Abstract
Heterocyst differentiation in the cyanobacterium Anabaena sp. strain PCC 7120 requires NtcA, the global nitrogen regulator in cyanobacteria, and HetR, the master regulator of heterocyst differentiation. Expression of hetR is upregulated by nitrogen deprivation, and its upregulation depends on NtcA. However, it has not yet been revealed how NtcA regulates the expression of hetR. In the experiments presented here, it was confirmed that NrrA (All4312), a nitrogen-responsive response regulator, was required for the upregulation of hetR. The use of the nitrogen-responsive transcription initiation sites (TISs) for the hetR gene depended upon NrrA. NrrA bound specifically to the region upstream of TISs located at positions −728 and −696 in vitro. Overexpression of nrrA resulted in enhanced hetR expression and heterocyst formation. A molecular regulatory cascade is proposed whereby NtcA upregulates the expression of nrrA upon limitation of combined nitrogen in the medium and then NrrA upregulates the expression of hetR, leading to heterocyst differentiation.
Heterocysts are terminally differentiated cells of cyanobacteria that are capable of fixing atmospheric nitrogen. Upon limitation of combined nitrogen in the medium, particular vegetative cells change their form into heterocysts with a regular spacing of 10 to 15 cells (10, 12, 29). In the filamentous cyanobacterium Anabaena sp. strain PCC 7120, about 10% of genes on the chromosome are estimated to be upregulated during heterocyst development (6, 7). Heterocysts markedly differ from vegetative cells in physiological and morphological aspects (30). Heterocyst development provides an excellent model for studying cell differentiation and multicellular pattern formation.
The hetR gene is a master gene that regulates heterocyst differentiation. While mutation in the hetR gene blocks early steps in differentiation, extra copies of hetR result in the formation of multiple contiguous heterocysts (Mch phenotype) under nitrogen-depleted conditions (4). Expression of hetR from the petE promoter, which is regulated by copper, induces heterocyst differentiation irrespective of cellular nitrogen status (5). The level of the hetR transcripts increases shortly after nitrogen deprivation, and its expression is localized to heterocysts (2). The upregulation of hetR depends on ntcA (9).
The ntcA gene, which is required for heterocyst development (9, 28), encodes a transcriptional regulator that globally controls nitrogen metabolism in cyanobacteria (19, 27). The NtcA protein, belonging to the cyclic AMP receptor protein family of bacterial regulators, binds to promoter regions containing the sequence signature GTAN8TAC (12), and an eight-base palindromic sequence, TGTAN8TACA, is shown to be the optimal NtcA-binding sequence by in vitro selection (15). The DNA-binding activity of NtcA is enhanced by 2-oxoglutarate (2-OG) (17, 26), and NtcA-dependent transcription from the glnA promoter of Synechococcus sp. strain PCC 7942 increases in the presence of 2-OG in vitro (25). 2-OG is proposed to be a signaling molecule that transmits information regarding cellular nitrogen status (24) and that triggers heterocyst differentiation (17). The NtcA-dependent promoters of hetR do not contain the NtcA recognition sequence, and NtcA does not bind to DNA fragments corresponding to the hetR promoter regions (23). Thus, the molecular basis of the NtcA-dependent regulation of hetR remains unknown.
Recently we found that the nrrA gene (all4312), encoding a response regulator of the OmpR family, was involved in the regulation of heterocyst development (6). The nrrA gene is upregulated by nitrogen deprivation, but the induction of nrrA does not take place in the ntcA mutant strain (6, 22). The promoter sequence of the nrrA gene is consistent with the NtcA-activated promoter sequence, and NtcA binds to the promoter region of nrrA (22). The deletion of nrrA delays heterocyst development and diminished hetR induction under nitrogen deprivation (6).
In this study, we investigated the regulation of hetR expression by NrrA. The NrrA protein showed sequence-specific binding to the promoter region of hetR. In Anabaena sp. strain PCC 7120 having extra copies of the nrrA gene on a plasmid, the amount of the hetR transcript increased and heterocyst formation was enhanced. These results indicate that NrrA directly regulates hetR expression. A molecular cascade between activation of NtcA and upregulation of hetR in the early stage of heterocyst differentiation is linked by NrrA, i.e., NtcA upregulates the expression of nrrA, and then NrrA upregulates the expression of hetR during heterocyst differentiation.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Anabaena sp. strain PCC 7120 and its derivatives were grown in BG-11 medium as described previously (6). The Escherichia coli strains used as hosts were JM109 [recA1 endA1 gyrA96 thi hsdR17(rK− mK+) supE44 relA1 Δ(lac-proAB)/F′ (traD36 proAB lacIq lacZΔM15)] for cloning and BL21(DE3) [F− ompT hsdS(rB− mB−) dcm gal λ(DE3)] for the expression of recombinant proteins. E. coli strains were grown in Luria-Bertani medium (21).
Plasmid constructions.
A DNA fragment containing the nrrA promoter and coding regions was amplified by PCR using the primer pair 4312F2 (5′-CAGGCTGAAATGTGAAGGGTA-3′) and 4312R (5′-AACCAAGCCGATGAAGAATG-3′) and was cloned into the EcoRV site of pBluescript II SK+ (Stratagene, CA). This fragment was excised by digestion with BamHI and KpnI and was cloned between the BamHI and KpnI sites of pRA201 (8) to construct pRA4312.
