Abstract
The major peptidoglycan hydrolase of Enterococcus faecalis, AtlA, has been identified, but its enzyme activity remains unknown. We have used tandem mass spectrometry analysis of peptidoglycan hydrolysis products obtained using the purified protein to show that AtlA is an N-acetylglucosaminidase. To gain insight into the regulation of its enzyme activity, the three domains of AtlA were purified alone or in combination following expression of truncated forms of the atlA gene in Escherichia coli or partial digestion of AtlA by proteinase K. The central domain of AtlA was catalytically active, but its activity was more than two orders of magnitude lower than that of the complete protein. Partial proteolysis of AtlA was detected in vivo: zymograms of E. faecalis extracts revealed two catalytically active protein bands of 62 and 72 kDa that were both absent in extracts from an atlA null mutant. Limited digestion of AtlA by proteinase K in vitro suggested that the proteolytic cleavage of AtlA in E. faecalis extracts corresponds to the truncation of the N-terminal domain, which is rich in threonine and glutamic acid residues. We show that the truncation of the N-terminal domain from recombinant AtlA has no impact on enzyme activity. The C-terminal domain of the protein, which contains six LysM modules bound to highly purified peptidoglycan, was required for optimal enzyme activity. These data indicate that AtlA is not produced as a proenzyme and that control of the AtlA glucosaminidase activity is likely to occur at the level of LysM-mediated binding to peptidoglycan.
Peptidoglycan (or murein) is a major component of the bacterial cell wall. This molecule forms a bag-shaped exoskeleton enclosing the plasma membrane and protects the cell against internal osmotic pressure in hypoosmotic conditions (23). Peptidoglycan consists of glycan strands of alternating β-1,4-linked N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) residues cross-linked to each other by short peptides made of l- and d-amino acids (20). Throughout growth, the insertion of new precursors and separation of daughter cells requires limited cleavage of the peptidoglycan molecule (13). The enzymes responsible for this process are potentially lethal enzymes referred to as autolysins, as they cleave the high-molecular-weight polymer. In addition to their contribution to cell growth and division, some autolysins play a role in adhesion (8, 17) and in amplification of the inflammatory response by releasing muramyl peptides (6). Depending on the bond they cleave, autolysins are classified as lytic tranglycosylases, N-acetylmuramidases, N-acetylglucosaminidases, N-acetylmuramoyl l-alanine amidases, or endopeptidases.
In Enterococcus faecalis, two autolytic activities have been described (12). One of the corresponding proteins, designated AtlA in this report, has been identified (4, 18), but its activity has not been characterized. AtlA is a three-domain enzyme composed of an N-terminal threonine- and glutamic acid-rich (T/E-rich) domain of unknown function (domain I), a central putative catalytic domain (domain II), and a C-terminal cell wall binding domain consisting of six LysM modules (domain III) (3). In this study, we have identified the peptidoglycan bond cleaved by AtlA and analyzed the contribution of the domains of the protein to its enzyme activity.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All strains and plasmids used in this study are described in Table 1. The bacteria were grown at 37°C in brain heart infusion broth or agar (15 g/liter) (BHI; Difco laboratories, Detroit, Mich.). When required, the growth medium was supplemented with 100 μg/ml ampicillin and 50 μg/ml kanamycin.
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides
| Strain, plasmid, or oligonucleotide | Relevant property | Source or reference |
|---|---|---|
| Strains | ||
| Enterococcus faecalis | ||
| V583 | Sequenced strain (clinical isolate) | 18 |
| OG1RF | 17 | |
| OG1RF atlA | 17 | |
| Micrococcus lysodeikticus | ||
| ATCC 4698 | Pasteur Institute | |
| Escherichia coli | ||
| BL21(DE3)(pREP4GroESL) | Expression strain | 1 |
| XL1-blue | Cloning strain | Stratagene |
| Plasmids | ||
| pET28/16 | pET28a derivative | 7 |
| pET2818 | pET28/16 variant for C-terminal histidine tag fusion | Lab stock |
| pML118 | pET2818 carrying an atlA fragment encoding residues 54 to 737 | This work |
| pML318 | pET2818 carrying an atlA fragment encoding residues 54 to 335 | This work |
| pML418 | pET2818 carrying an atlA fragment encoding residues 182 to 335 | This work |
| pML518 | pET2818 carrying an atlA fragment encoding residues 335 to 737 | This work |
| Oligonucleotides | Sequence (5′→3′) | |
| EF0799-1 | TTCCATGGGGACAGAAGAGCAGCCAACAAATGC | |
| EF0799-2 | TTGGATCAGAAGATGGTGTATCATATTG | |
| EF0799-3 | TTCCATGGGGTCAGAATTTATTGCCGAGTTAGC | |
| EF0799-4 | TTGGATCCACCAACTTTTAAAGTTTGACCAA | |
| EF0799-5 | AAACCATGGGAACGAACACGTACTATACTGTAAAATC |
Plasmid construction.
