Abstract
Escherichia coli σ70-dependent promoters have typically been characterized as either −10/−35 promoters, which have good matches to both the canonical −10 and the −35 sequences or as extended −10 promoters (TGn/−10 promoters), which have the TGn motif and an excellent match to the −10 consensus sequence. We report here an investigation of a promoter, Pminor, that has a nearly perfect match to the −35 sequence and has the TGn motif. However, Pminor contains an extremely poor σ70 −10 element. We demonstrate that Pminor is active both in vivo and in vitro and that mutations in either the −35 or the TGn motif eliminate its activity. Mutation of the TGn motif can be compensated for by mutations that make the −10 element more canonical, thus converting the −35/TGn promoter to a −35/−10 promoter. Potassium permanganate footprinting on the nontemplate and template strands indicates that when polymerase is in a stable (open) complex with Pminor, the DNA is single stranded from positions −11 to +4. We also demonstrate that transcription from Pminor incorporates nontemplated ribonucleoside triphosphates at the 5′ end of the Pminor transcript, which results in an anomalous assignment for the start site when primer extension analysis is used. Pminor represents one of the few −35/TGn promoters that have been characterized and serves as a model for investigating functional differences between these promoters and the better-characterized −10/−35 and extended −10 promoters used by E. coli RNA polymerase.
Recognition and binding of DNA promoter elements by RNA polymerase set the start site for transcription initiation. In Escherichia coli, these elements are recognized by a σ factor when σ is present in RNA polymerase holoenzyme, (σ plus core [β, β′, α, α, and ω]) (15, 36). E. coli encodes several σ factors that can be part of the holoenzyme, which are used under various conditions of growth and stress (39). Each σ factor interacts with different DNA sequences, and thus the recognition and usage of a given promoter is dependent upon the σ present in the holoenzyme. The presence of specific σ factors allows bacteria to coordinate the expression of gene sets and is one of the major ways bacteria regulate expression in response to changing growth conditions.
The primary σ factor of E. coli, σ70, is used during exponential growth and belongs to a large family of prokaryotic primary σ factors related to each other by sequence, structure, and function (15, 39). Primary σ factors have four regions of similarity. It is known that residues in region 2 recognize a −10 element (TATAAT) (37), residues in region 3 recognize an extended TGn −10 motif (positions −15 to −13) (1), and residues in region 4 recognize a −35 element (TTGACA) (4). However, not all three of these promoter elements need to be present for promoter function. E. coli σ70-dependent promoters have typically been characterized as either −10/−35 promoters, which have good matches to both the canonical −10 and −35 sequences and do not require the TGn motif (32), or as extended −10 promoters (TGn/-10 promoters), which have the TGn motif and an excellent match to the −10 consensus sequence and do not require a −35 element (2, 23, 26).
In addition to the sequence elements themselves, the distance between them is important for promoter recognition. Because σ70 regions 4 and 2 simultaneously contact the −35 and the −10 elements, polymerase structure dictates the distance between the elements in a −10/−35 promoter (3, 10, 38). The −35 and −10 elements are ideally separated by a spacer length of 17 bp. Although this spacer length may vary, transcription is affected by a change of even one base pair (35). Likewise, the distance between the −10 element and the transcription start site is determined by the polymerase structure. The transcriptional start site is typically located seven nucleotides downstream of the −10 element (−12 TATAAT −7), with a preference for A as the incoming nucleotide (24, 28).
The consensus sequences for σ70-dependent promoters have been studied extensively and are well defined (16, 27, 34). Promoter elements for σ70-dependent promoters are initially assigned based on sequences that match −10/−35 or extended −10 consensus sequences at the appropriate distance from the +1 transcription start. However, the bacteriophage T4 Pminor promoter (50), which was identified by its activity in vitro, is an example of a promoter that does not readily fit into either the −10/−35 or the extended −10 promoter categories. Examination of the Pminor promoter region failed to locate good matches to any of the typical σ70 DNA elements at proper positions relative to the transcriptional start site, which had been determined by primer extension. Nonetheless, recognition of Pminor is specific for polymerase containing σ70, since it is not recognized by polymerase containing the closely related stationary phase σ factor, σ38 (50). In addition, Pminor is of interest because the formation of stable polymerase/Pminor complexes increases when σ70 lacks the N-terminal 99 residues (region 1.1); other tested promoters have been either unaffected or negatively affected by the lack of σ70 region 1.1 (50, 53).
To understand how Pminor is recognized and used by polymerase containing σ70, we have investigated how specific mutations within the Pminor promoter region affect transcription from this promoter. Here we define the minimal Pminor promoter and show that it functions in vivo as well as in vitro. We demonstrate that transcription from Pminor incorporates nontemplated ribonucleoside triphosphates (NTPs) at the 5′ end of the Pminor transcript, which results in an anomalous assignment of the start site when primer extension analysis is used. The correct assignment of the start site suggests that Pminor has both a good −35 element and a TGn motif but has an extremely poor −10 element. Our mutational analysis indicates that both the −35 element and the TGn motif are required for efficient transcription and that these elements compensate for the poor −10 element. Pminor represents one of only a few characterized −35/TGn promoters and is useful for comparing the properties of this class of promoter with the well-characterized −10/−35 and TGn/-10 classes.
MATERIALS AND METHODS
Constructs and DNA templates.
pFW11-Pnull, pFW11-P1, and pFW11-P2 are pACYC184 derivatives that have a EcoRI/SalI fragment without a promoter sequence (Pnull), with a good promoter (P1), or with a very good promoter (P2) upstream of the 5′ end of the lacZ gene (52). These plasmids also contain the 3′ end of the lacI gene and the kanamycin resistance gene upstream of the promoter insert. Thus, homologous recombination will transfer the antibiotic resistance and the promoter sequence into an F′ plasmid that contains the entire lac operon. To construct the Pminor-lacZ transcriptional fusion plasmid pXBJ203, a PCR product was first obtained using pDKT90 (29) as a template, Pfu polymerase (Stratagene), and primers designed to create restriction sites. The upstream primer annealed to pDKT90 starting at position −63 relative to the start of Pminor and included a 5′ EcoRI site, and the downstream primer annealed to pDKT90 starting at position +4 and included a 5′ SalI site. After isolation and digestion of this product with EcoRI and SalI, the fragment was ligated with pFW11-null that had been previously digested with EcoRI and SalI. The resulting plasmid is pXBJ203. Constructs pXBJ302, pXBJ402, and pXBJ503, which include the Pminor region from positions −44 to +4, −35 to +4, or −29 to +4, respectively, were created by inserting synthesized oligomers that contained the desired region of Pminor with upstream EcoRI and downstream SalI sites into pFW11-Pnull that had been previously digested with EcoRI and SalI. pIH4021(P−33A), pIH4022 (P−31C), pIH4023 (P−14A−13A−12T), pIH4024 (P−14A−13A−12T−31C), pIH4025 (P−14A), pIH4026 (P−16G), pIH4027 (P−12T−10T−7T), pIH4028 (P+1C), and pIH4029 (P−11G), which include the Pminor region from the +4 to −35 positions with specific promoter mutations, were constructed in the same manner using the appropriate oligomers to clone the promoter region. DNA sequence analyses (performed by the CBR-DNA Sequencing Facility of the University of Maryland or the Facility for Biotechnology Resources of the Food and Drug Administration) confirmed the promoter sequences in these various constructs.