For construction of an expression plasmid for the histidine-tagged NrrA (His-NrrA) protein, a DNA fragment containing the nrrA gene was amplified by PCR using the primer pair NrrA-1F (5′-GACATATGGGTTCGGTTTGTATTGAA-3′) and NrrA-R (5′-ATGTCGACTCCCTCTGCCTCCTCTACAC-3′). The amplified DNA fragment was cloned between the NdeI and SalI sites of the pET-28a expression vector (Merck KGaA, Darmstadt, Germany). The resulting plasmid, pENrrA1, contains the nrrA gene fused to the His tag sequence.
RNA gel blot analysis and primer extension analysis.
Extraction of total RNA and analysis of RNA gel blots were carried out as described previously (6).
Two fluorescence-labeled primers, IRD-hetR-120 (5′-GGTCAAGATGCTCATTCCTC-3′) and IRD-hetR-603 (5′-TGCTCTTATGGCAGTGTAGG-3′), were used for primer extension analysis as described previously (6). DNA sequencing was carried out with a SequiTherm EXCELII DNA sequencing kit LC (EPICENTRE, WI) using a plasmid pBhetRu containing the region from −967 to +25 with respect to the translation start site of the hetR gene as a template. The cDNA and sequencing products were analyzed with a model 4200 DNA sequencer (LI-COR Biosciences, NE).
Expression and purification of His-NrrA.
E. coli BL21(DE3) cells harboring pENrrA1 were grown at 37°C in l liter of Luria-Bertani medium supplemented with kanamycin (30 μg/ml). The recombinant gene was expressed in exponentially growing cells (optical density at 600 nm of 0.6) by adding 1 mM isopropyl-β-d-thiogalactopyranoside. After 1 h of incubation, the cells were harvested by centrifugation, washed with buffer A (20 mM Tris-HCl [pH 8.0], 0.3 M NaCl, 10% glycerol), resuspended in 30 ml of buffer A, and disrupted with a French pressure cell (20,000 lb/in2). The cell extract was centrifuged at 16,000 × g for 10 min, and the supernatant was further centrifuged at 150,000 × g for 60 min. The 150,000 × g supernatant was loaded onto a HiTrap chelating HP column (GE Healthcare Bio-Science, NJ) equilibrated with buffer A, and the proteins were eluted using a gradient of 30, 60, 100, and 250 mM imidazole in buffer A. The 250 mM imidazole fraction containing purified His-NrrA was loaded onto a PD-10 column (GE Healthcare Bio-Science) equilibrated with buffer B (20 mM Tris-HCl [pH 8.0], 0.1 M NaCl, 10% glycerol), and the protein was eluted with buffer B.
Gel mobility shift assay.
Two DNA fragments, F1 and F2, were generated by PCR using the primer pair hetRF-876 (5′-AACTCTGGACTTCTGGCTCA-3′) and hetRR-659 (5′-GACAGGTAATTCCAACTTAGCAA-3′) and primer pair hetRF-440 (5′-CACCTCAAGGAGAGATTGTGC-3′) and hetRR-182 (5′-CGTGCTTGTATCTTACCGAAA-3′), respectively. The digoxigenin (DIG)-labeled primers, hetRF-876 and hetR-F440, were purchased from Japan Bio Services (Saitama, Japan). The DIG-labeled DNA fragments (0.1 ng) were incubated with His-NrrA in 20 μl of the binding buffer [20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol, 10% glycerol, 0.5 μg/ml of poly(dI-dC)] for 30 min at room temperature. The mixtures were subjected to electrophoresis on a native 5% polyacrylamide gel and transferred to a nylon membrane (Hybond-N+; GE Healthcare Bio-Science). The probes were detected with an alkaline phosphatase-conjugated anti-DIG Fab fragment and a chemiluminescence reagent CDP-Star (Roche Diagnostics, Basel, Switzerland) according to the recommendation of the supplier.
DNase I footprinting analysis.
A fluorescence-labeled DNA fragment was generated by PCR using the primer pair hetRF-876 and IRD-hetR-603. The fluorescence-labeled DNA fragment (500 ng) was incubated with His-NrrA in 25 μl of the binding buffer containing 40 μg/ml of poly(dI-dC) for 30 min at room temperature. Then, 0.2 U of DNase I (RNase-free) (TaKaRa Bio, Shiga, Japan) was added to the reaction mixture, which was then incubated for 10 seconds at 30°C. The reaction was terminated by the addition of 25 μl of 20 mM EDTA. The reaction mixture was extracted once with phenol-chloroform-isoamyl alcohol (25:24:1), and DNA was collected by ethanol precipitation. The partially digested DNA fragments were analyzed with a model 4200 DNA sequencer (LI-COR Biosciences).
Other analytical procedures.
Protein concentration was determined by the method of Bradford (3) using bovine serum albumin as the standard. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out in polyacrylamide gels containing 0.1% SDS by the method of Laemmli (16).
RESULTS
NrrA upregulates the hetR gene under nitrogen deprivation.
