Abstract
Iron deprivation in bacteria causes the derepression of genes controlled by the ferric uptake regulator (Fur). The present microarray analysis of iron-starved Bacillus subtilis cells grown in minimal medium unveils additional physiological effects on a large number of genes linked to stringent-response regulation and to genes involved in amino acid biosynthesis associated with pathways essential for bacillibactin production.
Iron is an essential cofactor in various biosynthetic and bioenergetic pathways. In many bacteria, the Fur-dependent derepression of iron acquisition genes is a common strategy used to overcome iron starvation (13). The Fur regulon of B. subtilis comprises 39 genes coding mainly for siderophore biosynthesis and several iron transporters (1). The Escherichia coli Fur protein controls an even larger regulon of similar constitution and was shown to regulate the cellular iron-protein content (17). This study investigated the consequences of permanent iron depletion under nonrich growth conditions on global gene expression and shows a complex physiological response beyond Fur regulation.
For our analyses, Bacillus subtilis strain ATCC 21332 (sfp+) (7) was grown in a defined medium without (iron-depleted cultures) or with (iron-replete cultures) the addition of 10 μM FeSO4 as described previously (18). The growth rates of iron-depleted and iron-replete cultures were similar until the late exponential phase. The growth of the iron-depleted culture then declined and, in contrast to that of the iron-replete culture, catechol siderophore secretion greatly increased (see Fig. S1 in the supplemental material), confirming iron as the limiting nutrient in the late exponential phase. At this time point (optical density at 600 nm, ∼0.35), the mRNA populations of iron-depleted and iron-replete cultures were compared by microarray analysis (essentially performed as described previously [15; see also technical details in the supplemental material]). Genes of several prominent functional and regulatory classes exhibited iron-dependent repression or induction (see Tables S1 and S2 in the supplemental material). As expected, iron starvation led to induction of the Fur regulon, although there were slight differences from previous studies of Fur regulation carried out with B. subtilis 168 (sfp0) in broth medium (1). Interestingly, among the Fur-dependent ABC transporters, the feuABC genes for ferri-bacillibactin uptake (18, 20) showed the strongest induction. In contrast, the fhuBGC-fhuD genes for ferrichrome uptake (24) were only slightly induced and the yfmCDEF and yfiYZ-yfhA ferric iron transporter genes were not induced. This might indicate a hierarchical expression of iron transporters when dominant iron chelators such as bacillibactin are present. Additionally, 24 genes were upregulated that were reported to be regulated by the transcriptional repressor CodY (19). It has been suggested that derepression of GTP-activated CodY is mediated by a decreasing cellular GTP pool upon RelA-dependent formation of (p)ppGpp, the second messenger of the stringent response (21). To investigate this relationship more closely, we compared our transcriptome data with a global stringent-response analysis (8). Indeed, there was a high coincidence of gene regulation between the two studies. Among the genes earlier described to be either positively or negatively RelA regulated, we found 19 and 28 genes to be up- and downregulated during iron starvation, respectively. Furthermore, among the genes that were reported to be affected independently of RelA during the stringent response, we found 26 genes showing a similar iron-dependent repression or induction. These genes mainly belonged to the functional categories of amino acid, purine, and pyrimidine biosynthesis. To confirm these and further results of the microarray analysis, iron-dependent repression or induction of selected genes was subsequently compared by an independent dot blot analysis (Fig. 1; see technical details in the supplemental material). The specific transcript detection showed the same expression pattern for both the RelA-dependent genes rpsP and ald and the CodY-dependent genes lpdV and yurO, as revealed by the transcriptome analysis.
FIG. 1.
Dot blot analysis of selected genes. Shown are the relative transcriptional levels of cells cultured in minimal medium under iron-starved (−) or iron-replete (+) conditions. Panels: A, Fur-regulated genes (as controls); B, amino acid biosynthesis genes of the threonine (yclM, hom), serine/glycine (yoaD), and glutamate (gltA) pathways; C, stringent-response-regulated genes; D, CodY-regulated genes; E, genes of the tricarboxylic acid cycle. The arrows indicate increased (↑), decreased (↓), or equal (→) transcript amounts that were detected during iron starvation in comparison with iron-replete conditions.