Using the procedure of Whipple (52), pFW11-Pnull, pFW11-P1, pFW11-P2, pXBJ203, pXBJ302, pXBJ402, pXBJ503, and pIH4028 (P+1C) promoter constructs were transferred to single copy F′ plasmids by homologous recombination. The recombinant F′ plasmids were then transferred to the streptomycin-resistant E. coli strain FW102 by conjugation.
In some cases, templates for in vitro transcriptions were prepared by digesting plasmids, which had been previously isolated and purified, with BglI. This digestion linearized the plasmids at position +209, relative to the +1 position of P2. Linear templates, which were used in transcriptions to identify the Pminor +1 (see Fig. 2A), were 120-bp PCR products containing only the Pminor promoter. PCR was carried out with pIH4022, Pfu polymerase (Stratagene), and primers chosen to produce a fragment from position −99 to position +21 relative to the Pminor +1 start site.
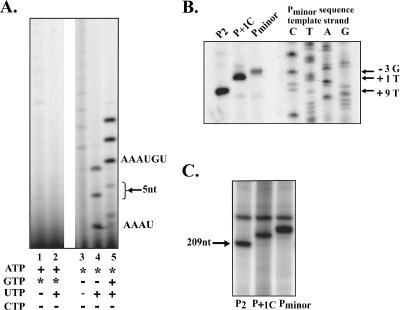
FIG. 2.
The Pminor transcriptional start is identified. In vitro transcription reactions were assembled by adding 1.95 μl of a solution containing reconstituted polymerase (0.2 pmol core plus 0.5 pmol of σ70) in protein buffer I to 0.02 pmol of linear DNA in 2.05 μl of DNA buffer I. (A) Transcription was initiated by adding 1 μl of NTP mix to the protein-DNA mix containing Pminor DNA. As indicated, each NTP was added at the following concentrations: 1 mM UTP and 0.25 mM each ATP and GTP. The specific activity of [γ-32P]GTP or [γ-32P]ATP (where indicated by the asterisk) was 7 × 105 dpm/pmol. Assignments of labeled RNA products are shown. The black arrow signifies 5-nt products, consistent with the migration of the pppACN3 marker (not shown) kindly provided by N. Nossal. (B) Denaturing acrylamide gel showing the products of primer extension assays next to a Pminor DNA sequencing ladder. The unlabeled Pminor, P+1C, and P2 transcripts were generated by multiple-round transcription assays, which were initiated by the addition of 1 μl of NTP mix II. (C) Single-round transcription was initiated by adding 1 μl of NTP mix I with heparin. Transcripts arising from the Pminor, P+1C, and P2 promoters are indicated.
The labeled templates used in KMnO4 footprinting were 156-bp PCR products containing only the Pminor promoter. PCR was carried out with either pXBJ402 (Pminor) or pIH4023 (P−14A−13A−12A) template, Pfu polymerase (Stratagene), and primers chosen to produce a fragment from position −99 to position +57, relative to the Pminor +1 start. Primers were 5′ end labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs) prior to PCR. Each reaction contained labeled primer that annealed to one strand, and unlabeled primer that annealed to the other strand. The γ-32P-labeled PCR product was purified by gel electrophoresis.
Buffers and proteins.
σ70 with an N-terminal His6 tag was purified from E. coli BL21(DE3)/pLysE (46) cultures containing the rpoD plasmid (pETσfl [20, 53]), as previously described (50), by denaturation of inclusion bodies containing the protein, Ni2+ resin affinity chromatography under denaturing conditions, followed by a slow renaturation of the protein. E. coli RNA polymerase core was purchased from Epicenter Technologies. Protein buffer I contained 27 mM Tris-Cl (pH 7.9), 54 mM Tris-acetate (pH 7.9), 52 mM NaCl, 40% (vol/vol) glycerol, 0.9 mM EDTA, 0.007% Triton X-100, 0.24 mM dithiothreitol, 154 mM potassium glutamate, 4.1 mM magnesium acetate, and 102.6 μg of bovine serum albumin/ml. DNA buffer I contained 21.9 mM Tris-Cl (pH 7.9), 43.4 mM Tris-acetate (pH 7.9), 71 mM NaCl, 3.4% (vol/vol) glycerol, 0.5 mM EDTA, 0.15 mM dithiothreitol, 219 mM potassium glutamate, 5.8 mM magnesium acetate, 146 μg of bovine serum albumin/ml, and 0.34 mM 2-mercaptoethanol. NTP mix I contained 1 mM each of ATP, GTP, and CTP and 25 μM [α-32P]UTP (7 × 105 dpm/pmol). NTP mix II contained 1 mM each ATP, GTP, CTP, and UTP. A set of NTP mixes was used for determining the Pminor +1. The NTPs present in each mix are indicated in Fig. 2.
β-Galactosidase assays.
The level of β-galactosidase activity in Miller units in FW102 lysates containing the single-copy F′ plasmids with the various promoter-lacZ constructs was determined as described previously (41) except that cells were grown in the presence of 30 μg of kanamycin and 100 μg of streptomycin/ml and no IPTG (isopropyl-β-d-thiogalactopyranoside) was added.
In vitro transcription assays.