Figure 1 shows the result of primer extension analyses of the hetR transcripts using RNA isolated from Anabaena sp. strain PCC 7120 and the nrrA deletion mutant strain DR4312S (6). In Anabaena sp. strain PCC 7120, four putative transcription initiation sites (TISs) were detected as reported previously (5, 23). Under the present experimental conditions, the TISs reported at positions −271 and −184 were detected at positions −273 and −185, respectively. The use of the TIS located at −185 was independent of nitrogen status, while the use of the other three TISs (−728, −696 and −273) was activated by nitrogen deprivation (Fig. 1, lanes 1 and 2). Deletion of nrrA blocked induction of transcription from the nitrogen-responsive TISs after 3 h of nitrogen deprivation (lanes 3 and 4), indicating that the use of these TISs depended on NrrA. The constitutive expression from position −185 was not affected by deletion of nrrA, suggesting that NrrA was not required for transcription from this TIS.
FIG. 1.
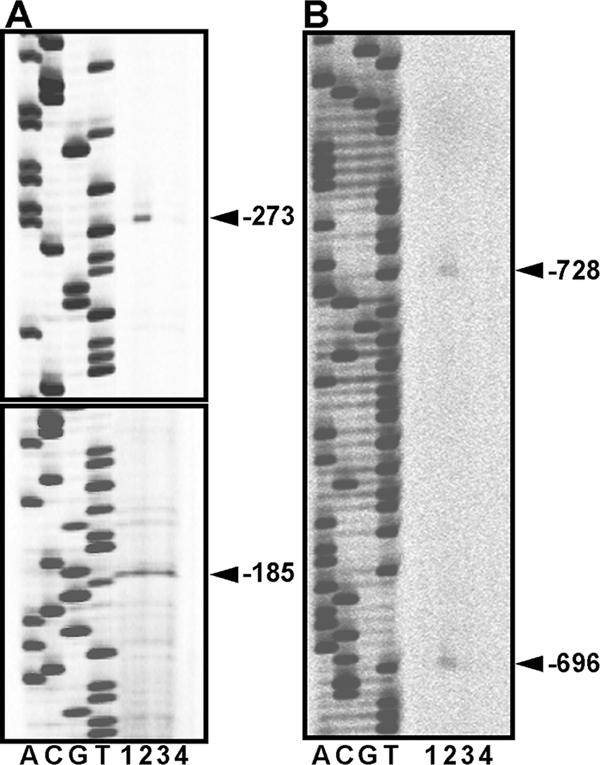
Primer extension analyses of the expression of hetR in Anabaena sp. strain PCC 7120 and the nrrA deletion mutant DR4312S. RNA was isolated from Anabaena sp. strain PCC 7120 (lanes 1 and 2) and DR4312S (lanes 3 and 4) cells grown with nitrate (lanes 1 and 3) or cells subjected to nitrogen deficiency for 3 h (lanes 2 and 4). Oligonucleotides used as primers were IRD-hetR-120 (A) and IRD-hetR-603 (B). Sequencing ladders were generated with the corresponding primers and pBhetRu. The arrowheads indicate the positions of the putative transcription initiation sites.
NrrA binds to the region upstream of the hetR gene.
The NrrA protein was expressed as a recombinant protein fused to the His tag sequence at the amino terminus (His-NrrA) in E. coli cells bearing plasmid pENrrA1 (Fig. 2A). Two bands of affinity-purified His-NrrA were observed by SDS-PAGE (Fig. 2A, lane 2) and both of them had the His tag sequence (lane 4). The molecular mass of a major band was estimated to be 30 kDa, corresponding closely to the theoretical value. The minor band observed just below the major band had probably been cleaved at the carboxyl terminus.
FIG. 2.
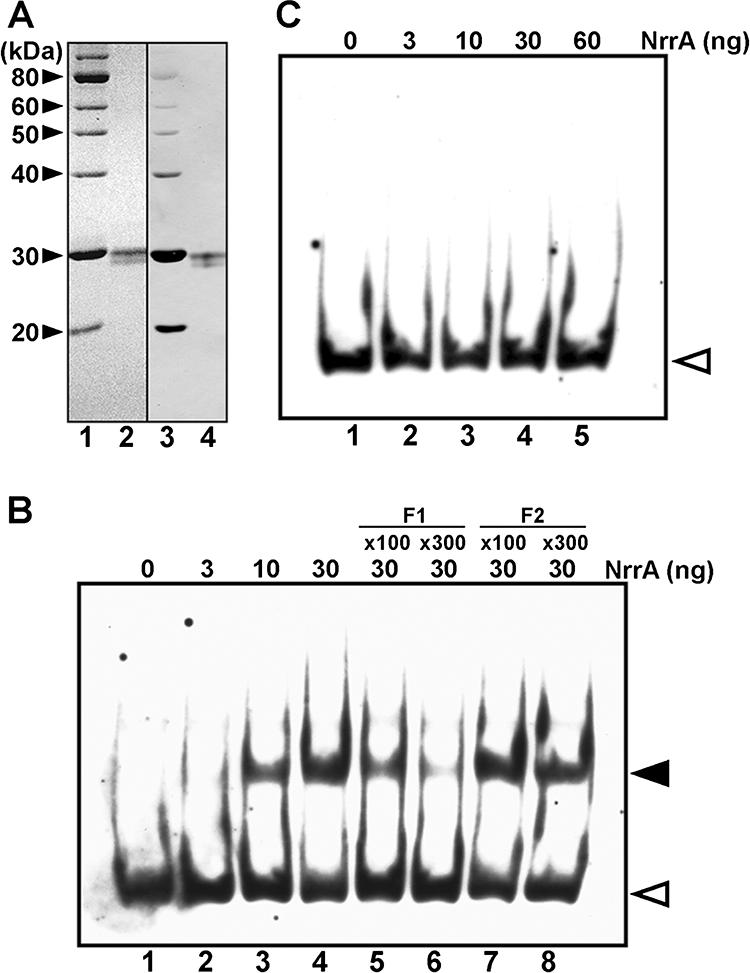
(A) Purification of His-tagged NrrA. SDS-PAGE was carried out using a 12% polyacrylamide gel. Lanes 1 and 3, His-tagged molecular size markers; lanes 2 and 4, His-tagged NrrA (1 μg). The gel was stained with Coomassie brilliant blue (lanes 1 and 2). His-tagged proteins were detected with horseradish peroxidase-conjugated Ni-nitrilotriacetic acid (lanes 3 and 4). (B and C) Gel mobility shift assays with His-NrrA and the regions upstream of the hetR gene. The binding of His-NrrA to fragment F1 (B) and fragment F2 (C), corresponding to the region from −876 to −659 and from −440 to −182 with respect to the translation start site of the hetR gene, respectively, was examined. His-NrrA was added in the amounts indicated above each lane. In panel B, nonlabeled F1 (lanes 5 and 6) and F2 (lanes 7 and 8) fragments were added at the indicated excess amounts. Open arrowheads indicate the positions of the free probe, and a closed arrowhead indicates the position of the NrrA-F1 complex.