Several enzymes in amino acid biosynthesis pathways are iron dependent, and iron limitation may subsequently cause amino acid starvation. The most abundant amino acid in both gram-positive and gram-negative bacteria is glutamate. Especially Bacillus spp. need a large intracellular glutamate pool (40 to >100 mM) for vegetative growth and adaptational processes (25). Since B. subtilis lacks an anabolic glutamate dehydrogenase (3), glutamate synthesis is strictly iron dependent as iron is needed to assemble the iron-sulfur cluster bound to glutamate synthase as a cofactor (26). The gltA and gltB genes coding for the large and small chains of the B. subtilis glutamate synthase, respectively, were downregulated during iron depletion (see also Fig. 1). The expression of the gltAB operon, which is triple regulated by TnrA (4), GltC (6), and CcpA (9), depends on a sufficient supply of ammonium and glucose (5, 28). Since both ammonium and glucose were present at nonlimiting concentrations in the minimal medium used, the underexpression of gltAB seems to be a direct result of low iron availability. Furthermore, we found that the citB gene coding for the iron-dependent B. subtilis aconitase involved in substrate supply for GltAB was also downregulated by iron depletion, as observed in previous work (1). citB repression seems to be directly iron dependent, since further genes of the tricarboxylic acid cycle coding for non-iron-dependent enzymes were not affected, such as citZ, which is regulated by the same carbon and nitrogen sources as citB (see also Fig. 1) (14, 22). Altogether, gltAB and citB downregulation indicates an iron-dependent bottleneck of glutamate synthesis. A recent study demonstrating SpoT-dependent (p)ppGpp accumulation during iron limitation in E. coli (27) might indicate similar effects in gram-negative bacteria. However, the stringent response observed in B. subtilis during iron starvation is most likely the consequence of deficiencies in iron-dependent amino acid biosynthesis.
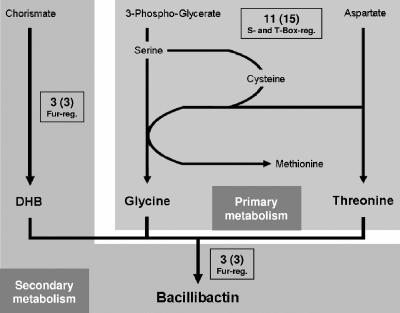
Recently, ferri-bacillibactin was shown to be the major endogenously derived iron source of B. subtilis during iron starvation (18). The utilization of this nonribosomally produced catecholic trilactone (2,3-dihydroxybenzoate-glycine-threonine)3 siderophore is mediated by the Fur-regulated FeuABC uptake system and the BesA trilactone hydrolase (18, 20). While the dhbACEBF operon coding for 2,3-dihydroxybenzoate synthesis (23) and bacillibactin assembly (16) is also controlled by Fur (1), iron-dependent regulation of primary metabolic genes involved in bacillibactin synthesis was not reported. In total, our transcriptome study revealed 11 amino acid biosynthesis genes that were more than 40% upregulated during iron starvation. Strikingly, all of them code for enzymes that are essential for the synthesis of the bacillibactin precursors threonine and glycine, as shown schematically in Fig. 2. Threonine synthesis starting from aspartate needs five enzymatic activities. Five genes coding for four of these activities were upregulated: lysC (aspartokinase II) and yclM (aspartokinase III, thrD), encoding two isozymes for the initial reaction (2, 10), as well as hom, thrB, and thrC, coding for homoserine dehydrogenase, homoserine kinase, and threonine synthase, respectively. In the synthesis pathway leading from 3-phosphoglycerate via serine to glycine, the yoaD gene, coding for a putative paralog of the initial enzyme SerA, was upregulated. The genes yclM, hom, and yoaD were selected for dot blot analysis (Fig. 1). In the amino acid biosynthesis network, the threonine, serine/glycine, and cysteine/methionine pathways are interdependent. In total, there are seven specific enzymatic activities needed for cysteine/methionine synthesis. The genes yjcI, yjcJ, yitJ (12), and cysE, coding for four of these activities, were upregulated. Additionally, yxjG, coding for a protein similar to the methionine synthase MetE, possibly provides a further activity to this pathway. Altogether, 9 out of the 11 genes are either S box (yjcI, yjcJ, yitJ, yoaD, and yxjG) or T box (hom, thrB, thrC, and cysE) regulated (11, 12). Furthermore, yjcI, yjcJ, yitJ, and yoaD were shown to be induced RelA independently during the stringent response (8). Thus, in addition to these regulatory mechanisms, it is tempting to speculate that there could be a specific link between iron starvation and/or bacillibactin synthesis and the threonine, serine/glycine, and cysteine/methionine pathways. However, because of the moderate induction of the precursor biosynthesis genes, a more obvious explanation might be the occurrence of S- and T-box-dependent feedback regulation(s) caused by the consumption of threonine, glycine, and serine (as glycine precursor) in bacillibactin synthesis, thus leaving a “regulatory footprint” in the primary metabolism. However, this is the first time that siderophore synthesis-dependent regulation in the primary metabolism was observed, underlining the importance of bacillibactin as a major iron deficiency rescue system in B. subtilis.