Transcription reactions were assembled as indicated in the figure legends. Polymerase and DNA were incubated at 37°C for 10 min, NTPs were added, and reactions were incubated at 37°C for an additional 8 min. When indicated, single-round reactions were performed by including heparin (0.5 μl of 1 mg/ml) with the NTPs. Gel load solution (1× Tris-borate-EDTA, 7 M urea, 0.1% bromophenol blue, 0.1% xylene cyanol FF) was added at a volume three times that of the reaction aliquots, and the reactions were collected on ice. Each reaction solution was heated at 95°C for 2 min before electrophoresis on 6% polyacrylamide-7 M urea denaturing gels run in 1× Tris-borate-EDTA. After autoradiography, the films were scanned by using a Powerlook 2100XL densitometer and QuantityOne software from Bio-Rad, Inc.
Primer extension analyses.
Primer extension reactions were carried out as described previously (50). Primer IGH107 is complementary to Pminor, P+1C, and P2 transcripts from 155 to 171 nucleotides (nt) downstream of the SalI cut site, which is located from position +159 to position +175 relative to the Pminor +1A.
Potassium permanganate footprinting.
Reactions were assembled as indicated in the legend of Fig. 5. DNA-protein complexes were treated with potassium permanganate as described previously (29, 44). Potassium permanganate (0.5 μl of a 50 mM solution) was added to each 4-μl reaction mixture, and the mixture was incubated for 2 min at 37°C. After the addition of 5 μl of stop solution (0.69 M sodium acetate, 1 M 2-mercaptoethanol, 200 μg of salmon sperm DNA/ml), the DNA was precipitated with ethanol and dried. The pellets were resuspended in 100 μl of 1 M piperidine and incubated at 90°C for 30 min. The DNA was then precipitated with butanol, dried, and resuspended in gel loading solution. The DNA products were separated on denaturing gels as described above. As controls, G+A ladders were obtained using PCR product labeled on either the top or the bottom strand (31).
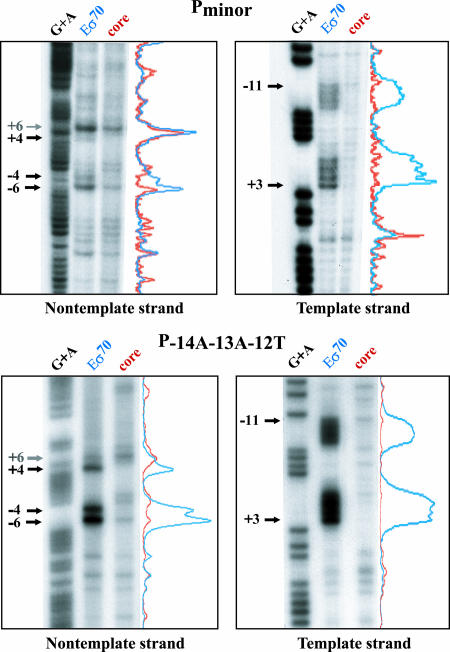
FIG. 5.
Potassium permanganate footprints of Pminor (A) and P−14A−13A−12T (B). Reactions were assembled by adding 1.95 μl of a solution containing reconstituted polymerase (Eσ70; 0.4 pmol core plus 1.0 pmol of σ70) or core alone (0.4 pmol) in protein buffer I to 0.2 pmol of the indicated DNA in 2.05 μl of DNA buffer I. Eσ70-dependent bands are marked by arrows and are numbered relative to the Pminor transcriptional +1 site. Nontemplate strand results for Pminor are consistent with the findings of Vuthoori et al. (50), but bands −6 and −4 are referred to as −2 and +1 in that study. Traces for the lanes are shown.
RESULTS
The 5′ end of Pminor RNA has nontemplated nucleotides.
Previously, Pminor was identified as a σ70-dependent promoter active in vitro (50). Although this promoter is present within T4 DNA, located about 70 bp downstream of the T4 middle promoter PuvsX, Pminor RNA has not been observed after T4 infection (17), suggesting that it is not active for T4 under typical growth conditions. In a previous study (50), the +1 transcription start for Pminor was identified as G (now position −3 in Fig. 1A). This assignment was based on primer extension experiments and the migration of the Pminor RNA on denaturing gels. However, typical σ70 elements are not observed at the correct positions upstream of this assignment. Although an excellent −35 element (TTGAAA) is seen from 27 to 32 bp upstream of this start site, such a position would be highly unusual. Thus, we considered the possibility that the actual Pminor transcriptional start is 3 bp downstream of where the primer extension analyses indicated (+1A, Fig. 1). Primer extension acts as a measuring tape to determine the distance from a fixed point to the 5′ end of a transcript, which typically corresponds to the transcriptional +1 position. However, any extra nucleotides added to the transcript would result in a false measure of the transcriptional +1 position. This seemed plausible for the Pminor RNA since an A-rich sequence surrounds the start of Pminor transcription (GGAAAAT, positions −3 to +4 in Fig. 1). Such an A-rich sequence could facilitate slippage of the polymerase during initiation, resulting in the incorporation of extra A's at the start of the Pminor transcript. To determine whether the Pminor RNA starting nucleotide is a G, as assigned by primer extension, or an A, consistent with promoter element spacing, we carried out in vitro transcription with either [γ-32P]GTP or [γ-32P]ATP (Fig. 2A). During transcription only the initiating nucleotide retains the γ-phosphate; therefore, the labeled nucleotide must be incorporated at position +1 to result in detectable product. Reactions in which [γ-32P]GTP was added yielded no observable product (Fig. 2A, lanes 1 and 2). In contrast, reactions that included [γ-32P]ATP produced bands (Fig. 2A, lanes 3 to 5). This indicates that in vitro Pminor transcription begins with an A rather than a G.
FIG. 1.
Promoter constructs. Sequences between EcoRI (GAATTC) and SalI (GTCGAC) cloning sites (enclosed in boxes) in pFW11 are shown. (A) Pnull (pFW11-null), P1 (pFW11-P1), and P2 (pFW11-P2) are as described elsewhere (52). A 67-bp Pminor fragment is from pDKT90 (29). Promoter elements (−35, TGn, and −10) and the +1 start site are noted in red above the sequence. Note that the +1A starts of P1 and P2 are located within the SalI site, whereas the Pminor +1 is 8 nt upstream. Arrows indicate 5′ end of the Pminor fragment before the EcoRI site in the P-63, P-44, P-35, and P-29 clones. (B) The P-35 construct, used as wild-type Pminor promoter in the present study, is shown. Derivative promoters with the indicated changes are listed below Pminor. The dotted line indicates that the sequence is identical to Pminor.