Gel mobility shift assays were carried out with purified His-NrrA and DNA fragments of the region upstream of hetR (Fig. 2B and C). His-NrrA reduced the electrophoretic mobility of the DNA probe F1 that included the TISs located at positions −728 and −696, and the amount of the NrrA-F1 complex increased in proportion to the concentration of His-NrrA (Fig. 2B, lanes 1 to 4). However, His-NrrA did not form a complex with probe F2 that included the TISs at positions −273 and −185 (Fig. 2C). The band intensity of the NrrA-F1 complex was reduced by the addition of a nonlabeled F1 fragment but was not affected by the addition of a nonlabeled F2 fragment (Fig. 2B, lanes 5 to 8). These results show that His-NrrA binds to the probe F1, corresponding to the region from −876 to −659 with respect to the translation start site of the hetR gene, in a sequence-specific manner.
Figure 3 shows the result of a DNase I footprinting assay performed using His-NrrA and the region upstream of hetR. The His-NrrA protein protected a region of 27 bp, covering −844 to −818 with respect to the translation start site of the hetR gene, from DNase I digestion. The region protected by NrrA includes tandemly repeated sequences CTT(A/G)AT(G/T)T. The NrrA protein belongs to the OmpR family, and response regulators of the OmpR family recognize tandemly repeated sequences (13, 20).
FIG. 3.
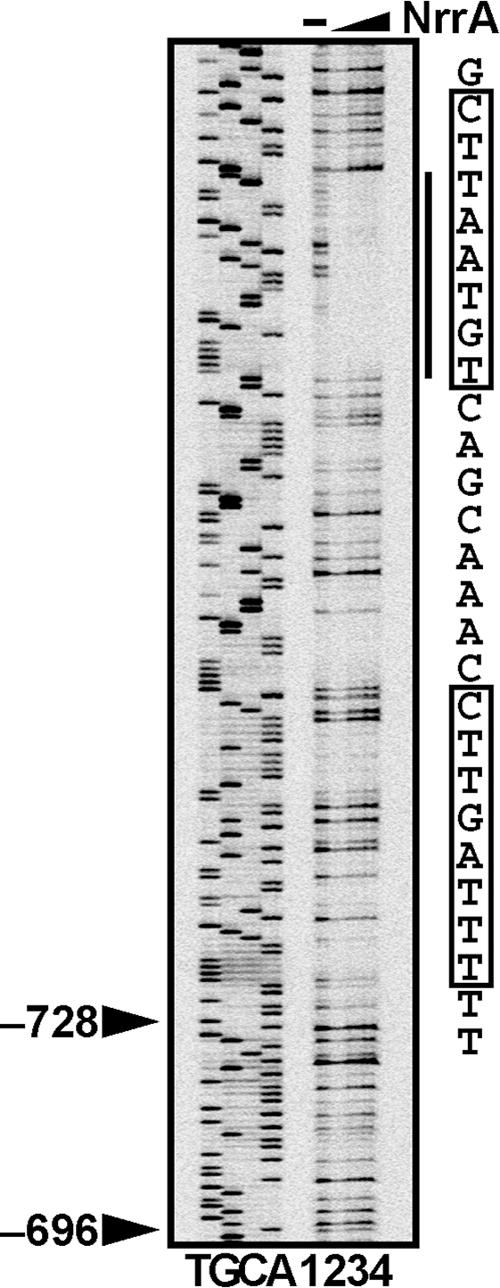
DNase I footprinting assay with His-NrrA and the region upstream of the hetR gene. The patterns of fragments resulting from digestion with DNase I of the fluorescence-labeled DNA fragment containing the region from −876 to −603 with respect to the translation start site of the hetR gene are shown. His-NrrA was added in the concentration of 0 (lane 1), 0.25 (lane 2), 0.5 (lane 3), and 1 μM (lane 4) in each reaction. Sequencing ladders were generated with primer IRD-hetR-603 and pBhetRu. Arrowheads indicate the positions of the transcription initiation sites. A region protected from DNase I digestion is shown by a solid line, and its sequence is shown on the right. Candidates for the NrrA recognition sequence are indicated by boxes.
