Abstract
Pseudomonas syringae pv. tabaci 6605 possesses a genetic region involved in flagellin glycosylation. This region is composed of three open reading frames: orf1, orf2, and orf3. Our previous study revealed that orf1 and orf2 encode glycosyltransferases; on the other hand, orf3 has no role in posttranslational modification of flagellin. Although the function of Orf3 remained unclear, an orf3 deletion mutant (Δorf3 mutant) had reduced virulence on tobacco plants. Orf3 shows significant homology to a 3-oxoacyl-(acyl carrier protein) synthase III in the fatty acid elongation cycle. The Δorf3 mutant had a significantly reduced ability to form acyl homoserine lactones (AHLs), which are quorum-sensing molecules, suggesting that Orf3 is required for AHL synthesis. In comparison with the wild-type strain, swarming motility, biosurfactant production, and tolerance to H2O2 and antibiotics were enhanced in the Δorf3 mutant. A scanning electron micrograph of inoculated bacteria on the tobacco leaf surface revealed that there is little extracellular polymeric substance matrix surrounding the cells in the Δorf3 mutant. The phenotypes of the Δorf3 mutant and an AHL synthesis (ΔpsyI) mutant were similar, although the mutant-specific characteristics were more extreme in the Δorf3 mutant. The swarming motility of the Δorf3 mutant was greater than that of the ΔpsyI mutant. This was attributed to the synergistic effects of the overproduction of biosurfactants and/or alternative fatty acid metabolism in the Δorf3 mutant. Furthermore, the amounts of iron and biosurfactant seem to be involved in biofilm development under quorum-sensing regulation in P. syringae pv. tabaci 6605.
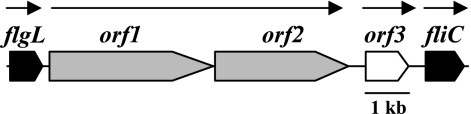
Pseudomonas syringae pv. tabaci 6605, an phytopathogenic bacterial isolate, causes wildfire disease on host tobacco plants and induces a hypersensitive reaction (HR) on nonhost plants. In previous studies, we demonstrated that flagellin, a component of the flagellar filament, is a major elicitor of HR by P. syringae pv. tabaci 6605 (30, 35). Flagellin of P. syringae pv. tabaci 6605 induces HR on nonhost plants but not on its host tobacco plant. Although the deduced amino acid sequence of P. syringae pv. glycinea race 4 flagellin (FliC) is identical to that of P. syringae pv. tabaci 6605 flagellin, the flagellin of P. syringae pv. glycinea does induce HR on tobacco plants, suggesting that posttranslational modification of flagellin determines the specificity to induce HR (34). We recently reported that genes existing upstream of fliC, which encodes flagellin protein, are involved in the glycosylation of flagellin in these two pathovars (33, 36). There are three open reading frames, namely, orf1, orf2, and orf3, in the glycosylation islands of P. syringae pv. tabaci and pv. glycinea (Fig. 1). Because the proteins encoded by orf1 and orf2 showed homology to a putative glycosyltransferase and the molecular mass of the flagellin of each orf1 and orf2 deletion mutant (Δorf1 and Δorf2 mutants) was decreased, these genes are thought to encode the glycosyltransferases. Inoculation of these mutants into each host plant confirmed that glycosylation of flagellin proteins plays an important role in their virulence (33, 36). While the Δorf3 mutants of P. syringae pv. tabaci also had significantly reduced virulence on host tobacco plants, mass spectrometry analysis indicated that orf3 is not involved in posttranslational modification (33).
FIG. 1.
Schematic organization of the glycosylation island in the flagellum gene cluster of Pseudomonas syringae pv. tabaci and pv. glycinea. flgL, gene encoding HAP3; orf1 and orf2, genes encoding glycosyltransferases for flagellin glycosylation; orf3, putative gene encoding 3-oxoacyl-(acyl carrier protein) synthase III, fliC, gene encoding flagellin. Arrows indicate putative transcripts and directions of transcription.
A homology search revealed that orf3 is highly homologous to the putative 3-oxoacyl-(acyl carrier protein [ACP]) synthase III (β-ketoacyl-ACP synthase III) genes of Escherichia coli, Salmonella enterica serovar Typhimurium, and Pseudomonas putida strain KT2440 (24, 36). It has been reported that a 3-oxoacyl-ACP synthase III called FabH is one of the enzymes in the type II fatty acid synthesis system (20). Most bacteria and plant plastids use this cycle, with each enzyme catalyzing an individual reaction to produce long-chain fatty acids. In this elongation cycle, 3-oxoacyl-ACP synthase III first catalyzes a condensation reaction to supply intermediates of short-chain fatty acids (14, 20).
Recently, the expression of many virulence factors has been reported to be regulated by a cell density-dependent system called quorum sensing. Several studies revealed that quorum sensing by gram-negative bacteria involves N-acyl homoserine lactones (AHLs) that differ in the structures of their N-linked acyl side chains as signal molecules (23, 29). AHLs are synthesized by the coupling of homoserine lactone rings from S-adenosylmethionine and acyl chains from the acyl-ACP pool in cells by the enzymes LuxI in Vibrio fischeri (27) and PsyI in P. syringae (12). Because 3-oxoacyl-ACP synthase III correlates with fatty acid biosynthesis, we speculated that orf3 has a critical role in AHL production. In addition, it was reported that other biosynthetic pathways, such as the synthesis of phospholipids and lipopolysaccharides, also use acyl-ACP intermediates (14), suggesting an important role for Orf3 in fatty acid cellular metabolism.
In this study, the role of the orf3 gene in the glycosylation island of the flagellin gene cluster of P. syringae pv. tabaci 6605 was investigated by analysis of the AHL production and some virulence factors under regulation of quorum sensing in the Δorf3 mutant. In addition, we generated the AHL-defective ΔpsyI mutant and compared its characteristics with those of the Δorf3 mutant. The physiological role of the orf3 gene in biofilm formation on the tobacco leaf surface was also examined.
MATERIALS AND METHODS
Bacterial strains and growth condition.
All bacterial strains used in this study are shown in Table 1. Pseudomonas syringae pv. tabaci 6605 strains were maintained as described previously (35). E. coli strains were grown at 37°C in Luria-Bertani medium (LB). Chromobacterium violaceum CV026 as the AHL biosensor strain was maintained at 30°C in LB with a final concentration of 50 μg/ml kanamycin (22).