The pattern of products seen in Fig. 2A suggests that stuttering occurs at Pminor. The addition of [γ-32P]ATP alone (lane 3) resulted in a ladder of products, which is consistent with reiterative incorporation of NTPs (42). The addition of [γ-32P]ATP and the next templated base, UTP, produced three distinct bands (lane 4). These bands migrate as four-, five-, and six-nucleotide (nt) products, which is consistent with a templated product (AAAU) plus products with one or two additional nucleotides. We assign these products as AAAAU and AAAAAU. The addition of [γ-32P]ATP, UTP, and GTP in the reaction (lane 5) resulted again in three major bands consistent with the 6-nt templated product (AAAUGU) plus products with one or two additional nucleotides. We conclude that the added transcript length of Pminor is due to the incorporation of nontemplated A nucleotides. The sizes of the transcription products (≥4) also suggest that the first A (in the sequence 5′-GGAAAAUGU-3′) is not the +1 nt. Rather, transcription appears to begin at one of the other A's.
To further examine the Pminor transcriptional start, we changed the putative +1 nucleotide from an A to a C. C is the least favored nucleotide for beginning transcription, and changing the +1 nt to a C will often result in selection of the +2 nt as the transcriptional start (24, 28). We reasoned that, if the second A is the transcriptional start, then the +1 C mutation will disrupt the run of A's and prevent stuttering. To investigate the effect of this mutation, we used a primer that annealed 159 to 175 nt downstream of the assigned +1 start site of the Pminor transcript. As seen previously (50), after primer extension the product from Pminor RNA comigrated with the −3 position of the Pminor DNA sequence. This is because of the additional nontemplated nucleotides at the 5′ end of the Pminor RNA. In contrast, the primer extension product using the (P+1C) RNA was slightly shorter. Similar results were observed in a transcription gel (Fig. 2C), in which the P+1C transcript migrated slightly faster than Pminor RNA. We conclude that interrupting the run of A's at the Pminor start site results in a loss of stuttering, and thus we assign the Pminor transcription start as the +1A depicted in Fig. 1. As a control for these analyses, we used the P2 transcript (Fig. 1 and see below). As expected, in the primer extension assay (Fig. 2B) the P2 product migrated correctly, with the +9 position of the Pminor DNA. (Note in Fig. 1 that the Pminor DNA construct has an extra 8 bp relative to the P2 DNA.) In addition, in the transcription gel (Fig. 2C) the P2 RNA migrated as a smaller product, which is consistent with its expected size of 209 nt.
Defining the minimal Pminor promoter.
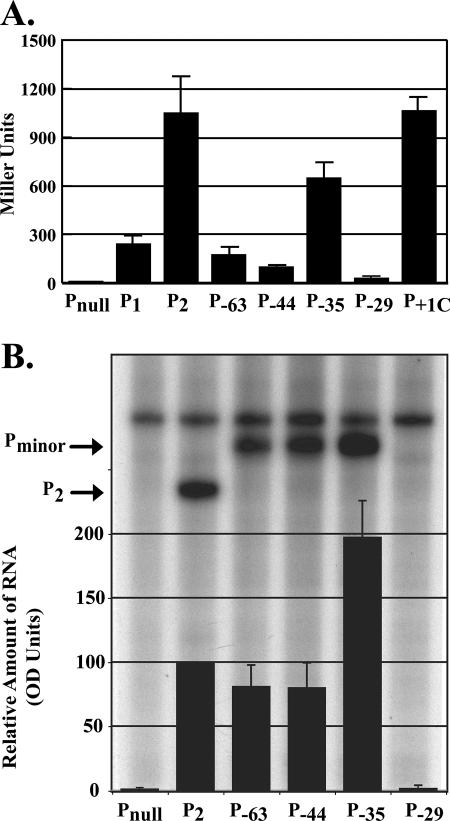
To precisely define the salient sequence features needed for the recognition of Pminor, a set of lacZ transcriptional fusions was designed based on the system of Whipple (52). A 67-bp fragment known to contain the Pminor promoter (−63 to +4) was first cloned into pFW11 and designated P−63 (Fig. 1). Similar constructs were then made (P−44, P−35, and P−29) with smaller promoter fragments, resulting in 5′ nested deletion constructs. pFW11-P2 and pFW11-P1, which contain the PlacUV5-derived −10/−35 promoters P2 and P1, respectively, represented the positive controls. In vivo, P2 and P1 produce high and moderate levels of transcription, respectively (52) (see also Fig. 3A). pFW11-Pnull, which lacks a promoter, was used as a negative control. The lacZ transcriptional fusions were moved into F′ plasmids, so promoter activity could be measured in vivo by β-galactosidase activity. The positive and negative controls behaved as expected (Fig. 3A), yielding significant or negligible activity, respectively. Strains carrying either P−63 or P−44 were less active than P1, whereas P−35 and its derivative P+1C yielded even more activity than P1. However, the removal of the next six base pairs, containing the sequence TTGAAA, significantly reduced Pminor activity (P−29). We conclude that Pminor is active in vivo and that sequences downstream of position −36 are sufficient for this activity. In addition, these results suggest that the sequence from −35 to −30 is required for significant Pminor activity in vivo.
FIG. 3.
Minimal Pminor promoter defined. (A) Graph showing the β-galactosidase assay activity (in Miller units) determined in vivo for each promoter. (B) A denaturing acrylamide gel showing the products of multiple-round in vitro transcription reactions is overlaid with a graph of the quantitation data. Transcription reactions were assembled as described in Fig. 2C. Multiple-round transcription was initiated by adding 1 μl of NTP mix I (without heparin). The amounts of RNA were determined by densitometry and are shown relative to P2, which is set at 100. The values represent the average of three or more transcriptions.
In multiple-round in vitro transcription assays, we found that the relative activities of the promoters differed somewhat from that observed in vivo. However, deletion of the TTGAAA sequence (P−29, Fig. 3B) was again deleterious, in this case eliminating promoter activity. Thus, because P−35 contained the minimum functional Pminor promoter both in vivo and in vitro, this construct was designated and used as wild-type Pminor for the rest of the present study.
Positions −35 to −30 of Pminor, TTGAAA, define the Pminor −35 element.