Extra copies of the nrrA gene enhance the expression of hetR.
A plasmid carrying the nrrA gene, pRA4312, and the control plasmid pRA201 were transferred into Anabaena sp. strain PCC 7120 by conjugation, and transformants were selected for resistance to neomycin. In Anabaena sp. strain PCC 7120 bearing pRA201, the nrrA transcript was not detected in the presence of nitrate and was induced within 3 h after nitrogen deprivation (Fig. 4A). In the strain bearing pRA4312, the nrrA transcript was detected even in the presence of nitrate and markedly increased after nitrogen deprivation (Fig. 4A). Overexpression of the nrrA gene increased the transcript level of hetR, while the increase was less than that of the nrrA transcript (Fig. 4B). The amount of the hetR transcripts increased by twofold after 3 and 8 h of nitrogen deprivation and by threefold after 24 h of nitrogen deprivation in the strain bearing pRA4312.
FIG. 4.
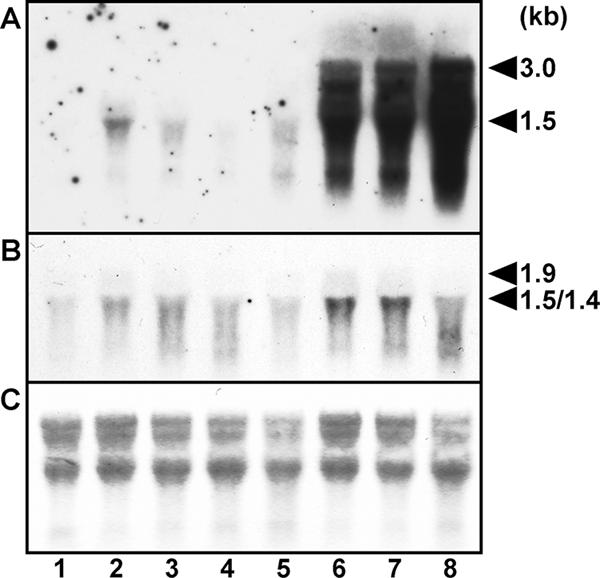
RNA gel blot analysis in Anabaena sp. strain PCC 7120 having extra copies of the nrrA gene on a plasmid. Total RNA was isolated from nitrate-grown cells (lanes 1 and 5) and cells subjected to nitrogen deficiency for 3 h (lanes 2 and 6), 8 h (lanes 3 and 7), and 24 h (lanes 4 and 8). Lanes 1 to 4, Anabaena sp. strain PCC 7120 bearing the control plasmid pRA201; lanes 5 to 8,: Anabaena sp. strain PCC 7120 bearing pRA4312, a plasmid carrying the nrrA gene. The filters were hybridized to an nrrA probe (A) and a hetR probe (B). RNA was stained with methylene blue (C). The sizes of transcripts are shown to the right of the panels.
Figure 5 shows the effect of overexpression of the nrrA gene on heterocyst formation. At least 1,000 cells each from two independently grown cultures were scored to determine the frequency of heterocysts. The heterocyst frequency in Anabaena sp. strain PCC 7120 bearing pRA201 grown with nitrate was 0.8%, while it was 2.1% in the strain bearing pRA4312 (Fig. 5A and B). After nitrogen deprivation, heterocyst differentiation was induced and a multiple contiguous heterocyst (Mch) phenotype with pairs of heterocysts and occasionally groups of three or four heterocysts was observed in the multicopy nrrA strain (Fig. 5C). Many free heterocysts detached from filaments, probably resulting from breakage of labile connections between contiguous heterocysts, were also observed (Fig. 5C). Heterocyst formation was completely repressed by the addition of ammonium in both strains (data not shown).
FIG. 5.