To construct pML118 encoding amino acids 54 to 737 of AtlA (domains I, II, and III), V583 genomic DNA (19) was PCR amplified using Vent DNA polymerase (Biolabs) and oligonucleotides EF0799-1 and EF0799-4 (Table 1). The resulting fragment was cloned in frame with the hexahistidine sequence of pET2818, a pET2816b derivative (9), using NcoI and BamHI. The same cloning procedure was used to obtain pML318 (encoding domains I and II of AtlA; amino acids 54 to 335) with primers EF0799-1 and EF0799-2; pML418 (encoding domain II; amino acids 182 to 335) with EF0799-3 and EF0799-2; and pML518 (encoding domain III; amino acids 335 to 737) with EF0799-5 and EF0799-4.
Production and purification of histidine-tagged AtlA and its derivatives.
Escherichia coli BL21(DE3)(pREP4GroESL) (1) harboring recombinant plasmids was grown at 37°C in BHI broth containing kanamycin and ampicillin. When the cultures had reached an optical density at 600 nm of 0.7, production of the recombinant protein was induced by addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and incubation was continued for 12 h at 16°C. The cells were harvested, washed, and resuspended in buffer A (50 mM Tris-HCl, pH 7.5, 300 mM NaCl). Crude lysates were obtained by sonication (six times for 30 s; 20% output; Branson Sonifier 450). Proteins were loaded onto Ni2+-nitrilotriacetate agarose resin (QIAGEN GmbH, Hilden, Germany) and eluted with stepwise-increasing concentrations of imidazole (25, 50, 100, and 250 mM in buffer A). AtlA eluting at 100 mM imidazole was further purified by anion exchange chromatography (MonoQ column; Amersham Biosciences, Uppsala, Sweden) using a 0 to 1 M NaCl gradient in 25 mM ethanolamine (pH 9.25). The concentration of purified proteins was determined using the Bio-Rad Protein Assay (Bio-Rad Laboratories GmbH, Postfach, Germany).
AtlA derivatives were purified by the same method, except that a single affinity chromatography step was carried out.
Proteolysis of AtlA.
Purified AtlA (10 μg) was incubated with 10 ng of proteinase K (Boehringer GmbH, Ingelheim, Germany) in 20 μl of 25 mM Tris-HCl, pH 7.5, 25 mM NaCl, 0.5 mM MgCl2, and 2 mM CaCl2 (buffer B). After various incubation times at 37°C (1 to 10 min), aliquots were withdrawn and digestion was stopped by adding phenylmethylsulfonyl fluoride to a final concentration of 2 mM. The samples were analyzed by sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE). For N-terminal sequencing, proteins were transferred onto polyvinylidene difluoride membranes by passive absorption and sequenced using a Perkin-Elmer Procise 494 HT protein sequencer as described elsewhere (10).
To prepare AtlA with a truncated domain I, 400 μg of purified protein was digested with 12.5 ng of proteinase K for 3 h at 37°C in 500 μl of buffer B. The digestion products were separated by size exclusion chromatography on a Superdex75 HR 10/30 column (Amersham Biosciences, Uppsala, Sweden) equilibrated with 20 mM Tris-HCl, pH 7.5, 100 mM NaCl. The fractions containing undigested AtlA and partially digested AtlA were analyzed by SDS-PAGE, pooled separately, and tested for activity.
Cell wall purification and peptidoglycan structural analysis.
Bacteria were grown in 500 ml of BHI broth at 37°C to an optical density at 650 nm of 0.7. Peptidoglycan was extracted by treating the bacterial pellet with 14 ml of 4% SDS at 100°C for 30 min. Peptidoglycan was washed five times by centrifugation (12,000 × g for 10 min at 20°C) with 20 ml of water. Peptidoglycan was serially treated overnight at 37°C with Pronase (200 μg/ml) in 1 ml of Tris-HCl (10 mM, pH 7.4) and with trypsin (200 μg/ml) in 1 ml of phosphate buffer (20 mM, pH 7.8). Peptidoglycan was washed twice with 20 ml of water and digested overnight with mutanolysin (45 μg/ml; Sigma-Aldrich) or AtlA (200 μg/ml) at 37°C in 1 ml of phosphate buffer (25 mM, pH 6.0) containing MgCl2 (0.1 mM). Soluble disaccharide peptides were recovered by ultracentrifugation (100,000 × g for 30 min at 20°C). For reduction of MurNAc to N-acetylmuramitol or GlcNAc to N-acetylglucosaminitol, equal volumes (200 μl) of the solution of disaccharide peptides and of borate buffer (250 mM, pH 9.0) were mixed. Two milligrams of sodium borohydride was added, and the solution was incubated for 20 min at room temperature. The pH of the solution was adjusted to 4.0 with 20% orthophosphoric acid.
