levels. IC regulation may occur at the level of pyrR2 expression or the gene product activity. PyrR2* represents the functional PyrR2 regulator.
PyrR2 positively regulates pyr gene expression in response to a decrease in IC concentrations. The involvement of a regulator belonging to the PyrR regulator family in IC-responding regulation has never been described. It is not clear how L. plantarum PyrR2 senses the IC level or how the IC response is mediated. A first hypothesis would be that PyrR2 modulates PyrR1 activity by forming heteromers, as proposed for the other regulators found at two copies in L. plantarum, ArgR1 and ArgR2. Both ArgR1 and ArgR2 are required for the arginine-responding repression of the arg regulon, probably by forming a heterohexamer (18). PyrR homologs are known to repress gene expression by binding to RNA loops, leading to transcriptional termination. PyrR from Bacillus caldolyticus, B. subtilis, Thermus thermophilus, or Mycobacterium tuberculosis forms dimers with identically concave, basic surfaces, presumably the RNA-binding sites (6). Thus, it could be speculated that L. plantarum PyrR1/PyrR2 form a heterodimer that is unable to promote transcriptional termination, leading to increased expression of the pyr operon. This hypothesis predicts that a pyrR2 mutant would have reduced pyr gene transcription, as more PyrR1 homodimer would be available to favor termination. Another prediction is that at low IC levels, this mutant would have altered pyrimidine-mediated regulation, but such phenotype was not observed. Reciprocally, in a pyrR1 mutant, a loss of the IC response would be expected but was not observed. Since an important clue came from the observation that the IC dependent regulation is independent of PyrR1, if a PyrR1/PyrR2 heterodimer were formed, it would have no major role in IC-mediated or pyrimidine-mediated regulation. PyrR2 is highly similar to PyrR1, suggesting that they both recognize the mRNA. Since PyrR2 acts as an activator, one could speculate that PyrR2, instead of stabilizing the anti-antiterminator loop as PyrR1, prevents the formation of the anti-antiterminator, thus favoring antitermination.
Our model proposes that PyrR1 and PyrR2 regulate the expression levels of pyr genes by two independent mechanisms (Fig. 5). Two possible mechanisms are proposed to explain the contribution of PyrR2 to the increased expression of the pyr regulon at low IC levels. In addition to the above-proposed model in which PyrR2 directly prevents the formation of the anti-antiterminator, PyrR2 could indirectly modulate the transcription of the pyr genes, by modulating the expression of a regulator specifically dedicated to IC response, which in turn would regulate pyr gene transcription. To date, gene transcription in response to IC in bacteria involves transcription activators: regulators of the LysR-family, such as CbbR (14) and AtxA (8, 16), or two-component signal transducing systems, such as MgA (15). Since PyrR homologs have been described as repressors (25), PyrR2 may be a repressor that controls the expression of another repressor when IC is present at low concentrations.
Based on this model, a double pyrR1 pyrR2 mutant would have the HCR phenotype with reduced pyr gene expression independently of the IC concentration in the media. Moreover, pyr genes would be expressed even in the presence of uracil. As previously stated, residual pyrimidine regulation is observed in the pyrR1 mutant. Therefore, the levels of pyr gene expression in the pyrR1 pyrR2 double mutant would be similar in the presence and in the absence of IC, whereas the residual pyrimidine regulation would still be present. Further experiments are required to identify the molecular mechanism of PyrR2-mediated regulation in response to IC concentration.
Is IC-mediated regulation of the pyr operon impaired in HCR auxotrophs?