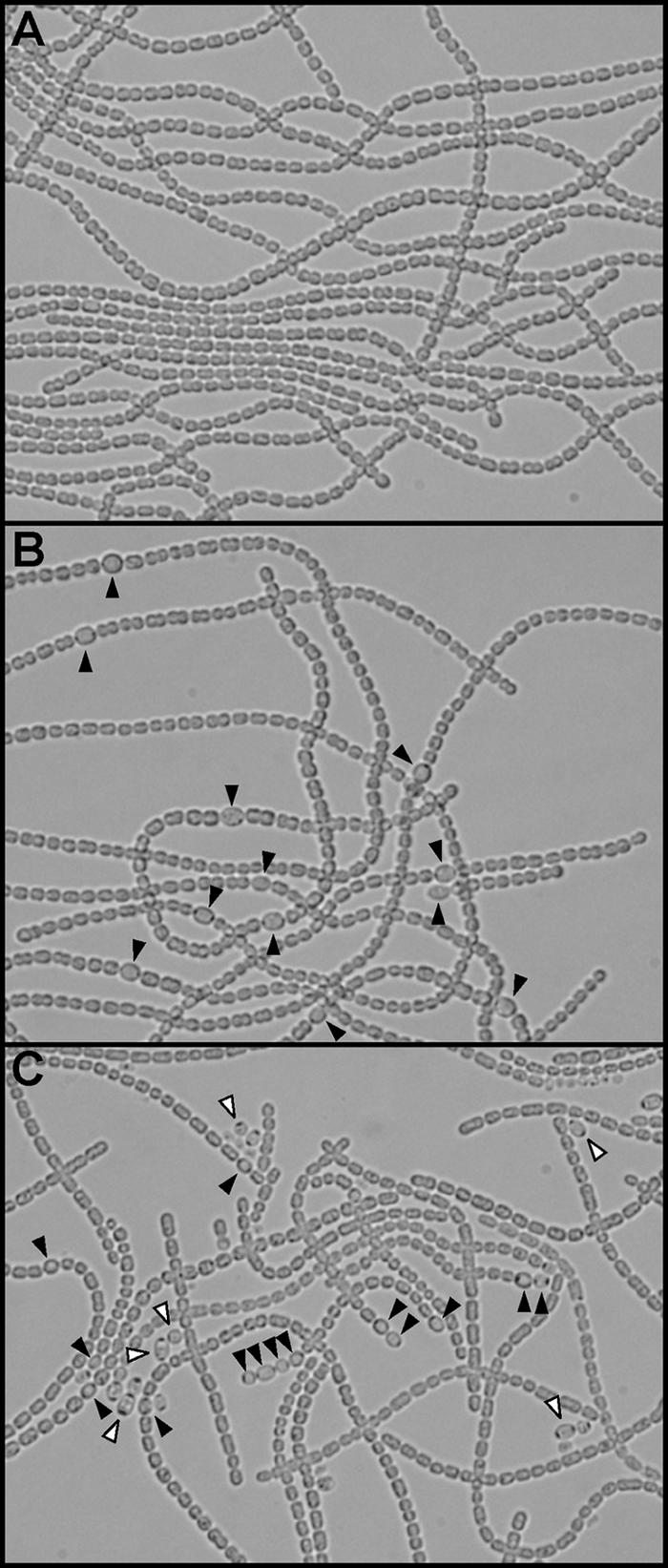
Light microscopy of Anabaena sp. strain PCC 7120 having extra copies of the nrrA gene. Anabaena sp. strain PCC 7120 bearing pRA201 was grown with nitrate (A). Anabaena sp. strain PCC 7120 bearing pRA4312 was grown with nitrate (B) or without combined nitrogen (C). Closed arrowheads show heterocysts. Open arrowheads show heterocysts detached from filaments.
DISCUSSION
NtcA and HetR play the main regulatory roles in the early stage of heterocyst development. The induction of hetR expression during heterocyst differentiation requires NtcA, whereas molecular mechanisms of regulation of the hetR expression by NtcA have not been revealed (9, 23). In this study, we showed that the nrrA gene was required for the upregulation of hetR by nitrogen deprivation (Fig. 1) and that NrrA specifically bound to the region upstream of hetR in vitro (Fig. 2 and 3). In Anabaena sp. strain PCC 7120 having extra copies of the nrrA gene, the amount of hetR transcripts and the frequency of heterocysts were increased (Fig. 4 and 5). Excessive NrrA could increase the level of the expression of hetR in vivo and lead to supernumerary heterocysts. These results indicate that NrrA directly regulates the expression of hetR during heterocyst differentiation.
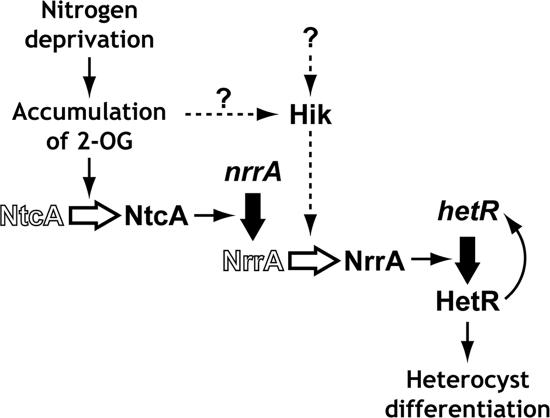
Figure 6 shows a model for sequential activation of gene expression in the early stage of heterocyst differentiation. Limitation of combined nitrogen in the medium causes accumulation of 2-oxoglutarate within the cells of Anabaena sp. strain PCC 7120 (17, 18). It has been proposed that accumulated 2-OG serves as the nitrogen limitation signal triggering the signal pathways that lead to heterocyst differentiation (17, 18). 2-OG enhances the DNA-binding activity of NtcA (17). NtcA directly regulates the expression of genes involved in heterocyst maturation, such as hetC and devBCA, as well as genes involved in nitrogen assimilation, such as glnA and the nir operon (11). NtcA also regulates the expression of nrrA (22). The nrrA gene is upregulated by nitrogen deprivation and is involved in regulation of heterocyst differentiation. In the nrrA deletion mutant, heterocyst formation after nitrogen deprivation is delayed, and the hetR induction by nitrogen deprivation is reduced up to 24 h (6). It was shown that the nrrA gene was required for the initiation of transcription from the nitrogen-responsive TISs of hetR located at positions −728, −696, and −273 (Fig. 1). The 1.9-kb transcript of hetR originates from positions −728 and −696, and the 1.5-kb transcript originates from position −273 (5). In the nrrA mutant, the 1.9-kb transcript is not detected, and the level of induction of the 1.5-kb transcript is reduced after nitrogen deprivation (6). NrrA bound to the region upstream of the TISs at positions −728 and −696 (Fig. 2B and 3), suggesting that transcription from these TISs is directly regulated by NrrA. Use of the TIS at position −728 also depends on NtcA (23). Thus, the NtcA-dependent regulation of hetR must be mediated by NrrA. HetR is a DNA-binding protein and binds to the region around the TISs at positions −728 and −696 (14). However, HetR is not required for transcription from these TISs (23). Functions of HetR in transcription from these TISs remain unknown. Expression of hetR is subjected to autoregulation, a key aspect of the regulation of heterocyst differentiation (2). Transcription from the TIS at position −273 requires HetR itself (23), and HetR binds to the DNA fragment around this TIS (14). NrrA was also required for transcription from this TIS, but NrrA did not bind to the DNA fragment that includes this TIS (Fig. 1 and 2C). In the nrrA deletion mutant, absence of the induction of hetR from the TISs at positions −728 and −696 would diminish the autoregulatory induction from the TIS at position −273. It has been reported that the hetF gene is involved in the regulation of hetR in Nostoc punctiforme (31). The hetF gene is required for the quick induction of hetR after nitrogen deprivation and localization of the HetR protein in heterocysts. The HetF protein is suggested to be a protease (1), but its role in a regulatory cascade of heterocyst differentiation has not been revealed yet.