The reduced muropeptides were separated by reverse-phase high-performance liquid chromatography (rp-HPLC) on a C18 column (3 μm; 4.6 by 250 mm; Interchrom, Montluçon, France) at a flow rate of 0.5 ml/min with a 0 to 20% gradient applied between 10 and 90 min (buffer A, 0.05% [vol/vol] trifluoroacetic acid in water; buffer B, 0.035% [vol/vol] trifluoroacetic acid in acetonitrile). Mass spectral data were collected with an electrospray time-of-flight mass spectrometer operating in the positive mode (Qstar Pulsar I; Applied Biosystems, Courtaboeuf, France). The data were acquired with a capillary voltage of 5,200 V and a declustering potential of 20 V. The mass scan range was from m/z 350 to 1,500, and the scan cycle was 1 s. Tandem mass spectrometry (MS/MS) was carried out as previously described (2).
Determination of peptidoglycan hydrolase activity.
Hydrolysis of purified cell walls (200 μg/ml) was measured using an Ultrospec 2000 spectrophotometer (Amersham Biosciences, Uppsala, Sweden) and following the decrease in turbidity at 450 nm for 1 h at 37°C in 25 mM Tris-HCl, pH 7.5, 100 mM NaCl buffer. Various dilutions of AtlA and its derivatives were tested to identify conditions in which the velocity of hydrolysis was proportional to enzyme concentration. Enzymatic activity was expressed as A450 units per minute per millimole of protein.
To determine the optimal pH for AtlA activity, a buffer containing 30 mM malonic acid, 30 mM sodium phosphate, 30 mM Tris-HCl, and 30 mM ethanolamine was prepared, and the pH was adjusted as required.
To determine whether partial proteolysis stimulated AtlA activity, 25 ng of proteinase K was added to the reaction mixture containing 1 μg of AtlA in a final volume of 1 ml. In these experiments, aliquots were analyzed by SDS-PAGE to monitor partial digestion of AtlA.
For zymogram analysis, crude extracts were separated by SDS-PAGE using gels containing 0.2% autoclaved Micrococcus lysodeikticus cells. After electrophoresis, the proteins were renatured by incubating the gel for 24 h in 25 mM Tris (pH 8.0) buffer containing 0.1% Triton at 37°C. Lytic activities could be visualized as clear bands on the opaque SDS-PAGE gel.
Analysis of the LysM-peptidoglycan interaction.
Purified peptidoglycan (100 μg) was incubated with purified LysM domain III (10 μg) in 20 mM Tris-HCl (pH 8.0), 500 mM NaCl in a final volume of 125 μl for 30 min at 4°C under agitation. The suspension was centrifuged for 10 min at 15,000 × g, and the supernatant (soluble fraction) was kept for further analyses. The pellet was washed twice with 250 μl of buffer and resuspended in 125 μl of buffer (insoluble fraction). Unbound proteins in the soluble fractions and bound proteins in the insoluble fractions were analyzed by SDS-12% PAGE.
RESULTS AND DISCUSSION
Enzymatic activity and purification of recombinant AtlA.
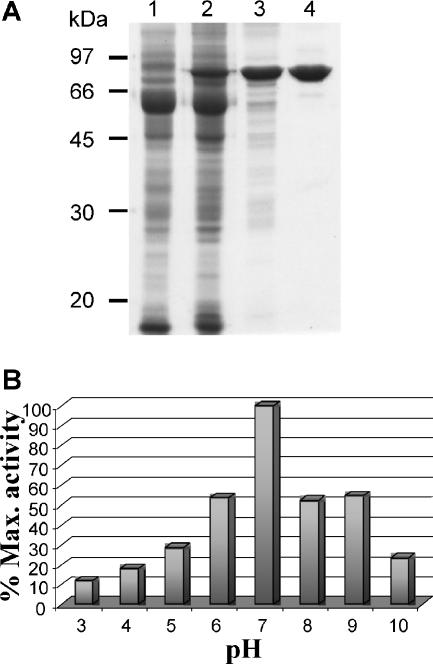
Mature AtlA (residues 54 to 737; EF0799 at www.tigr.org; or ALYS_ENTFA, accession no. P37710 in Swissprot) was produced in E. coli as a C-terminally histidine-tagged protein and purified using affinity and anion exchange chromatography (Fig. 1A). The purified recombinant protein migrated as a 72-kDa polypeptide band on SDS-PAGE, in agreement with the predicted molecular mass of 72.540 kDa. A faint polypeptide band of approximately 62 kDa (Fig. 1A, lane 4) was present in all the purification steps. Storage of the recombinant enzyme at 4°C for 8 weeks led to an increase in the abundance of this 62-kDa polypeptide, indicating that it resulted from proteolysis (data not shown).