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Escherichia coli | ||
| DH5a | F− λ− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) supE44 thi-1 gyrA relA1 | Takara, Kyoto, Japan |
| S17-1 | thi pro hsdR hsdM+recA [chr::RP4-2-Tc::Mu-Km::Tn7] | 26 |
| Chromobacterium violaceum CV026 | Double mini-Tn5 mutant from C. violaceum ATCC 31532; AHL biosenser | 22 |
| Pseudomonas syringae pv. tabaci | ||
| 6605 | Wild type; Nalr | 30 |
| 6605-d3 | 6605 Δorf3 | 33 |
| 6605-dpsyI | 6605 ΔpsyI | This study |
| Plasmids | ||
| pGEM-T Easy | 3.015-kb cloning vector for PCR product; Ampr | Promega, Tokyo, Japan |
| pK18mobsacB | Small mobilizable vector; Kmr; sucrose sensitive (sacB) | 26 |
| pDSK519 | Broad-host-range cloning vector; Kmr | 16 |
| pM3 | 1.69-kb chimeric PCR product deleting orf3 cloned into pK18mobsacB at EcoRI site; Kmr | 33 |
| pDSKGI | 9-kb HindIII fragment containing orf1, orf2, and orf3 genes from P. syringae pv. tabaci 6605 in pDSK519; Kmr | 33 |
Ampr, ampicillin resistance; Kmr, = kanamycin resistance; Nalr, nalidixic acid resistance.
Plant material and inoculation procedure.
Tobacco plants (Nicotiana tabacum L. cv. Xanthi NC) were grown at 25°C with a 12-h photoperiod. For the inoculation experiments, bacterial strains were suspended in 10 mM MgSO4 and 0.02% Silwet L77 (OSI Specialties, Danbury, CT) at a density of 2 × 108 CFU/ml. After spray inoculation on both surfaces of tobacco leaves, the leaves were incubated in a growth cabinet for 8 days at 23°C.
Construction of mutants.
Generation of the Δorf3 mutant and its complementary strain by pDSKGI (Table 1) from P. syringae pv. tabaci 6605 was described previously (33). To generate an AHL synthesis-defective (ΔpsyI) mutant, the genetic region of psyI and psyR was first isolated by using a TA cloning system (pGEMT-Easy; Promega, Tokyo, Japan). PCR primers (PsyI5′ [5′-ATGTCGAGCGGGTTTGAGTTTCAG-3′] and PsyR5′ [5′-ATGGAGGTTCGTACCGTGAAAGCC3′]) were designed based on the registered sequences of psyI and psyR of P. syringae pv. tabaci (accession number AF110468). The ΔpsyI mutant was constructed by the replacement of the codon AAG for Lys148 with a TAG stop codon by using a QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). Two complementary oligonucleotides containing a stop codon, PsyI-S (5′-CACGGTGGTCAGCTAGGCAATGGCGCGGAT-3′) and PsyI-AS (5′-ATCCGCGCCATTGCCTAGCTGACCACCGTG-3′), were synthesized. Mutation was introduced according to the manufacturer's protocol and confirmed by sequencing.
Detection of AHL by TLC analysis.
Bacterial strains were grown in King's B (KB) medium for 24 h at 25°C. After removal of cells by centrifugation, AHLs were extracted from the supernatant with an equal volume of ethyl acetate, and the organic phases were evaporated. Residues were dissolved in 1/500 the original volume of ethyl acetate, and 10 μl of the solution was subjected to C18 reverse-phase thin layer chromatography (TLC) (20 cm by 20 cm, RP-18F254S; Merck, Darmstadt, Germany) with a solvent system of methanol and water (60:40, vol/vol). After development, the dried TLC plate was overlaid with 50 ml of semisolid LB agar medium containing 6 ml of an overnight culture of C. violaceum CV026. After 24 h of incubation at 30°C, AHL was visualized as violet spots by the induction of violacein production. Chemically synthesized N-hexanoyl-l-homoserine lactone (HHL) and N-(3-oxohexanoyl)-l-homoserine lactone (OHHL) as the standard molecules were gifts from T. Ikeda (Utsunomiya University). All samples were stored at −20°C in ethyl acetate.
Northern blot analysis.
Pseudomonas strains were incubated in LB with 10 mM MgCl2 for 24 h at 25°C. The cells were transferred to new minimal medium (MM) [50 mM potassium phosphate buffer, 7.6 mM (NH4)2SO4, 1.7 mM MgCl2, and 1.7 mM NaCl, pH 5.7] supplemented with 10 mM each of mannitol and fructose (MMMF) (30) and incubated to an optical density at 600 nm (OD600) of 0.3. Total RNA was extracted using a High Pure RNA isolation Kit (Roche, Mannheim, Germany), and 10 μg of RNA was used for Northern blot analysis. The probes were labeled with a PCR digoxigenin synthesis kit (Roche). The conditions for hybridization and detection were as described by Shimizu et al. (30).
Scanning electron and optical microscopy.
Leaves inoculated with the wild-type (WT) strain or mutant strains for 8 days at 23°C were observed by scanning electron microscopy. The detailed procedure has been described by Taguchi et al. (33). Colony morphologies of the WT and each mutant on 1.5% KB agar plates after 2 days of incubation were observed under an optical microscope (Olympus IX70, Tokyo, Japan).
Swarming assay.
Bacteria cultured overnight in LB containing 10 mM MgCl2 at 25°C were resuspended in 10 mM MgSO4 and adjusted to an OD600 of 0.1. Three-microliter aliquots were inoculated in the center of the medium for swarming (SWM plate [0.5% peptone, 0.3% yeast extract, and 0.5% agar; Difco, Detroit, MI]) (18) or MMMF (MMMF plate with 0.5% agar) (33). The swarming motility was observed after 24 h of incubation at 27°C for SWM and at 23°C for MMMF agar plates.
Biosurfactant assay.
Overnight cultures of WT and mutant strains in LB with 10 mM MgCl2 were subcultured into MMMF and incubated for 24 h at 23°C. After centrifugation, aliquots of 10 μl of supernatant were spotted on the film (Parafilm; Alcan Packaging, Neenah, WI) to detect the drop-collapsing activity (21).
For biosurfactant detection, overnight cultures in LB containing 10 mM MgCl2 at 25°C were centrifuged and adjusted to an OD600 of 0.1 with 10 mM MgSO4. Five microliters of the bacterial suspension was placed on an MMMF agar plate with 0.0005% methylene blue and 0.02% hexadecyltrimetyl ammonium bromide, as described by Kohler et al. (19). The plate was incubated for 48 h at 27°C until the appearance of a blue halo.