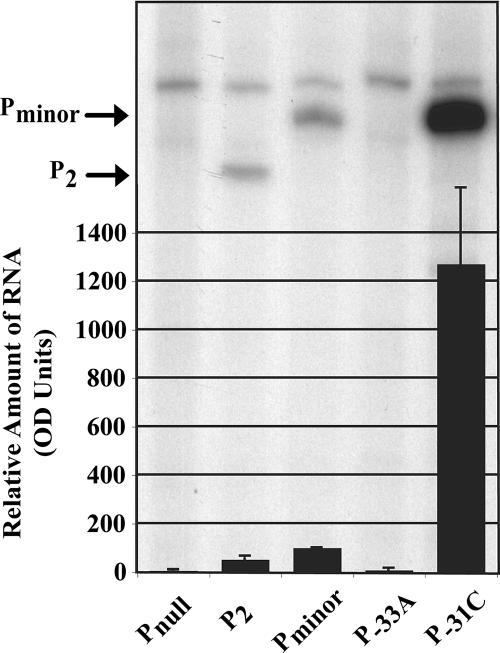
To determine whether the TTGAAA sequence of Pminor (positions −35 to −30) functions as the −35 element, we investigated how specific mutations in this sequence affected transcription. Altering the −33G:C base pair within a σ70 −35 element (TTGACA) has been shown to significantly reduce transcription from promoters because the C moiety is directly contacted by σ70 region 4 residues (22). The −33A mutation eliminated the production of Pminor RNA (Fig. 4), whereas the −31C mutation, which makes the −35 element a perfect match to a consensus, dramatically increased transcription (Fig. 4). These results indicate that the TTGAAA sequence of Pminor functions as the −35 promoter element and confirm that this sequence is required for Pminor activity. In addition, other work has shown that the anti-sigma factor, AsiA (19), which blocks σ70 region 4 recognition of −35 elements, inhibits transcription from Pminor (20), further confirming the requirement for a −35 recognition element at Pminor.
FIG. 4.
TTGAAA functions as −35 element and is required for Pminor transcription. A denaturing acrylamide gel showing the products of single-round in vitro transcription reactions is overlaid with a graph of the quantitation data. Transcription reactions were assembled and carried out as described in Fig. 2C. The amounts of RNA were determined by densitometry and are shown relative to Pminor, which is set at 100. The values represent the average of three or more transcriptions.
The transcription bubble at Pminor ranges from position −11 to position +4.
Identification of the Pminor −35 element and +1A suggested that GAAAAC should function as the −10 element based on location. However, such a sequence deviates substantially from the canonical σ70 −10 sequence, TATAAT (positions −12 to −7) (26). Previous work has shown that the nontemplate strand −11A is crucial for DNA melting and that transcription bubbles typically extend from this −11A to position +3 (11, 25, 30). Thus, KMnO4 footprinting, which reveals single-stranded thymines, is a reliable way to identify the −10 element because it can identify the single-stranded T on the template strand opposite the nontemplate −11A.
We performed KMnO4 experiments with DNA containing Pminor and either Eσ70 or core alone (Fig. 5). When the nontemplate strand was 5′ end labeled (Fig. 5, top left), reactive bands occurred at positions +4, −4, and −6. A band was also visible at position +6 but was σ70 independent. Footprinting using labeled template strand produced eight bands representing every thymine from positions −11 to +3 (Fig. 5, top right). Therefore, in Pminor, the transcription bubble extends from −11A to about position +4, as is typical (5, 7, 44), and the polymerase-DNA interaction within the −10 element appears to be normal as judged by this assay. We conclude that GAAAAC (positions −12 to −7) is the functional −10 element, a finding consistent with data discussed above assigning the +1A as the transcriptional start site.
The TGn motif of Pminor compensates for its noncanonical −10 element.
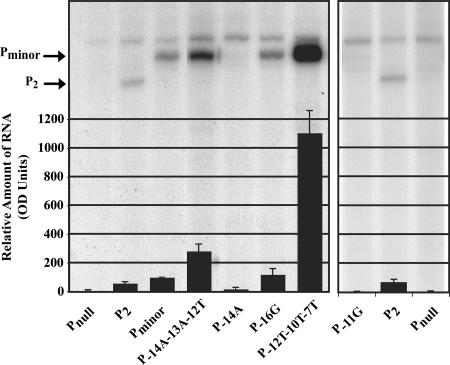
Although the Pminor −10 element (GAAAAC) deviates substantially from the consensus −10 sequence (TATAAT), Pminor has a TGn motif located just upstream at positions −15 to −13. To address whether the TGn was important for Pminor activity, we made a set of constructs to examine the roles of the TGn motif and the −10 element (Fig. 1 and 6). When the −14G was changed to −14A, transcription was significantly reduced, a finding consistent with the idea that the TGn motif is required for Pminor function. In contrast, the multiple mutation, −14A−13A−12T, had a positive effect on transcription. In this construct, the TGn motif was changed to TAn, but the −12G was also changed to T. Improving the −10 element allowed transcription in the absence of the TGn motif. These results suggest that the TGn motif of Pminor compensates for the poor −10 consensus.
FIG. 6.
Pminor requires TGn motif to compensate for a weak −10 element. A denaturing acrylamide gel showing the products of single-round in vitro transcription reactions is overlaid with a graph of the quantitation data. Transcription reactions were assembled and carried out as described in Fig. 2C. The amounts of RNA transcript were determined by densitometry and are shown relative to Pminor, which is set at 100. The values represent the average of three or more transcriptions.
Another consequence of the −14A−13A−12T mutation is that it creates a perfect −10 consensus sequence (TATAAT) at positions −17 to −13 relative to the Pminor start. The presence of this sequence could create a situation in which this perfect −10 sequence would be favored over the weaker −10 element of TAAAAC, present in this mutant. However, this is not the case since migration of the transcriptional product is unchanged (Fig. 6), and KMnO4 footprints with this mutant show a transcription bubble in the same location as that seen with wild-type Pminor (Fig. 5, bottom). Thus, the functional −10 element is not changed in this mutant. Combining the −14A−13A−12T mutations with a −31C mutation created a promoter with a perfect −35 element and the improved −10 element. As expected, transcription from this promoter was significantly higher than from wild-type Pminor (data not shown) and roughly the same as that seen with the −31C mutation alone. Finally, when the −12, −10, and −7 nucleotides were all changed to T, creating a −10 element that perfectly matches consensus (P−12T−10T−7T), transcription again was greatly improved (Fig. 6). As a control, we found that changing the −16A of Pminor, which is upstream of the TGn motif, to a G had no effect.
As discussed above, the nontemplate A at position −11, which is also highly conserved among σ70-dependent promoters, is thought to lie at the transition between double-stranded DNA (upstream and including position −12) and the open (single-stranded) DNA within the transcription bubble (from position −11 to position +3) (11, 25, 30). To investigate the importance of the −11A in Pminor, we changed the −11A to a G. This change was particularly deleterious, resulting in a significant decrease in transcription (Fig. 6). We conclude that the −11A is a crucial determinant for Pminor activity.