FIG. 1.
Purification of AtlA and determination of the optimal pH for activity. (A) Purification of AtlA. Lane 1, crude extract of BL21(DE3)(pREP4GroESL) transformed with pET2818 after IPTG induction (15 μg of protein); lane 2, crude extract of BL21 (DE3)(pREP4GroESL) transformed with pML118 after IPTG induction (15 μg); lane 3, protein fraction eluting from metal affinity chromatography with 100 mM imidazole (5 μg); lane 4, protein fraction eluted with 100 to 200 mM NaCl from the anion exchange column (4 μg). (B) pH activity profile of AtlA. Enzymatic activity was assayed on M. lysodeikticus cell walls at 37°C. Max., maximum.
The optimal pH for the activity of recombinant AtlA was 7.0 at 37°C (Fig. 1B). Preincubation of the enzyme (at a concentration of 15 nM) in 10 mM EDTA did not inhibit its activity, indicating that divalent cations are not essential for AtlA activity. AtlA was more active on M. lysodeikticus (1,900 ± 290 U), a reference substrate for autolysins, than on E. faecalis peptidoglycan (350 ± 20 U). The fact that M. lysodeikticus peptidoglycan is more susceptible to AtlA than homologous peptidoglycan could be due to an unusually large amount of unsubstituted MurNAc residues leading to a low-cross-linked molecule therefore quickly solubilized by the enzyme (14). Alternatively, a lower degree of O acetylation of the M. lysodeikticus peptidoglycan could explain this difference, as O acetylation has been shown to modulate autolysin activity (5, 22).
Determination of AtlA hydrolytic bond specificity.
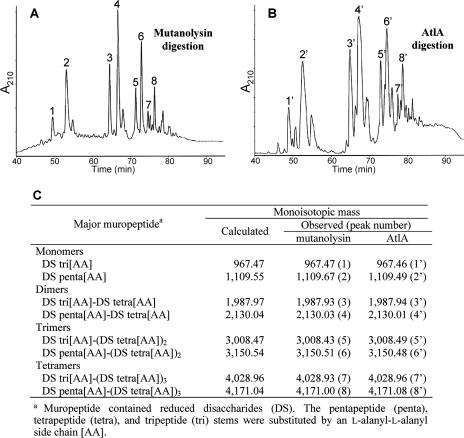
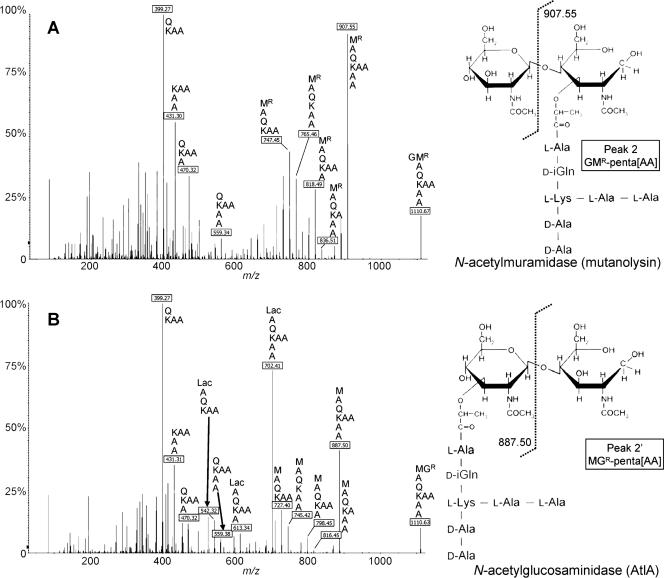
To identify the peptidoglycan bond cleaved by AtlA, we compared the structure of the muropeptides obtained after hydrolysis of E. faecalis OG1RF peptidoglycan by the purified AtlA protein and a commercially available muramidase (mutanolysin). After digestion and reduction, the muropeptides were separated by rp-HPLC on a C18 column (Fig. 2A and B), and the peaks containing the main monomers, dimers, trimers, and tetramers were analyzed by mass spectrometry (MS) (Fig. 2C). The major muropeptides obtained with mutanolysin (peaks 1 to 8) had the same mass as their counterparts obtained after digestion with the purified AtlA protein (peaks 1′ to 8′), confirming that AtlA cleaves the glycan moiety of the peptidoglycan. N-acetylglucosaminidases and N-acetylmuramidases generate muropeptides carrying GlcNAc or MurNAc at the reducing end of the disaccharide, respectively. To discriminate between these two activities, tandem mass spectrometry (MS/MS) was performed on the major muropeptide monomer generated by mutanolysin (Fig. 3A) and AtlA (Fig. 3B). Fragmentation of the ion at m/z 1110.6, corresponding to the [M+H]+ form of a reduced disaccharide pentapeptide replaced by an l-alanyl-l-alanyl side chain (DS-penta[AA]), led to different patterns for the two enzymes. For mutanolysin, loss of unreduced GlcNAc gave an ion at m/z 907.55 as previously described (24). For AtlA, loss of reduced