Glycolipid detection by TLC.
Each bacterial strain was grown in 200 ml of KB for 24 h at 25°C, and cells were removed by centrifugation. The supernatant was filtered through a 0.45-μm-pore-size filter, and glycolipids were extracted with an equal volume of ethyl acetate. The organic phase was evaporated to dryness and redissolved in 400 μl of ethyl acetate. Fifteen microliters of sample was subjected to TLC on a precoated silica gel glass plate (Silica Gel 60, Whatman Inc., Clifton, NJ) with a solvent system of chloroform-methanol-water (95:20:2, vol/vol/vol). After development, glycolipids were visualized by spraying with 0.2% orcinol reagent dissolved in 11.2% H2SO4 (final concentrations) and heating at 110°C for 15 min (31).
CAS assay.
The siderophore production by bacterial strains in the culture supernatant was determined by the methods of Schwyn and Neilands (28). Each strain was incubated in LB containing 10 mM MgCl2 for 24 h at 25°C and was adjusted to an OD600 of 0.001, 0.01, or 0.1 with MMMF. After 24 h of incubation, each culture was filtered through a 0.45-μm-pore-size filter (Millipore). Fifty microliters of each sample or MMMF medium as a reference was added to 150 μl of chromoazurol S (CAS) solution (0.6 mM hexadecyltrimetyl ammonium bromide, 0.015 mM FeCl3, 0.15 mM HCl, and 0.15 mM CAS in MMMF). The absorbance of each sample at 630 nm was measured after 30 min of incubation at room temperature. The amount of siderophore was calculated by subtracting the absorbance value of the reference.
Tolerance to H2O2 and antibiotics.
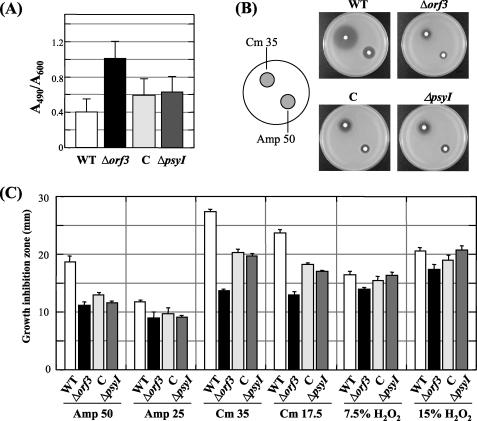
P. syringae pv. tabaci strains were grown for 24 h in KB. Two milliliters of bacterial suspension was diluted in 15 ml of new KB medium containing 0.3% agar and overlaid on a KB plate. A paper disc (3MM paper; Whatman plc, Brentford, United Kingdom) containing antibiotic (ampicillin at 50 μg/μl or 25 μg/μl or chloramphenicol at 35 μg/μl or 12.5 μg/μl) was placed on the plate. For the H2O2 tolerance assay, a nitrocellulose membrane (Millipore Corporation, Bedford, MA) with 20 μl of H2O2 solution (7.5% or 15%) was put on the plate. After incubation for 24 h at 27°C, the diameter of the growth inhibition zone was measured.
Quantitative analysis of EPS.
Each bacterial strain was grown on an MMMF plate containing 1.5% agar for 48 h at 27°C. The bacterial cells were then harvested and suspended in 200 μl of distilled water. After centrifugation at 8,000 × g for 3 min, the supernatant was mixed with 3 volumes of chilled 95% ethanol for 24 h at −20°C, and extracellular polysaccharide (EPS) was precipitated by centrifugation at 8,000 × g for 10 min. Quantification of the purified extracellular polysaccharide was carried out by the phenol-sulfate method (15). Total protein in the sample was determined by a Bradford protein assay (Bio-Rad protein assay; Bio-Rad, Hercules, CA). Total extracellular polysaccharide was calculated as a relative value per total cellular protein.
Nucleotide sequence accession numbers.
The nucleotide sequence of the psyI and psyR genes has been deposited in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession number AB257774.
RESULTS
Detection of AHLs and Northern analysis.
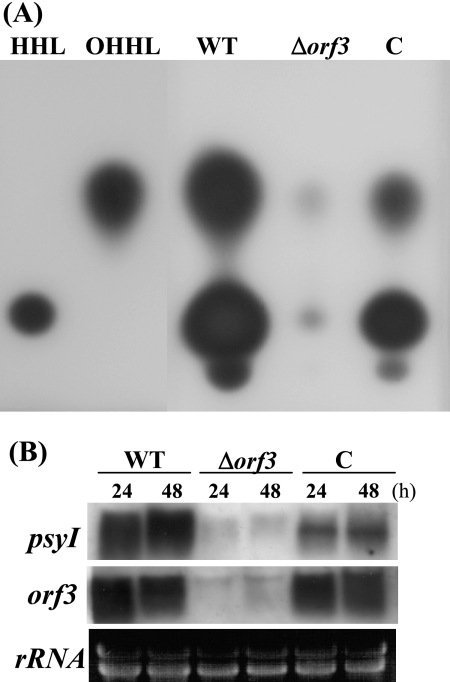
To examine whether AHLs are produced by P. syringae pv. tabaci 6605, TLC analysis with a C18 reverse-phase plate and C. violaceum CV026 as the AHL biosensor strain was carried out (Fig. 2A). The production of OHHL and HHL by P. syringae pv. tabaci 2024 has been reported, and an OHHL structure was confirmed by mass spectrometry (29). In the WT supernatant from overnight culture in KB, two major signals and one minor signal for violacein were detected. Because the mobilities of the two major signals were consistent with those of the standard molecules, OHHL and HHL, P. syringae pv. tabaci 6605 also produces both AHLs (Fig. 2A). The concentration of AHLs in the WT culture medium was estimated at about 0.05 μg/ml for OHHL and at about 0.1 μg/ml for HHL. In contrast, a small amount of AHLs was detected in the Δorf3 mutant. Complementation of the strain almost restored production both of HHL and OHHL under these experimental conditions.
FIG. 2.
Detection of AHLs produced by P. syringae pv. tabaci and Northern blot analysis. (A) AHL TLC assay. Ten-microliter aliquots of 500-fold-concentrated extracts in ethyl acetate were developed by C18 reverse-phase TLC. (B) Expression of orf3 and psyI genes in each strain after 24 and 48 h of incubation in MMMF. HHL and OHHL (0.1 μg each) were used as standards. C, orf3-complemented strain.