IC availability is a limiting parameter for pyrimidine and arginine metabolism in lactobacilli since HCR prototrophs were commonly isolated in natural environments (5). These isolates require high CO2 levels in the absence of arginine and pyrimidine for their growth. Under low-IC conditions (ordinary air), it was assumed that the CPS-A activity was too low to produce enough carbamoyl phosphate for arginine or pyrimidine biosynthesis pathways, and the CPS-P produced the majority of carbamoyl phosphate for both pathways (20). Recently, mutations were introduced in the regulatory region of the pyr operon, encoding the CPS-P. These mutations led to constitutive repression of pyr gene expression (19). The resulting strain (strain AE1023) had an HCR phenotype. Therefore, it was proposed that constitutively low pyr gene expression prevented growth at low IC concentrations. In this work, we have shown that pyr gene expression is higher when IC concentration is low. One could suppose that under this condition, the level of the CPS-P enzymatic reaction is lower because the concentration of one of its substrates is reduced. Therefore, the amount of the CPS-P enzyme required to produce enough carbamoyl phosphate would be larger under conditions of low IC than under those of enriched IC. Reduced pyr gene expression under low-IC conditions (as, for example, in mutant AE1023) reduces CPS-P amounts, resulting in a shortage of the carbamoyl phosphate required for arginine and pyrimidine biosynthesis, which confers the HCR phenotype. In this work, we have shown that PyrR2 is required to reach maximal transcriptional efficiency when the IC level is low. Moreover, the ΔpyrR2 mutant has the HCR phenotype with a reduced amount of CPS-P compared to that in the wild type strain when the IC level is low. All these observations are in agreement with the hypothesis that reduced carbamoyl phosphate pools due to low pyr gene expression would lead to the HCR phenotype. Therefore, the impaired integrity of the pyrR2 gene as well as the activity of the PyrR2 protein in the HCR auxotrophs may be the molecular basis that confers the HCR phenotype in some auxotrophs isolated from natural environments (5).
In this work, we demonstrated that the two copies of PyrR homologs in L. plantarum were functional and responded to different stimuli. To unravel microbial adaptation to IC environment, future work should investigate the role of PyrR-like proteins in microorganisms with several PyrR homologs, as detected not only in the genome of L. plantarum but also in a few other gram-positive species (data not shown), all found so far to belong to the Lactobacillus genus, such as Lactobacillus acidophilus, Lactobacillus delbrueckii, Lactobacillus gasseri, and Lactobacillus johnsonii.
Acknowledgments
This work was supported by CNRS. 2D equipment was purchased with Université Louis Pasteur funding to Marie-Claire Lett. Jan Martinussen received a fellowship under the OECD cooperative research program for biological resource management of sustainable agriculture systems.
We thank Christelle Guillier, Philippe Hamman, and the Plateforme proteomique laboratory as well as Hélène Diemer and Alain Van Dorsselaer for the mass spectrometry analysis. We thank Brett Johnson for his help in writing the manuscript.
Footnotes
Published ahead of print on 13 October 2006.
REFERENCES
- 1.Arsène-Ploetze, F., and F. Bringel. 2004. Role of inorganic carbon on lactic acid bacteria metabolism. Lait 84:45-59. [Google Scholar]
- 2.Arsène-Ploetze, F., H. Nicoloff, B. Kammerer, J. Martinussen, and F. Bringel. 2006. Uracil salvage pathway in Lactobacillus plantarum: transcription and genetic studies. J. Bacteriol. 188:4777-4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Bringel, F., L. Frey, S. Boivin, and J. C. Hubert. 1997. Arginine biosynthesis and regulation in Lactobacillus plantarum: the carA gene and the argCJBDF cluster are divergently transcribed. J. Bacteriol. 179:2697-2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bringel, F., and J. C. Hubert. 2003. Extent of genetic lesions of the arginine and pyrimidine biosynthetic pathways in Lactobacillus plantarum, L. paraplantarum, L. pentosus, and L. casei: prevalence of CO2 dependent auxotrophs and characterization of deficient arg genes in L. plantarum. Appl. Environ. Microbiol. 69:2674-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chander, P., K. M. Halbig, J. K. Miller, C. J. Fields, H. K. Bonner, G. K. Grabner, R. L. Switzer, and J. L. Smith. 2005. Structure of the nucleotide complex of PyrR, the pyr attenuation protein from Bacillus caldolyticus, suggests dual regulation by pyrimidine and purine nucleotides. J. Bacteriol. 