FIG. 6.
A model for a regulatory cascade in the early stage of heterocyst differentiation in Anabaena sp. strain PCC 7120. Thick black arrows represent upregulation of gene expression. Open arrows represent activation of proteins. A hitherto unidentified pathway that regulates the NrrA activity is indicated by broken arrows. See the text for details. Hik, histidine kinase.
NrrA is a response regulator belonging to the OmpR family, and thus, its activity is regulated by a sensory histidine kinase. An aspartate residue that corresponds to the phosphorylation site of the OmpR protein in E. coli is conserved in NrrA (6). However, incubation of NrrA with 100 mM acetyl phosphate did not enhance the DNA-binding activity of NrrA (data not shown). The other OmpR-type response regulator ManR from Anabaena sp. strain PCC 7120 and Synechocystis sp. strain PCC 6803 is also not activated by incubation with acetyl phosphate (13, 32). It is possible that recombinant NrrA proteins have already been phosphorylated inside the E. coli cells or that nonphosphorylated NrrA is able to bind to the upstream region of hetR and phosphorylation of NrrA might be required for the transcriptional activation of hetR.
Why does NtcA not directly regulate the expression of hetR but does indirectly via NrrA? One idea is that NrrA ensures the more strict regulation of the hetR expression than NtcA alone. The 2-OG signal regulates the expression of nrrA via NtcA, and some signal other than 2-OG could also regulate the activity of NrrA via a hitherto unidentified histidine kinase (Fig. 6). The possibility that the 2-OG signal is transmitted to NrrA cannot be excluded. Multiple signals could be integrated into NrrA to prevent inappropriate expression of hetR.
Acknowledgments
This work was supported by a Grant-in-Aid for General Scientific Research (15370019) to M.O. from the Ministry of Education, Culture, Sports, Science and Technology of Japan and partly by a grant to M.O. from the Research Institute of Innovative Technology for the Earth and partly by a Sasakawa Scientific Research Grant to S.E. from the Japan Science Society.
Footnotes
Published ahead of print on 13 October 2006.
REFERENCES
- 1.Aravind, L., and E. V. Koonin. 2002. Classification of the caspase-hemoglobinase fold: detection of new families and implications for the origin of the eukaryotic separins. Proteins 46:355-367. [DOI] [PubMed] [Google Scholar]
- 2.Black, T. A., Y. Cai, and C. P. Wolk. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 9:77-84. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Buikema, W. J., and R. Haselkorn. 1991. Characterization of a gene controlling heterocyst differentiation in the cyanobacterium Anabaena 7120. Genes Dev. 5:321-330. [DOI] [PubMed] [Google Scholar]
- 5.Buikema, W. J., and R. Haselkorn. 2001. Expression of the Anabaena hetR gene from a copper-regulated promoter leads to heterocyst differentiation under repressing conditions. Proc. Natl. Acad. Sci. USA 98:2729-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ehira, S., and M. Ohmori. 2006. NrrA, a nitrogen-responsive response regulator facilitates heterocyst development in the cyanobacterium Anabaena sp. strain PCC 7120. Mol. Microbiol. 59:1692-1703. [DOI] [PubMed] [Google Scholar]
- 7.Ehira, S., M. Ohmori, and N. Sato. 2003. Genome-wide expression analysis of the responses to nitrogen deprivation in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 10:97-113. [DOI] [PubMed] [Google Scholar]
- 8.Ehira, S., M. Ohmori, and N. Sato. 2005. Role of the 5′-UTR in accumulation of the rbpA1 transcript at low temperature in the cyanobacterium Anabaena variabilis M3. FEMS Microbiol. Lett. 251:91-98. [DOI] [PubMed] [Google Scholar]
- 9.Frias, J. E., E. Flores, and A. Herrero. 1994. Requirement of the regulatory protein NtcA for the expression of nitrogen assimilation and heterocyst development genes in the cyanobacterium Anabaena sp. PCC 7120. Mol. Microbiol. 14:823-832. [DOI] [PubMed] [Google Scholar]
- 10.Golden, J. W., and H. S. Yoon. 2003. Heterocyst development in Anabaena. Curr. Opin. Microbiol. 6:557-563. [DOI] [PubMed] [Google Scholar]
- 11.Herrero, A., A. M. Muro-Pastor, and E. Flores. 2001. Nitrogen control in cyanobacteria. J. Bacteriol. 183:411-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herrero, A., A. M. Muro-Pastor, A. Valladares, and E. Flores. 2004. Cellular differentiation and the NtcA transcription factor in filamentous cyanobacteria. FEMS Microbiol. Rev. 28:469-487. [DOI] [PubMed] [Google Scholar]