To ascertain the expression of the orf3 and psyI genes, which encode the AHL synthesis protein in P. syringae pv. tabaci 6605, Northern blot analysis was carried out using RNA after 24 and 48 h of incubation in MMMF. As shown in Fig. 2B, the expression of both genes was strong in the WT but was hardly detectable in the Δorf3 mutant. In the complemented strain, the expression of orf3 was recovered to previous levels and that of psyI was partially restored.
Biosurfactant detection and colony morphology.
The results shown in Fig. 2 suggest that Orf3 is involved in the quorum-sensing system. In some pathogenic bacteria, it has been reported that biosurfactant production is regulated by quorum sensing (7). Therefore, the characteristics of the WT and Δorf3 mutant were examined. Further, to examine whether alternation of the phenotype in the Δorf3 mutant is due to a defect in AHL production, a psyI-deficient (ΔpsyI) mutant was generated as a typical mutant defective in AHL production. Loss of ability to produce AHLs by the ΔpsyI mutant was confirmed by TLC analysis using C. violaceum CV026 (data not shown).
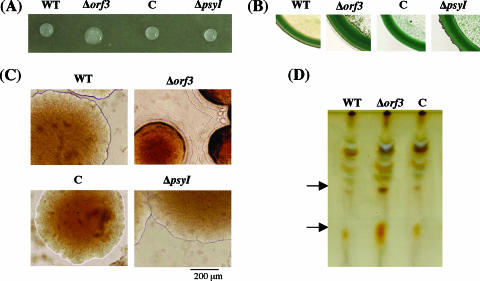
The production of biosurfactants was examined by the drop-collapsing test and standard methylene blue plate assay. In the drop-collapsing test, the ability of the Δorf3 mutant to produce biosurfactants seemed to be enhanced in comparison with those in WT and complemented strains (Fig. 3A). In the standard methylene blue plate assay, the dark blue ring around the colony of the Δorf3 mutant was significantly greater than those produced by WT and complemented strains (Fig. 3B). Although the production of biosurfactants by the ΔpsyI mutant did not differ from that by the WT in the drop-collapsing test, the ΔpsyI mutant exhibited enhanced production of biosurfactants in the methylene blue plate assay.
FIG. 3.
Detection of biosurfactants, colony morphology, and separation of glycolipids. (A) Drop-collapsing test of bacterial suspension. (B) Methylene blue plate assay. (C) Colony morphology of the WT and each mutant observed by optical microscopy on 1.5% KB agar plate after 2 days of incubation at 27°C. (D) TLC analysis of glycolipids. Arrows indicate orcinol-positive spots with higher intensity in the Δorf3 mutant. Bacterial strains are indicated as: C, orf3-complemented strain.
The colony morphology of each strain on KB plates with 1.5% agar was also observed with an optical microscope. The Δorf3 mutant showed a somewhat diffused colony whose periphery was surrounded with translucent bilayers, suggesting an overproduction of biosurfactants (Fig. 3C). On the other hand, there was no significant difference in colony morphology among the WT, the complemented strain, and the ΔpsyI mutant.
The most common class of biosurfactants are glycolipids, consisting of carbohydrates and long-chain aliphatic acids or hydroxyaliphatic acids. Because rhamnolipid, one of the best-studied glycolipids produced in Pseudomonas aeruginosa was not detected in the supernatants of overnight cultures of WT P. syringae pv. tabaci and the Δorf3 mutant in KB medium by the procedure previously reported (31), P. syringae pv. tabaci seems not to produce rhamnolipids, at least under these culture conditions. Therefore, glycolipids were extracted from the supernatant with a different solvent, ethyl acetate, and analyzed by TLC. Figure 3D showed detection of glycolipids on the TLC plate with orcinol reagent. The WT, the Δorf3 mutant, and its complement strain produced many kinds of glycolipids with different Rf values. Among them, two spots are remarkable in the Δorf3 mutant and are candidates for the biosurfactants of this strain.
Swarming motility.
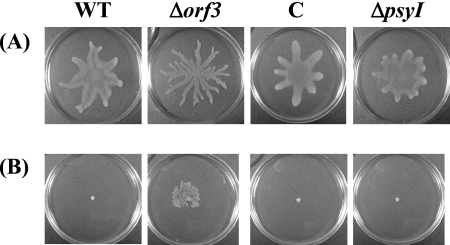
Because biosurfactants have been reported to enhance swarming motility (7), the motility of the Δorf3 mutant was investigated on 0.5% agar SWM and MMMF plates. As shown in Fig. 4A, the WT, the ΔpsyI mutant, and the orf3-complemented strain showed similar swarming patterns on SWM agar plates. On the other hand, the Δorf3 mutant showed a more irregular and branched swarming pattern on SWM plates. Furthermore, only the Δorf3 mutant had swarming ability on 0.5% MMMF agar plates (Fig. 4B). The doubling times of the WT and the complemented strain of P. syringae pv. tabaci 6605 are 1.03 ± 0.16 h and 1.17 ± 0.14 h, respectively, whereas that of the Δorf3 mutant is longer (1.44 ± 0.08 h) and the ΔpsyI mutant has a slightly longer doubling time (1.27 ± 0.11 h) than the WT, indicating that enhanced swarming ability in the Δorf3 mutant was not caused by rapid growth. These results suggest that overproduction of biosurfactants in the Δorf3 mutant is one of the causes for the hyperswarming phenotype.
FIG. 4.
Swarming motility of P. syringae pv. tabaci 6605. Swarming patterns on SWM plates with 0.5% agar after 24 h of incubation at 27°C (A) and on MMMF plates with 0.5% agar after 24 h of incubation at 23°C (B) are shown. C, orf3-complemented strain.
EPS production and tolerance to H2O2 and antibiotics.
Previous papers have reported that the production of EPS is regulated by quorum sensing and that an EPS-deficient mutant of P. syringae is hypersensitive to environmental stresses (7, 17, 25). To compare extracellular polysaccharide production on an MMMF plate with 1.5% agar after 48 h of incubation at 27°C, each mutant was scraped off and the amounts of extracellular polysaccharide and extracellular proteins as an internal control were quantified (Fig. 5A). The results demonstrated that the amount of extracellular polysaccharide in the Δorf3 mutant was about twice that in the WT. The extracellular polysaccharide production in the ΔpsyI mutant was also increased, but weakly. These results suggest that the quorum-sensing system might repress the production of extracellular polysaccharide by P. syringae pv. tabaci 6605 under less-nutritional conditions.