GlcNAc gave an ion at m/z 887.50. Additional loss of alanyl residues from the C terminus of the pentapeptide or the N terminus of the side chain gave additional ions characteristic of the muropeptides generated by mutanolysin (Fig. 3A) and AtlA (Fig. 3B) carrying either unreduced or reduced GlcNAc, respectively. As expected, ions corresponding to peptides resulting from the loss of both sugars were found in the two fragmentation patterns. These data show that MS/MS is a powerful method for discriminating between muropeptides generated by N-acetylmuramidases and those generated by N-acetylglucosaminidases and clearly demonstrate that AtlA displays the latter specificity. Characterization by other techniques of autolysins related to AtlA indicated that the protein family includes both N-acetylmuramidases, such as Mur-2 from E. hirae (11), and N-acetylglucosaminidases, such as AcmA from Lactococcus lactis (21) and LytG from Bacillus subtilis (15).
FIG. 2.
Digestion of E. faecalis peptidoglycan by mutanolysin and AtlA. rp-HPLC muropeptide profiles of OG1RF peptidoglycan digested by mutanolysin (A) or AtlA (B). (C) The mass and predicted structures for peaks 1 to 8 and 1′ to 8′.
FIG. 3.
Determination of AtlA cleavage specificity by MS/MS. The major muropeptide monomers generated by mutanolysin (peak 2) and AtlA (peak 2′) were analyzed by MS/MS yielding fragmentation patterns shown in panels A and B, respectively. The m/z values of the most informative ions are boxed, and the inferred structures are indicated with a one-letter code: M, MurNAc; MR, reduced MurNAc; G, GlcNAc; GR, reduced GlcNAc; A, l-Ala or d-Ala; Lac, d-lactate; K, l-Lys; Q, d-iso-Gln. Percentages on the ordinates show percentages of intensity.
Domain organization of AtlA.
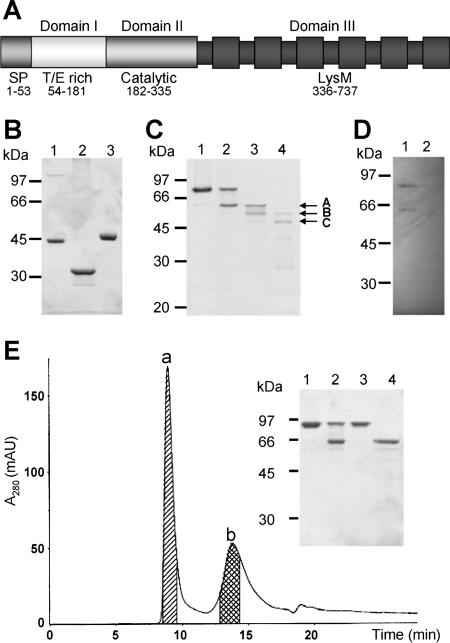
Sequence comparison (data not shown) revealed that the three domains of the AtlA protein are present in different combinations in proteins from various databases, allowing approximate boundaries to be defined as depicted in Fig. 4A. The T/E-rich region is found in E. faecalis AtlA homologs (EF0252 and EF1823; www.tigr.org) as well as in Enterococcus faecium AtlA homologs (contigs 643 and 533; http://genome.jgi-psf.org, database released June 2004). No function has been assigned to this low-complexity region. The central domain is similar to the catalytic domain of several autolysins from gram-positive bacteria, including B. subtilis, L. lactis, and E. hirae (see above) as well as autolysins from E. faecium, Staphylococcus aureus, Streptococcus pyogenes, and Listeria monocytogenes. Finally, the LysM domain is composed of six LysM modules of approximately 50 amino acids. These modules form βααβ secondary structures separated by intervening sequences of 15 to 20 residues (3). LysM modules occur most often in cell wall-degrading enzymes but are also present in many other bacterial proteins (3). The LysM modules bind to peptidoglycan, but the nature of the interaction remains to be characterized.