DISCUSSION
Within promoter DNA, specific sequences provide the recognition elements needed to define the start of transcription. E. coli σ70-dependent promoters have three well-characterized recognition elements: −35 sequences, TGn, and −10 sequences. Previous work has shown that base determinants within these elements are recognized by specific contact with σ70 residues. The σ70 region 4.2 residues R584 and E585 contact the −31G and −33C, respectively, on the template (bottom) strand of the double-stranded −35 element (TTGACA) (4, 14, 45), whereas recognition of the TGn element is thought to arise by the interaction of region 3 residues E458 and H455 (1, 43) with the −14G:C base pair. For the −10 element, σ70 residues recognize and interact both with double-stranded DNA and with the single-stranded base determinants formed at the transcription bubble. Region 2.4 residues T440 (45) and Q437 (51) are required for recognition of the −12 T:A base pair in double-stranded DNA, facilitating closed complex formation (8, 9, 12, 37, 54). Aromatic residues Y425, Y430, W433, and W434 in region 2.3 promote DNA strand separation beginning at −11 and interact with the nontemplate strand of the −10 element in the open transcription bubble (12, 21, 37, 40, 48, 49).
Despite the multiple contacts between σ70 and various base determinants in the promoter DNA, recognition of all of these elements is not biologically optimal because maximum promoter binding works against the need to leave the promoter once transcription has been initiated (reference 13 and references therein). In addition, deviation from consensus allows transcription to be conditionally regulated by transacting factors, allowing the best level of expression for the environment rather than the highest level of expression. Recognition of just the −10 and −35 elements (−10/−35 promoters) or recognition of just the extended −10 motif TGnTATAAT (TGn/-10 promoters) is sufficient for excellent promoter activity (2, 26, 34). In fact, the absence of σ70 region 4 is nonessential for transcription from an extended promoter, indicating that the total loss of the σ70/−35 contacts can be tolerated if both the TGn and canonical −10 sequences are present (23).
Although −35/-10 and TGn/-10 promoters have been established as σ70 promoter classes, previous work has suggested that perhaps any combination of the −35, TGn, and −10 modules might be acceptable for recognition (13, 33). A study of more than 500 promoters (34) revealed that promoters with poorer matches to the canonical −10 sequences were more likely to have the TGn motif than those with consensus −10 sequences. This finding led to the speculation that the TGn motif might be able to compensate for a less canonical −10 sequence. Recent work with the gapA P1 promoter of E. coli (47) has demonstrated that the −35/TGn promoter is indeed a viable promoter architecture. Thus, the Pminor promoter, described here, and the gapA P1 promoter represent the first well-characterized members of a −35/TGn promoter class. Both of these promoters have excellent −35 sequences and the TGn motif. However, both of these promoters lack the conserved T at position −12 within the −10 element. It is this contact in particular for which −14G seems to compensate at Pminor, as seen by the activity of P−14A versus P−14A−13A−12T. In addition, Pminor also has a C rather than a T at the highly conserved position −7. With both promoters, mutation of either the −35 or the TGn motif away from consensus essentially eliminates transcription, suggesting that at these promoters, the −35 and TGn elements are the primary elements for promoter recognition. Thus, it seems that having a minimal number of strong contacts is required, but these contacts can occur in any combination. Despite their deviations from the −10 consensus sequence, both gapA P1 and Pminor retain the A nucleotide at position −11, and this base determinant is indeed crucial for Pminor activity. Thus, the presence of the −11A, which lies at the upstream edge of the single-stranded transcription bubble in the stable polymerase-promoter complex may be important for promoter melting despite whatever promoter modules are used for recognition.
Promoter elements are frequently assigned by an inspection of sequences upstream of the identified +1 sequence. Because the −35/TGn class has not been previously appreciated, other members of this class may have escaped notice and instead have been assigned as −35/-10 promoters with less than optimal spacer distances. Indeed, gapA P1 was not originally identified as a −35/TGn promoter and was not included in the study by Mitchell et al. (34).
Previous work in our lab failed to identify the correct promoter elements of Pminor in part because it is a member of the −35/TGn class but also because Pminor RNA contains nontemplated NTPs at the 5′ end. Consequently, primer extension analyses were misleading, and the +1 of Pminor was incorrectly assigned (50). Although several promoters have been identified that generate transcripts with nontemplated NTPs at the 5′ end due to polymerase slippage (6, 18, 42), in most of these cases, there is not a well-defined number of nontemplated bases incorporated. Rather, primer extension reveals a “stutter” stop as the primer is extended along RNAs of various lengths. However, with Pminor, primer extension gives a well-defined stop because of the addition of (mostly) three nontemplated bases. If this phenomenon occurs at other promoters, then the incorrect assignment of the transcriptional start may be more common than is currently realized. Indeed, several promoters align with a distance between the −7 and +1 that is less than the standard 6 bp (see the study by Mitchell et al. [34]). However, studies examining start site selection found that the 6-bp distance between the −10 element and the start of transcription was strongly preferred, such that decreasing the distance by 1 bp moved the start site downstream (24, 28). This makes sense because start site selection is dictated by polymerase structure, with the −10 nontemplate strand DNA held by region 2.3, whereas the template +1 is in the active site. Shorter distances between the transcriptional start site and −10 element might not be tolerated. However, polymerase slippage that specifically incorporates two to three nontemplated NTPs could be interpreted as a short distance between the −7 and +1 positions. Our study suggests that a reconsideration of the DNA elements of promoters with short distances between the −10 element and the transcriptional start is warranted.
Acknowledgments
We thank N. Nossal for the pppACn3 marker and F. Whipple for providing the in vivo lacZ system. We are grateful to R. Bonocora, N. Nossal, and C. Turnbough for helpful discussions and to J. Lee for purification of the His6-tagged σ70 protein.
This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
Published ahead of print on 29 September 2006.