FIG. 5.
Extracellular polysaccharide quantification and growth inhibition test. (A) Relative amount of extracellular polysaccharide in bacteria grown on MMMF plate for 48 h at 27°C. (B) Photographs of growth inhibition on plates by 50 μg/μl ampicillin (Amp) and 35 μg/μl chloramphenicol (Cm). (C) Growth inhibition of each bacterial strain by 7.5% and 15% H2O2, 50 μg/μl and 25 μg/μl ampicillin, and 35 μg/μl and 17.5 μg/μl chloramphenicol. C, orf3-complemented strain. Error bars indicate standard deviations.
The tolerance to H2O2 and antibiotics among strains was compared by a growth inhibition test using H2O2, ampicillin, or chloramphenicol. The growth inhibition by both ampicillin and chloramphenicol was significantly decreased in the Δorf3 mutant in comparison with that in the WT strain, indicating that the Δorf3 mutant had increased tolerance to them (Fig. 5B). The degrees of growth inhibition by H2O2 and antibiotics in each strain are shown in Fig. 5C. These results indicate that the Δorf3 mutant shows higher tolerance to not only H2O2 but also to antibiotics and that the ΔpsyI mutant also has slightly increased tolerance to these substances. Overproduction of EPS and/or biosurfactants may contribute to enhanced tolerance in these mutants.
Production of siderophore.
Many bacteria secrete iron-chelating molecules to acquire iron for their own growth. Siderophores are low-molecular-weight iron-chelating compounds, and their production is reportedly controlled by quorum sensing (13, 32). To examine whether siderophore synthesis in P. syringae pv. tabaci 6605 is regulated by AHL molecules, the amounts of siderophores in the WT and each mutant were compared using a CAS solution assay.
Figure 6A shows the result of color change due to iron-siderophore complex formation. Because siderophores in the culture supernatant had a high affinity for iron, an orange iron-siderophore complex was formed instead of the blue iron-dye complex. Although all strains produced no or little siderophore at lower cell density, the WT and the complemented strain increased siderophore production in proportion to the increase in cell density (Fig. 6). On the other hand, both the Δorf3 and ΔpsyI mutants produced significantly less siderophore, even at higher cell density, suggesting that iron acquisition is regulated by quorum sensing via AHL molecules.
FIG. 6.
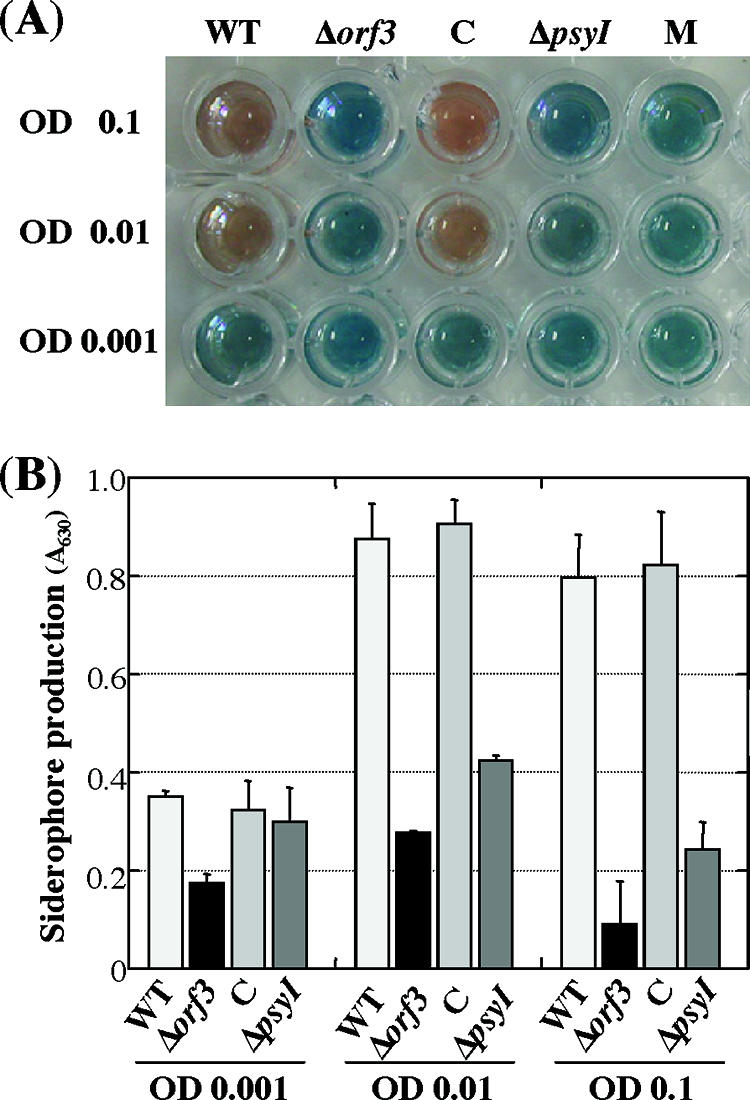
Siderophore production. (A) Color change of CAS solution containing culture supernatant from each strain. CAS solution with 50 μl of each bacterial supernatant or MMMF as a control (M) were mixed. (B) Quantitative analysis of siderophore production. C, orf3-complemented strain. Error bars indicate standard deviations.
Virulence of mutants on host tobacco leaves and observation of biofilm by scanning electron microscopy.
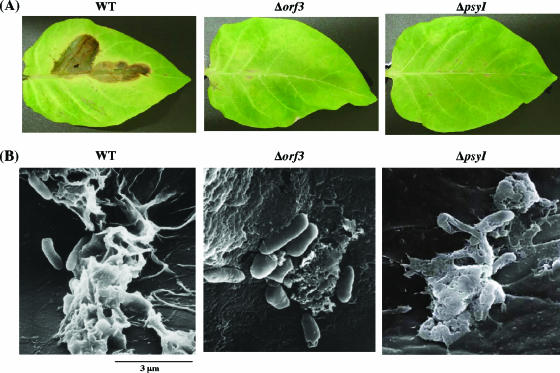
The ability of the WT and the Δorf3 and ΔpsyI mutants to cause disease on host tobacco leaves was examined. The results demonstrate that the Δorf3 mutant was less virulent than the WT, as previously reported (33). The ΔpsyI mutant also showed decreased virulence against tobacco (Fig. 7A).
FIG. 7.