187:1773-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devroede, N., N. Huysveld, and D. Charlier. 2006. Mutational analysis of intervening sequences connecting the binding sites for integration host factor, PepA, PurR, and RNA polymerase in the control region of the Escherichia coli carAB operon, encoding carbamoylphosphate synthase. J. Bacteriol. 188:3236-3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drysdale, M., A. Bourgogne, and T. M. Koehler. 2005. Transcriptional analysis of the Bacillus anthracis capsule regulators. J. Bacteriol. 187:5108-5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han, X., and C. L. Turnbough, Jr. 1998. Regulation of carAB expression in Escherichia coli occurs in part through UTP-sensitive reiterative transcription. J. Bacteriol. 180:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herbert, S., S. W. Newell, C. Lee, K. P. Wieland, B. Dassy, J. M. Fournier, C. Wolz, and G. Doring. 2001. Regulation of Staphylococcus aureus type 5 and type 8 capsular polysaccharides by CO2. J. Bacteriol. 183:4609-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kandler, O., and N. Weiss. 1986. Regular, nonsporing gram-positive rods, p. 1208-1260. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. The Williams and Wilkins Co., Baltimore, Md. [Google Scholar]
- 12.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koehler, T. M. 2002. Bacillus anthracis genetics and virulence gene regulation. Curr. Top. Microbiol. Immunol. 271:143-164. [DOI] [PubMed] [Google Scholar]
- 14.Kusian, B., and B. Bowien. 1997. Organization and regulation of cbb CO2 assimilation genes in autotrophic bacteria. FEMS Microbiol. Rev. 21:135-155. [DOI] [PubMed] [Google Scholar]
- 15.McIver, K. S., and R. L. Myles. 2002. Two DNA-binding domains of Mga are required for virulence gene activation in the group A Streptococcus. Mol. Microbiol. 43:1591-1601. [DOI] [PubMed] [Google Scholar]
- 16.Mignot, T., E. Couture-Tosi, S. Mesnage, M. Mock, and A. Fouet. 2004. In vivo Bacillus anthracis gene expression requires PagR as an intermediate effector of the AtxA signalling cascade. Int. J. Med. Microbiol. 293:619-624. [DOI] [PubMed] [Google Scholar]
- 17.Molenaar, D., F. Bringel, F. H. Schuren, W. M. de Vos, R. J. Siezen, and M. Kleerebezem. 2005. Exploring Lactobacillus plantarum genome diversity using microarrays. J. Bacteriol. 187:6119-6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicoloff, H., F. Arsène-Ploetze, C. Malandain, M. Kleerebezem, and F. Bringel. 2004. Two arginine repressors regulate arginine biosynthesis in Lactobacillus plantarum. J. Bacteriol. 186:6059-6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicoloff, H., A. Elagöz, F. Arsène-Ploetze, B. Kammerer, J. Martinussen, and F. Bringel. 2005. Repression of the pyr operon in Lactobacillus plantarum prevents its ability to grow at low carbon dioxide levels. J. Bacteriol. 187:2093-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicoloff, H., J. C. Hubert, and F. Bringel. 2000. In Lactobacillus plantarum, carbamoyl phosphate is synthesized by two carbamoyl-phosphate synthetases (CPS): carbon dioxide differentiates the arginine-repressed from the pyrimidine-regulated CPS. J. Bacteriol. 182:3416-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwarz, R., J. Lieman-Hurwitz, M. Hassidim, and A. Kaplan. 1992. Phenotypic complementation of high CO2-requiring mutants of the Cyanobacterium Synechococcus sp. strain PCC 7942 by inosine 5′ monophosphate. Plant Physiol. 100:1987-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stretton, S., and A. E. Goodman. 1998. Carbon dioxide as a regulator of gene expression in microorganisms. Antonie Leeuwenhoek 73:79-85. [DOI] [PubMed] [Google Scholar]
- 23.Switzer, R. L., R. J. Turner, and Y. Lu. 1999. Regulation of the Bacillus subtilis pyrimidine biosynthetic operon by transcriptional attenuation: control of gene expression by an mRNA-binding protein. Prog. Nucleic Acid Res. Mol. Biol. 62:329-367. [DOI] [PubMed] [Google Scholar]
- 24.Takayama, M., T. Ohyama, K. Igarashi, and H. Kobayashi. 1994. Escherichia coli cad operon functions as a supplier of carbon dioxide. Mol. Microbiol. 11:913-918. [DOI] [PubMed] [Google Scholar]
- 25.Tomchick, D. R., R. J. Turner, R. L. Switzer, and J. L. Smith. 1998. Adaptation of an enzyme to regulatory function: structure of Bacillus subtilis PyrR, a pyr RNA-binding attenuation protein and uracil phosphoribosyltransferase. Structure 6:337-350. [DOI] [PubMed] [Google Scholar]
- 26.Wang, H. L., B. L. Postier, and R. L. Burnap. 2004. Alterations in global patterns of gene expression in Synechocystis sp. PCC 6803 in response to inorganic carbon limitation and the inactivation of NdhR, a LysR family regulator. J. Biol. Chem. 279:5739-5751. [DOI] [PubMed] [Google Scholar]