FIG. 2.
Scheme of bacillibactin synthesis. The bacillibactin precursors glycine and threonine derive from the primary metabolism. The synthesis of 2,3-dihydroxybenzoate (DHB) and the nonribosomally catalyzed bacillibactin assembly are Fur regulated in the secondary metabolism. The numbers shown indicate the numbers of genes that were upregulated during iron starvation. The total numbers of activities involved in the presented pathway sections are in parentheses. reg., regulation.
In conclusion, the results of this study show the relevance of both culture medium composition and the capability of siderophore production to global gene expression during iron starvation and establish novel iron-dependent functional and regulatory connections between differentially classified genes.
Supplementary Material
Acknowledgments
We thank the group of E. Bremer (Marburg) for help with RNA preparation and chemiluminescence detection. Anne de Jong, Siger Holsappel, and Anne Sadewasser are acknowledged for technical assistance.
This work was supported by EC grant LSHG-CT-2004-503468.
Footnotes
Published ahead of print on 29 September 2006.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Baichoo, N., T. Wang, R. Ye, and J. D. Helmann. 2002. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 45:1613-1629. [DOI] [PubMed] [Google Scholar]
- 2.Belitsky, B. R. 2002. Biosynthesis of amino acids of the glutamate and aspartate families, alanine, and polyamines, p. 203-231. In A. L. Sonenshein (ed.), Bacillus subtilis and its closest relatives. ASM Press, Washington, D.C.
- 3.Belitsky, B. R., and A. L. Sonenshein. 1998. Role and regulation of Bacillus subtilis glutamate dehydrogenase genes. J. Bacteriol. 180:6298-6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belitsky, B. R., L. V. Wray, Jr., S. H. Fisher, D. E. Bohannon, and A. L. Sonenshein. 2000. Role of TnrA in nitrogen source-dependent repression of Bacillus subtilis glutamate synthase gene expression. J. Bacteriol. 182:5939-5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohannon, D. E., M. S. Rosenkrantz, and A. L. Sonenshein. 1985. Regulation of Bacillus subtilis glutamate synthase genes by the nitrogen source. J. Bacteriol. 163:957-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohannon, D. E., and A. L. Sonenshein. 1989. Positive regulation of glutamate biosynthesis in Bacillus subtilis. J. Bacteriol. 171:4718-4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper, D. G., C. R. Macdonald, S. J. Duff, and N. Kosaric. 1981. Enhanced production of surfactin from Bacillus subtilis by continuous product removal and metal cation additions. Appl. Environ. Microbiol. 42:408-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eymann, C., G. Homuth, C. Scharf, and M. Hecker. 2002. Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis. J. Bacteriol. 184:2500-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faires, N., S. Tobisch, S. Bachem, I. Martin-Verstraete, M. Hecker, and J. Stülke. 1999. The catabolite control protein CcpA controls ammonium assimilation in Bacillus subtilis. J. Mol. Microbiol. Biotechnol. 1:141-148. [PubMed] [Google Scholar]
- 10.Graves, L. M., and R. L. Switzer. 1990. Aspartokinase III, a new isozyme in Bacillus subtilis 168. J. Bacteriol. 172:218-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grundy, F. J., and T. M. Henkin. 1998. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol. Microbiol. 30:737-749. [DOI] [PubMed] [Google Scholar]
- 12.Grundy, F. J., and T. M. Henkin. 2002. Synthesis of serine, glycine, cysteine, and methionine, p. 245-254. In A. L. Sonenshein (ed.), Bacillus subtilis and its closest relatives. ASM Press, Washington, D.C.