Bacterial cells on the surfaces of tobacco leaves inoculated with WT P. syringae pv. tabaci 6605 and the Δorf3 and ΔpsyI mutants. Leaves were photographed after 8 days of incubation at 23°C (A) and the inoculated tobacco surface was observed by scanning electron microscopy (B).
Bacterial cells on the surfaces of tobacco leaves inoculated with each strain were observed by scanning electron microscopy (Fig. 7B). The WT bacteria were fully embedded in the adhesive EPS matrix, which promoted adhesion to the leaf surface. In contrast, there was little material around the bacterial surface of the Δorf3 and ΔpsyI mutants, and only dried material was observed around them. This result suggests an intimate relationship between AHL production and biofilm formation.
DISCUSSION
In this study, we investigated functions of the orf3 gene in the glycosylation island of the flagellar gene cluster in P. syringae pv. tabaci 6605 by using the orf3 deletion mutant. Based on homology research, orf3 was predicted to encode a 3-oxoacyl-ACP synthase III homologue. In the fatty acid biosynthesis pathway of P. aeruginosa, acetyl-ACP is derived from acetyl coenzyme A and ACP by FabH (14). In Lactococcus lactis subsp. lactis IL1403, the homologue of this enzyme was reported to produce not only acetyl-ACP but also acetoacetyl-ACP (20). Acetyl-ACP is one of the initiators of the fatty acid elongation cycle, and acetoacetyl-ACP is thought to be a precursor of 3-oxo-acyl homoserine lactones. However, because there are several pathways to produce them, FabH is reported to be a dispensable enzyme for biosyntheses of both fatty acids and AHLs in P. aeruginosa (14). In contrast, Lai and Cronan (20) reported that FabH is essential for bacterial fatty acid biosynthesis in E. coli and L. lactis subsp. lactis IL-1403 and that FabH-defective mutants failed to grow without exogenous supplementation of long-chain fatty acids.
As shown in Fig. 2, WT P. syringae pv. tabaci 6605 synthesized OHHL and HHL as major AHLs, whereas the Δorf3 mutant had a significantly reduced ability to produce AHLs. The quorum-sensing system using AHL signals is a recently well-studied bacterial mechanism that unicellular organisms use to communicate with each other and act like multicellular organisms by monitoring their own population density (7). The orf3 gene is thought to participate in this system by supplementation of AHL precursors. However, the Δorf3 mutant produced a small amount of detectable AHLs, suggesting that a minor pathway to produce AHLs may exist in P. syringae pv. tabaci 6605, as shown in the previous report for P. aeruginosa (14). 3-oxoacyl-ACP synthase I or II may compensate for the function of Orf3. Furthermore, the Δorf3 mutant was able to grow in MMMF, suggesting that this enzyme is not indispensable for fatty acid biosynthesis.
Biosurfactants are wetting agents produced by some bacteria to reduce surface tension. The major biosurfactants reported previously are rhamnolipid in Pseudomonas, surfactin in Bacillus, and serrawettin in Serratia, which are glycolipids or lipopeptides. Rhamnolipid production in P. aeruginosa is regulated via a quorum-sensing system (4). In the biosynthetic pathway, the 3-ketoacyl-ACP derived from the fatty acid biosynthesis cycle becomes a primer for the subsequent complex steps. Although the structure and de novo biosynthesis pathway of biosurfactants of P. syringae pv. tabaci 6605 have not been elucidated, we detected biosurfactant production by this bacterium (Fig. 3). In particular, the production of biosurfactants in the Δorf3 mutant was significantly higher than those in the WT and the ΔpsyI mutant. Although some candidates for biosurfactants were detected (Fig. 3D), their structures have not yet been determined. Structural analysis of these compounds by mass spectrometry and nuclear magnetic resonance will be needed in near future.
Why is the production of biosurfactants facilitated in the Δorf3 mutant? The answer to this question is not clear at present. However, if Orf3 catalyzes the rate-limiting step of fatty acid biosynthesis, short-chain ACPs such as malonyl-ACP, substrates for Orf3, will be accumulated in the Δorf3 mutant. In this case, the enzymes for biosurfactant production may be able to use these short-chain ACPs without competition.
Previously, it was reported that swarming motility is enhanced by the addition of biosurfactants and that mutants of Serratia liquefaciens defective in biosurfactant production have no swarming ability (19, 21). Indeed, swarming motility in the Δorf3 mutant was enhanced in both SWM and MMMF plates with 0.5% agar (Fig. 4). The ΔpsyI mutant showed the same level of swarming ability as the WT, although AHL-deficient mutants of P. syringae pv. syringae B728a exhibited high motility (25). Because this mutant is defective in both the ahlI and ahlR genes, which encode an AHL synthetic enzyme and a transactivator of AHL-responsive genes, the phenotype might be different from that caused by our single mutation of psyI in P. syringae pv. tabaci 6605. Further investigation of the regulation of swarming ability is required.
It was reported that a P. syringae mutant defective in EPS production was hypersensitive to H2O2 (17, 25). In P. syringae pv. tabaci 6605, tolerance to H2O2 and antibiotics was enhanced in the Δorf3 mutant, probably owing to the overproduction of biosurfactants and/or EPS (Fig. 3 and 5). Our results suggest that EPS and/or biosurfactants are important for swarming motility and tolerance to environmental stresses. Thus, the quorum-sensing system in P. syringae pv. tabaci 6605 is thought to negatively regulate their production.
Iron uptake using siderophores has been reported to be regulated by the quorum-sensing system in P. aeruginosa (32). Because environmental iron is almost insoluble at biological pH, many bacteria have developed systems to acquire iron by using siderophores, which show high affinity for iron(III) (28). As shown in Fig. 6, both the Δorf3 and the ΔpsyI mutants showed a drastically reduced ability to produce siderophores even at high bacterial density. This result suggests that siderophore production is positively regulated via the quorum-sensing system in P. syringae pv. tabaci 6605.
Many species of pseudomonads produce a fluorescent yellow-green siderophore called pyoverdine. Fluorescent pseudomonads are also able to produce other, minor siderophores such as pyochelin, pseudomonine, quinolobactin, and corrugatin (6). Recently, it was reported that pyoverdine is generally detected by many pathovars of P. syringae, although the spectral characteristics are different from those of typical pyoverdine in animal pathogens (5). WT P. syringae pv. tabaci 6605 has greater ability to produce fluorescence under UV light than the Δorf3 and ΔpsyI mutants (data not shown), suggesting that pyoverdine may be the major siderophore in this bacterium. Indeed, there are highly homologous genes for pyoverdine side chain peptide synthase in P. syringae pv. tabaci 6605 (data not shown).