FIG. 4.
Domain organization of AtlA. (A) Domain organization of AtlA deduced from sequence analysis. SP, signal peptide; T/E-rich, threonine- and glutamic acid-rich region (domain I); Catalytic, catalytic domain (domain II); LysM, LysM domain (domain III). (B) Purification of AtlA and its derivatives overexpressed in E. coli. Lane 1, domains I and II (2 μg); lane 2, domain II alone (2 μg); lane 3, domain III alone (4 μg). (C) Limited digestion of AtlA by proteinase K. Full-length AtlA (lane 1) was digested with proteinase K, and aliquots were withdrawn after 1 min (lane 2), 5 min (lane 3), or 10 min (lane 4). The three polypeptides A, B, and C (indicated by an arrow) were subjected to N-terminal sequencing. (D) Zymogram showing cell wall lytic activity of AtlA. Proteins were separated by SDS-PAGE in a gel containing 0.2% autoclaved M. lysodeikticus cells, renatured in situ, and incubated at 37°C. Lane 1, crude extract of OG1RF (20 μg); lane 2, crude extract of OG1RF atlA (20 μg) (18). (E) Purification of truncated AtlA lacking the N-terminal domain. The full-length AtlA was subjected to limited digestion by proteinase K. The digestion products were loaded on a size-exclusion column to separate undigested AtlA (domains I, II, and III) from AtlA devoid of its N-terminal region (domains II and III). Inset: lane 1, undigested AtlA; lane 2, partial digest; lane 3, purified peak a; lane 4, purified peak b. The unexpected high retention time of domains II and III (peak b) could result from nonspecific interaction of this polypeptide with the Sephadex matrix of the column. AU, absorbance units.
In this study, we have developed two approaches to gain insights into the role of the three domains of AtlA in its enzyme activity. First, fragments of the atlA open reading frame were cloned in an E. coli expression vector, and the corresponding polypeptides were purified. Second, AtlA was partially digested by proteinase K to experimentally probe its domain organization and identify sites sensitive to proteolytic cleavage that might be involved in activation of a putative proenzyme.
Purification of AtlA domains produced in E. coli.
The polypeptides corresponding to the different domains of AtlA were produced in E. coli and purified by affinity chromatography as described in Materials and Methods. Domain II alone, domain III alone, and domains I and II together were successfully purified to homogeneity (Fig. 4B). As domains II and III were produced at a very low level in E. coli, this fragment of AtlA was generated by partial digestion of the mature protein (see below).
Probing of the structural organization of AtlA by limited proteinase K digestion.
The first cleavage by proteinase K generated a protease-resistant core (Fig. 4C, polypeptide A) with an estimated molecular mass of 62 kDa. A polypeptide with a similar apparent molecular mass displaying lytic activity against M. lysodeikticus cells was detected in crude extracts of E. faecalis (Fig. 4D, lane 1). Since no autolytic activity is detected in crude extracts of the E. faecalis OG1RF atlA mutant (Fig. 4D, lane 2), these results suggested that AtlA is cleaved in vivo in the original host. The N-terminal sequence of the 62-kDa polypeptide obtained in vitro was SALSPT, indicating a cleavage between Phe 171 and Ser 172, near the transition between domains I and II as deduced from sequence analysis (Fig. 4A, Ser 181-Glu 182). The corresponding fragment was purified by size-exclusion chromatography (Fig. 4E) for enzymatic analyses. Further proteolysis events (Fig. 4C) gave rise to polypeptides B (56 kDa) and C (50 kDa). These polypeptides had the same N-terminal sequence, suggesting that they resulted from the sequential loss of one or two LysM modules (ca. 6 kDa) from the C terminus of the protein.
The central domain of AtlA is catalytically active.
Domain II alone displayed enzymatic activity, although it was much less active than AtlA (4.85 ± 0.4 U versus 1,900 ± 290 U). This result confirmed that domain II is the catalytic domain of AtlA and indicated that one or both of the N-terminal and the C-terminal domains are required for optimal activity.
The N-terminal T/E-rich domain does not function as a propeptide.