- 13.Huang, W., and Q. Wu. 2004. The ManR specifically binds to the promoter of a Nramp transporter gene in Anabaena sp. PCC 7120: a novel regulatory DNA motif in cyanobacteria. Biochem. Biophys. Res. Commun. 317:578-585. [DOI] [PubMed] [Google Scholar]
- 14.Huang, X., Y. Dong, and J. Zhao. 2004. HetR homodimer is a DNA-binding protein required for heterocyst differentiation, and the DNA-binding activity is inhibited by PatS. Proc. Natl. Acad. Sci. USA 101:4848-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang, F., S. Wisen, M. Widersten, B. Bergman, and B. Mannervik. 2000. Examination of the transcription factor NtcA-binding motif by in vitro selection of DNA sequences from a random library. J. Mol. Biol. 301:783-793. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 17.Laurent, S., H. Chen, S. Bedu, F. Ziarelli, L. Peng, and C. C. Zhang. 2005. Nonmetabolizable analogue of 2-oxoglutarate elicits heterocyst differentiation under repressive conditions in Anabaena sp. PCC 7120. Proc. Natl. Acad. Sci. USA 102:9907-9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, J. H., S. Laurent, V. Konde, S. Bedu, and C. C. Zhang. 2003. An increase in the level of 2-oxoglutarate promotes heterocyst development in the cyanobacterium Anabaena sp. strain PCC 7120. Microbiology 149:3257-3263. [DOI] [PubMed] [Google Scholar]
- 19.Luque, I., E. Flores, and A. Herrero. 1994. Molecular mechanism for the operation of nitrogen control in cyanobacteria. EMBO J. 13:2862-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makino, K., H. Shinagawa, M. Amemura, and A. Nakata. 1986. Nucleotide sequence of the phoB gene, the positive regulatory gene for the phosphate regulon of Escherichia coli K-12. J. Mol. Biol. 190:37-44. [DOI] [PubMed] [Google Scholar]
- 21.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Muro-Pastor, A. M., E. Olmedo-Verd, and E. Flores. 2006. All4312, an NtcA-regulated two-component response regulator in Anabaena sp. strain PCC 7120. FEMS Microbiol. Lett. 256:171-177. [DOI] [PubMed] [Google Scholar]
- 23.Muro-Pastor, A. M., A. Valladares, E. Flores, and A. Herrero. 2002. Mutual dependence of the expression of the cell differentiation regulatory protein HetR and the global nitrogen regulator NtcA during heterocyst development. Mol. Microbiol. 44:1377-1385. [DOI] [PubMed] [Google Scholar]
- 24.Muro-Pastor, M. I., J. C. Reyes, and F. J. Florencio. 2001. Cyanobacteria perceive nitrogen status by sensing intracellular 2-oxoglutarate levels. J. Biol. Chem. 276:38320-38328. [DOI] [PubMed] [Google Scholar]
- 25.Tanigawa, R., M. Shirokane, S. Maeda, T. Omata, K. Tanaka, and H. Takahashi. 2002. Transcriptional activation of NtcA-dependent promoters of Synechococcus sp. PCC 7942 by 2-oxoglutarate in vitro. Proc. Natl. Acad. Sci. USA 99:4251-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vázquez-Bermúdez, M. F., A. Herrero, and E. Flores. 2002. 2-Oxoglutarate increases the binding affinity of the NtcA (nitrogen control) transcription factor for the Synechococcus glnA promoter. FEBS Lett. 512:71-74. [DOI] [PubMed] [Google Scholar]
- 27.Vega-Palas, M. A., E. Flores, and A. Herrero. 1992. NtcA, a global nitrogen regulator from the cyanobacterium Synechococcus that belongs to the Crp family of bacterial regulators. Mol. Microbiol. 6:1853-1859. [DOI] [PubMed] [Google Scholar]
- 28.Wei, T.-F., T. S. Ramasubramanian, and J. W. Golden. 1994. Anabaena sp. strain PCC 7120 ntcA gene required for growth on nitrate and heterocyst development. J. Bacteriol. 176:4473-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolk, C. P. 2000. Heterocyst formation in Anabaena, p. 83-104. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. American Society for Microbiology, Washington, D.C.
- 30.Wolk, C. P., A. Ernst, and J. Elhai. 1994. Heterocyst metabolism and development, p. 769-823. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 31.Wong, F. C., and J. C. Meeks. 2001. The hetF gene product is essential to heterocyst differentiation and affects HetR function in the cyanobacterium Nostoc punctiforme. J. Bacteriol. 183:2654-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamaguchi, K., I. Suzuki, H. Yamamoto, A. Lyukevich, I. Bodrova, D. A. Los, I. Piven, V. Zinchenko, M. Kanehisa, and N. Murata. 2002. A two-component Mn2+-sensing system negatively regulates expression of the mntCAB operon in Synechocystis. Plant Cell 14:2901-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]