Recently, gene expression profiles of S. enterica serovar Typhimurium during swarming were compared with those in liquid medium by microarray analysis (37). The results demonstrated that genes for iron metabolism were strongly induced in bacteria grown on swarming agar plates with less-nutritional conditions. It was reported that excess iron prevents swarming motility, and less-nutritional conditions may induce swarming and biosurfactant production in P. aeruginosa (7). From these reports, a reduced ability to acquire iron may relate to the hyperswarming motility in the Δorf3 mutant.
Iron acquisition and biosurfactant production were reported to influence biofilm formation (2, 3). Normal biofilm is composed of bacterial cells and EPS, with large amounts of water in its structure (10). However, when a WT strain of P. aeruginosa was incubated with lactoferrin, an iron chelator, a thick, mushroom-like structured biofilm was not observed by confocal scanning laser microscopy (2). The siderophore-defective mutant also formed only a thin uniform layer (2). Furthermore, overproduction of biosurfactants inhibited biofilm development in P. aeruginosa (8), probably because bacterial detachment from the biofilm occurred earlier and more extensively (3). As shown in Fig. 7, both the Δorf3 and ΔpsyI mutants had reduced virulence toward the host tobacco leaves, and the biofilm formation by each strain seemed not to develop normally. To elucidate iron functions as a signal for swarming and biofilm development, we now plan to construct a mutant of P. syringae pv. tabaci 6605 that is defective in iron acquisition by disruption of related genes of pyoverdine synthesis. In addition, further characterization of the genes responsible for biosurfactant synthesis is required.
We investigated the orf3-mediated regulation of virulence factors in P. syringae pv. tabaci 6605. Why is orf3, a gene concerned with quorum sensing, located in the glycosylation island of the flagellar gene cluster? There are two stages in the process of bacterial attachment: the primary docking stage and the secondary locking stage (11). In the primary docking stage, bacteria must approach the surface against electrostatic and hydrophobic forces via swimming motility and chemotaxis. During this stage, at lower cell density, flagellum-mediated motility is thought to play an important role; however, EPS, if any, may obstruct active flagellar motility. In the subsequent secondary locking stage, loosely bound bacteria attach firmly to the surface by producing EPS for biofilm maturation. In this stage, EPS becomes an essential factor, but flagellum-mediated motility might not be necessary. After sufficient maturation of the biofilm, bacteria begin to escape from the old biofilm and colonize other surfaces. The flagellum-mediated swarming motility might be induced at this detachment process owing to the limited availability of nutrients, including iron, in the mature biofilm. Recent papers have suggested that each process of biofilm formation might be regulated by expression of quorum-sensing genes (1, 9). These dynamic alternations in gene expression for biofilm formation might be regulated by the orf3 gene in relation to flagellum expression.
Acknowledgments
We thank the Leaf Tobacco Research Laboratory of Japan Tobacco Inc. for providing P. syringae pv. tabaci 6605. We are also grateful to P. Williams (Nottingham University, United Kingdom) and T. Ikeda (Utsunomiya University, Japan) for providing Choromobacterium violaceum CV026 and the chemically synthesized AHLs, respectively.
This work was supported in part by Grants-in-Aid for Scientific Research (S) (no. 15108001) and (B) (no. 18380035) from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by the Okayama University COE program “Establishment of Plant Health Science.”
Footnotes
Published ahead of print on 6 October 2006.
REFERENCES
- 1.Allison, D. G., B. Ruiz, C. Sanjose, A. Jaspa, and P. Gilbert. 1998. Extra-cellular products as mediators of the formation and determination of Pseudomonas fluorescens biofilm. FEMS Microbiol. Lett. 167:179-184. [DOI] [PubMed] [Google Scholar]
- 2.Banin, E., M. L. Vasil, and E. P. Greenberg. 2005. Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. USA 102:11076-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boles, B. R., M. Thoendel, and P. K. Singh. 2005. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol. Microbiol. 57:1210-1223. [DOI] [PubMed] [Google Scholar]
- 4.Brint, J. M., and D. E. Ohman. 1995. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of rhlR-rhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive luxR-luxI family. J. Bacteriol. 177:7155-7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bultreys, A., I. Gheysen, H. Maraite, and E. D. Hoffmann. 2001. Characterization of fluorescent and nonfluorescent peptide siderophores produced by Pseudomonas syringae strain and their potential use in strain identification. Appl. Environ. Microbiol. 67:1718-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cornelis, P., and S. Matthijs. 2002. Diversity of siderophore-mediated iron uptake systems in fluorescent pseudomonas: not only pyoverdines. Environ. Microbiol. 4:787-798. [DOI] [PubMed] [Google Scholar]
- 7.Daniels, R., J. Vanderleyden, and J. Michiels. 2004. Quorum sensing and swarming migration in bacteria. FEMS Microbiol. Rev. 28:261-289. [DOI] [PubMed] [Google Scholar]
- 8.Davey, M. E., N. C. Caiazza, and G. A. O'Toole. 2003. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185:1027-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies, D. G., M. R. Parsek, J. P. Person, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 10.Donlan, R. M. 2002. Biofilms: microbial life on surfaces. Emerg. Infect. Dis. 8:881-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunne, W. M. Jr. 2002. Bacterial adhesion: seen any good biofilms lately? Clin. Microbiol. Rev. 15:155-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elasri, M., S. Delorme, P. Lemanceau, G. Stewart, B. Laue, E. Glickmann, P. M. Oger, and Y. Dessaux. 2001. Acyl-homoserine lactone production is more common among plant-associated Pseudomonas spp. than among soilborne Pseudomonas spp. Appl. Environ. Microbiol. 67:1198-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fong, K. P., L. Gao, and D. R. Demuth. 2003. luxS and arcB control aerobic growth of Actinobacillus actinomycetemcomitans under iron limitation. Infect. Immun. 71:298-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoang, T. T., S. A. Sullivan, J. K. Cusick, and H. P. Schweizer. 2002. β-Ketoacyl acyl carrier protein reductase (FabG) activity of the fatty acid biosynthetic pathway is a determining factor of 3-oxo-homoserine lactone acyl chain lengths. Microbiology 148:3849-3856. [DOI] [PubMed] [Google Scholar]