REFERENCES
- 1.Barne, K. A., J. A. Bown, S. J. Busby, and S. D. Minchin. 1997. Region 2.5 of the Escherichia coli RNA polymerase sigma70 subunit is responsible for the recognition of the “extended-10” motif at promoters. EMBO J. 16:4034-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Browning, D. F., and S. J. Busby. 2004. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2:57-65. [DOI] [PubMed] [Google Scholar]
- 3.Callaci, S., E. Heyduk, and T. Heyduk. 1999. Core RNA polymerase from Escherichia coli induces a major change in the domain arrangement of the sigma 70 subunit. Mol. Cell 3:229-238. [DOI] [PubMed] [Google Scholar]
- 4.Campbell, E. A., O. Muzzin, M. Chlenov, J. L. Sun, C. A. Olson, O. Weinman, M. L. Trester-Zedlitz, and S. A. Darst. 2002. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol. Cell 9:527-539. [DOI] [PubMed] [Google Scholar]
- 5.Chan, B., A. Spassky, and S. Busby. 1990. The organization of open complexes between Escherichia coli RNA polymerase and DNA fragments carrying promoters either with or without consensus −35 region sequences. Biochem. J. 270:141-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, Y., S. M. Dylla, and C. L. Turnbough, Jr. 2001. A long T.A tract in the upp initially transcribed region is required for regulation of upp expression by UTP-dependent reiterative transcription in Escherichia coli. J. Bacteriol. 183:221-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craig, M. L., O. V. Tsodikov, K. L. McQuade, P. E. Schlax, Jr., M. W. Capp, R. M. Saecker, and M. T. Record, Jr. 1998. DNA footprints of the two kinetically significant intermediates in formation of an RNA polymerase-promoter open complex: evidence that interactions with start site and downstream DNA induce sequential conformational changes in polymerase and DNA. J. Mol. Biol. 283:741-756. [DOI] [PubMed] [Google Scholar]
- 8.Daniels, D., P. Zuber, and R. Losick. 1990. Two amino acids in an RNA polymerase sigma factor involved in the recognition of adjacent base pairs in the −10 region of a cognate promoter. Proc. Natl. Acad. Sci. USA 87:8075-8079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dombroski, A. J. 1997. Recognition of the −10 promoter sequence by a partial polypeptide of sigma70 in vitro. J. Biol. Chem. 272:3487-3494. [PubMed] [Google Scholar]
- 10.Dombroski, A. J., B. D. Johnson, M. Lonetto, and C. A. Gross. 1996. The sigma subunit of Escherichia coli RNA polymerase senses promoter spacing. Proc. Natl. Acad. Sci. USA 93:8858-8862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fenton, M. S., and J. D. Gralla. 2001. Function of the bacterial TATAAT −10 element as single-stranded DNA during RNA polymerase isomerization. Proc. Natl. Acad. Sci. USA 98:9020-9025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fenton, M. S., S. J. Lee, and J. D. Gralla. 2000. Escherichia coli promoter opening and −10 recognition: mutational analysis of sigma70. EMBO J. 19:1130-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grana, D., T. Gardella, and M. M. Susskind. 1988. The effects of mutations in the ant promoter of phage P22 depend on context. Genetics 120:319-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gregory, B. D., B. E. Nickels, S. A. Darst, and A. Hochschild. 2005. An altered-specificity DNA-binding mutant of Escherichia coli σ70 facilitates the analysis of σ70 function in vivo. Mol. Microbiol. 56:1208-1219. [DOI] [PubMed] [Google Scholar]
- 15.Gruber, T. M., and C. A. Gross. 2003. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 57:441-466. [DOI] [PubMed] [Google Scholar]
- 16.Hawley, D. K., and W. R. McClure. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 11:2237-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hinton, D. M. 1989. Transcript analyses of the uvsX-40-41 region of bacteriophage T4: changes in the RNA as infection proceeds. J. Biol. Chem. 264:14432-14439. [PubMed] [Google Scholar]
- 18.Hinton, D. M. 1991. Transcription from a bacteriophage T4 middle promoter using T4 MotA protein and phage-modified RNA polymerase. J. Biol. Chem. 266:18034-18044. [PubMed] [Google Scholar]
- 19.Hinton, D. M., S. Pande, N. Wais, X. B. Johnson, M. Vuthoori, A. Makela, and I. Hook-Barnard. 2005. Transcriptional takeover by sigma appropriation: remodeling of the sigma70 subunit of Escherichia coli RNA polymerase by the bacteriophage T4 activator MotA and coactivator AsiA. Microbiology 151:1729-1740. [DOI] [PubMed] [Google Scholar]
- 20.Hinton, D. M., S. Vuthoori, and R. Mulamba. 2006. The bacteriophage T4 inhibitor and coactivator AsiA inhibits Escherichia coli RNA Polymerase more rapidly in the absence of sigma70 region 1.1: evidence that region 1.1 stabilizes the interaction between sigma70 and core. J. Bacteriol. 188:1279-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Juang, Y. L., and J. D. Helmann. 1994. A promoter melting region in the primary sigma factor of Bacillus subtilis: identification of functionally important aromatic amino acids. J. Mol. Biol. 235:1470-1488. [DOI] [PubMed] [Google Scholar]
- 22.Keener, J., and M. Nomura. 1993. Dominant lethal phenotype of a mutation in the −35 recognition region of Escherichia coli sigma 70. Proc. Natl. Acad. Sci. USA 90:1751-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar, A., R. A. Malloch, N. Fujita, D. A. Smillie, A. Ishihama, and R. S. Hayward. 1993. The minus 35-recognition region of Escherichia coli sigma 70 is inessential for initiation of transcription at an “extended minus 10” promoter. J. Mol. Biol. 232:406-418. [DOI] [PubMed] [Google Scholar]
- 24.Lewis, D. E., and S. Adhya. 2004. Axiom of determining transcription start points by RNA polymerase in Escherichia coli. Mol. Microbiol. 54:692-701. [DOI] [PubMed] [Google Scholar]
- 25.Lim, H. M., H. J. Lee, S. Roy, and S. Adhya. 2001. A “master” in base unpairing during isomerization of a promoter upon RNA polymerase binding. Proc. Natl. Acad. Sci. USA 98:14849-14852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lisser, S., and H. Margalit. 1993. Compilation of Escherichia coli mRNA promoter sequences. Nucleic Acids Res. 21:1507-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lisser, S., and H. Margalit. 1994. Determination of common structural features in Escherichia coli promoters by computer analysis. Eur. J. Biochem. 223:823-830. [DOI] [PubMed] [Google Scholar]