As described above, the zymogram analysis of E. faecalis crude extracts indicated that AtlA is cleaved by endogenous proteases (Fig. 4D). To test whether domain I functions as a propeptide, we compared (i) the activity of domains I, II, and III with that of domains II and III and (ii) the activity of domains I and II with that of domain II alone. The activity of AtlA was similar to that of domains II and III (1,900 ± 290 U versus 2,830 ± 420 U, respectively). Similarly, the activity of domains I and II was similar to that of domain II alone (6.97 ± 0.9 U versus 4.85 ± 0.4 U, respectively). In agreement with these results, the rate of hydrolysis of M. lysodeikticus peptidoglycan by AtlA did not increase upon addition of proteinase K to the reaction mixture (data not shown). Since the addition of exogenous proteases increases the autolysis rate in E. faecalis (18), it is likely that another autolysin (different from AtlA) is activated by proteolysis in this bacterium. This putative autolysin could be related to the E. hirae Mur-1 enzyme, which is also activated by proteolysis (16). Further experiments are required to identify the role of the T/E-rich region, which may be involved in posttranslational modification of AtlA, subcellular targeting, or interaction with protein(s) modulating its activity.
The LysM domain is critical for AtlA activity.
As expected, domain III (consisting of six LysM modules) displayed no enzymatic activity and was able to bind peptidoglycan in vitro (data not shown). The impact of the LysM domain on AtlA activity was tested by comparing (i) the activity of domains I, II, and III with that of domains I and II and (ii) the activity of domains II and III with that of domain II alone. Truncation of domain III from the full-length protein led to a 270-fold reduction in activity. Similarly, the truncation of domain III from domains II and III led to a 580-fold reduction of activity. Altogether, our results show that cell wall binding is critical for full AtlA activity. Zymogram analyses have been used to investigate the activity of AcmA from L. lactis, which is made of a catalytic domain fused to a C-terminal LysM domain. Deletion of the LysM modules of AcmA led to an inactive protein, indicating that the peptidoglycan-binding domain is also important for the activity of this autolysin (21). The critical role of LysM modules suggests that the activity of autolysins may be controlled at the level of binding of the enzymes to their substrate. The binding onto the cell wall may increase the local concentration of the enzyme or may provide proper positioning of the catalytic domain towards its substrate. Alternatively, the LysM domain may be required to induce a proper conformation of the catalytic domain as described for the Streptococcus pneumoniae LytA autolysin (7).
Acknowledgments
We thank Barbara Murray (University of Texas Medical School, Houston) for providing E. faecalis OG1RF and OG1RF atlA and Jean-Claude Huet (INRA, Jouy en Josas, France) for N-terminal sequencing. Sophie Magnet and Laurent Gutmann are acknowledged for constructive comments on the manuscript.
Footnotes
Published ahead of print on 13 October 2006.
REFERENCES
- 1.Amrein, K. E., B. Takacs, M. Stieger, J. Molnos, N. A. Flint, and P. Burn. 1995. Purification and characterization of recombinant human p50csk protein-tyrosine kinase from an Escherichia coli expression system overproducing the bacterial chaperones GroES and GroEL. Proc. Natl. Acad. Sci. USA 92:1048-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arbeloa, A., J. E. Hugonnet, A. C. Sentilhes, N. Josseaume, L. Dubost, C. Monsempes, D. Blanot, J. P. Brouard, and M. Arthur. 2004. Synthesis of mosaic peptidoglycan cross-bridges by hybrid peptidoglycan assembly pathways in gram-positive bacteria. J. Biol. Chem. 279:41546-41556. [DOI] [PubMed] [Google Scholar]
- 3.Bateman, A., and M. Bycroft. 2000. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J. Mol. Biol. 299:1113-1119. [DOI] [PubMed] [Google Scholar]
- 4.Beliveau, C., C. Potvin, J. Trudel, A. Asselin, and G. Bellemare. 1991. Cloning, sequencing, and expression in Escherichia coli of a Streptococcus faecalis autolysin. J. Bacteriol. 173:5619-5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bera, A., S. Herbert, A. Jakob, W. Vollmer, and F. Gotz. 2005. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol. Microbiol. 55:778-787. [DOI] [PubMed] [Google Scholar]