- 15.Hodge, J. E., and B. T. Hofreiter. 1962. Determination of reducing sugars and carbohydrates, p. 388. In R. L. Whistler and M. L. Wolfrom (ed.), Methods in carbohydrate chemistry, vol. 1. Academic Press, New York, N.Y. [Google Scholar]
- 16.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 17.Keith, I. M., and C. L. Bender. 1999. AlgT (σ22) controls alginate production and tolerance to environmental stress in Pseudomonas syringae. J. Bacteriol. 181:7176-7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kinscherf, T. G., and D. K. Willis. 1999. Swarming by Pseudomonas syringae B728a requires gacS (lemA) and gacA but not the acyl-homoserine lactone biosynthetic gene ahlI. J. Bacteriol. 181:4133-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohler, T., L. K. Curty, F. Barja, C. van Delden, and J. C. Pechere. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pilli. J. Bacteriol. 182:5990-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai, C.-Y., and J. E. Cronan. 2003. β-Keroacyl-acyl carrier protein synthase III (FabH) is essential for bacterial fatty acid synthesis. J. Biol. Chem. 278:51494-51503. [DOI] [PubMed] [Google Scholar]
- 21.Lindum, P. W., U. Anthoni, C. Christophersen, L. Eberi, S. Molin, and M. Givskov. 1998. N-Acyl-l-homoserine lactone autoinducers control production of an extracellular lipopeptide biosurfactant required for swarming motility of Serratia liquefaciens MG1. J. Bacteriol. 180:6384-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McClean, K. H., M. K. Winson, L. Fish, A. Taylor, S. R. Chhabra, M. Camara, M. Daykin, J. H. Lamb, S. Swift, B. W. Bycroft, G. S. Stewart, and P. Williams. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143:3703-3711. [DOI] [PubMed] [Google Scholar]
- 23.More, M. I., L. D. Finger, J. L. Stryker, C. Fuqua, A. Eberhard, and S. C. Winans. 1996. Enzymatic synthesis of a quorum-sensing autoinducer through use of defined substrates. Science 272:1655-1658. [DOI] [PubMed] [Google Scholar]
- 24.Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, H. Hilbert, V. A. Martins dos Santos, D. E. Fouts, S. R. Gill, M. Pop, M. Holmes, L. Brinkac, M. Beanan, R. T. DeBoy, S. Daugherty, J. Kolonay, R. Madupu, W. Nelson, O. White, J. Peterson, H. Khouri, I. Hance, P. C. Lee, E. Holtzapple, D. Scanlan, K. Tran, A. Moazzez, T. Utterback, M. Rizzo, K. Lee, D. Kosack, D. Moestl, H. Wedler, J. Lauber, D. Stjepandic, J. Hoheisel, M. Straetz, S. Heim, C. Kiewitz, J. A. Eisen, K. N. Timmis, A. Dusterhoft, B. Tummler, and C. M. Fraser. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4:799-808. [DOI] [PubMed] [Google Scholar]
- 25.Quinones, B., G. Dulla, and S. E. Lindow. 2005. Quorum sensing regulates exopolysaccharide production, motility, and virulence in Pseudomonas syringae. Mol. Plant-Microbe Interact. 18:682-693. [DOI] [PubMed] [Google Scholar]
- 26.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 27.Schäfer, A. L., D. L. Val, B. L. Hanzelka, J. E. Cronan, Jr., and E. P. Greenberg. 1996. Generation of cell-to-cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc. Natl. Acad. Sci. USA 93:9505-9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Ana. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 29.Shaw, P. D., G. Ping, S. L. Daly, C. Cha, J. E. Cronan, Jr., K. L. Rinehart, and S. K. Farrand. 1997. Detecting and characterizing N-acyl-homoserine lactone signal molecule by thin-layer chromatography. Proc. Natl. Acad. Sci. USA 94:6036-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimizu, R., F. Taguchi, M. Marutani, T. Mukaihara, Y. Inagaki, K. Toyoda, T. Shiraishi, and Y. Ichinose. 2003. The ΔfliD mutant of Pseudomonas syringae pv. tabaci, which secretes flagellin monomers, induces a strong hypersensitive reaction (HR) in non-host tomato cells. Mol. Genet. Genomics 269:21-30. [DOI] [PubMed] [Google Scholar]
- 31.Sim, L., O. P. Ward, and Z-Y. Li. 1997. Production and characterization of a biosurfactant isolated from Pseudomonas aeruginosa UW-1. J. Ind. Microbiol. 19:232-238. [DOI] [PubMed] [Google Scholar]
- 32.Stintzi, A., K. Evans, J. M. Meyer, and K. Poole. 1998. Quorum-sensing and siderophore biosynthesis in Pseudomonas aeruginosa: lasR/lasI mutants exhibit reduced pyoverdine biosynthesis. FEMS Microbiol. Lett. 166:341-345. [DOI] [PubMed] [Google Scholar]
- 33.Taguchi, F., K. Takeuchi, E. Katoh, K. Murata, T. Suzuki, M. Marutani, T. Kawasaki, M. Eguchi, S. Katoh, H. Kaku, C. Yasuda, Y. Inagaki, K. Toyoda, T. Shiraishi, and Y. Ichinose. 2006. Identification of glycosylation genes and glycosylated amino acids of flagellin in Pseudomonas syringae pv. tabaci. Cell. Microbiol. 8:923-938. [DOI] [PubMed] [Google Scholar]
- 34.Taguchi, F., R. Shimizu, Y. Inagaki, K. Toyoda, T. Shiraishi, and Y. Ichinose. 2003. Post-translational modification of flagellin determines the specificity of HR induction. Plant Cell Physiol. 44:342-349. [DOI] [PubMed] [Google Scholar]
- 35.Taguchi, F., R. Shimizu, R. Nakajima, K. Toyoda, T. Shiraishi, and Y. Ichinose. 2003. Differential effects of flagellins from Pseudomonas syringae pv. tabaci, tomato and glycinea on plant defense response. Plant Physiol. Biochem. 41:165-174. [Google Scholar]
- 36.Takeuchi, K., F. Taguchi, Y. Inagaki, K. Toyoda, T. Shiraishi, and Y. Ichinose. 2003. Flagellin glycosylation island in Pseudomonas syringae pv. glycinea and its role in host specificity. J. Bacteriol. 185:6658-6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, Q., J. G. Frye, M. McClelland, and R. M. Harshey. 2004. Gene expression patterns during swarming in Salmonella typhimurium: genes specific to surface growth and putative new motility and pathogenicity genes. Mol. Microbiol. 52:169-187. [DOI] [PubMed] [Google Scholar]