- 28.Liu, J., and C. L. Turnbough, Jr. 1994. Effects of transcriptional start site sequence and position on nucleotide-sensitive selection of alternative start sites at the pyrC promoter in Escherichia coli. J. Bacteriol. 176:2938-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.March-Amegadzie, R., and D. M. Hinton. 1995. The bacteriophage T4 middle promoter PuvsX: analysis of regions important for binding of the T4 transcriptional activator MotA and for activation of transcription. Mol. Microbiol. 15:649-660. [DOI] [PubMed] [Google Scholar]
- 30.Matlock, D. L., and T. Heyduk. 2000. Sequence determinants for the recognition of the fork junction DNA containing the −10 region of promoter DNA by Escherichia coli RNA polymerase. Biochemistry 39:12274-12283. [DOI] [PubMed] [Google Scholar]
- 31.Maxam, A. M., and W. Gilbert. 1980. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 65:499-560. [DOI] [PubMed] [Google Scholar]
- 32.McClure, W. R., D. K. Hawley, P. Youderian, and M. M. Susskind. 1983. DNA determinants of promoter selectivity in Escherichia coli. Cold Spring Harbor Symp. Quant. Biol. 47(Pt. 1):477-481. [DOI] [PubMed] [Google Scholar]
- 33.Miroslavova, N. S., and S. J. Busby. 2006. Investigations of the modular structure of bacterial promoters. Biochem. Soc. Symp. 73:1-10. [DOI] [PubMed] [Google Scholar]
- 34.Mitchell, J. E., D. Zheng, S. J. Busby, and S. D. Minchin. 2003. Identification and analysis of “extended-10” promoters in Escherichia coli. Nucleic Acids Res. 31:4689-4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mulligan, M. E., J. Brosius, and W. R. McClure. 1985. Characterization in vitro of the effect of spacer length on the activity of Escherichia coli RNA polymerase at the TAC promoter. J. Biol. Chem. 260:3529-3538. [PubMed] [Google Scholar]
- 36.Murakami, K. S., and S. A. Darst. 2003. Bacterial RNA polymerases: the whole story. Curr. Opin. Struct. Biol. 13:31-39. [DOI] [PubMed] [Google Scholar]
- 37.Murakami, K. S., S. Masuda, E. A. Campbell, O. Muzzin, and S. A. Darst. 2002. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science 296:1285-1290. [DOI] [PubMed] [Google Scholar]
- 38.Murakami, K. S., S. Masuda, and S. A. Darst. 2002. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science 296:1280-1284. [DOI] [PubMed] [Google Scholar]
- 39.Paget, M. S., and J. D. Helmann. 2003. The sigma70 family of sigma factors. Genome Biol. 4:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panaghie, G., S. E. Aiyar, K. L. Bobb, R. S. Hayward, and P. L. de Haseth. 2000. Aromatic amino acids in region 2.3 of Escherichia coli sigma 70 participate collectively in the formation of an RNA polymerase-promoter open complex. J. Mol. Biol. 299:1217-1230. [DOI] [PubMed] [Google Scholar]
- 41.Pande, S., A. Makela, S. L. Dove, B. E. Nickels, A. Hochschild, and D. M. Hinton. 2002. The bacteriophage T4 transcription activator MotA interacts with the far-C-terminal region of the sigma70 subunit of Escherichia coli RNA polymerase. J. Bacteriol. 184:3957-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qi, F., C. Liu, L. S. Heath, and C. L. Turnbough, Jr. 1996. In vitro assay for reiterative transcription during transcriptional initiation by Escherichia coli RNA polymerase. Methods Enzymol. 273:71-85. [DOI] [PubMed] [Google Scholar]
- 43.Sanderson, A., J. E. Mitchell, S. D. Minchin, and S. J. Busby. 2003. Substitutions in the Escherichia coli RNA polymerase sigma70 factor that affect recognition of extended −10 elements at promoters. FEBS Lett. 544:199-205. [DOI] [PubMed] [Google Scholar]
- 44.Sasse-Dwight, S., and J. D. Gralla. 1989. KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. J. Biol. Chem. 264:8074-8081. [PubMed] [Google Scholar]
- 45.Siegele, D. A., J. C. Hu, W. A. Walter, and C. A. Gross. 1989. Altered promoter recognition by mutant forms of the sigma 70 subunit of Escherichia coli RNA polymerase. J. Mol. Biol. 206:591-603.2661828 [Google Scholar]
- 46.Studier, F. W. 1991. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J. Mol. Biol. 219:37-44. [DOI] [PubMed] [Google Scholar]
- 47.Thouvenot, B., B. Charpentier, and C. Branlant. 2004. The strong efficiency of the Escherichia coli gapA P1 promoter depends on a complex combination of functional determinants. Biochem. J. 383:371-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomsic, M., L. Tsujikawa, G. Panaghie, Y. Wang, J. Azok, and P. L. deHaseth. 2001. Different roles for basic and aromatic amino acids in conserved region 2 of Escherichia coli sigma(70) in the nucleation and maintenance of the single-stranded DNA bubble in open RNA polymerase-promoter complexes. J. Biol. Chem. 276:31891-31896. [DOI] [PubMed] [Google Scholar]
- 49.Tsujikawa, L., O. V. Tsodikov, and P. L. deHaseth. 2002. Interaction of RNA polymerase with forked DNA: evidence for two kinetically significant intermediates on the pathway to the final complex. Proc. Natl. Acad. Sci. USA 99:3493-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vuthoori, S., C. W. Bowers, A. McCracken, A. J. Dombroski, and D. M. Hinton. 2001. Domain 1.1 of the sigma(70) subunit of Escherichia coli RNA polymerase modulates the formation of stable polymerase/promoter complexes. J. Mol. Biol. 309:561-572. [DOI] [PubMed] [Google Scholar]
- 51.Waldburger, C., and M. M. Susskind. 1994. Probing the informational content of Escherichia coli sigma 70 region 2.3 by combinatorial cassette mutagenesis. J. Mol. Biol. 235:1489-1500. [DOI] [PubMed] [Google Scholar]
- 52.Whipple, F. W. 1998. Genetic analysis of prokaryotic and eukaryotic DNA-binding proteins in Escherichia coli. Nucleic Acids Res. 26:3700-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson, C., and A. J. Dombroski. 1997. Region 1 of sigma70 is required for efficient isomerization and initiation of transcription by Escherichia coli RNA polymerase. J. Mol. Biol. 267:60-74. [DOI] [PubMed] [Google Scholar]
- 54.Zuber, P., J. Healy, H. L. Carter III, S. Cutting, C. P. Moran, Jr., and R. Losick. 1989. Mutation changing the specificity of an RNA polymerase sigma factor. J. Mol. Biol. 206:605-614. [DOI] [PubMed] [Google Scholar]