- 6.Boneca, I. G. 2005. The role of peptidoglycan in pathogenesis. Curr. Opin. Microbiol. 8:46-53. [DOI] [PubMed] [Google Scholar]
- 7.Briese, T., and R. Hakenbeck. 1985. Interaction of the pneumococcal amidase with lipoteichoic acid and choline. Eur. J. Biochem. 146:417-427. [DOI] [PubMed] [Google Scholar]
- 8.Cabanes, D., O. Dussurget, P. Dehoux, and P. Cossart. 2004. Auto, a surface associated autolysin of Listeria monocytogenes required for entry into eukaryotic cells and virulence. Mol. Microbiol. 51:1601-1614. [DOI] [PubMed] [Google Scholar]
- 9.Chastanet, A., J. Fert, and T. Msadek. 2003. Comparative genomics reveal novel heat shock regulatory mechanisms in Staphylococcus aureus and other gram-positive bacteria. Mol. Microbiol. 47:1061-1073. [DOI] [PubMed] [Google Scholar]
- 10.Da Costa, B., C. Chevalier, C. Henry, J. C. Huet, S. Petit, J. Lepault, H. Boot, and B. Delmas. 2002. The capsid of infectious bursal disease virus contains several small peptides arising from the maturation process of pVP2. J. Virol. 76:2393-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dolinger, D. L., L. Daneo-Moore, and G. D. Shockman. 1989. The second peptidoglycan hydrolase of Streptococcus faecium ATCC 9790 covalently binds penicillin. J. Bacteriol. 171:4355-4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fontana, R., M. Boaretti, A. Grossato, E. A. Tonin, M. M. Lleo, and G. Satta. 1990. Paradoxical response of Enterococcus faecalis to the bactericidal activity of penicillin is associated with reduced activity of one autolysin. Antimicrob. Agents Chemother. 34:314-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster, S. J., and D. L. Popham. 2001. Structure and synthesis of cell wall, spore cortex, teichoic acids, S-layers, and capsules, p. 21-41. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 14.Ghuysen, J. M., E. Bricas, M. Lache, and M. Leyh-Bouille. 1968. Structure of the cell walls of Micrococcus lysodeikticus. 3. Isolation of a new peptide dimer, N-alpha-[L-alanyl-gamma-(alpha-D-glutamylglycine)]-L-lysyl-D-alanyl-N-alpha-[L-alanyl-gamma-(alpha-D-glutamylglycine)]-L-lysyl-D-alanine. Biochemistry 7:1450-1460. [DOI] [PubMed] [Google Scholar]
- 15.Horsburgh, G. J., A. Atrih, M. P. Williamson, and S. J. Foster. 2003. LytG of Bacillus subtilis is a novel peptidoglycan hydrolase: the major active glucosaminidase. Biochemistry 42:257-264. [DOI] [PubMed] [Google Scholar]
- 16.Kawamura, T., and G. D. Shockman. 1983. Purification and some properties of the endogenous, autolytic N-acetylmuramoylhydrolase of Streptococcus faecium, a bacterial glycoenzyme. J. Biol. Chem. 258:9514-9521. [PubMed] [Google Scholar]
- 17.Milohanic, E., R. Jonquieres, P. Cossart, P. Berche, and J. L. Gaillard. 2001. The autolysin Ami contributes to the adhesion of Listeria monocytogenes to eukaryotic cells via its cell wall anchor. Mol. Microbiol. 39:1212-1224. [DOI] [PubMed] [Google Scholar]
- 18.Qin, X., K. V. Singh, Y. Xu, G. M. Weinstock, and B. E. Murray. 1998. Effect of disruption of a gene encoding an autolysin of Enterococcus faecalis OG1RF. Antimicrob. Agents Chemother. 42:2883-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sahm, D. F., J. Kissinger, M. S. Gilmore, P. R. Murray, R. Mulder, J. Solliday, and B. Clarke. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33:1588-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steen, A., G. Buist, G. J. Horsburgh, G. Venema, O. P. Kuipers, S. J. Foster, and J. Kok. 2005. AcmA of Lactococcus lactis is an N-acetylglucosaminidase with an optimal number of LysM domains for proper functioning. FEBS J. 272:2854-2868. [DOI] [PubMed] [Google Scholar]
- 22.Strating, H., and A. J. Clarke. 2001. Differentiation of bacterial autolysins by zymogram analysis. Anal. Biochem. 291:149-154. [DOI] [PubMed] [Google Scholar]
- 23.Weidel, W., and H. Pelzer. 1964. Bagshaped macromolecules - a new outlook on bacterial cell walls. Adv. Enzymol. Relat. Areas Mol. Biol. 26:193-232. [DOI] [PubMed] [Google Scholar]
- 24.Xu, N., Z. H. Huang, B. L. de Jonge, and D. A. Gage. 1997. Structural characterization of peptidoglycan muropeptides by matrix-assisted laser desorption ionization mass spectrometry and postsource decay analysis. Anal. Biochem. 248:7-14. [DOI] [PubMed] [Google Scholar]