- 13.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 14.Jourlin-Castelli, C., N. Mani, M. M. Nakano, and A. L. Sonenshein. 2000. CcpC, a novel regulator of the LysR family required for glucose repression of the citB gene in Bacillus subtilis. J. Mol. Biol. 295:865-878. [DOI] [PubMed] [Google Scholar]
- 15.Lulko, A. T., G. Buist, J. Kok, and O. P. Kuipers. Transcriptome analysis of temporal regulation of carbon-metabolism by CcpA in Bacillus subtilis. J. Mol. Biol. Biotechnol., in press. [DOI] [PubMed]
- 16.May, J. J., T. M. Wendrich, and M. A. Marahiel. 2001. The dhb operon of Bacillus subtilis encodes the biosynthetic template for the catecholic siderophore 2,3-dihydroxybenzoate-glycine-threonine trimeric ester bacillibactin. J. Biol. Chem. 276:7209-7217. [DOI] [PubMed] [Google Scholar]
- 17.McHugh, J. P., F. Rodriguez-Quinones, H. Abdul-Tehrani, D. A. Svistunenko, R. K. Poole, C. E. Cooper, and S. C. Andrews. 2003. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J. Biol. Chem. 278:29478-29486. [DOI] [PubMed] [Google Scholar]
- 18.Miethke, M., O. Klotz, U. Linne, J. J. May, C. L. Beckering, and M. A. Marahiel. 2006. Ferri-bacillibactin uptake and hydrolysis in Bacillus subtilis. Mol. Microbiol. 61:1413-1427. [DOI] [PubMed] [Google Scholar]
- 19.Molle, V., Y. Nakaura, R. P. Shivers, H. Yamaguchi, R. Losick, Y. Fujita, and A. L. Sonenshein. 2003. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J. Bacteriol. 185:1911-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ollinger, J., K. B. Song, H. Antelmann, M. Hecker, and J. D. Helmann. 2006. Role of the Fur regulon in iron transport in Bacillus subtilis. J. Bacteriol. 188:3664-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ratnayake-Lecamwasam, M., P. Serror, K. W. Wong, and A. L. Sonenshein. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 15:1093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenkrantz, M. S., D. W. Dingman, and A. L. Sonenshein. 1985. Bacillus subtilis citB gene is regulated synergistically by glucose and glutamine. J. Bacteriol. 164:155-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rowland, B. M., T. H. Grossman, M. S. Osburne, and H. W. Taber. 1996. Sequence and genetic organization of a Bacillus subtilis operon encoding 2,3-dihydroxybenzoate biosynthetic enzymes. Gene 178:119-123. [DOI] [PubMed] [Google Scholar]
- 24.Schneider, R., and K. Hantke. 1993. Iron-hydroxamate uptake systems in Bacillus subtilis: identification of a lipoprotein as part of a binding protein-dependent transport system. Mol. Microbiol. 8:111-121. [DOI] [PubMed] [Google Scholar]
- 25.Schreier, H. J. 1993. Biosynthesis of glutamine and glutamate and the assimilation of ammonia, p. 281-298. In A. L. Sonenshein (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, D.C.
- 26.van den Heuvel, R. H., D. Ferrari, R. T. Bossi, S. Ravasio, B. Curti, M. A. Vanoni, F. J. Florencio, and A. Mattevi. 2002. Structural studies on the synchronization of catalytic centers in glutamate synthase. J. Biol. Chem. 277:24579-24583. [DOI] [PubMed] [Google Scholar]
- 27.Vinella, D., C. Albrecht, M. Cashel, and R. D'Ari. 2005. Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli. Mol. Microbiol. 56:958-970. [DOI] [PubMed] [Google Scholar]
- 28.Wacker, I., H. Ludwig, I. Reif, H. M. Blencke, C. Detsch, and J. Stülke. 2003. The regulatory link between carbon and nitrogen metabolism in Bacillus subtilis: regulation of the gltAB operon by the catabolite control protein CcpA. Microbiology 149:3001-3009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.